Abstract
Flower color is most often conferred by colored flavonoid pigments. Aurone flavonoids confer a bright yellow color on flowers such as snapdragon (Antirrhinum majus) and dahlia (Dahlia variabilis). A. majus aureusidin synthase (AmAS1) was identified as the key enzyme that catalyzes aurone biosynthesis from chalcones, but transgenic flowers overexpressing AmAS1 gene failed to produce aurones. Here, we report that chalcone 4′-O-glucosyltransferase (4′CGT) is essential for aurone biosynthesis and yellow coloration in vivo. Coexpression of the Am4′CGT and AmAS1 genes was sufficient for the accumulation of aureusidin 6-O-glucoside in transgenic flowers (Torenia hybrida). Furthermore, their coexpression combined with down-regulation of anthocyanin biosynthesis by RNA interference (RNAi) resulted in yellow flowers. An Am4′CGT-GFP chimeric protein localized in the cytoplasm, whereas the AmAS1(N1-60)-RFP chimeric protein was localized to the vacuole. We therefore conclude that chalcones are 4′-O-glucosylated in the cytoplasm, their 4′-O-glucosides transported to the vacuole, and therein enzymatically converted to aurone 6-O-glucosides. This metabolic pathway is unique among the known examples of flavonoid, including anthocyanin biosynthesis because, for all other compounds, the carbon backbone is completed before transport to the vacuole. Our findings herein not only demonstrate the biochemical basis of aurone biosynthesis but also open the way to engineering yellow flowers for major ornamental species lacking this color variant.
Keywords: Antirrhinum, chalcone, flavonoid, glucosyltransferase, flower coloration
Flavonoids and their color class, anthocyanins, are the principal color determinants in most flowers, and their biosynthetic pathways are well established (1, 2). Biosyntheses, including modifications such as glucosylation and acylation, are completed before products are transported to the vacuole. Genetic engineering of a flavonoid biosynthetic pathway in floriculture would provide a powerful method for obtaining novel flower colors beyond genetic constraints that conventional breeding is not able to overcome (1). Transgenic blue/violet carnations already are now being marketed, and transgenic blue roses have been reported (1). Both were obtained by expressing heterologous flavonoid 3′,5′-hydroxylase genes necessary to produce blue anthocyanins, a gene function that was missing in both carnations and roses. Such important ornamentals as geranium, sweet pea, cyclamen, saintpaulia, and morning glory lack yellow varieties, and, although considerable efforts have been made to obtain yellow colors through genetic engineering, only very pale yellow has been reported on buds (3).
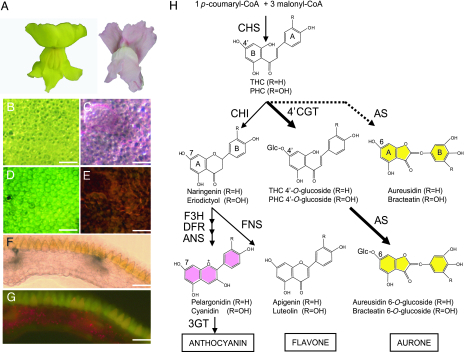
Aurones, represented by aureusidin, are a class of flavonoids that confer a bright yellow color with characteristic fluorescence on some ornamental flowers, such as the snapdragon (Scrophulariaceae, Antirrhinum majus) (Fig. 1A–G) (4, 5). Aurones as well as anthocyanins in flowers are considered to provide a nectar guide for pollinating bumblebees; their biosyntheses therefore is an evolutionally important issue concerning the plant–pollinator interaction (6). Previous genetic studies of Antirrhinum had identified some loci regulating yellow flower coloration (7, 8); structural genes in aurone biosynthesis, however, remained to be identified. To reveal the aurone biosynthesis, we previously identified a polyphenol oxidase (PPO) homolog from A. majus that encodes aureusidin synthase (AmAS1) (9). AmAS1 catalyzes aureusidin biosynthesis from 2′,4′,6′,4-tetrahydroxychalcone (THC) and 2′,4′,6′,3,4-pentahydroxychalcone (PHC) (9–11). It also converts substrates PHC, THC 4′-O-glucoside, and PHC 4′-O-glucoside to bracteatin, aureusidin 6-O-glucoside, and bracteatin 6-O-glucoside in vitro as diagrammed in Fig. 1H (9–11). AmAS1 is a glycoprotein and does not carry a typical N-terminal sequence (NTPP) for plastid targeting that is highly conserved in plant PPOs (9). Moreover, its peak activity at the low pH of 5.4 toward THC suggests that it is localized in the vacuole (9–11). In support of this finding, we showed experimentally that AmAS1 is conveyed from endoplasmic reticulum (ER) to vacuole via Golgi body (12). This finding demonstrated that aurone biosynthesis occurs in vacuole but not in cytoplasm.
Fig. 1.
Flavonoids and their biosynthetic pathway in A. majus. (A) Flower colors in A. majus cultivar (cv.) Snap Yellow (Left) and cv. Merryland Pink (Right). Shown are petal color on the adaxial side of cv. Snap Yellow (B) and of cv. Merryland Pink (C). Fluorescent microcopies in cv. Snap Yellow (D) and Merryland Pink (E). Cross sections show that fluorescence is restricted to the pigmented adaxial epidermis of cv. Snap Yellow (F and G). (Scale bar: 100 μm.) (H) Flavonoid biosynthetic pathway. The 4′ position of chalcones corresponds to the 6 position of aurones and the 7 position of flavanone (naringenin). Dotted arrow, aurone biosynthetic activity of AmAS1 in vitro; thick arrows, the aurone biosynthetic pathway in vivo reported in this study; thin arrows, the anthocyanin biosynthetic pathway; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanindin synthase; 3GT, anthocyanin 3-O-glucosyltransferase; FNS, flavone synthase; AS, aureusidin synthase; 4′CGT, chalcone 4′-O-glucosyltransferase; THC, tetrahydroxychalcone; PHC, pentahydroxychalcone; Glc, glucose.
In this report, we show that glucosylation of chalcone by cytosolic chalcone 4′-O-glucosyltransferase followed by oxidative cyclization by vacuolar aureusidin synthase is the biochemical basis of aurone 6-O-glucosides in vivo. Moreover, we open molecular breeding strategies to generate monotonous yellow flowers that produce aurone 6-O-glucoside dominantly.
Results and Discussion
Transgenic Plants Overexpressing AmAS1 Fail to Produce Aurones.
To obtain yellow flowers that accumulate aurone, a binary vector (pSFL211) to overexpress the AmAS1 gene by a constitutive cauliflower mosaic virus 35S promoter was introduced into Scrophulariaceae, Torenia hybrida (cv. Summer Wave Blue). This cultivar has blue flowers conferred by a malvidin class of anthocyanin but does not accumulate aurones (Table 1). Although 48 independent transgenic plants carrying the AmAS1 gene were obtained, their petals produced no aurones, and no flower color changes occurred (Table 1 and Figs. 5 and 6, which are published as supporting information on the PNAS web site).
Table 1.
Flavonoid analysis of transgenic flowers
| (n) | Genotype | Flavonoid content, mg/g fresh petal weight | ||||
|---|---|---|---|---|---|---|
| THC 4′-O-glucoside | Aureusidin 6-O-glucoside | Anthocyanidins | Flavones | Phenotype | ||
| NT (3) | cv. Summer Wave Blue | ND | ND | 0.656 ± 0.178 | 4.416 ± 1.687 | Blue |
| SFL211 (5) | AmAS1OX | ND | ND | 0.639 ± 0.030 | 5.051 ± 0.175 | Blue |
| SFL209 (3) | Am4′CGTOX | 0.467 ± 0.261 | ND | 0.727 ± 0.112 | 4.999 ± 0.174 | Blue |
| SFL210 (5) | ThF3HRNAi | ND | ND | 0.014 ± 0.008 | 5.194 ± 0.475 | White |
| SFL201 (3) | Am4′CGTOX + AmAS1OX | 0.200 ± 0.129 | 0.422 ± 0.161 | 0.581 ± 0.330 | 3.007 ± 0.869 | Blue/Yellow |
| SFL307 (3) | Am4′CGTOX + AmAS1OX + ThDFRRNAi | ND | 0.749 ± 0.157 | 0.065 ± 0.021 | 2.287 ± 0.625 | Yellow |
| SFL308 (10) | Am4′CGTOX + AmAS1OX + ThF3HRNAi | ND | 0.757 ± 0.157 | 0.032 ± 0.010 | 3.172 ± 0.453 | Yellow |
| A. majus (4) | cv. Snap Yellow (Endogenous Am4′CGT and AmAS1) | 0.030 ± 0.001 | 0.411 ± 0.003 | ND | 6.767 ± 0.101 | Yellow |
Flavonoids in all the transgenic lines and nontransformants were identified by HPLC, and the data were summarized. Standards were eluted at R.T. 6.2 (aureusidin 6-O-glucoside), R.T. 14.2 (THC 4′-O-glucoside) and R.T. 24.0 minutes (THC). The amounts of THC 4′-O-glucoside and aureusidine 6-O-glucoside in the transgenic flowers were compared with the amounts in Antirrhinum flower (cv. Snap Yellow at flower stage 6). NT, nontransformant (Standard Torenia hybrida cv. Summer Wave Blue); cv., cultivar; ND, not detected. Anthocyanidins: delphinidin, cyanidin, peonidin, petunidin, and malvidin. Flavones: luteolin and apigenin.
This puzzling result prompted us to hypothesize that THC is glucosylated at its 4′-hydroxyl group in the cytoplasm, then transported to the vacuole and there converted to aureusidin 6-O-glucoside by AmAS1; i.e., THC 4′-O-glucoside is the authentic substrate for AmAS1 in vivo.
UGT88D3 Has Chalcone 4′-O Glucosylating Activity.
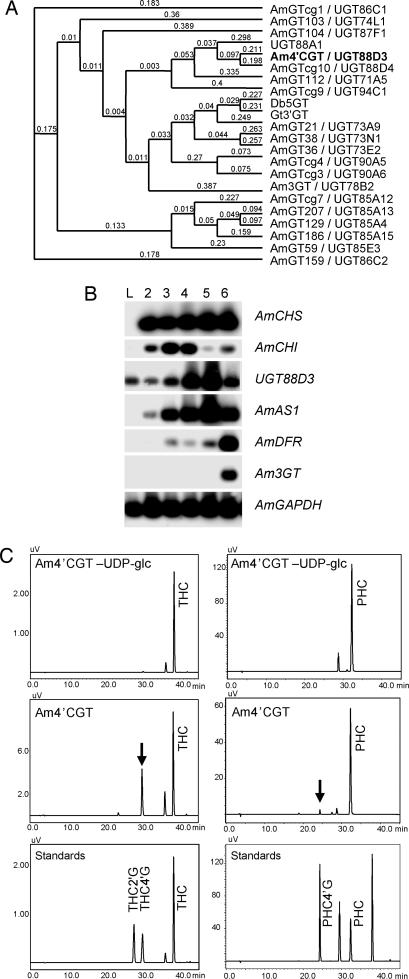
Plants harbor a wide variety of glucosyltransferases that glucosylate various molecules, including secondary metabolites, phytohormones, and biotic and abiotic environmental toxins (13). Glucosylation is a key mechanism to modify the bioactivity and location of small molecules (13). Yet, so far, no glucosyltransferases that catalyze glucosylation at the 4′-hydroxyl group of chalcones have been identified. We detected UDP-glucose:chalcone 4′-O-glucosyltransferase (4′CGT) activity in a crude extract of yellow Antirrhinum petals that accumulated aureusidin 6-O-glucoside. That activity was not adsorbed to a Hi-Trap Q column in 25 mM potassium-phosphate buffer (pH 7.0); therefore, the enzyme corresponding to 4′CGT activity has a relatively high pI for flavonoid-related glucosyltransferases.
To identify an A. majus glucosyltransgerase gene responsible for the 4′CGT activity, we screened 5 × 105 plaques of an A. majus petal cDNA library with labeled C-terminal portions of five flavonoid glucosyltransferases; these C-terminal portions, including the PSPG-box, are well conserved (14). We obtained 18 independent molecular species of glucosyltransferase homologues (Fig. 2A), one of which was a known gene, anthocyanidin 3-O-glucosyltransferase (15). Glucosyltransferase homologues were analyzed by semiquantitative RT-PCR to determine their developmental expression profiles (Fig. 2B). They also were expressed in Escherichia coli, and crude bacterial lysates were assessed by enzymatic assay in vitro (Fig. 2C). One clone, designated UGT88D3 by the committee for naming UDP Glucuronosyltransferase (http://som.flinders.edu.au/FUSA/ClinPharm/UGT/), had a developmental gene expression profile similar to that of AmAS1, indicative of a temporal correlation with the accumulation pattern of THC 4′-O-glucoside/aureusidin 6-O-glucoside in Antirrhinum flower development (Fig. 2B) (10). The crude fraction from bacteria expressing UGT88D3 had UDP-glucose-dependent 4′CGT activity toward THC (Fig. 2C). Moreover, it catalyzed glucosylation of the 4′-hydroxyl group of PHC (Fig. 2C). The predicted sequence of this glucosyltransferase consists of 457-aa residues and has a calculated pI of 6.82, consistent with native 4′CGT behavior in ion exchange column chromatography. The calculated pIs of the other cloned Antirrhinum glucosyltransferase homologs ranged from 4.73 to 5.89. Antirrhinum majus 4′CGT (Am4′CGT) has the highest amino acid sequence identity (40%) to a functionally unknown Arabidopsis glucosyltransferase, UGT88A1 (Fig. 2A). We isolated the genomic Am4′CGT gene from A. majus genomic DNA by PCR. The genomic gene has no intron, whereas UGT88A1 has one (Fig. 7, which is published as supporting information on the PNAS web site). The genomic gene was detected as multiple bands by Southern blotting analysis, suggesting that other glucosyltransferase genes similar to Am4′CGT exist in A. majus (Fig. 7).
Fig. 2.
Functional characterizations of Am4′CGT. (A) A phylogenic tree for the flavonoid-related and 18 A. majus glucosyltransferases constructed by a clustal-w program (32), macvector 7.2.2 software (Accelrys, San Diego). The number on the branches indicates sequence difference (0.05 corresponds to a 5% change). UGT88A1 (AAN28841) is an Arabidopsis, function unknown, glucosyltransferase; Am3GT is an Antirrhinum anthocyanidin 3-O-GT; Db5GT (Y18871) is a Dorotheanthus betanidin 5-O-GT; and Gt3′GT (AB076697) is a Gentiana anthocyanin 3′-O-GT. (B) Expression patterns of Antirrhinum flavonoid-related structural genes during flower development. L, leaves; numbers above the lanes, a previously defined developmental flower stage (10). (C) Enzymatic activity of recombinant Am4′CGT protein against THC and PHC. Chromatograms show the absorption at 360 nm in each reaction mixture. HPLC condition is described in Materials and Methods. Standards were eluted at retention times (R.T.) 24.3 (PHC 4′-O-glucoside), 27.7 (THC 2′-O-glucoside), 30.0 (THC 4′-O-glucoside), 32.4 (PHC), and 38.2 min (THC). Arrows indicate the THC 4′-O-glucoside and PHC 4′-O-glucoside synthesized, respectively, by Am4′CGT from THC and PHC.
UGT88D3 Functions as a 4′CGT in Vivo.
Six series of transgenic Torenia plants were generated to clarify whether 4′CGT activity is required for aurone 6-O-glucoside biosynthesis in vivo. The six binary vectors for transformation are shown in Fig. 5. Although 40 independent transgenic plants carrying pSFL209 (expression of Am4′CGT) were obtained, flower color was not altered. Flavonoids were extracted from petals and analyzed by HPLC. Results are shown in Table 1. Petals of transgenic line SFL209 accumulated significant amounts of THC 4′-O-glucoside, evidence that this glucosyltransferase functions as a 4′CGT in vivo. Although chalcones themselves are pale yellow pigments, flower color was still blue in all transformants of the SFL209 line, indicating that anthocyanins are visually dominant in flower color over chalcones (Table 1 and Fig. 6).
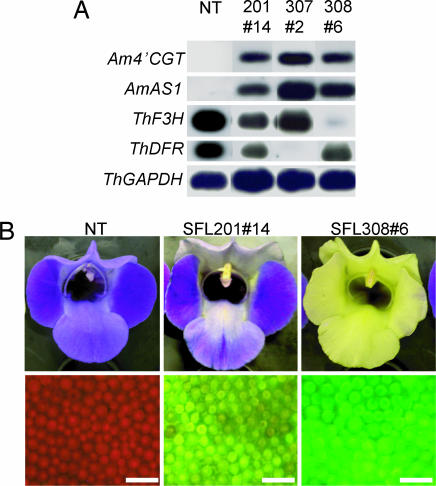
Coexpression of Am4′CGT and AmAS1 Is Sufficient for Aurone Biosynthesis in Vivo.
Fifty-nine independent transgenic plants carrying pSFL201 (coexpression of Am4′CGT and AmAS1 as verified by RT-PCR, Fig. 3A) were obtained, of which 24 had a yellow hue on their stamens, on their carpels, and in the region between the petal lobes and throat (Fig. 3B). There was significant cellular fluorescence in the petals, unlike transgenic line SFL209, which expresses the Am4′CGT gene solely. Flavonoid analysis clearly showed that coexpression of Am4′CGT and AmAS1 is sufficient to synthesize aureusidin 6-O-glucoside via THC 4′-O-glucoside (Table 1). Aureusidin 6-O-glucoside accumulation in transgenic flowers expressing cellular fluorescence was correlated to the reduction of flavones and anthocyanidins, the precursors to blue anthocyanins (Table 1). In the early step of flavonoid biosynthesis, chalcone isomerase (CHI) efficiently catalyzes THC to a flavanone, naringenin (Fig. 1H) (1, 16). Moreover some varieties of A. majus synthesize simultaneously anthocyanins, flavones, and aurones in the colored cells of petals (7, 8). Taken together, we surmise that 4′CGT competes with CHI and all three pathways are derived from the same substrate pool of chalcone.
Fig. 3.
Phenotypic and expression analyses of transgenic Torenia Flowers. (A) Expression analysis of each transgenic line by RT-PCR/Southern blotting. Total RNAs extracted from nontransformant and transgenic flowers were reverse-transcribed then amplified by PCR with the gene-specific primers in Table 2. (B) Phenotypes of transgenic Torenia flowers. (Upper) Flower color under white light. (Lower) Cellular fluorescence from adaxial side of petal in each line. (Scale bars: 100 μm.) NT, nontransformant (T. hybrida cv. Summer Wave Blue).
Molecular Breeding of Yellow Flowers.
In Antirrhinum, the yellow phenotype is associated genetically with a breakdown of anthocyanin biosynthesis, represented by the incolorata mutant, which lacks flavanone 3-hydroxylase (F3H) activity (Fig. 1H) (7, 8). These reports indicate that anthocyanins disturb the monotonous yellow flowers by aurones. To obtain yellow flowers by genetic engineering, the production of aurone 6-O-glucoside was combined with depletion of anthocyanins by RNA interference. Blue Torenia was transformed with pSFL307 [coexpression of Am4′CGT and AmAS1, and knockdown of Torenia DFR by RNA interference (RNAi)] or pSFL308 (coexpression of Am4′CGT and AmAS1, and knockdown of Torenia F3H by RNAi) (Fig. 5). Fifteen of 30 (SFL307: DFR RNAi) and 25 of 41 (SFL308: F3H RNAi) plants in these transgenic lines had bright yellow flowers accompanied by enhanced cellular fluorescence (Fig. 3B and Fig. 8A, which is published as supporting information on the PNAS web site). Expression of two heterologous genes and the knockdown of endogenous DFR or F3H were confirmed by RT-PCR (Figs. 3A and 8B). Compared with flowers of transgenic line SFL201, the yellow flowers of both triply transgenic lines accumulated abundant aureusidin 6-O-glucoside and almost no anthocyanins (Table 1). Furthermore, because no chalcone 4′-O-glucoside was detected, the aureusidin biosynthetic reaction seems to be efficient in both transgenic lines (Table 1). Down-regulation of anthocyanin biosynthesis, therefore, efficiently promotes a flavonoid metabolic flux toward aureusidin 6-O-glucoside. In contrast, transgenic lines carrying pSFL209 (Am4′CGT expression), 210 (F3H RNAi), or 211 (AmAS1 expression) did not synthesize detectable aureusidin 6-O-glucoside in their flowers (Table 1 and Figs. 5 and 6). Because chalcone, the first compound in flavonoid biosynthesis, is widespread throughout the plant kingdom, our strategies to generate yellow flowers by production of aureusidin 6-O-glucoside are widely applicable to most plant species producing chalcone.
Biochemical Basis of Aurone Biosynthesis.
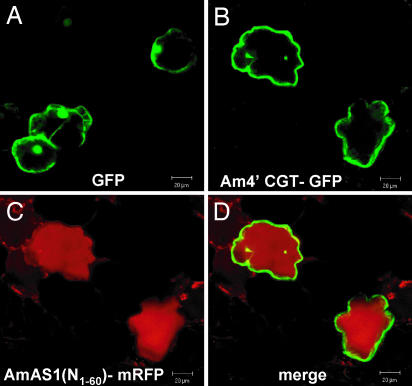
Subcellular localization of both Am4′CGT and AmAS1 was analyzed by transient expression (12). The C-terminal end (Arg-457) of a full-length Am4′CGT was fused to an N terminus of GFP (17), resulting in the Am4′CGT-GFP chimeric protein gene. This fusion gene was coexpressed with AmAS1(N1–60)-monomeric red fluorescent protein (mRFP) (12, 18) in Antirrhinum petal cells. Confocal laser scanning microscopy confirmed that Am4′CGT-GFP and AmAS1(N1–60)-mRFP were distinctly localized in the cytoplasm and vacuole lumen, respectively (Fig. 4). Am4′CGT therefore functions in the cytoplasm where CHS synthesizes chalcones (19). These results suggests that glucosylation of chalcones facilitates vacuolar transport of chalcones by modulating metabolic channeling (20). A previous structural study revealed that there is hydrogen-bonding interaction between the Thr-190 residue of alfalfa CHI and the 7-hydroxyl group of naringenin, which corresponds to the 4′-hydroxyl group of chalcones (Fig. 1) (16). Therefore, 4′-O-glucosylation of chalcones is likely to inhibit the interaction. Here, we show that 4′CGT plays a critical role in aurone biosynthesis by providing chalcone 4′-O-glucosides, the in vivo substrates of vacuolar AmAS1, and likely by avoiding channeling to anthocyanins/flavones.
Fig. 4.
Subcellular localization of Am4′CGT in Antirrhinum petal cells. (A) Control sGFP localizes in the cytoplasm and nucleus. (B) Am4′CGT-GFP fusion protein localizes in the cytoplasm. (C) AmAS1(N1–60)-mRFP protein localizes in the vacuole lumen. (D) Merged image of B and C. (Scale bar: 20 μm.)
Evolution of Aurone Biosynthesis and Other Yellow Pigment.
The 6-O-glycosylated forms of other aurones, such as bracteatin, sulfuretin, and maritimetin, also have been identified in the Angiosperm genera Antirrhinum, Dahlia, Oxalis, Linaria, Limonium, Coreopsis, and Bidens (2). Therefore, the biochemical basis of aurone biosynthesis also may be applicable to these species. Notably, to obtain bright color yellow required a depletion of anthocyanin biosynthesis in our transgenic experiments as predicted from previous genetic studies (Fig. 3 and Fig. 9, which is published as supporting information on the PNAS web site) (7, 8). Similarly, anthocyanin biosynthesis should be lowered in these species. Interestingly, distribution of aurone 6-O-glucosides expands to the Bryophyte such as Marchantia and Conocephalum (2), suggesting that aurone 6-O-glucosides-biosynthetic machinery occurred in the predate of floral evolution. It is of particular interest to unravel physiological roles of aurones in nonflowering plants in which aurones are unlikely to function as a guide for pollinators.
Carotenoid is known as a major hydrophobic class of pigments ranging from yellow, orange to red in flowers and fruits such as Chrysanthemum, Tulipa, and Lycopersicum (21). It is synthesized and accumulated in plastids and is therefore totally different from water-soluble pigment, flavonoid. Recent advanced studies reported that transgenic rice grains producing the carotenoid β-carotene (Golden Rice) show yellow pigmentation (22, 23). Two different classes of yellow pigments (flavonoid and carotenoid) therefore are available to modify flower color by genetic engineering.
Materials and Methods
Am4′CGT Cloning.
A cDNA library derived from A. majus yellow petals (9) was screened with a mixture of five UGTs involved in flavonoid biosynthesis; Ipomoea nil anthocyanin 3GGT (AB192314), Petunia hydrida anthocyanidin 3GT (AB027454), Verbena hybrida anthocyanin 5GT (AB013598), Scutellaria baicalensis baicalen 7GT (AB031274), and Gentiana triflora anthocyanin 3′GT (AB076697) (24). All were labeled with a DIG dye and detection kit (Roche, Mannheim, Germany) by means of the gene-specific primer set given in Table 2, which is published as supporting information on the PNAS web site. The procedure for screening and detection was basically the same as described elsewhere (24, 25).
RT-PCR.
Semiquantitative RT-PCR followed by Southern blotting (25) was performed for Antirrhinum and Torenia genes with the gene-specific primer sets Antirrhinum CHS (CAA27338), Antirrhinum CHI (M68326), Antirrhinum 4′CGT, AmAS1 (AB044884), Antirrhinum DFR (P14721), Antirrhinum anthocyanidin 3GT (15), Antirrhinum glyceraldehyde 3-phosphate dehydrogenase (GAPDH: X59517), Torenia F3H (AB211958), Torenia DFR (AB012924), and Torenia GAPDH (AB106523) (Table 2).
Enzymatic Assay.
cDNAs of cloned Antirrhinum glucosyltransferase homologues were inserted in the pQE61 expression vector (Qiagen, Valencia, CA) for expression in E. coli. Bacterial soluble fractions were prepared as described (26). Because the THC and PHC substrates spontaneously isomerized to flavanones at a neutral pH, they were immobilized in a resin. One milligram of chalcone dissolved in 1 ml of H2O containing 5% ethanol was applied to the resin (TOYO Pearl HW-40F, TOSO, Tokyo) in H2O for the immobilization. To detect 4′CGT activity, 100 μl of the chalcone-immobilized resin, 100 μl of the soluble fraction, and 10 μl of 10 mM UDP-glucose were gently mixed together and kept at 30°C for 30 min. After precipitation, the supernatant was removed, and the resin was washed with H2O. Chalcone then was extracted from the resin by adding 70% acetonitrile containing 0.1% (vol/vol) trifluoroacetic acid (TFA). The extract was analyzed by HPLC under the following conditions (condition 1). Reversed-phase HPLC was performed with an LC-2010A HT system (Shimadzu, Kyoto) equipped with a photodiode array detector, SPD-M10A VP (250–600 nm, Shimadzu), and a YMC-pack ODS-A-312 (150 × 6.0 mm, YMC, Kyoto) column. The extract first was eluted in a linear gradient of 15–40% solvent B (100% methanol) in solvent A [H2O containing 2% (vol/vol) acetic acid] at a flow rate of 1 ml/min for 22 min, then eluted with 40% solvent B for 5 min more. It was further eluted in a linear gradient of 40–62% solvent B for 14 min and then eluted with 62% solvent B for 2 min. Chalcones were monitored at A at 360 nm.
Torenia Transformation.
Torenia hybrida cv. Summer Wave Blue (Suntory Flowers Ltd.) was the host plant. Construction of the binary vector shown in Fig. 5 was essentially the same as in Fukuchi-Mizutani et al. (26). As previously reported (27–30), Torenia fresh leaves were transformed via Agrobacterium tumefaciens-mediated transformation.
Flavonoid Analyses of Transgenic Flowers.
Freeze-dried petals from the transgenic Torenia were dissolved in 50% acetonitrile containing 0.1% (vol/vol) TFA, final concentration 0.1 g/ml (fresh weight/vol). The extracts were analyzed by HPLC with a Shim-pack FC-ODS (50 × 4.6 mm, Shimadzu) column under the following condition (condition 2). Linear gradient elution using 10–23% solvent B [acetonitrile containing 0.05% (vol/vol) TFA] at a flow rate of 0.8 ml/min for 3 min in solvent A [H2O containing 0.05% (vol/vol) TFA], next with 23% solvent B for 17 min more, followed by linear gradient elution of 23–80% solvent B for 2 min, then with 80% solvent B for 3 min more. Chalcones, aurones, anthocyanidins, and flavones were monitored respectively at A at 360 nm, A at 400 nm, A at 520 nm, and A at 360 nm.
Subcellular Localization of the Am4′CGT Protein.
Full-length Am4′CGT cDNA was amplified with primer set 20 shown in Table 2 to introduce the restriction sites for SalI and NcoI. To construct the N-Am4′CGT-sGFP-C fusion gene, Am4′CGT cDNA was inserted at the sites of SalI and NcoI in the sGFP/pUC vector (17). Construction of the AmAS1 (N1–60)-mRFP fusion gene has been described (12). These fusion genes were cotransformed into an Antirrhinum petal (a white cultivar) by particle bombardment with a Helios gene gun system (Bio-Rad) (31), incubated at 20°C for 36 h in the dark, and then observed under a fluorescent microscope (Axioplan 2 imaging, Carl Zeiss) as described in ref. 9.
Supplementary Material
Acknowledgments
We thank Dr. H. Okuhara for advice on the Am4′CGT enzymatic assay; Drs. I. Hara-Nishimura (Kyoto University, Kyoto, Japan) and K. Tamura (Kyoto University) for providing the fluorescent microscope equipped with a confocal system; Drs. H. Satake (SUNBOR, Osaka, Japan) and R. Y. Tsien (University of California, San Diego) for providing the mRFP vector; Dr. Y. Niwa (University of Shizuoka, Shizuoka, Japan) for the sGFP vector; Dr. Y. Ohashi (National Institute of Agrobiological Sciences, Ibaraki, Japan) for the modified El2-CaMV35S promoter; and Dr. R. A. Ludwig (University of California, Santa Cruz) for the A. tumefaciens strain Agl0 strain. Dr. P. Mackenzie named the A. majus glucosyltransferase homologues. Drs. Y. Yamada, P. Yamada, and M. Nakao participated in discussions; Dr. K. Suzuki, Y. Katsumoto, C. Mizutani, Y. Kobayashi, and M. Akagi helped with the transgenic experiments; and N. Kitai, T. Matsuo, and Y. Takeuchi provided experimental support in Am4′CGT cloning.
Abbreviations
- AmAS1
Antirrhinum majus aureusidin synthase 1
- 4′CGT
chalcone 4′-O-glucosyltransferase
- Am4′CGT
Antirrhinum majus 4′CGT
- mRFP
monomeric red fluorescent protein
- THC
2′,4′,6′,4-tetrahydroxychalcone
- PHC
2′,4′,6′,3,4-pentahydroxychalcone
- CHI
chalcone isomerase
- F3H
flavanone 3-hydroxylase
- TFA
trifluoroacetic acid
- OX
overexpression.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The sequence for Am4′CGT/UGT88D3 reported in this paper has been deposited in the DNA Data Bank of Japan (DDBJ) database (accession no. AB198665).
References
- 1.Tanaka Y., Katsumoto Y., Brugliera F., Mason J. Plant Cell Tissue Organ Cult. 2005;80:1–24. [Google Scholar]
- 2.Harborne J. B., Baxter H. The Handbook of Natural Flavonoids 2. London: Wiley; 1999. pp. 196–201. [Google Scholar]
- 3.Davies K. M., Bloor S. J., Spiller G. B., Deroles S. C. Plant J. 1998;13:259–266. [Google Scholar]
- 4.Schwarz-Sommer Z., Davies B., Hudson A. Nat. Rev. Genet. 2003;4:655–664. doi: 10.1038/nrg1127. [DOI] [PubMed] [Google Scholar]
- 5.Asen S., Norris K. H., Stewart R. N. Phytochemistry. 1972;11:2739–2741. [Google Scholar]
- 6.Lunau K., Wacht S., Chittka L. J. Comp. Physiol. 1996;A178:477–489. [Google Scholar]
- 7.Forkmann G., Stotz G. Z. Naturforsch. 1981;36c:411–416. [Google Scholar]
- 8.Spribille R., Forkmann G. Phytochemistry. 1982;21:2231–2234. [Google Scholar]
- 9.Nakayama T., Yonekura-Sakakibara K., Sato T., Kikuchi S., Fukui Y., Fukuchi-Mizutani M., Ueda T., Nakao M., Tanaka Y., Kusumi T., et al. Science. 2000;290:1163–1166. doi: 10.1126/science.290.5494.1163. [DOI] [PubMed] [Google Scholar]
- 10.Sato T., Nakayama T., Kikuchi S., Fukui Y., Yonekura-Sakakibara K., Ueda T., Nishino T., Tanaka Y., Kusumi T. Plant Sci. 2001;160:229–236. doi: 10.1016/s0168-9452(00)00385-x. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama T., Sato T., Fukui Y., Yonekura-Sakakibara K., Hayashi H., Tanaka Y., Kusumi T., Nishino T. FEBS Lett. 2001;499:107–111. doi: 10.1016/s0014-5793(01)02529-7. [DOI] [PubMed] [Google Scholar]
- 12.Ono E., Hatayama M., Isono Y., Sato T., Watanabe R., Yonekura-Sakakibara K., Fukuchi-Mizutani M., Tanaka Y., Kusumi T., Nishino T., et al. Plant J. 2006;45:133–143. doi: 10.1111/j.1365-313X.2005.02625.x. [DOI] [PubMed] [Google Scholar]
- 13.Lim E.-K., Bowles D. J. EMBO J. 2003;23:2915–2922. doi: 10.1038/sj.emboj.7600295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogt T., Jones P. Trends Plant Sci. 2000;5:380–386. doi: 10.1016/s1360-1385(00)01720-9. [DOI] [PubMed] [Google Scholar]
- 15.Martin C. R., Prescott A., Mackey S., Bartlett J., Vrijlandt E. Plant J. 1991;1:37–49. doi: 10.1111/j.1365-313x.1991.00037.x. [DOI] [PubMed] [Google Scholar]
- 16.Jez J. M., Bowman M. E., Dixon R. A., Noel J. P. Nat. Struct. Biol. 2000;7:786–791. doi: 10.1038/79025. [DOI] [PubMed] [Google Scholar]
- 17.Chiu W.-L., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. Curr. Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 18.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. Proc. Natl. Acd. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burbulis I. E., Winkel-Shirley B. Proc. Natl. Acad. Sci. USA. 1999;96:12929–12934. doi: 10.1073/pnas.96.22.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jørgensen K., Rasmussen A. V., Morant M., Nielsen A. H., Bjarnholt N., Zagrobelny M., Bak S., Møller B. L. Curr. Opin. Plant Biol. 2005;8:280–291. doi: 10.1016/j.pbi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 21.Giovanni G., Bartley G. E., Scolnik P. A. Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye X., Al-Babili S., Klöti A., Zhang J., Lucca P., Beyer P., Potrykus I. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 23.Paine J. A., Shipton C. A., Chaggar S., Howells R. M., Kenndy M. J., Vernon G., Wright S. Y., Hinchliffe E., Adams J. L., Silverstone A. L., et al. Nat. Biotech. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- 24.Yonekura-Sakakibara K., Tanaka Y., Fukuchi-Mizutani M., Fujiwara H., Fukui Y., Ashikari T., Murakami Y., Yamaguchi M., Kusumi T. Plant Cell Physiol. 2000;41:495–502. doi: 10.1093/pcp/41.4.495. [DOI] [PubMed] [Google Scholar]
- 25.Morita Y., Hoshino A., Kikuchi Y., Okuhara H., Ono E., Tanaka Y., Fukui Y., Saito N., Nitasaka E., Noguchi H., et al. Plant J. 2005;42:353–363. doi: 10.1111/j.1365-313X.2005.02383.x. [DOI] [PubMed] [Google Scholar]
- 26.Fukuchi-Mizutani M., Okuhara H., Fukui Y., Nakao M., Katsumoto Y., Yonekura-Sakakibara K., Kusumi T., Tanaka Y. Plant Physiol. 2003;132:1652–1663. doi: 10.1104/pp.102.018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Engelen F. A., Molthoff J. W., Conner A. J., Nap J. P., Pereira A., Stiekema W. J. Transgenic Res. 1995;4:288–290. doi: 10.1007/BF01969123. [DOI] [PubMed] [Google Scholar]
- 28.Mitsuhara I., Ugaki M., Hirochika H., Ohshima M., Murakami T., Gotoh Y., Katayose Y., Nakamura S., Honkura R., Nishimiya S., et al. Plant Cell Physiol. 1996;37:49–59. doi: 10.1093/oxfordjournals.pcp.a028913. [DOI] [PubMed] [Google Scholar]
- 29.Aida R., Kishimoto S., Tanaka Y., Shibata M. Plant Sci. 2000;153:33–42. doi: 10.1016/s0168-9452(00)00364-2. [DOI] [PubMed] [Google Scholar]
- 30.Lazo G. R., Pascal A. S., Ludwig R. A. Bio/Technology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- 31.Noji M., Inoue K., Kimura N., Gouda A., Saito K. J. Biol. Chem. 1998;273:32739–32745. doi: 10.1074/jbc.273.49.32739. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J. D., Higgins D. G., Gibson T. J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.