Abstract
Adenocarcinomas of the prostate can be categorized into tumor grades based on the extent to which the cancers histologically resemble normal prostate glands. Because grades are surrogates of intrinsic tumor behavior, characterizing the molecular phenotype of grade is of potential clinical importance. To identify molecular alterations underlying prostate cancer grades, we used microdissection to obtain specific cohorts of cancer cells corresponding to the most common Gleason patterns (patterns 3, 4, and 5) from 29 radical prostatectomy samples. We paired each cancer sample with matched benign lumenal prostate epithelial cells and profiled transcript abundance levels by microarray analysis. We identified an 86-gene model capable of distinguishing low-grade (pattern 3) from high-grade (patterns 4 and 5) cancers. This model performed with 76% accuracy when applied to an independent set of 30 primary prostate carcinomas. Using tissue microarrays comprising >800 prostate samples, we confirmed a significant association between high levels of monoamine oxidase A expression and poorly differentiated cancers by immunohistochemistry. We also confirmed grade-associated levels of defender against death (DAD1) protein and HSD17β4 transcripts by immunohistochemistry and quantitative RT-PCR, respectively. The altered expression of these genes provides functional insights into grade-associated features of therapy resistance and tissue invasion. Furthermore, in identifying a profile of 86 genes that distinguish high- from low-grade carcinomas, we have generated a set of potential targets for modulating the development and progression of the lethal prostate cancer phenotype.
Keywords: carcinoma, monoamine oxidase A, microarray, expression profile
The prognosis and choice of therapy for prostate cancer is based primarily on three parameters obtained at the time of diagnosis: clinical stage, serum prostate-specific antigen (PSA), and the Gleason score of the cancer (1). The Gleason grading system, which is based on microscopic tumor architecture, consists of five histological patterns that annotate cancers into categories exhibiting well differentiated (pattern 1) to poorly differentiated (pattern 5) features (2, 3). A number from 1 to 5 is assigned to the most prevalent pattern. A second number, also from 1 to 5, is assigned to the second most prevalent pattern. The Gleason grade, which is the sum of these two numbers, has a value between 2 and 10. In current practice, the vast majority of prostate cancers have a Gleason score of ≥6 (4, 5). Hence, tumors composed of patterns 3, 4, and/or 5 are considered clinically significant. The reporting of individual Gleason patterns is not a trivial distinction, because the amount of pattern 4 and the presence of any pattern 5 has been highly correlated with probability of cancer dissemination, response to therapy, disease outcome, patient-management decisions, and clinical-trial enrollment (6, 7). Numerous studies have demonstrated a direct correlation between Gleason score and clinical measurements of disease outcome, including death due to tumor within 15 years and likelihood of remaining free of biochemical evidence of disease recurrence after either definitive, potentially curative radical prostatectomy or radiation therapy (8). Although different Gleason patterns are histologically distinctive, the molecular features underlying these tumor phenotypes are not precisely defined. In this study, we sought to characterize the molecular profile of prostate carcinomas of specific Gleason patterns. Tumor cell transcript levels were used to develop a model capable of distinguishing the low-grade (pattern 3) from high-grade (patterns 4 and 5) tumors. We validated the predictive power of the model by using an independent set of primary prostate cancers and confirmed grade-associated differences in protein-expression levels using a third sample cohort. Together, these results identify both consistent and divergent features of the molecular framework that underlies the histological classification of cancer grade.
Results and Discussion
Gene-Expression Profiles Associated with Prostate Cancer Grades.
To identify molecular alterations correlating with histological tumor grade, we used laser-capture microdissection to exclusively acquire the epithelial cell component of prostate carcinoma foci corresponding to individual Gleason pattern 3, 4, or 5 cancers. After linear amplification, gene-expression alterations in tumor cells were measured by cDNA microarray hybridizations in a head-to-head fashion against patient-matched microdissected benign secretory epithelial cells. A total of 121 benign and neoplastic samples from 59 radical prostatectomies and 30 prostate needle core biopsies contributed to the analysis.
To assess the generalized applicability and consistency of the methods, we first sought to identify consistent prostate cancer-associated transcript alterations irrespective of grade. Using microdissected epithelium from radical prostatectomy samples, we identified 736 genes with altered expression levels between benign and neoplastic epithelium (false discovery rate <0.01%). This cohort included several genes previously reported to be differentially expressed in prostate cancers, such as hepsin, AMACR, and CAMKK2 (Fig. 1A) (9–12). These findings provided validation that our methods replicated results of earlier expression-profiling studies, despite using the very small sample quantities obtained by laser microdissection and subjecting the RNA from these samples to amplification and transcript quantitation by microarray analysis.
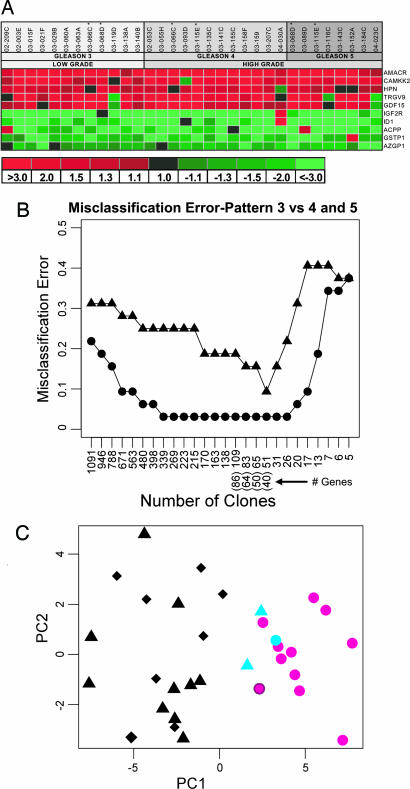
Fig. 1.
Gene-expression changes associated with prostate cancer grade. (A) Genes differentially expressed between microdissected benign and neoplastic prostate epithelium were identified in a supervised analysis of transcript profiles of 32 matched samples from 29 individuals. A Statistical Analysis Of Microarrays (SAM) one-sample t test comparing cancerous epithelium to matched benign epithelium across all Gleason patterns identified 736 differentially expressed genes (false discovery rate ≤0.05%). Cancer-associated transcript alterations for several genes with previously described alterations in prostate carcinoma are shown. (B) Identification of gene-expression alterations predictive of Gleason pattern. PAM analysis of gene-expression profiles generated from microdissected Gleason pattern 3, 4, and 5 cancers. Circles represent the training error. Triangles represent the leave-one-out cross-validation error. The x axis shows the number of array clones (and corresponding unique genes) used for classification. (C) Principal-components analysis of grade-specific prostate cancers using the 40-gene (51-clone) Gleason pattern classifier. For each sample, the score on the first (x axis) and second (y axis) principal component is plotted. Gleason pattern 3 (circle) samples are generally distinct from Gleason pattern 4 and 5 sample space (triangle and diamond, respectively). Pattern 4 and 5 samples were intermixed, indicating a high degree of molecular similarity. Three misclassified samples (under cross-validation of the 40-gene model) between pattern 3 and pattern 4/5 cancers are denoted in blue.
To identify gene-expression alterations that associated with specific Gleason patterns, we used a supervised-learning approach, Prediction Analysis for Microarrays (PAM) that represents a modification of standard nearest-neighbor classification (13). Models comprising 40, 64, and 86 genes were able to partition Gleason 3 vs. Gleason 4 and 5 cancers with 90%, 84%, and 81% accuracy (under leave-one-out cross-validation) (Fig. 1B). We were unable to identify a cohort of genes that could distinguish between pattern 4 and 5 cancers with sufficiently high accuracy to be useful, suggesting a high degree of similarity between these cancer histologies or substantial molecular heterogeneity in one or both of these groups. Therefore, we grouped pattern 4 and 5 cancers together for subsequent comparisons. Principal-components analysis of the 40-gene model demonstrated segregation of cancers according to histological grade, with only one pattern 4/5 cancer misclassified as a pattern 3, and two pattern 3 cancers misclassified as pattern 4/5 (Figs. 1C and 2A). Rereview of the histology and the specific composition of cells acquired by laser-capture microdissection did not alter the histological classification of these samples. Although Gleason patterns can be reproducibly categorized (14), there are clear examples of heterogeneity in clinical behavior within each histological category (15). Further studies are required to determine whether molecular signatures within a specific cancer pattern exhibit prognostic power.
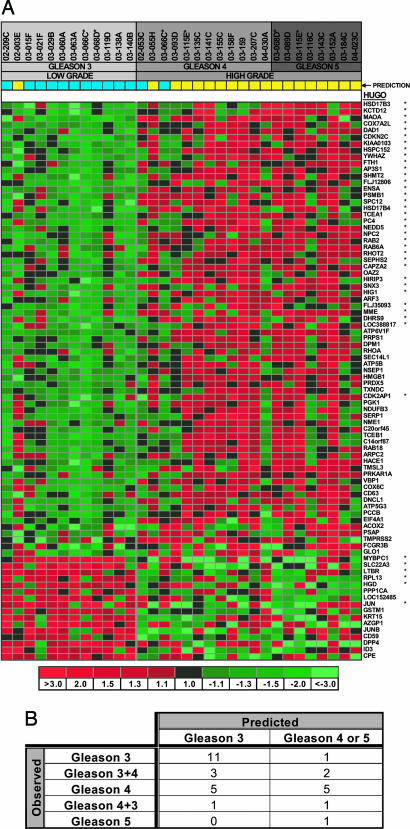
Fig. 2.
Genes associated with specific Gleason pattern prostate cancers. (A) Shown are the 86 genes exhibiting the greatest discriminatory power to partition cancer grades using an independent set of prostate cancer tissues. Genes are ordered according to t test score. The 40 genes used in the original PAM model are denoted by asterisks. (B) Application of the 86-gene Gleason classification model to an independent test set of 23 prostate cancers of singular pattern (e.g., 3 + 3) or exclusively high grade (4 + 4 or 4 + 5) resulted in an overall classification accuracy of 74%. The predicted pattern of 7 additional cancers of mixed low grade and high grade (3 + 4 and 4 + 3) varied between pattern 3 and pattern 4/5. The functional properties of grade-classifying genes are annotated in Table 1, which is published as supporting information on the PNAS web site.
Performance Characteristics of the Gleason Grade Classifier.
To determine the general applicability of the Gleason pattern classifier, we generated and analyzed an independent prostate cancer gene-expression data set derived from microdissected matched benign and neoplastic epithelium from 30 prostate needle biopsies. We measured expression alterations by microarray hybridization and assessed the models that provided the greatest discriminatory power in the original prostatectomy samples. The classifier comprised 86 genes (109 clones), including the 40 genes with 90% discriminatory power in the original radical prostatectomy cases performed with highest accuracy, producing a 76% overall correct classification rate (P = 0.056, Fisher’s exact test) (Fig. 2B) on this independent sample set. Of the 12 cancers histologically called Gleason pattern 3, all but one was correctly classified. Of the cancers with a histological classification of 4 or 4 + 5, 6 of 11 were correctly identified (Fig. 2B). As expected, microdissected samples recognized to contain mixed grades of 3 + 4 or 4 + 3 cancer cells were divided between pattern 3 or pattern 4 molecular categories. These results suggest that pattern 3 cancers exhibit relatively consistent molecular alterations, whereas cancers with histological features of patterns 4 and 5 are more diverse and, in some cases, exhibit molecular features common to pattern 3 cancers.
We next attempted to compare our results with other reports correlating prostate tumor gene expression with grade. These comparisons were problematic, because all publicly available prostate cancer data sets generated expression profiles and classifiers based on Gleason-sum scores rather than individual patterns. For example, a given Gleason-sum score 8 cancer could be composed of patterns 5 + 3 or 4 + 4. Reports by Singh et al. (11) and LaPointe et al. (12) identified 29 and 42 genes, respectively, that statistically associated with Gleason-sum score. However, only three genes, SPARC, BGN, and COL1A2, were in common between these analyses. None of the genes comprising our classifier were present in the published grade-associated gene sets. An additional contributor to these discrepancies concerns the use of samples that, although enriched for tumor, also contain variable amounts of tumor-adjacent stroma, benign glands, and inflammatory cells. Alterations in the expression of genes in tumor-adjacent stroma is well recognized (16). In the context of these studies, the expression of SPARC has been shown to be up-regulated in the stroma adjacent to non-small-cell lung cancers (17) and pancreatic carcinomas (18) and may be similarly overexpressed in prostate cancer-adjacent stroma. A final source of discrepancies between studies involves technical variables, attributable, for example, to the use of different platforms (cDNA arrays vs. Affymetrix chips) and the necessity of amplifying RNA before hybridization.
Although the grade-discriminatory gene sets derived from our analyses and those of prior studies exhibited no concordance, we explored the ability of the Gleason grade classifier to partition prostate cancers in available data sets that were composed of singular grade combinations such as 3 + 3 and 4 + 4. Of 86 genes used in our classifier, 77 were present in the study by LaPointe et al., with sufficient data to allow for grade-associated analyses. Unsupervised hierarchical clustering of samples using these 77 genes produced two major clusters generally partitioned according to grade. Of 26 tumors with Gleason 3 + 3 histology, 19 (73%) were correctly classified, as were 11 of 15 tumors (73%) with 4 + 4 or 4 + 5 Gleason patterns.
Characteristics and Confirmation of Gleason Grade-Associated Molecular Alterations.
Several genes that we found to be associated with prostate cancer cell differentiation patterns have been linked to cancer grade or stage classifications. A study of breast carcinomas found a statistical relationship between increased expression of the antioxidant enzyme peroxiredoxin 5 (PRDX5) and larger tumor size, positive lymph node status, and shorter survival (19). Elevated levels of the transcription elongation factor TCEB1/Elongin C are associated with advanced androgen-independent prostate cancers (20), and changes in the subcellular localization of the membrane metalloendopeptidase MME/CD10 expression are linked to prostate cancer grade (21). Immunohistochemical analysis of the Akt-regulated transcriptional repressor NSEP/YB-1 demonstrates a strong positive correlation with Gleason grade (22). NSEP/YB-1 may influence cancer progression through multiple mechanisms that include enhancement of anchorage-independent cell growth and up-regulation of the p-glycoprotein multidrug-resistance protein (22, 23).
Many genes exhibiting altered expression in high- relative to low-grade prostate cancers possess characteristics supporting potential roles in cancer cell survival, invasion, or metastasis. Of the 86 genes we used for grade classification, 6 are located on chromosome 8. Of these, 4 map to 8q21–23, a region shown to be amplified in >40% of primary prostate cancers in association with higher cancer grades (24). The expression of 4 genes involved in aspects of sex hormone metabolism or regulation were increased in higher-grade cancers, including two members of the 17β-hydroxysteroid dehydrogenase (HSD) family, HSD17β3 and HSD17β4. The 3-α-hydroxysteroid dehydrogenase DHRS9 is capable of catalyzing the conversion of 3-α-androstanediol to dihydrotestosterone (25) and also contributes to the β-oxidation of fatty acids. The expression of HSD17β4, also known as D-bifunctional protein, was recently shown to be up-regulated in neoplastic prostate epithelium (26). We confirmed this result using quantitative RT-PCR analysis of microdissected prostate epithelium and further confirmed the grade-associated elevation of HSD17β4 transcripts found in the microarray-based analyses (see Supporting Materials and Methods and Fig. 4, which are published as supporting information on the PNAS web site). Together, these results suggest the emergence of mechanisms that favor intracrine utilization of alternative androgen precursors by tumor cells. Five members of the ras homologue gene family RHOT2, RHOA, RAB2, RAB6A, and RAB18 were up-regulated in high-grade cancers. The RAB18-related gene, RAB25, was recently shown to influence the malignant potential of breast and ovarian cancers, suggesting that members of this gene family may be general mediators of epithelial cancer progression (27). The products of RHOA, CAPZA2, HMGB1, NM23H1, CD63, and Saposin C have all been shown to directly modulate tumor progression through enhanced cell motility, invasion, or metastasis.
Immunohistochemical (IHC) Analysis of Gleason Grade-Associated Protein Expression.
In current clinical practice, diagnostic and prognostic cancer markers are most commonly assessed by using IHC quantitation. We sought to determine whether grade-associated alterations identified through transcript profiling would have corresponding changes in protein levels discernable by IHC. Although immunoperoxidase histochemistry is an imprecise method of protein quantification, it is the only technique currently available for assessing protein levels in small tissue samples and localizing expression to specific cell types. We focused on two pathways known to be associated with prostate cancer behavior: neuroendocrine effectors and apoptosis (28, 29–31). Of the 80 genes in the grade classifier with known biological functions, 5 are reported to modulate neuropeptide or amine metabolism: monoamine oxidase A (MAOA), YWHAZ/14–3-3-ζ, OAZ2, CPE, and SLC22A3. MAOA is encoded by a polymorphic gene located on the X chromosome that catalyzes the oxidative deamination of biogenic amines, such as catecholamines and indolamine transmitters, throughout the body. Gene variants conferring up to 50-fold differences in MAOA activity have been associated with a variety of neurological and psychosocial disorders, but few studies of MAOA in the context of cancer biology have been reported. The physiological functions of amine oxidases remain to be completely established, but amine metabolism is linked to essential cellular processes such as cell growth and differentiation (32). The byproducts of amine metabolism include H202 and hydroxyl radicals that may contribute to aspects of tumorigenesis, including redox-sensitive pathways such as MAP-kinase signaling (33).
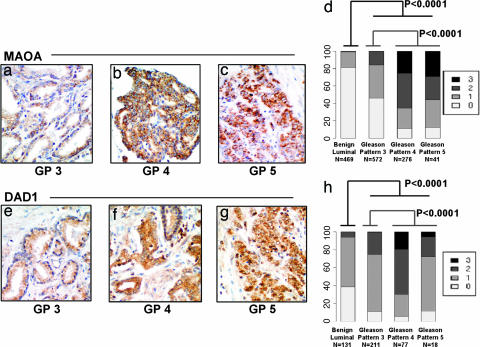
To further characterize the association between MAOA expression and the differentiation state of prostate cancers, we measured MAOA protein levels by IHC on panels of tissue microarrays (TMAs) representing independent patient cohorts not evaluated in the initial analyses of transcript levels. A comparison of 469 benign and 889 cancerous samples demonstrated that MAOA protein expression was elevated in cancerous epithelium relative to benign secretory epithelium (P < 0.0001, proportional odds-regression analysis) (34) (Fig. 3a–d), and MAOA expression was significantly elevated in Gleason 4 or 5 samples relative to Gleason 3 samples (P < 0.0001, proportional odds-regression analysis). The finding that high levels of expression of MAOA protein characterizes high-grade prostate carcinoma raises the possibility that patients on long-term MAOA inhibitors might have lower frequencies of high-grade prostate cancer if MAOA plays a mechanistic role in the development of high-grade prostate cancer.
Fig. 3.
IHC analysis of Gleason grade-associated gene expression. Representative immunohistochemical staining of Gleason pattern 3 (a and e), 4 (b and f), and 5 (c and g) prostate adenocarcinoma for MAOA (a–c) and DAD1 (e–g) expression. Magnifications are ×400. Summary of MAOA (1,358 scoreable samples) (d) and DAD1 (437 scoreable samples) (d and h) expression quantitation in tissue cores comprised of benign secretory epithelium and specific Gleason pattern adenocarcinomas.
Six genes functionally linked to the regulation of apoptosis were present in the cohort of grade-classifying genes. Of these, the gene encoding defender against death (DAD1) provides an intriguing link between apoptosis and the influence of tumor survival factors associated with perineural invasion (PNI). DAD1 is a downstream target of the NFkB survival pathway and exhibits an antiapoptotic function (35). In vitro studies modeling PNI-associated prostate cancer growth measured increased proliferation, reduced apoptosis, and elevated expression of NFkB and DAD1 in tumor cells located in proximity to ganglia and nerve tissue (36). We evaluated the expression of DAD1 protein by IHC in TMAs of formalin-fixed radical prostatectomy cores that, together, comprised 131 benign and 306 cancerous samples. High DAD1 expression was significantly associated with cancerous epithelium relative to benign secretory epithelium (P < 0.0001). In agreement with the transcript analyses, DAD1 protein levels also exhibited a strong association with Gleason pattern. Cancers of patterns 4 and 5 were more likely to stain intensely than low-grade cancer of pattern 3 (P < 0.0001) (Fig. 3 e–h).
Conclusions
In this study, we have identified a panel of molecular alterations that associate with the histological interpretation of prostate cancer grades. The panel performed with high accuracy across three independent panels of prostate adenocarcinomas processed and analyzed by using divergent techniques. Gleason patterns 4 and 5 cancers were virtually indistinguishable at the molecular level with our model. This finding is in accord with the clinical observation that Gleason patterns 4 and 5 tumors are associated with similar prognoses and, thus, are managed identically (37). If demonstrated to be mechanistically involved in cancer progression, the proteins encoded by grade-discriminating genes could serve as targets for pharmaceutical inhibition. Grade-discriminatory proteins may also have utility as serum markers for identifying high-grade prostate cancer. In this context, the association of MAOA expression with prostate cancer and grade is a finding that further implicates neuroendocrine features as mediators of prostate carcinogenesis. This finding has clinical importance, because the presence and extent of a neuroendocrine component in a prostate carcinoma correlates with tumor aggressiveness (37). The ready availability of monamine oxidase inhibitors offers an immediate opportunity to determine the clinical relevance of this finding.
Materials and Methods
Tissue Acquisition.
All materials were acquired and used in conformity with Institutional Review Board-approved protocols at the University of Washington and Oregon Health & Science University. Two types of tissue were used for this study: fresh, nonfixed tissue as a source of RNA for laser microdissection and formalin-fixed, paraffin-embedded tissue for IHC studies. The nonfixed tissue consisted of frozen tissue blocks from radical prostatectomies and an independent sample set of frozen tissue blocks containing prostate needle core biopsies. Details of tissue handling are provided in Supporting Materials and Methods. Fixed tissue samples consisted of two types: blocks corresponding to the fresh tissue samples that were used for the laser microdissection preparations and TMAs.
Laser-Capture Microdissection (LCM) and RNA Preparation.
Frozen sections (8 μm) were cut from optimal cutting temperature medium (OCT) blocks and immediately fixed in cold 95% ethanol. After brief (5–10 seconds) staining with hematoxylin using the HistoGene staining solution (Arcturus Engineering Mountain View, CA), the sections were dehydrated in 100% ethanol, followed by xylenes (per the manufacturer’s protocol). Epithelial cells (≈5,000) from both histologically benign glands and cancer glands were separately laser-capture microdissected by using the Arcturus PixCell II instrument. Only one Gleason pattern was included in each laser-captured cancer sample. A total of 32 different Gleason patterns were captured from the 29 radical prostatectomy samples: 12 Gleason pattern 3, 12 Gleason pattern 4, and 8 Gleason pattern 5 samples. A total of 30 Gleason patterns were captured from the 30 needle core biopsy samples, with some samples comprising combinations of Gleason patterns. Matched benign epithelium was captured for each cancer sample, for a total of 121 samples. Digital photographs were taken of tissue sections before, during, and after LCM and assessed independently by two investigators to confirm the Gleason patterns of the laser-captured cells. RNA extraction and amplification were performed by using standard procedures described in Supporting Materials and Methods.
Microarray Hybridization, Data Acquisition, and Analysis.
Prostate Expression Database cDNA microarrays were prepared on polyL-lysine-coated glass microscope slides by using a robotic spotting tool as described in ref. 38. cDNA probes were made from 2 μg of amplified RNA and randomly labeled with either Cy3 or Cy5 dye to account for dye bias. Patient-matched normal and cancer probes were combined, filtered, and competitively hybridized to microarrays under a coverslip for 14 h at 63°C. Further details are provided in Supporting Materials and Methods.
Fluorescent array images were collected for both Cy3 and Cy5 emissions by using a GenePix 4000B fluorescent scanner (Axon Instruments, Foster City, CA). The image-intensity data were gridded and extracted by using genepix pro 4.1 software. The specifics of microarray data processing are provided in Supporting Materials and Methods. Microarray data sets from this study are deposited in the GEO respository under accession no. GSE5132.
To compare the overall expression patterns of all radical prostatectomy cancer samples to their patient-matched normal samples, the filtered log-ratio measurements were analyzed by using the Significance Analysis of Microarrays (SAM) procedure (39) (www-stat.stanford.edu/∼tibs/SAM). In this analysis, a one-sample t test was used to determine which genes were significantly differentially expressed between cancer samples and their patient-matched normal samples. We call the set of significant genes the expression profile associated with prostate cancer.
To identify gene-expression alterations associated with specific Gleason patterns, we used Prediction Analysis for Microarrays (PAM) (13), a supervised classification method. Full technical details are provided on the PAM web site (www-stat.stanford.edu/∼tibs/PAM). We divided the radical prostatectomy samples into two classes (Gleason pattern 3 and Gleason pattern 4 or 5) and applied PAM to identify several small gene cohorts that classified the samples with low error rates under leave-out-one cross-validation. To evaluate the predictive properties of these gene sets, we classified an independent sample set of Gleason pattern cancers composed of prostate needle core biopsy samples. An additional visual assessment of the degree to which our gene model partitioned the radical prostatectomy samples by Gleason pattern was undertaken by using principal-components analysis of the samples (40).
Quantitative (q)RT-PCR.
cDNA was generated from 1 μg of aRNA by using 2 μg of random hexamers for priming reverse transcription by SuperScript II (200 units per reaction; Invitrogen). qRT-PCRs were done in triplicate, by using ≈5 ng of cDNA, 0.2 mM each primer, and SYBR green PCR master mix (Applied Biosystems) in a 20-μl reaction volume. Reactions were carried out and analyzed by using an Applied Biosystems 7900 sequence detector. Samples were normalized to the cycle threshold value obtained during the exponential amplification of RPL13A. The expression level of HSD17B4 was calculated. Values were reported as the ratio of gene expression in neoplastic to normal epithelium. Additional details and primer sequences are provided in Supporting Materials and Methods.
TMAs.
Eight TMAs were used for these studies. All samples in all arrays were provided in duplicate. Two arrays of predominantly primary prostate cancers (of 159 and 234 samples, respectively) have been described (41). Six arrays represented a range of Gleason grades, a mix of prostate cancer tissue of different biologic states [normal, atrophy, benign prostatic hyperplasia (BPH), prostatic intraepithelial neoplasia (PIN), primary prostate carcinoma, and metastatic prostate carcinoma], and a mix of different normal and neoplastic tissues. Altogether, 469 unique samples of benign prostate glands and 889 unique samples of primary prostate carcinoma (572 Gleason pattern 3, 276 Gleason pattern 4, and 41 Gleason pattern 5) were used for MAOA immunostaining. And 131 unique samples of benign prostate glands and 306 unique samples of primary prostate carcinoma (211 Gleason pattern 3, 77 Gleason pattern 4, and 18 Gleason pattern 5) were used for DAD1 immunostaining.
IHC.
Antibodies recognizing MAOA (sc-20156; Santa Cruz Biotechnology) and DAD1 (sc-25557; Santa Cruz Biotechnology) were used to stain TMAs composed of benign and neoplastic prostate tissues. Specificity of labeling was confirmed by both omission of the primary antibody and immunostaining the sections with a primary antibody against an irrelevant antigen. Immunolocalization was done by using a three-step avidin–biotin–peroxidase method. The sections were counterstained with hematoxylin. Further details are provided in Supporting Materials and Methods.
IHC stains were evaluated by using the following categorical compositional scale: 0, no expression; 1, ≤5% of the cells express the antigen; 2, 5–20% of the cells express the antigen; and 3, 20–100% of cells express the antigen. The following cell types were evaluated: secretory and basal epithelial, high-grade PIN, and Gleason pattern 3, pattern 4, and pattern 5 tumor cells. When a section had several Gleason patterns, each pattern was scored.
To test for differences in the staining intensity of different cell types, we used a proportional-odds model and included the covariates Gleason grade and tissue source. The model was fit in sas (SAS Institute, Cary, NC), implementing a generalized estimating-equations approach to account for multiple sections from the same patient. Further details of the analytical methods are provided in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mahul Amin for helpful suggestions and assistance with study design; Devon Felise and Julie Hahn for tissue collection; members of the immunohistochemistry laboratories at the University of Washington (supervisor, Farinaz Shokri) and Fred Hutchinson Cancer Research Center (Kimberly Adolphson and Linda Cherepow); Ruth Dumpit for assistance with microarray hybridizations; members of the Nelson laboratory for helpful discussions; Andrew Glass and Lukas Bubendorf for TMA construction; and Ying Zhang of the Biostatistics and Bioinformatics Shared Resource of the Lombardi Comprehensive Cancer Center for statistical support. This work was supported by Department of Defense Grants DAMD17-03-2-033 and PC041158 and National Institutes of Health Grants P01CA85859, DK65204, and the Pacific Northwest Prostate Cancer SPORE Grant P50CA97186.
Abbreviations
- IHC
immunohistochemical
- MAOA
monoamine oxidase A
- PAM
prediction analysis for microarrays
- TMA
tissue microarray.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The microarray data sets reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE5132).
References
- 1.Partin A. W., Kattan M. W., Subong E. N., Walsh P. C., Wojno K. J., Oesterling J. E., Scardino P. T., Pearson J. D. J. Am. Med. Assoc. 1997;277:1445–1451. [PubMed] [Google Scholar]
- 2.Gleason D. F., Mellinger G. T. J. Urol. 1974;111:58–64. doi: 10.1016/s0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 3.Epstein J. I., Allsbrook W. C., Jr., Amin M. B., Egevad L. L. Am. J. Surg. Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 4.Epstein J. I. Am. J. Surg. Pathol. 2000;24:477–478. doi: 10.1097/00000478-200004000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Albertsen P. C., Hanley J. A., Barrows G. H., Penson D. F., Kowalczyk P. D., Sanders M. M., Fine J. J. Natl. Cancer Inst. 2005;97:1248–1253. doi: 10.1093/jnci/dji248. [DOI] [PubMed] [Google Scholar]
- 6.Chan T. Y., Partin A. W., Walsh P. C., Epstein J. I. Urology. 2000;56:823–827. doi: 10.1016/s0090-4295(00)00753-6. [DOI] [PubMed] [Google Scholar]
- 7.Rasiah K. K., Stricker P. D., Haynes A. M., Delprado W., Turner J. J., Golovsky D., Brenner P. C., Kooner R., O’Neill G. F., Grygiel J. J., et al. Cancer. 2003;98:2560–2565. doi: 10.1002/cncr.11850. [DOI] [PubMed] [Google Scholar]
- 8.Albertsen P. C., Fryback D. G., Storer B. E., Kolon T. F., Fine J. J. Am. Med. Assoc. 1995;274:626–631. [PubMed] [Google Scholar]
- 9.Magee J. A., Araki T., Patil S., Ehrig T., True L., Humphrey P. A., Catalona W. J., Watson M. A., Milbrandt J. Cancer Res. 2001;61:5692–5696. [PubMed] [Google Scholar]
- 10.Dhanasekaran S. M., Barrette T. R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K. J., Rubin M. A., Chinnaiyan A. M. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 11.Singh D., Febbo P. G., Ross K., Jackson D. G., Manola J., Ladd C., Tamayo P., Renshaw A. A., D’Amico A. V., Richie J. P., et al. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 12.Lapointe J., Li C., Higgins J. P., van de Rijn M., Bair E., Montgomery K., Ferrari M., Egevad L., Rayford W., Bergerheim U., et al. Proc. Natl. Acad. Sci. USA. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tibshirani R., Hastie T., Narasimhan B., Chu G. Proc. Natl. Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allsbrook W. C., Jr., Mangold K. A., Johnson M. H., Lane R. B., Lane C. G., Amin M. B., Bostwick D. G., Humphrey P. A., Jones E. C., Reuter V. E., et al. Hum. Pathol. 2001;32:74–80. doi: 10.1053/hupa.2001.21134. [DOI] [PubMed] [Google Scholar]
- 15.Graefen M., Karakiewicz P. I., Cagiannos I., Quinn D. I., Henshall S. M., Grygiel J. J., Sutherland R. L., Stricker P. D., Klein E., Kupelian P., et al. J. Clin. Oncol. 2002;20:3206–3212. doi: 10.1200/JCO.2002.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Cunha G. R., Hayward S. W., Wang Y. Z. Differentiation. 2002;70:473–485. doi: 10.1046/j.1432-0436.2002.700902.x. [DOI] [PubMed] [Google Scholar]
- 17.Koukourakis M. I., Giatromanolaki A., Brekken R. A., Sivridis E., Gatter K. C., Harris A. L., Sage E. H. Cancer Res. 2003;63:5376–5380. [PubMed] [Google Scholar]
- 18.Sato N., Fukushima N., Maehara N., Matsubayashi H., Koopmann J., Su G. H., Hruban R. H., Goggins M. Oncogene. 2003;22:5021–5030. doi: 10.1038/sj.onc.1206807. [DOI] [PubMed] [Google Scholar]
- 19.Karihtala P., Mantyniemi A., Kang S. W., Kinnula V. L., Soini Y. Clin. Cancer Res. 2003;9:3418–3424. [PubMed] [Google Scholar]
- 20.Porkka K., Saramaki O., Tanner M., Visakorpi T. Lab. Invest. 2002;82:629–637. doi: 10.1038/labinvest.3780457. [DOI] [PubMed] [Google Scholar]
- 21.Albrecht M., Mittler A., Wilhelm B., Lundwall A., Lilja H., Aumuller G., Bjartell A. Eur. Urol. 2003;44:415–422. doi: 10.1016/s0302-2838(03)00322-1. [DOI] [PubMed] [Google Scholar]
- 22.Gimenez-Bonafe P., Fedoruk M. N., Whitmore T. G., Akbari M., Ralph J. L., Ettinger S., Gleave M. E., Nelson C. C. Prostate. 2004;59:337–349. doi: 10.1002/pros.20023. [DOI] [PubMed] [Google Scholar]
- 23.Sutherland B. W., Kucab J., Wu J., Lee C., Cheang M. C., Yorida E., Turbin D., Dedhar S., Nelson C., Pollak M., et al. Oncogene. 2005;24:4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 24.Tsuchiya N., Slezak J. M., Lieber M. M., Bergstralh E. J., Jenkins R. B. Genes Chromosomes Cancer. 2002;34:363–371. doi: 10.1002/gcc.10064. [DOI] [PubMed] [Google Scholar]
- 25.Chetyrkin S. V., Belyaeva O. V., Gough W. H., Kedishvili N. Y. J. Biol. Chem. 2001;276:22278–22286. doi: 10.1074/jbc.M102076200. [DOI] [PubMed] [Google Scholar]
- 26.Zha S., Ferdinandusse S., Hicks J. L., Denis S., Dunn T. A., Wanders R. J., Luo J., De Marzo A. M., Isaacs W. B. Prostate. 2005;63:316–323. doi: 10.1002/pros.20177. [DOI] [PubMed] [Google Scholar]
- 27.Cheng K. W., Lahad J. P., Kuo W. L., Lapuk A., Yamada K., Auersperg N., Liu J., Smith-McCune K., Lu K. H., Fishman D., et al. Nat. Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- 28.Puccetti L., Supuran C. T., Fasolo P. P., Conti E., Sebastiani G., Lacquaniti S., Mandras R., Milazzo M. G., Dogliani N., De Giuli P., Fasolis G. Eur. Urol. 2005;48:215–221. doi: 10.1016/j.eururo.2005.03.018. and Discussion, pp 221–223. [DOI] [PubMed] [Google Scholar]
- 29.Weinstein M. H., Partin A. W., Veltri R. W., Epstein J. I. Hum. Pathol. 1996;27:683–687. doi: 10.1016/s0046-8177(96)90398-6. [DOI] [PubMed] [Google Scholar]
- 30.Stapleton A. M., Zbell P., Kattan M. W., Yang G., Wheeler T. M., Scardino P. T., Thompson T. C. Cancer. 1998;82:168–175. doi: 10.1002/(sici)1097-0142(19980101)82:1<168::aid-cncr21>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 31.Krajewska M., Krajewski S., Banares S., Huang X., Turner B., Bubendorf L., Kallioniemi O. P., Shabaik A., Vitiello A., Peehl D., et al. Clin. Cancer Res. 2003;9:4914–4925. [PubMed] [Google Scholar]
- 32.Pietrangeli P., Mondovi B. Neurotoxicology. 2004;25:317–324. doi: 10.1016/S0161-813X(03)00109-8. [DOI] [PubMed] [Google Scholar]
- 33.Tonks N. K. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 34.McCullagh P., Nelder J. Generalized Linear Models. London: Chapman and Hall; 1989. [Google Scholar]
- 35.Patel N. M., Nozaki S., Shortle N. H., Bhat-Nakshatri P., Newton T. R., Rice S., Gelfanov V., Boswell S. H., Goulet R. J., Jr., Sledge G. W., Jr., Nakshatri H. Oncogene. 2000;19:4159–4169. doi: 10.1038/sj.onc.1203768. [DOI] [PubMed] [Google Scholar]
- 36.Ayala G. E., Dai H., Ittmann M., Li R., Powell M., Frolov A., Wheeler T. M., Thompson T. C., Rowley D. Cancer Res. 2004;64:6082–6090. doi: 10.1158/0008-5472.CAN-04-0838. [DOI] [PubMed] [Google Scholar]
- 37.Abrahamsson P. A. Prostate. 1999;39:135–148. doi: 10.1002/(sici)1097-0045(19990501)39:2<135::aid-pros9>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 38.Clegg N., Eroglu B., Ferguson C., Arnold H., Moorman A., Nelson P. S. J. Steroid Biochem. Mol. Biol. 2002;80:13–23. doi: 10.1016/s0960-0760(01)00167-4. [DOI] [PubMed] [Google Scholar]
- 39.Tusher V. G., Tibshirani R., Chu G. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raychaudhuri S., Stuart J. M., Altman R. B. Pac. Symp. Biocomput. 2000:455–66. doi: 10.1142/9789814447331_0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zellweger T., Ninck C., Mirlacher M., Annefeld M., Glass A. G., Gasser T. C., Mihatsch M. J., Gelmann E. P., Bubendorf L. Prostate. 2003;55:20–29. doi: 10.1002/pros.10209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.