Abstract
Pioneer oral bacteria, including Streptococcus gordonii, initiate the formation of oral biofilms on tooth surfaces, which requires differential expression of genes that recognize unique environmental cues. An S. gordonii::Tn917-lac biofilm-defective mutant was isolated by using an in vitro biofilm formation assay. Subsequent inverse PCR and sequence analyses identified the transposon insertion to be near the 3′ end of an open reading frame (ORF) encoding a protein homologous to a Streptococcus pneumoniae repressor, AdcR. The S. gordonii adc operon, consisting of the four ORFs adcR, adcC, adcB, and adcA, is homologous to the adc operon of S. pneumoniae, which plays a role in zinc and/or manganese transport and genetic competence in S. pneumoniae. AdcR is a metal-dependent repressor protein containing a putative metal-binding site, AdcC contains a consensus-binding site for ATP, AdcB is a hydrophobic protein with seven hydrophobic membrane-spanning regions, and AdcA is a lipoprotein permease with a putative metal-binding site. The three proteins (AdcC through -A) are similar to those of the binding-lipoprotein-dependent transport system of gram-positive bacteria. Reverse transcriptase PCR confirmed that adcRCBA are cotranscribed as an operon in S. gordonii and that the transposon insertion in S. gordonii adcR::Tn917-lac had resulted in a polar mutation. Expression of adcR, measured by the β-galactosidase activity of the adcR::Tn917-lac mutant, was growth phase dependent and increased when the mutant was grown in media with high levels of manganese (>1 mM) and to a lesser extent in media with zinc, indicating that AdcR may be a regulator at high levels of extracellular manganese. A nonpolar inactivation of adcR generated by allelic replacement resulted in a biofilm- and competence-defective phenotype. The biofilm-defective phenotype observed suggests that AdcR is an active repressor when synthesized and acts at a distant site(s) on the chromosome. Thus, the adc operon is involved in manganese acquisition in S. gordonii and manganese homeostasis and appears to modulate sessile growth in this bacterium.
Oral streptococci (viridans streptococci) initiate the formation of oral biofilms known as dental plaque and are ubiquitous primary colonizers that are the predominant cultivable bacteria found in supragingival plaque (45). Viridans streptococci are also frequent etiologic agents of bacterial endocarditis (21, 64) and are becoming significant opportunistic pathogens and a major cause of bacteremia in immunocompromised patients, accounting for 40% of infections in neutropenic patients (25, 32). Fully developed, surface-attached oral biofilms are highly structured, with distinct architectural and physiochemical properties commonly observed with other biofilm communities (12). Sessile bacteria found in biofilms represent a distinct mode of growth that differs from planktonic cells, characterized by differences in gene expression and cellular physiology and an increased resistance to antibiotics.
The multistep process of biofilm formation has just begun to be understood (12). This complex biofilm developmental process is initiated by the attachment of the colonizing bacteria to a surface. Subsequent accumulation and growth of attached bacteria result in microcolonies that increase in size and eventually appear as towering pillar- and mushroom-shaped biofilms. The growth of this complex, multilayered, cellular matrix-embedded community and its eventual dispersal and spread require the coordinated expression of an array of genes (57, 59). These include general housekeeping genes and some global regulatory genes, which have been identified by using genetic analyses of biofilm-defective phenotypes, DNA microarrays, and proteomic analysis (4, 38, 46, 61, 63, 68, 69). Thus, the transition between planktonic and sessile phenotypes does not appear to arise from the expression of unique genes but probably requires a differential pattern of gene expression as the bacteria enter a distinct physiological state. The same attached bacteria can revert to planktonic cells by switching off this coordinated gene expression (59).
Assays that measure the ability of bacteria to form adherent biofilms on abiotic surfaces by staining attached bacterial biofilm with crystal violet or safarinin have been used extensively to screen for biofilm-defective mutants. Specific genes involved in single-species biofilms have been identified in a variety of bacteria, including Escherichia coli (24, 52, 54), Staphylococcus aureus (13), Staphylococcus epidermidis (43), Streptococcus gordonii (42), Streptococcus mutans (68, 71), Pseudomonas aeruginosa (48), Pseudomonas fluorescens (49), Salmonella enterica (44), and Vibrio cholerae (67). Identifying and analyzing biofilm-defective mutants resulted in the identification of a number of biofilm-associated genetic loci. These include genetic competence (39, 40, 42), quorum sensing and signaling (4, 39, 40, 42), exopolysaccharide synthesis (27, 70), and specific bacterial surface proteins (22, 36, 52, 55, 67).
A number of transcriptional regulators that are stress inducible and growth phase dependent have been found to modulate bacterial biofilm formation. Biofilm formation of S. epidermidis was found to require the expression of RsbU, a positive regulator of σB, an alternative sigma factor that modulates biofilm formation during environmental challenges by altering the expression of the polysaccharide intercellular adhesin (33). A transcriptional repressor in S. epidermidis encoded by icaR is involved in both the environmental regulation of the ica operon and biofilm formation (10). The E. coli OmpR/EnvZ two-component regulatory system modulates biofilm formation in response to changes in osmolarity through a complex regulatory network that controls expression of curli (53), whereas the Cpx two-component signal transduction system is involved in the attachment of stationary-phase E. coli cells to hydrophobic abiotic surfaces (18, 50). PvrR, a putative sensor regulator homolog, modulates antibiotic resistance and biofilm formation of P. aeruginosa during phase variation (19). Crc, the global carbon metabolism regulator in P. aeruginosa (47), and CcpA, the catabolite control protein in S. mutans (68), are necessary during biofilm formation on abiotic surfaces. CytR, a transcriptional repressor that inhibits polysaccharide synthesis in V. cholerae, also modulates biofilm development (26). A ToxR homolog that regulates its own synthesis in Vibrio anguillarum serotype 01 has been implicated in biofilm formation on glass surfaces (66). The availability of microbial genome sequences has enabled investigators to mutate global transcriptional regulators identified in silico and to identify several biofilm-associated genetic loci among these regulators in the oral pathogen S. mutans (4, 38-40, 68). Taken together, these studies demonstrated that certain transcriptional regulators play a crucial role in biofilm formation by regulating the expression of biofilm-related genes and reinforced the concept that coordinated gene expression may modulate the transition between the planktonic and sessile phenotypes.
A mutant generated by transcriptional fusion with a reporter gene would represent an important advantage in the study of genes involved in biofilm formation, as it is an important tool for facilitating studies of gene regulation. In order to investigate factors that may regulate the expression of biofilm-associated genes, the gram-positive transposon Tn917, containing the easily detectable promoterless E. coli lacZ reporter gene, was used to generate insertion-mediated transcriptional fusions in S. gordonii and the mutants generated were screened to identify genes associated with biofilm formation. An in vitro biofilm formation assay (42) was used to screen for biofilm-defective mutants. The use of Tn917-lac (Eryr, Gal+) (14) enabled the detection of the levels of gene expression under various environmental conditions. The aims of this study were to identify a biofilm-associated gene(s) in S. gordonii and to determine the influence of environmental factors on the expression of the gene(s) identified.
MATERIALS AND METHODS
Bacteria, media, and chemicals.
S. gordonii Challis 2, the rifamycin-resistant (500 μg/ml) parent strain of S. gordonii Challis (42), was used as the wild-type (WT) strain. Unless otherwise indicated, bacteria were subcultured and maintained routinely on brain heart infusion (BHI) agar (BBL; Becton Dickinson, Cockeysville, Md.) or Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.2% yeast extract (THBYE) at 37°C under anaerobic conditions (VWR brand anaerobic chamber; VWR, Plainfield, N.J.). E. coli strain XL1-Blue MRF' (Stratagene, Cedar Creek, Tex.) was grown at 37°C in Luria-Bertani broth or on Luria-Bertani agar under aerobic conditions.
All chemicals were purchased from Sigma (St. Louis, Mo.) or Fisher Scientific (Pittsburgh, Pa.). All enzymes for DNA manipulations were purchased from Promega (Madison, Wis.) or Fisher Scientific unless stated otherwise. Oligonucleotide primers were from Invitrogen Life Technologies (Rockville, Md.), and their sequences are available on request.
Tn917-lac mutagenesis of S. gordonii Challis.
S. gordonii Challis 2 was transformed with 2 to 3 μg of pTV32-OK (14) by the method described previously (42). The transformants were then plated on BHI agar containing 10 μg of erythromycin (ERY)/ml and 500 μg of kanamycin (KAN)/ml at 30°C and were incubated for 3 to 5 days under anaerobic conditions. Single-colony transformants that were recovered on ERY and KAN containing BHI agar at 30°C were used for marker rescue of interrupted genes of S. gordonii by using the methods described by Cvitkovitch et al. (14). Briefly, single colonies were inoculated into 5 ml of THBYE containing ERY and KAN and were grown anaerobically for 48 h at 30°C. Independent pools of Tn917 insertions into host chromosomal DNA were generated by a temperature shift to 40°C. Aliquots (5, 10, 25, and 50 μl) of culture were then inoculated into 5 ml of fresh THBYE with no antibiotics and were incubated overnight at 40°C. Following the temperature shift, overnight aliquots were plated on BHI plates containing ERY alone or both ERY and KAN or no antibiotics to estimate the relative efficiencies of transpositions. S. gordonii cells in which the Tn917 had transposed into the chromosome and had lost the plasmid were erythromycin resistant (Eryr) and kanamycin sensitive (Kans). An equal volume of sterile glycerol was added to the rest of the culture and mixed, and aliquots were frozen at −80°C.
A Tn917-lac library was made by plating the frozen cultures onto BHI plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 100 μg/ml) and ERY at the selective concentration (10 μg/ml) and was grown at 37°C. After 36 to 48 h of growth, blue (Lac+) and white (Lac−) colonies were observed on the plates. Each blue, Eryr colony was picked and transferred into a well on a 96-well microtiter plate containing 200 μl of BHI and ERY. After mixing of the inoculum, 100 μl of the BHI was transferred to another well on a new microtiter plate, thus creating a duplicate, and plate contents were incubated anaerobically at 37°C for 24 to 48 h. After glycerol was added to a final concentration of 30%, this library was stored at −80°C.
Biofilm assay.
The in vitro biofilm formation assay was performed in biofilm medium (BM) as previously described (42). In addition to the microtiter plate assay, biofilm formation on borosilicate glass coverslips was visualized directly by using phase-contrast microscopy (47). Cells from overnight colonies grown on BHI agar plates were inoculated into BM in 50-ml Falcon tubes at an A600 of ∼0.04. Biofilms were formed at 37°C on borosilicate coverslips placed in the Falcon tubes. At chosen time points, the coverslip was removed and rinsed with BM to remove nonadherent cells and biofilm bacteria were examined by phase-contrast microscopy with a Micromaster phase-contrast microscope (Fisher Scientific). Images were captured with a Nikon Coolpix 950 digital camera.
Southern hybridization.
Chromosomal DNA was isolated from the S. gordonii WT and putative biofilm-defective mutants by the method described by Ganeshkumar et al. (23). DNA was digested with KpnI, separated by agarose gel electrophoresis, and transferred onto a nitrocellulose membrane for Southern hybridization. The probe used for hybridization was pTV32-OK (14) labeled with digoxigenin (DIG) by using the DIG DNA labeling system (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the manufacturer's instructions. After hybridization, the membrane was developed by enzyme immunoassay with the DIG nucleic acid detection kit (Roche Molecular Biochemicals).
Localization of transposon insertion site and sequence analyses.
The location of the transposon insertion was determined by sequence analysis of the region flanking the transposon. Initially, pBluescript vector and chromosomal DNA from the mutant were digested with HindIII, purified by using the Qiagen Nucleotide Removal kit (Qiagen, Valencia, Calif.), ligated with each other, and used as the PCR template. Inverse PCR with the appropriate primers was then performed under standard conditions. The PCR products were analyzed by agarose gel electrophoresis, purified by using the Qiagen PCR purification kit (Qiagen), and sequenced at the Genetics Core Sequencing Facility at Boston University by using a Model 377 Automated Sequencer (Applied Biosystems, Foster City, Calif.). The sequence obtained was compared with sequences in GenBank by using the BLASTX and TBLASTN programs (1) to identify homologous bacterial sequences. Amino acid sequence alignments and phylogenetic analyses were performed by using the AlignX program in Vector NTI (Informax Inc., Bethesda, Md.), which utilizes the neighbor-joining algorithm (56). Transmembrane domains were predicted with TMpred software (http://www.isrec.isb-sib.ch/software/TMPRED_form.html).
RT-PCR of S. gordonii WT RNA.
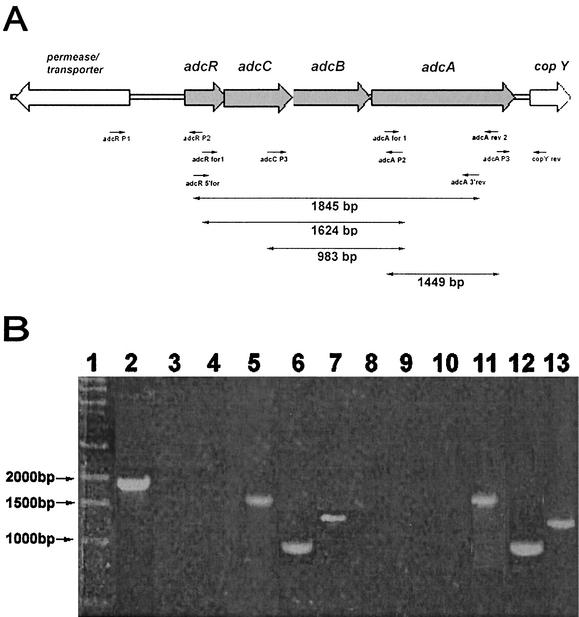
In order to determine the genes that constitute the adc operon, reverse transcriptase PCR (RT-PCR) was performed with total RNA extracted from an S. gordonii WT strain grown to mid-log phase (A600 of 0.3 to 0.4) by using a Qiagen RNeasy mini kit (Qiagen). Primers used for RT-PCR are adcR 5′ for and adcA 3′ rev (specific for a 1.845-kb region that spans adcR to adcA), primers adcR P1 and adcR P2 (specific for an 855-bp region that extends from the open reading frame [ORF] encoding a putative transporter to adcR) and primers adcA P3 and copY rev (specific for a 770-bp region that spans adcA to copY). Locations of the primers are shown in Fig. 5A.
FIG. 5.
RT-PCR analysis of RNA extracted from S. gordonii WT and adcR mutants. (A) Organization of adc operon and adjacent genes, location of primers used, and predicted size of RT-PCR products. Primers that successfully produced an amplicon and their predicted sizes are in boldface. (B) RT-PCR products were studied by using total RNA extracted from WT (lanes 2 to 7), adcR::Tn917-lac (lanes 8 to 10), and adcR::Specr (lanes 11 to 13) strains as the template. Lanes: 1, 1-kb DNA marker; 2, primers adcR 5′ for and adcA 3′ rev; 3, primers adcR P1 and adcR P2; 4, primers adcA P3 and copY rev; 5, primers adcR for 1 and adcA P2; 6, primers adcC P3 and adcA P2; 7, primers adcA for 1 and adcA rev 2; 8, primers adcR for 1 and adcA P2; 9, primers adcC P3 and adcA P2; 10, primers adcA for 1 and adcA rev 2; 11, primers adcR for 1 and adcA P2; 12, primers adcC P3 and adcA P2; 13, primers adcA for 1 and adcA rev 2.
RT-PCR was also performed with RNA isolated from both WT S. gordonii and an adcR::Tn917-lac mutant to determine whether the mutant generated was polar or nonpolar. This was carried out by using primers adcR for 1 and adcA P2, specific for a 1,624-bp region spanning adcR to adcA; primers adcC P3 and adcA P2, specific for a 983-bp region spanning adcC to adcA; and primers adcA for 1 and adcA rev 2, specific for a 1,449-bp region within adcA.
The reaction mixture (25 μl) contained 0.1 μg of template RNA, a 1.2 μM concentration of each primer, 0.2 mM deoxynucleoside triphosphates, 1 mM MgSO4, 2.5 U of avian myeloblastosis virus RT for first-strand DNA synthesis, and 2.5 U of thermostable Tfl DNA polymerase (from Thermus flavus) for second-strand cDNA synthesis and DNA amplification (Access RT-PCR system; Promega). The RT reaction was performed at 37°C for 60 min and was then stopped by raising the temperature to 95°C for 2 min. Subsequent PCR amplification was performed under standard conditions, and the RT-PCR products were visualized after 1% agarose gel electrophoresis.
Expression of adcR in different environmental conditions.
The β-galactosidase activity of the biofilm-defective S. gordonii adcR::Tn917-lac mutant was determined by a fluorometric assay (28) by using methylumbelliferyl-β-glucuronide (MUG). Bacteria were grown in 10 ml of THBYE or BM containing ERY at 37°C (unless otherwise indicated) in the anaerobic chamber for 18 h. The conditions tested were BHI, THB, and THBYE without any supplement; THBYE without any supplement grown under aerobic conditions or at 30 or 42°C; THBYE without any supplement adjusted to various pHs (pH 5, 6, 7, 8, or 9); THBYE supplemented with zinc (1 or 10 mM), manganese (1 or 10 mM), FeSO4 (1 or 10 mM), NiCl2 (1 or 10 mM), 1 mM CoCl2, 10% clarified whole human saliva, horse serum (1 or 10%), human serum (1 or 10%), NaCl (0.1, 0.2, 0.3, 0.4, or 0.5 M), 0.8% (wt/vol) sugars (fructose, glucose, lactose, maltose, mannose, and sucrose), or 100 mM amino acid (alanine, arginine, aspartate, cysteine, glutamine, glutamate, glycine, histidine, leucine, lysine, methionine, phenylalanine, proline, serine, threonine, tyrosine, or valine). In a subsequent assay, the conditions tested were BM supplemented with various concentrations of manganese (0.1, 0.5, 1, 5, 10, 50, 100, and 200 μM and 1 and 2 mM).
Cells were centrifuged, resuspended in 10 ml of fresh, prewarmed appropriate liquid medium, and grown to log phase (A600 of 0.15 to 0.4). Fresh, prewarmed medium (100 ml) was inoculated with 2 ml of log-phase cultures and grown to a log-phase A600 of 0.15 to 0.4 (∼3 h). Bacterial growth was recorded (A600), and cells were washed twice with 5 ml of 10 mM Tris, 1 mM EDTA, and 150 mM NaCl (pH 8.0) and resuspended in 5 ml of it and sonicated for 2 min. Cells were placed on ice for 5 min and centrifuged, and the clear sonicate was transferred to a new tube. For each sample, 100 μl of the sonicate was transferred to an opaque microtiter plate, followed by 100 μl of 0.4 mg of MUG/ml in dimethyl sulfoxide. This was mixed well and incubated for 30 min at room temperature. After addition of 200 μl of 4-methyl-umbelliferone (MU) standards (ranging from 0.1 to 3.2 μM in dimethyl sulfoxide), the microtiter plate was read in a fluorescent plate reader (355-nm excitation and 460-nm emission). A Bradford protein assay (Bio-Rad Laboratories, Richmond, Calif.) was performed according to the manufacturer's instructions to determine protein concentration in each sonicate preparation. The activity was reported as micromolar concentration of MU per microgram of protein per minute.
Mutagenesis of the adcR gene in S. gordonii.
PCR ligation mutagenesis with vectorless intermediates (37) was used to construct adcR deletion mutants. The plasmids pDL151 (containing a KAN resistance gene, kan) and pDL152 (containing a spectinomycin resistance gene, spec) (62) were used as templates for amplifying kan and spec, respectively.
Initially, PCR amplifications of the two flanking regions and the antibiotic marker insert were performed with the appropriate primers, which incorporated MluI and XbaI, to enable directional ligation. After electrophoresis on a 1% agarose gel to confirm amplification, the PCR products were purified with the QIAquick PCR purification kit (Qiagen). The purified PCR products of the 5′ and 3′ flanking fragments were digested with MluI and XbaI, respectively, while the amplified kan and spec cassettes were digested with both enzymes by overnight double digestion at 37°C. After digestion, purification of the digested fragments was performed with the QIAquick Nucleotide Removal kit (Qiagen). Directional ligation was performed at room temperature for 18 h after mixing together 2 μl of the amplified 5′ flanking fragment, 2 μl of the amplified 3′ flanking fragment, 4 μl of the amplified antibiotic cassette, 1 μl of 10× ligase buffer, and 1 μl of T4 DNA ligase in a total volume of 10 μl.
After ligation, 4 μg of the ligated DNA was used for transformation of the S. gordonii WT by the method described previously (42). Transformants were then plated on BHI agar containing the appropriate antibiotics (either 350 μg of KAN/ml or 1,000 μg of spectinomycin/ml) and were incubated at 37°C anaerobically for 2 to 5 days. The competence of the S. gordonii WT strain and adcR::Tn917-lac, adcR::Specr, and adcR::Kanr mutants was assessed by transformation with 4 μg of DNA and by enumerating the CFU on solid media containing the appropriate antibiotics. Experiments were done in triplicate.
RT-PCR was performed with RNA isolated from both adcR::Specr and adcR::Kanr mutants to determine whether the mutant generated was polar or nonpolar. This was carried out using primers adcR for 1 and adcA P2, specific for a 1,624-bp region spanning adcR to adcA, primers adcC P3 and adcA P2, specific for a 983-bp region spanning adcC to adcA, and primers adcA for 1 and adcA rev 2, specific for a 1,449-bp region within adcA. The RT-PCR was carried out as described above. Locations of the primers are shown (see Fig. 5A).
The growth rates of the S. gordonii WT strain and the adcR::Tn917-lac, adcR::Specr, and adcR::Kanr mutants were assessed by inoculating the strains from an overnight THBYE culture into fresh 10 ml of THBYE and growing them at 37°C under anaerobic conditions. Growth was quantified by recording the A600 at regular intervals over 24 h.
Nucleotide sequence accession number.
The DNA sequence of the 4.175-kb fragment containing the adc operon has been assigned the GenBank accession number AY177418.
RESULTS
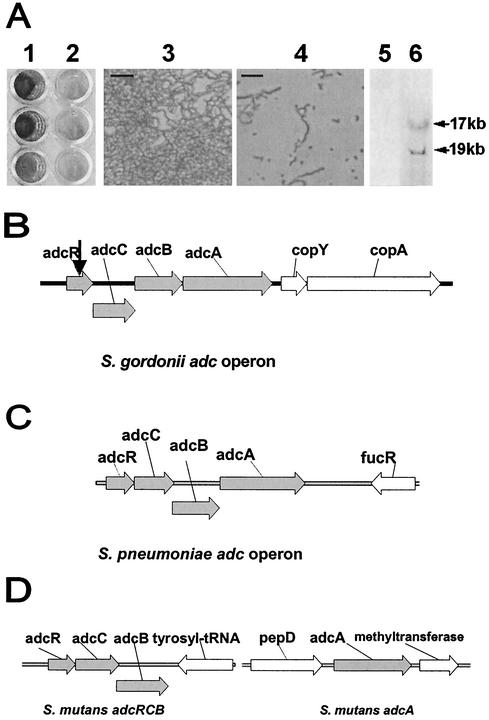
The transposition of Tn917-lac in S. gordonii Challis 2 occurred at a frequency of approximately 10−5, and approximately 10% of the transformants were also Kanr, indicating that plasmid cointegrants occurred at this frequency. A biofilm-defective Tn917-lac mutant was identified after a preliminary screening of 2,650 isogenic mutants generated by transposon mutagenesis with Tn917-lac (Fig. 1A, lanes 1 and 2). The biofilm assay showed that biofilm formation of this S. gordonii Tn917-lac mutant (A575 ± standard deviation, 1.03 ± 0.53) was reduced by 67% when compared to the S. gordonii WT strain (A575 ± standard deviation, 3.14 ± 0.33). In addition, biofilm development on borosilicate glass coverslips by both strains was examined microscopically at 1, 3, and 24 h after inoculation. After 1 h of incubation, a similar number of cells of the WT strain and Tn917-lac mutant had attached to the coverslip. Fewer cells of the Tn917-lac mutant were present after 3 h of incubation. After 24 h of incubation, the WT strain formed large chains of cells interspersed with areas of less densely packed cells. In contrast, only a few scattered Tn917-lac mutant cells had attached, with large, empty areas present (Fig. 1A, panels 3 and 4).
FIG. 1.
(A) Lanes 1 and 2, biofilm formation assay of two S. gordonii::Tn917-lac mutants. Lane 1, a biofilm-positive mutant (growth = A575 of 1.1); lane 2, biofilm-defective adcR::Tn917-lac mutant (growth = A575 of 1.1). Panels 3 and 4, phase-contrast micrographs of biofilm formation on borosilicate coverslips in BM after 24 h by WT (panel 3) and adcR::Tn917-lac mutant (panel 4). Bar = 10 μm. Images represent what was observed in multiple fields. Lanes 5 and 6, Southern hybridization of KpnI-digested DNA from WT (lane 5) and adcR::Tn917-lac mutant (lane 6). DIG-labeled pTV32-OK containing Tn917-lac (which has one KpnI site) was used as the probe. (B to D) Gene organization in the adc operon in various streptococci. Genes from the adc operon are shown in grey. (B) S. gordonii adc operon. adcR (positions 261 to 704) encodes a putative transcriptional repressor for Mn-responsive expression, adcC (positions 701 to 1411) encodes a putative ATP-binding cassette (ABC) transporter, adcB (positions 1404 to 2210) encodes a putative ABC transporter (membrane protein), and adcA (positions 2220 to 3722) encodes a metal-binding lipoprotein, an ABC transporter. The black vertical arrow indicates the position of Tn917-lac insertion in the biofilm-defective mutant. (C) S. pneumoniae adc operon, consisting of adcR, adcC, adcB, and adcA. (D) S. mutans adc operon, consisting of adcR, adcC, and adcB. The adcA gene is present on a different part of the chromosome.
The transposition was confirmed by Southern hybridization with Tn917-lac, which has a single KpnI site, and two hybridizing bands were predicted. Results from the Southern hybridization (Fig. 1A, lanes 5 and 6) show two bands that hybridized with the DIG-labeled Tn917-lac probe, at 17 and 19 kb. This confirmed that a single transposon insertion occurred in the biofilm-defective mutant.
Inverse PCR performed with genomic DNA that was digested with HindIII and ligated with pBluescript resulted in a PCR product, which was 2.8 kb in size. Sequence analysis of this PCR product containing the region 5′ to the transposon insertion revealed that transposition had occurred within an ORF that was homologous to adcR, which encodes a repressor in the adc operon of Streptococcus pneumoniae (16) and S. mutans (Fig. 1B to D). This gene was not identified when Tn916 mutagenesis was used in a previous screening of biofilm-defective mutants of S. gordonii Challis (42), suggesting that utilization of Tn917 has enabled us to identify an additional biofilm-associated gene.
Genetic organization of the adc operon.
The region flanking the transposition insertion in S. gordonii adcR::Tn917-lac was amplified by PCR. Appropriate primers were designed based on a homologous sequence found on an unfinished and nonannotated sequence of the S. gordonii chromosome (http://www.tigr.org), and the amplified products were sequenced.
Sequence similarity BLAST searches using the S. gordonii nucleotide sequence obtained found that the closest homologs of the deduced amino acid sequences were proteins encoded by genes of the S. pneumoniae adc operon, consisting of adcR, adcC, adcB, and adcA (Fig. 1B and C). This adc operon is involved in zinc and/or manganese transport and genetic competence of S. pneumoniae (9, 16). Four S. gordonii ORFs were identified and designated adcR, adcC, adcB, and adcA.
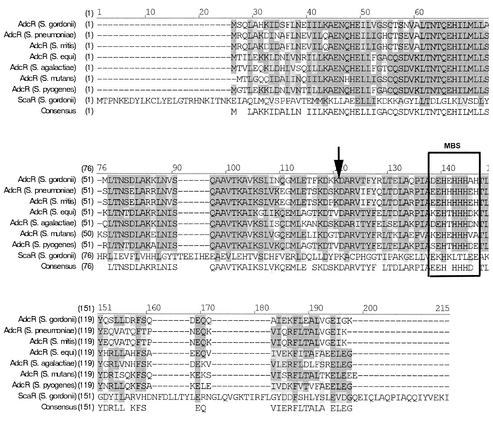
The 444-bp adcR ORF encodes a 148-amino-acid sequence with a predicted molecular mass of 16.7 kDa. The deduced amino acid sequence showed a high degree of homology to bacterial repressor proteins (Table 1; Fig. 2). Amino acid alignment of the AdcR of S. gordonii and all its homologs from S. pneumoniae, Streptococcus mitis, Streptococcus equi, Streptococcus agalactiae, S. mutans, and Streptococcus pyogenes and the ScaR of S. gordonii showed that ScaR was a larger protein that was not homologous to AdcR and did not contain a putative metal-binding region that is present in AdcR (Fig. 2).
TABLE 1.
Homology of proteins encoded by the S. gordonii adc operon and by those of other oral streptococci
| Species | % Identity for AdcR (% similarity) | % Identity for AdcC (% similarity) | % Identity for AdcB (% similarity) | % Identity for AdcA (% similarity) |
|---|---|---|---|---|
| S. pneumoniae | 69 (79) | 89 (91) | 87 (96) | 81 (86) |
| S. mitis | 70 (80) | 71 (73)a | NAc | 65 (68)b |
| S. equi | 60 (72) | 76 (83) | 71 (86) | 60 (71) |
| S. agalactiae | 61 (73) | 76 (82) | 73 (87) | 61 (72) |
| S. mutans | 61 (71) | 78 (81) | 69 (85) | 60 (71) |
| S. pyogenes | 58 (70) | 77 (83) | 74 (89) | 61 (72) |
Incomplete sequence. Homology was calculated over 189 amino acids only.
Incomplete sequence. Homology was calculated over 400 amino acids only.
NA, sequence not found in unfinished S. mitis genome database.
FIG. 2.
Multiple alignments of S. gordonii AdcR deduced amino acid sequence with AdcR homologs from various streptococci. S. gordonii AdcR was aligned with S. pneumoniae AdcR (GenBank accession no. CAA96184), the AdcR homologs from S. mitis and S. equi (identified from their respective unfinished genome sequences), S. agalactiae AdcR (NP687189), S. mutans AdcR (NP722293), S. pyogenes AdcR (AAL96906), and S. gordonii ScaR (AAF25184) through use of the AlignX program of Vector NTI (Informax). The black vertical arrow indicates the position of the transposon insertion in the S. gordonii adcR::Tn917-lac biofilm-defective mutant (pos 110). MBS, histidine-rich putative metal-binding site. Amino acids that are identical and conserved are highlighted in dark grey and light grey, respectively.
Analysis of the region 3′ from adcR revealed the presence of three other ORFs that were highly homologous to adcC, adcB, and adcA of S. pneumoniae. The adcC ORF was 711 bp, starting 1 bp 5′ of the adcR stop codon. The adcC ORF was predicted to encode a 237-amino-acid protein with a predicted molecular mass of 27.1 kDa that exhibited high levels of similarity to ATP-binding proteins in other streptococci (Table 1). It also contains a consensus site for binding to ATP. The adcB ORF was 807 bp in length, starting 5 bp 5′ of the adcC stop codon. It encodes a deduced 269-amino-acid sequence with a predicted molecular mass of 29.2 kDa, with homology to AdcB of S. pneumoniae (Table 1). S. gordonii AdcB contains seven hydrophobic transmembrane helices (at amino acid positions 13 to 32, 46 to 78, 89 to 107, 133 to 150, 180 to 210, 220 to 240, and 245 to 260). The adcB ORF was followed by adcA, which was 1,503 bp in length, starting 10 bp 3′ of the adcB stop codon. The adcA ORF has a deduced 501-amino-acid sequence, with a predicted molecular mass of 56.4 kDa, and a high level of homology to AdcA, a zinc-binding lipoprotein found in S. pneumoniae, and AdcA homologs in other streptococci (Table 1).
Further 3′ of adcA is another ORF, which starts 148 bp from the stop codon of adcA. This ORF encodes a protein with homology to CopY from S. mutans (37% identity; 60% similarity), which is a negative transcriptional repressor of the copYAZ operon in S. mutans (Fig. 1B). Nine base pairs 3′ of copY is copA, which encodes CopA, a 742-amino-acid protein that shares extensive homology with P-type ATPases (65). In S. mutans, the copYAZ operon encodes a conserved P-type ATPase, which enables the bacterium to tolerate high concentrations of extracellular copper. The expression of the ATPase appears to be regulated by copper via the activity of copY and copZ of the operon (65). Interestingly, in S. gordonii, there is no copZ homolog 3′ of copA. Located 5′ of the adcR is an ORF that is divergently transcribed, starting 567 bp 5′ from the start codon of adcR. This ORF has homology to bacterial transporter and/or permease genes (see Fig. 5A).
Sequence and phylogenetic analyses of S. gordonii adc operon.
The predicted amino acid sequences of the proteins encoded by the four adjacent ORFs in S. gordonii showed strong homology to the four proteins encoded by S. pneumoniae adcR, adcC, adcB, and adcA. Upstream of the adcR start site, a putative ribosomal binding site, putative −10 and −35 sites, and a 12-bp inverted repeat were found (Fig. 3). The inverted repeat region present in the 3′ region of adcR may be where the active repressor binds. A similar region has been observed in the ScaR repressor-binding domain of S. gordonii, which was localized to the 3′ region of scaC and is involved in positively regulating the sca operon in the presence of low levels of extracellular manganese (31). The presence of a histidine-rich putative metal-binding domain (DEHEHHHAH) in AdcR indicates that it may also require the binding of a metal for its function. No such histidine-rich metal-binding domain was found in ScaR (Fig. 2). These data suggest that AdcR may act as a metal-dependent repressor in vivo.
FIG. 3.
Alignment of the promoter regions of S. gordonii adcR with S. pneumoniae adcR (GenBank accession no. CAA96184), the adcR homologs from S. mitis and S. equi (identified from their respective unfinished genome sequences), S. agalactiae adcR (NP687189), S. mutans adcR (NP722293), and S. pyogenes adcR (AAL96906) by using the AlignX program of Vector NTI (Informax). Streptococcal sequences without accession numbers were obtained from the unfinished microbial genome database (http://www.ncbi.nlm.nih.gov). Nucleotides that are identical are highlighted in grey. Inverted repeat regions are indicated by arrows; region I is present in S. gordonii, S. equi, S. agalactiae, S. mutans, and S. pyogenes; and region II is present in S. pneumoniae and S. mitis. Putative ribosomal binding sites and potential −10 and −35 regions are underlined, and potential start codons are boxed.
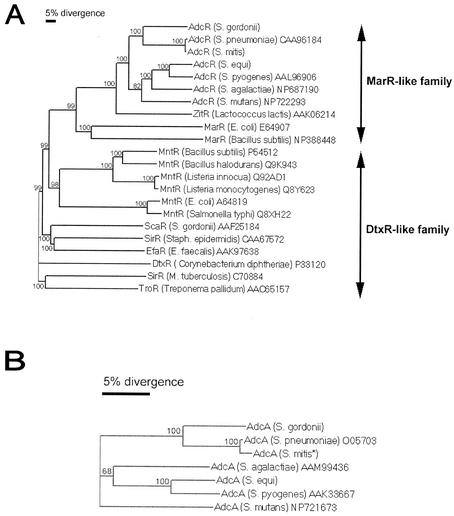
The genetic organization, phylogenetic analysis, and alignment of the deduced amino acid sequences of AdcR through -A were examined by identifying homologous sequences from streptococci deposited in the DNA databases (Fig. 4). Sequences encoding similar proteins were first identified with the TBLASTN algorithm (1) and were retrieved from the microbial genome databases (http://www.ncbi.nlm.nih.gov). The deduced amino acid sequences from these putative adcR through -A genes among the different streptococci were then aligned with the sequences from S. gordonii for comparison. Multiple-sequence alignment and the resulting phylogenetic tree of ABC metalloregulators indicated that ScaR belongs to the DtxR-like family of proteins, while AdcR belongs to the MarR-like family of proteins (Fig. 4A). Phylogenetic analyses demonstrate that S. gordonii AdcR is more closely related to repressors that modulate manganese transport and that AdcR is distantly homologous to other metalloregulators. Amino acid sequence alignments of AdcC and AdcB from S. gordonii, S. pneumoniae, S. mutans, S. equi, S. agalactiae, and S. pyogenes indicate that they share high levels of homology (data not shown), but the comparison among forms of AdcA from these streptococci showed distinct differences, even though they share high levels of homology (Fig. 4B).
FIG. 4.
Phylogenetic relatedness (dendrogram) constructed on the basis of amino acid sequence similarities by using the AlignX program in the Vector NTI software (Informax), which utilizes the neighbor-joining algorithm (56). The reliability of the topology was estimated by performing 100 bootstrap trials, and the bootstrap values are expressed in percentages at branch points (http://www.genebee.msu.su). Accession numbers follow the species names, and sequences without accession numbers were obtained from the unfinished microbial genome sequences (http://www.ncbi.nlm.nih.gov). (A) Phylogenetic tree of S. gordonii AdcR and other bacterial metalloregulatory proteins. (B) Phylogenetic tree of the metal-binding permease AdcA from S. gordonii and AdcA from other streptococcal species.
AdcA in S. gordonii contains a consensus sequence for the signal peptide cleavage site (LXXC) of the prolipoprotein signal peptide sequence (60). Notably, a histidine-rich motif (HEHGEEGHHHDYDPH) that is a putative metal-binding domain is present in AdcA but not in ScaA of S. gordonii (34). In contrast to ScaA, AdcA has an additional C-terminal region that is also present in AdcA proteins in other streptococci (9). These data suggested that the two distinct genetic systems, the adc and sca operons, are involved in metal transport in S. gordonii.
Phylogenetic analysis of the operon showed that there are significant differences in the organization of the adc operon in different streptococci, as determined by sequences from their completed and unfinished microbial genome databases (http://www.ncbi.nih.gov). The adcA genes in S. gordonii and S. pneumoniae are contiguous with adcRCB, whereas the adcA genes in S. mutans (Fig. 1B to D), S. pyogenes, S. equi, and S. agalactiae (data not shown) are noncontiguous with adcRCB. Also, S. gordonii AdcA and S. pneumoniae AdcA are lipoproteins and contain a consensus sequence for the signal peptide cleavage site in bacterial lipoproteins, whereas the AdcA proteins in S. pyogenes, S. mutans, S. equi, and S. agalactiae do not have the typical cleavage site for lipoproteins. However hydrophobicity analyses indicate that they are probably secreted proteins (data not shown). The presence of a lipoprotein signal peptide and phylogenetic analysis of the incomplete deduced AdcA protein sequence of S. mitis predict that these bacteria fall within the first group, but this can only be verified when the genome sequence of S. mitis is completed (Fig. 4B).
RT-PCR with RNA from the WT strain using primers adcR 5′ for and adcA 3′ rev (Fig. 5A), which are specific for a 1.845-kb region that spans the adcR and adcA intragenic region, yielded a product of approximately 1,845 bp (Fig. 5B, lane 2). This spans the ORFs adcR, adcC, adcB, and adcA and coincides with the predicted amplicon size of 1,845 bp. RT-PCR with primers adcR P1 and adcR P2 (Fig. 5A), which are specific for an 855-bp region that spans the putative transporter and/or permease 5′ of adcR to the adcR gene, and primers adcA P3 and copY rev, specific for a 770-bp region that spans adcA to copY, yielded no PCR products (Fig. 5B, lanes 3 and 4). All the primer pairs used in the RT-PCR above were also used in standard PCR amplifications with S. gordonii WT DNA, which yielded PCR products with the predicted sizes. These results demonstrate that adcR, adcC, adcB, and adcA are cotranscribed as a single operon, while copY and copA are probably transcribed as a separate operon. Therefore, the adc operon consists of the four ORFs adcR, adcC, adcB, and adcA only.
No amplified products were obtained from RT-PCR with primers spanning adcR to adcA or adcC to adcA or with primers within the adcA gene with RNA isolated from the S. gordonii adcR::Tn917-lac mutant, whereas all three primer pairs yielded an RT-PCR product of the predicted size when RNA from the WT strain was used, indicating that, as expected, the transposition resulted in a polar mutation and transcription did not arise from any alternative downstream promoter (Fig. 5B, lanes 5 to 10).
Regulation of adcR expression in response to changes in environmental conditions.
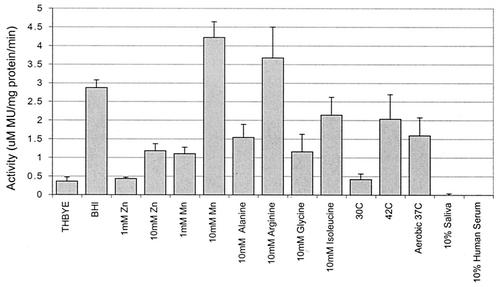
When S. gordonii adcR::Tn917-lac was grown on solid media containing X-Gal, a blue colony phenotype was produced, indicating that the Tn917-lac transposon was inserted as an in-frame fusion within adcR. Therefore, it was possible to identify and examine environmental factors that regulate the expression of adcR by measuring the β-galactosidase activity of adcR::Tn917-lac grown in medium supplemented with a variety of nutrients and under various conditions. Since the adcR in S. pneumoniae was reported to be involved in zinc and/or manganese transport (9, 15), the role of adcR in S. gordonii was examined by using the transcriptional fusion present in this adcR::Tn917-lac biofilm-defective mutant. Results from β-galactosidase assays show that the expression of adcR was increased significantly in the presence of 10 mM manganese and was increased to a lesser extent by the presence of 10 mM zinc (Fig. 6). Initial screening of the adcR::Tn917-lac mutant found that growth in BHI induced the expression approximately threefold compared to growth in THBYE. Expression was higher when cells were grown in THBYE at 42°C, under aerobic conditions, and in THBYE supplemented with manganese, zinc, alanine, arginine, glycine, or isoleucine (Fig. 6). These results identified manganese, zinc, arginine, glycine, alanine, and isoleucine and growth at 42°C and aerobic growth at 37°C as potential inducers of adcR expression.
FIG. 6.
Effect of various growth conditions on adcR expression. The β-galactosidase activities of the adcR-lacZ transcriptional fusion in the biofilm-defective S. gordonii adcR::Tn917-lac mutant are shown. Cultures were grown at 37°C under anaerobic conditions unless specified otherwise. The growth conditions examined were THBYE, BHI, and THBYE containing the following: whole saliva, human serum, Zn, Mn, and various amino acids. Cells were also grown in THBYE at 30 and 42°C and under aerobic conditions at 37°C. The β-galactosidase activities of sonic extracts from each of the cultures were quantified by using MUG (28). All assays were performed in triplicate, and mean values and standard deviations are shown.
There was minimal increase in β-galactosidase activity when cells were grown in media supplemented with 10% saliva, human serum, or THBYE. These conditions may simply reflect the concentration of extracellular manganese in saliva (50 to 176 μg/liter [20]), serum (0.1 to 2.9 μg/liter [35]), and THBYE (not known). The explanation for the enhanced β-galactosidase activity when adcR::Tn917-lac was grown in the presence of certain amino acids at 42°C and under aerobic conditions is unclear at this time.
Subsequent β-galactosidase activity assays were performed with adcR::Tn917-lac grown in the minimal, chemically defined BM supplemented with increasing concentrations of manganese. There was no increase in activity when the manganese concentrations were low (50 to 200 μM). However, when the concentration of manganese was increased to above 1 mM, adcR expression was induced, as indicated by an approximately twofold increase in β-galactosidase activity (data not shown). These results show that the adc operon is probably involved in the transport and recognition of high concentrations of extracellular manganese and suggest that adcR may be a transcriptional regulator. This is in contrast to the sca operon in S. gordonii, which is also involved in manganese homeostasis. The expression of scaR was induced at an extracellular manganese concentration of 0.1 μM and was repressed when the concentration was increased to 50 μM, suggesting that the scaR gene in the sca operon encodes a positive regulator in S. gordonii (31).
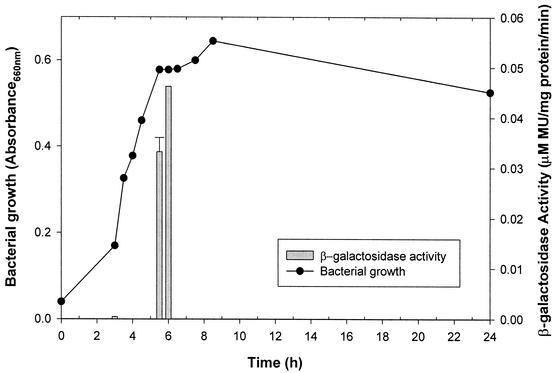
Several studies have suggested that the biofilm-associated phenotype is induced by slow growth and under duress (11, 41, 70). When the β-galactosidase activity of adcR::Tn917-lac was assayed at different growth phases, adcR was found to be expressed only during the late exponential phase of growth (Fig. 7).
FIG. 7.
Growth and the β-galactosidase activity of the biofilm-defective S. gordonii adcR::Tn917-lac mutant at 0, 3, 3.5, 4. 4.5, 5, 5.5, 6, 6.5, 7, 7.5, 8.5, and 24 h are shown. Cultures were grown at 37°C under anaerobic conditions in THBYE. The β-galactosidase activity of sonic extracts from each time point was quantified with MUG (28). All assays were performed in triplicate, and mean values and standard deviations are shown. MU, 4-methyl-umbelliferone.
Construction and characterization of an adcR insertion mutation.
In order to clarify the role of adcR in producing the biofilm-defective phenotype identified in adcR::Tn917-lac, a nonpolar insertional inactivation of adcR was done by allelic exchange with an antibiotic cassette using PCR ligation mutagenesis (37). The adcR gene (from amino acids 13 to 144) on the S. gordonii chromosome was replaced with either spec or kan, which encodes resistance to spectinomycin or KAN, respectively (see Materials and Methods). In both cases, transformation with the ligation mixture of PCR products resulted in viable mutations. Transformants were visible on BHI agar after 24 h of growth under anaerobic conditions for the adcR::Specr mutation, while transformants with the adcR::Kanr mutation became visible only after 48 h. PCR confirmed that integration of the spec or kan gene in adcR in the chromosome had occurred as predicted (data not shown).
The effects of insertional inactivations on biofilm formation and competence were assessed. Biofilm formation of the adcR::Specr mutant (A575 ± standard deviation = 1.07 ± 0.07) and the adcR::Kanr mutant (A575 ± standard deviation = 0.84 ± 0.10) was similar to that of the adcR::Tn917-lac mutant (A575 ± standard deviation = 1.03 ± 0.53), confirming that the adcR is involved in biofilm formation. All three adcR mutants were defective in biofilm formation when compared to the WT strain (A575 ± standard deviation = 3.13 ± 0.33). The biofilm formation of both the adcR::Specr and adcR::Kanr mutants observed with phase-contrast microscopy was similar to that of the adcR::Tn917-lac mutant (data not shown). Results from transformation of the WT, adcR::Tn917-lac, adcR::Specr, and adcR::Kanr strains with equal amounts of ligated DNA (amplified adcB flanking regions ligated with either the Specr or Kanr cassette) demonstrate that competence was defective in the adcR::Tn917-lac, adcR::Kanr, and adcR::Specr mutants. Therefore, a functional adc operon is probably required for competence in S. gordonii.
In growth experiments, the growth rates and final yields of the adcR::Specr and adcR::Kanr mutants were found to be similar to those of the WT strain, indicating that these mutations did not affect the growth rate or yield. In contrast, the adcR::Tn917-lac mutant grew at a lower rate than did the WT strain and the two nonpolar adcR mutants, indicating that a polar mutation in adcR reduced the growth rate. However, the final yield of the adcR::Tn917-lac mutant after 24 h appeared to be the same as that of the WT, adcR::Specr, and adcR::Kanr strains (data not shown).
When RT-PCRs with primers specific for regions spanning adcR to adcA, adcC to adcA, and within the adcA gene (Fig. 5A) were performed with RNA isolated from the WT or adcR::Specr mutant strains, RT-PCR products of approximately 1,624, 983, and 1,449 bp (Fig. 5B, lanes 5 to 7 and 11 to 13) were obtained. These span the ORFs adcR to -A, adcC to -A, and adcA and coincide with their respective predicted amplicon sizes. Amplicons of identical sizes were obtained when RT-PCR was performed with RNA isolated from the adcR::Kanr mutant (data not shown); however, no PCR products were obtained when RNA from adcR::Tn917-lac was used (Fig. 5B, lanes 8 to 10). All three primer pairs used in the RT-PCR above were also used in standard PCR amplifications with S. gordonii WT DNA, which produced PCR products with the predicted sizes (data not shown). These results indicate that both S. gordonii adcR::Specr and adcR::Kanr mutants are nonpolar. These results suggest that the repressor AdcR is probably synthesized as an active repressor de novo, because both polar and nonpolar mutations of adcR resulted in a biofilm-defective phenotype. If it was an inactive repressor, then there would not be a change in phenotype. Since the data suggest that AdcR is synthesized as an active repressor, it may also act on a remote region(s) on the S. gordonii chromosome and modulate biofilm formation on abiotic surfaces by an unknown mechanism.
DISCUSSION
In order to understand the environmental cues and the resulting phenotypic responses that influence S. gordonii biofilm formation and regulate biofilm-associated genes, the gram-positive transposon Tn917, which contains the promoterless E. coli lacZ reporter gene (14), was used to isolate and analyze a biofilm-associated gene in S. gordonii. Molecular analyses of a biofilm-defective S. gordonii::Tn917-lac mutant identified showed that a gene required for biofilm formation has a high degree of homology with adcR, part of the adc operon in S. pneumoniae (9, 16). The S. gordonii adc operon, consisting of adcR, adcC, adcB, and adcA, was homologous to the adc operon of S. pneumoniae, which belongs to the family of ABC transporters and plays a role in genetic competence and zinc and/or manganese transport in S. pneumoniae. RT-PCR was used to confirm that adcRCBA were also cotranscribed in S. gordonii.
Metal ion homeostasis in bacteria is generally controlled during uptake by transcriptional regulators and is mediated by ABC-type permeases (9, 29). When the possible regulation of adcR by various environmental factors was examined, two- to fourfold increases in β-galactosidase activity were observed when adcR::Tn917-lac was grown in the presence of 1 to 10 mM manganese when compared to growth with 100 μM manganese present in BM. Moderate increases in β-galactosidase activity were also observed when the adcR::Tn917-lac mutant was grown in the presence of zinc or some amino acids, in BHI, under aerobic conditions, or at 42°C. These results indicate that the adcR gene product plays an adaptive role in S. gordonii biofilm formation, probably by modulating adcR expression in response to specific environmental conditions. Moreover, the probable specificity for manganese is supported by the observation that, in the adcR::Tn917-lac mutant, manganese is a more potent activator of adcR expression than zinc.
Although the S. pneumoniae adc operon was reported to be involved in genetic competence and metal ion transport, specifically zinc (9, 16), our studies indicate that the adc operon in S. gordonii is mainly involved in the transport of manganese and competence, but actual transport experiments are required to support our hypothesis. Conclusions from studies of the S. pneumoniae adc operon were based on the slow growth of an adcC mutant in a chemically defined medium when compared to that of the WT strain and the restoration of growth of the mutant to WT levels by the addition of 0.8 μM ZnSO4 or ZnCl2. Interestingly, the growth of the adcC mutant was inhibited by the addition of 6 μM MnSO4, and this inhibition was suppressed by the addition of ZnSO4 or ZnCl2 (15). As the disruption of the adc operon in S. pneumoniae was made in adcC, the precise role of metal transport in the adcC mutant may have been obscured. On the other hand, the adc operon in S. gordonii appears to be involved in regulating gene expression primarily in the presence of high extracellular levels of manganese and to a lesser extent of zinc. More importantly, phylogenetic analysis demonstrates that AdcR is more closely related to repressors that modulate manganese transport, and these proteins are only distantly homologous to other metalloregulators.
Phylogenetic analysis, amino acid alignments, and the genetic organization of adc operons in a number of streptococci for which DNA sequences are available in the microbial genome databases indicate that there are at least two distinct groups. In one group, all four genes are contiguous and cotranscribed as an adcRCBA operon and the AdcA permease is a lipoprotein. S. gordonii and S. pneumoniae, both naturally transformable bacteria, are in this group. S. mitis may also be a member of this group, as AdcA is also a lipoprotein, but whether adcA is contiguous with the adc operon can only be determined when sequences of this region are determined. In the second group, adcA is noncontiguous with the adc operon and AdcA is not a lipoprotein but still contains a putative signal peptide. This group contains S. pyogenes, S. agalactiae, S. equi, and S. mutans.
Several studies have shown that the expression of biofilm-associated genes is growth phase dependent (11, 70). Similarly, expression of adcR was only observed during the later stages of the exponential growth phase in S. gordonii. In E. coli, RpoS mediates the synthesis of more than 50 proteins during the transition from the exponential to the stationary phase of growth, starvation, and osmotic and oxidative stress (41). Both adcR and rpoS were expressed during the late exponential phase, but the adcR mutant has a biofilm-defective phenotype, while a mutation in rpoS in E. coli resulted in enhanced biofilm formation (11). The data suggest that AdcR in S. gordonii is a positive regulator of biofilm-associated genes, in contrast to RpoS in E. coli, which is a negative regulator of biofilm-associated genes (11). This difference may explain the different phenotypes exhibited by these two mutants.
Bacteria have developed sophisticated acquisition systems to scavenge essential metals from the environment, including constitutively expressed or inducible low- and high-affinity transport systems for chelated or free metals (29). Manganese is an essential cofactor for a variety of enzymes, some of which are critical for bacterial growth and survival under oxidative stress, and is an important cofactor for enzymatic antioxidant defenses of bacteria, such as catalase and superoxide dismutase (8, 29, 30, 51). A number of proteins, such as catalases, oxidoreductases, and transferases have evolved a natural preference for manganese, using this metal for a regulatory or catalytic role. Manganese is also a tightly bound constituent of a few metalloenzymes involved in the metabolism of nitrogen and oxygen and in the modulation of signal transduction pathways (8, 29, 30).
S. gordonii has at least three different genes that are involved in manganese homeostasis. The sca and adc operons appear to be high-affinity manganese transport systems that recognize low and high concentrations of extracellular manganese, respectively, and are necessary for growth, genetic transformation, and biofilm formation (31, 34). Although both sca and adc operons are ABC solute-binding operons that share a high degree of homology, there are distinct differences between ScaA and AdcA, the lipoproteins responsible for solute binding. ScaA does not contain the histidine-rich metal-binding domain that is present in AdcA. The adcR gene is the first gene in the adc operon upstream of adcA, while the scaR gene is distant from the other genes of the sca operon elsewhere in the chromosome (31). Phylogenetic analysis revealed that scaR and adcR belong to two distinct classes of regulatory proteins that regulate manganese transport in S. gordonii. While there is clear evidence that Sca-like proteins are implicated in bacterial virulence in animal model studies (6), similar studies have not been done to demonstrate the role of Adc-like proteins in pathogenesis. Clearly these two operons are involved in biofilm formation and genetic competence. Although there is evidence to suggest that a third, unidentified manganese transport system exists in S. gordonii and is mediated by proton-dependent ATPase (34), whether this plays a role in biofilm formation, genetic competence, or virulence is yet to be determined. However, mutation in mntH, which encodes a P-type ATPase that is involved in manganese transport, does not affect virulence of Mycobacterium tuberculosis in a mouse model of tuberculosis (17), suggesting that not all manganese transport systems in bacteria are involved in virulence and perhaps in biofilm formation and competence.
The adcR gene is cotranscribed with the rest of the binding-lipoprotein-dependent transport genes adcCBA, and both a polar mutation and a nonpolar mutation of adcR have been shown to be biofilm defective, indicating that AdcR is an active repressor when synthesized de novo. Also, the biofilm-defective phenotype of the nonpolar mutation of adcR strongly suggests that the active adcR gene acting on a remote region(s) is essential to the biofilm phenotype observed on an abiotic surface. In addition an active repressor could abolish the transcription of the adc operon when a sufficient amount of AdcR is synthesized, thus regulating its own synthesis. Only when a sufficient amount of manganese is acquired by other manganese transport systems can the AdcR may be made inactive and can adc transcription be continued. Since the adc operon responds to high levels of extracellular manganese, S. gordonii can acquire the manganese through other transport genes (ScaA and P-type ATPase). Once AdcR is made inactive, the functional adc operon can respond to the high level of extracellular manganese.
Why would S. gordonii possess a system to sense a high level of manganese when its natural (saliva-bathed oral cavity) or its opportunistic habitat (human serum) is considered to be low in manganese (29, 30)? It may be that, when a bacterium like S. gordonii encounters high level of extracellular manganese, it might be an indication for it to switch off biofilm-associated genes through the inactivation of the AdcR repressor protein. Therefore, the AdcR/adc operon couple could be a sensory mechanism to enable the switch from a sessile to a planktonic phenotype, thus facilitating the dispersal and spread of the bacterium. This type of modulation by manganese may be involved in the pathogenicity of S. pneumoniae (3), Streptococcus parasanguis (6), S. enterica (5) and Yersinia pestis (2), all of which have conserved, ABC-type binding-protein-dependent, manganese acquisition systems that are implicated in virulence.
It is of interest that recent studies have identified metal ions as modulators of bacterial differentiation and growth. In addition to manganese, studies have shown that lactoferrin, a component of the innate immunity of mucosal surfaces in humans, chelates iron, which blocks biofilm development by the opportunistic pathogen P. aeruginosa (58). Likewise, the recently discovered autoinducer (AI-2) that is produced by numerous bacterial species allows bacterial populations to coordinate gene expression in a variety of developmental processes, such as bioluminescence, virulence, antibiotic production, and biofilm development, and is a furanosyl borate diester, indicating a potential role for boron in bacterial differentiation (7).
The role of manganese homeostasis in S. gordonii Challis biofilm formation provides insight into how metal-binding genes may be involved in biofilm differentiation and is a significant indicator of how changes in the environment can elicit a developmental change in bacteria. Since S. gordonii resides in environments such as human saliva or serum, which are low in manganese, it is conceivable that there may be other key extracellular cues, such as the presence of other metal ions that may regulate biofilm formation and virulence in S. gordonii. Blocking such transporter functions in bacteria may be an effective therapeutic strategy in controlling bacterial biofilms such as dental plaque and opens new avenues for therapy that do not involve immunological intervention such as vaccination.
Acknowledgments
This work was supported by Public Health Service grant RO1-DE13328 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 3.Berry, A. M., and J. C. Paton. 1996. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 64:5255-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, E., I. Bergevin, D. Malo, P. Gros, and M. F. Cellier. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnette-Curley, D., V. Wells, H. Viscount, C. L. Munro, J. C. Fenno, P. Fives-Taylor, and F. L. Macrina. 1995. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect. Immun. 63:4669-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 8.Christianson, D. W. 1997. Structural chemistry and biology of manganese metalloenzymes. Prog. Biophys. Mol. Biol. 67:217-252. [DOI] [PubMed] [Google Scholar]
- 9.Claverys, J. P. 2001. A new family of high-affinity ABC manganese and zinc permeases. Res. Microbiol. 152:231-243. [DOI] [PubMed] [Google Scholar]
- 10.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corona-Izquierdo, F. P., and J. Membrillo-Hernandez. 2002. A mutation in rpoS enhances biofilm formation in Escherichia coli during exponential phase of growth. FEMS Microbiol. Lett. 211:105-110. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 13.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cvitkovitch, D. G., J. A. Gutierrez, J. Behari, P. J. Youngman, J. E. Wetz, P. J. Crowley, J. D. Hillman, L. J. Brady, and A. S. Bleiweis. 2000. Tn917-lac mutagenesis of Streptococcus mutans to identify environmentally regulated genes. FEMS Microbiol. Lett. 182:149-154. [DOI] [PubMed] [Google Scholar]
- 15.Dintilhac, A., G. Alloing, C. Granadel, and J. P. Claverys. 1997. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and manganese resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25:727-739. [DOI] [PubMed] [Google Scholar]
- 16.Dintilhac, A., and J. P. Claverys. 1997. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res. Microbiol. 148:119-131. [DOI] [PubMed] [Google Scholar]
- 17.Domenech, P., A. S. Pym, M. Cellier, C. E. Barry III, and S. T. Cole. 2002. Inactivation of the Mycobacterium tuberculosis Nramp orthologue (mntH) does not affect virulence in a mouse model of tuberculosis. FEMS Microbiol. Lett. 207:81-86. [DOI] [PubMed] [Google Scholar]
- 18.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 19.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 20.Duggal, M. S., H. S. Chawla, and M. E. Curzon. 1991. A study of the relationship between trace elements in saliva and dental caries in children. Arch. Oral Biol. 36:881-884. [DOI] [PubMed] [Google Scholar]
- 21.Durack, D. T. 1995. Prevention of infective endocarditis. N. Engl. J. Med. 332:38-44. [DOI] [PubMed] [Google Scholar]
- 22.Froeliger, E. H., and P. Fives-Taylor. 2001. Streptococcus parasanguis fimbria-associated adhesin Fap1 is required for biofilm formation. Infect. Immun. 69:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganeshkumar, N., M. Song, and B. C. McBride. 1988. Cloning of a Streptococcus sanguis adhesin which mediates binding to saliva-coated hydroxyapatite. Infect. Immun. 56:1150-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genevaux, P., S. Muller, and P. Bauda. 1996. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 142:27-30. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Barca, E., A. Fernandez-Sevilla, J. Carratala, A. Granena, and F. Gudiol. 1996. Prospective study of 288 episodes of bacteremia in neutropenic cancer patients in a single institution. Eur. J. Clin. Microbiol. Infect. Dis. 15:291-296. [DOI] [PubMed] [Google Scholar]
- 26.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 28.Honeyman, A. L., C. K. Cote, and R. Curtiss III. 2002. Construction of transcriptional and translational lacZ gene reporter plasmids for use in Streptococcus mutans. J. Microbiol. Methods 49:163-171. [DOI] [PubMed] [Google Scholar]
- 29.Horsburgh, M. J., S. J. Wharton, M. Karavolos, and S. J. Foster. 2002. Manganese: elemental defence for a life with oxygen? Trends Microbiol. 10:496-501. [DOI] [PubMed] [Google Scholar]
- 30.Jakubovics, N. S., and H. F. Jenkinson. 2001. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology 147:1709-1718. [DOI] [PubMed] [Google Scholar]
- 31.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (manganese) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 32.Kern, W., E. Kurrle, and T. Schmeiser. 1990. Streptococcal bacteremia in adult patients with leukemia undergoing aggressive chemotherapy. Infection 18:138-145. [DOI] [PubMed] [Google Scholar]
- 33.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional rsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolenbrander, P. E., R. N. Andersen, R. A. Baker, and H. F. Jenkinson. 1998. The adhesion-asociated sca operon in Streptococcus gordonii encodes an inducible high-affinity ABC transporter for manganese uptake. J. Bacteriol. 180:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krachler, M., E. Rossipal, and D. Micetic-Turk. 1999. Concentrations of trace elements in sera of newborns, young infants, and adults. Biol. Trace Elem. Res. 68:121-135. [DOI] [PubMed] [Google Scholar]
- 36.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 37.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 38.Lemos, J. A., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, Y.-H., P. C. Y. Lau, N. Tang, G. Svensäter, R. P. Ellen, and D. G. Cvitkovitch. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184:6333-6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, Y.-H., N. Tang, M. B. Aspiras, P. C. Y. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon in Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 42.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of hexosamine-containing polysaccharide intracellular adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mireles, J. R., II, A. Toguchi, and R. M. Harshey. 2001. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183:5848-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24:267-272. [DOI] [PubMed] [Google Scholar]
- 46.Oosthuizen, M. C., B. Steyn, J. Theron, P. Cosette, D. Lindsay, A. Von Holy, and V. S. Brozel. 2002. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl. Environ. Microbiol. 68:2770-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 49.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 50.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signalling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Posey, J. E., J. M. Hardham, S. J. Norris, and F. C. Gherardini. 1999. Characterization of a manganese-dependent regulatory protein, TroR, from Treponema pallidum. Proc. Natl. Acad. Sci. USA 96:10887-10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 53.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejenne, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csdD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogers, J. D., R. J. Palmer, Jr., P. E. Kolenbrander, and F. A. Scannapieco. 2001. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69:7046-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 57.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552-555. [DOI] [PubMed] [Google Scholar]
- 59.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 60.Sutcliffe, I. C., and D. J. Harrington. 2002. Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology 148:2065-2077. [DOI] [PubMed] [Google Scholar]
- 61.Svensater, G., J. Welin, J. C. Wilkins, D. Beighton, and I. R. Hamilton. 2001. Protein expression by planktonic and biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 205:139-146. [DOI] [PubMed] [Google Scholar]
- 62.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 63.Tremoulet, F., O. Duche, A. Namane, B. Martinie, and J. C. Labadie. 2002. Comparison of protein patterns of Listeria monocytogenes grown in biofilm or in planktonic mode by proteomic analysis. FEMS Microbiol. Lett. 210:25-31. [DOI] [PubMed] [Google Scholar]
- 64.van der Meer, J. T., J. Thompson, H. A. Valkenburg, and M. F. Michel. 1992. Epidemiology of bacterial endocarditis in The Netherlands. I. Patient characteristics. Arch. Intern. Med. 152:1863-1868. [DOI] [PubMed] [Google Scholar]
- 65.Vats, N., and S. F. Lee. 2001. Characterization of a copper-transport operon, copYAZ, from Streptococcus mutans. Microbiology 147:653-662. [DOI] [PubMed] [Google Scholar]
- 66.Wang, S.-Y., J. Lauritz, J. Jass, and D. L. Milton. 2002. A ToxR homolog from Vibrio anguillarum serotype O1 regulates its own production, bile resistance, and biofilm formation. J. Bacteriol. 184:1630-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 70.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]