Abstract
The plant hormone cytokinin regulates numerous growth and developmental processes. A signal transduction pathway for cytokinin has been elucidated that is similar to bacterial two-component phosphorelays. In Arabidopsis, this pathway is comprised of receptors that are similar to sensor histidine kinases, histidine-containing phosphotransfer proteins, and response regulators (ARRs). There are two classes of response regulators, the type-A ARRs, which act as negative regulators of cytokinin responses, and the type-B ARRs, which are transcription factors that play a positive role in mediating cytokinin-regulated gene expression. Here we show that several closely related members of the Arabidopsis AP2 gene family of unknown function are transcriptionally up-regulated by cytokinin through this pathway, and we have designated these AP2 genes CYTOKININ RESPONSE FACTORS (CRFs). In addition to their transcriptional regulation by cytokinin, the CRF proteins rapidly accumulate in the nucleus in response to cytokinin, and this relocalization depends on the histidine kinases and the downstream histidine-containing phosphotransfer proteins, but is independent of the ARRs. Analysis of loss-of-function mutations reveals that the CRFs function redundantly to regulate the development of embryos, cotyledons, and leaves. Furthermore, the CRFs mediate a large fraction of the transcriptional response to cytokinin, affecting a set of cytokinin-responsive genes that largely overlaps with type-B ARR targets. These results indicate that the CRF proteins function in tandem with the type-B ARRs to mediate the initial cytokinin response. Thus, the evolutionarily ancient two-component system that is used by cytokinin branches to incorporate a unique family of plant-specific transcription factors.
Keywords: cell signaling, plant hormones
Cytokinins are N6 substituted adenine derivatives that were first identified by their ability to promote division in cultured plant cells together with a second hormone, auxin (1). Cytokinins have since been shown to play a role in diverse aspects of plant growth and development, including cell division, shoot initiation, apical meristem function, and vascular formation (2, 3). Recently, remarkable progress has been made in our understanding of cytokinin biosynthesis, metabolism, and perception. The genes encoding the key biosynthetic enzymes have been identified in plants, as have genes encoding several important cytokinin metabolic enzymes (4). A model for cytokinin perception and signaling has emerged that is similar to bacterial two-component phosphorelays (5). Binding of cytokinins to the Arabidopsis sensor histidine kinases (AHKs) initiates a phosphorelay in which the Arabidopsis histidine-containing phosphotransfer proteins (AHPs) are phosphorylated and then translocate into the nucleus where they likely transfer the phosphate to the Arabidopsis type-B response regulators (ARRs) (6–10). The type-B ARRs play a role in meditating the transcriptional response to cytokinin, including the induction of a second class of response regulators called the type-A ARRs (6, 11). The type-A ARRs are cytokinin primary response genes that act as highly redundant negative regulators of the primary signal transduction pathway (12–14). The pseudophosphotransfer protein AHP6 also acts as a negative regulator of cytokinin signaling, and plays a role in vascular development (15).
Gene expression in response to cytokinin has been extensively studied, and numerous genes have been identified that are transcriptionally up-regulated in response to cytokinin (16–18) including two members of the AP2/ERF superfamily of transcription factors within the ethylene response factor (ERF) family (17). Here, we demonstrate that this subgroup of AP2 transcription factors moves into the nucleus in response to cytokinin, and that they mediate, together with the type-B ARRs, the transcriptional response to cytokinin.
Results and Discussion
The CRF Genes Are Transcriptionally Induced by Cytokinin in a Type-B ARR-Dependent Manner.
The ERF family is comprised of 65 genes (19), several of which have been implicated in the response to the plant hormones ethylene, jasmonic acid, and cytokinin (20–22). Phylogenetic analysis of the predicted ERF-like proteins places the two cytokinin up-regulated genes in a subclade of six previously uncharacterized genes that fall into three pairs based on sequence similarity (Fig. 1B; see Figs. 5 and 6, which are published as supporting information on the PNAS web site). We named these six genes CYTOKININ RESPONSE FACTORS (CRFs) to reflect their similarity to ERFs and the observation that some members are transcriptionally up-regulated by cytokinin.
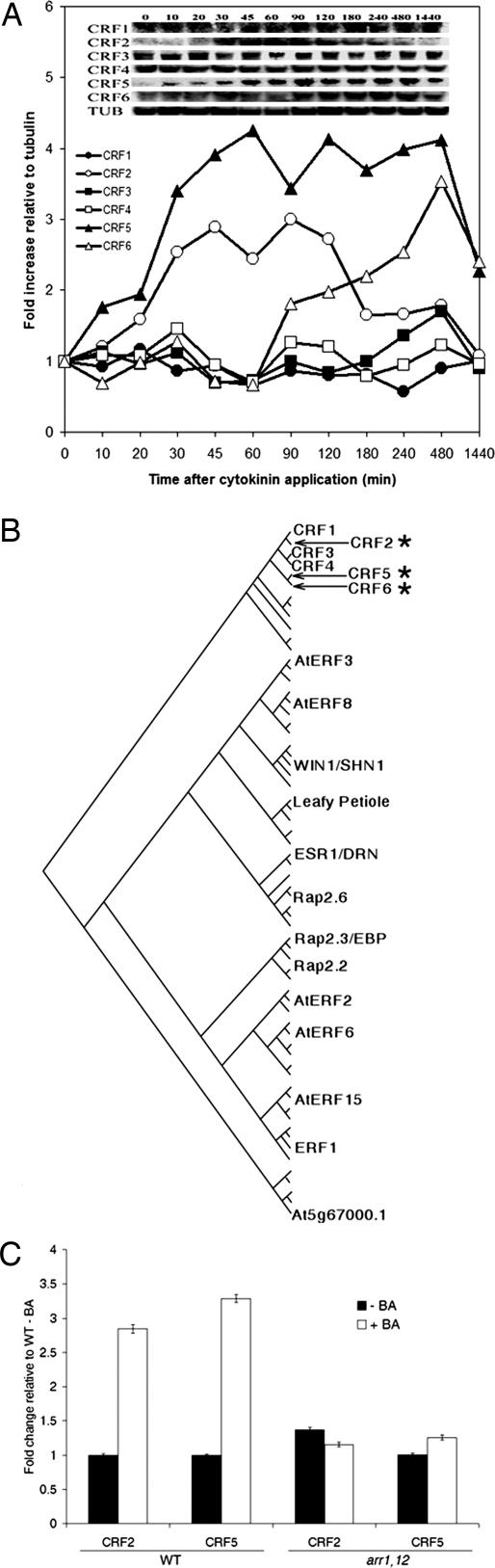
Fig. 1.
Induction of CRF genes by cytokinin. (A) Levels of CRF transcripts after cytokinin treatment were examined by using Northern blot analysis. (B) An unrooted phylogenic tree of the 65 ERF predicted proteins. Representative proteins are indicated; for a full list, see Fig. 5. The CRFs comprise a distinct subfamily with the cytokinin inducible members indicated by an asterisk. (C) Cytokinin induction of CRF transcripts requires type-B ARRs. Real-time PCR of CRF2 and CRF5 confirm that they are induced after cytokinin treatment (10 μM BA) for 1 h relative to a DMSO control in wild type, but are not induced in the type-B mutant arr1,12.
We examined the expression of the CRF genes in response to cytokinin by northern analysis. Consistent with previously reported microarray data (see Table 2, which is published as supporting information on the PNAS web site), the CRF2 and CRF5 transcripts are up-regulated 2- to 4-fold by cytokinin (Fig. 1A). The induction of both genes is rapid (<30 min), although CRF2 expression peaks at ≈60 min and then begins to decline, whereas induction of CRF5 is more sustained. CRF6 expression is also up-regulated by cytokinin, but more slowly. In contrast, the transcript levels of CRF1, CRF3, and CRF4 show little or no change in response to cytokinin (Fig. 1A and Table 2). The induction of CRF2 and CRF5 by cytokinin depends on the type-B ARRs, as an arr1,12 double mutant (23) severely reduces the response of these genes to cytokinin (Fig. 1C). Examination of the 5′ region of the CRF2 and CRF5 genes reveals the presence of multiple type-B consensus binding sites (17, 24), consistent with these CRFs being direct targets of type-B ARRs.
CRF Proteins Rapidly Relocalize to the Nucleus in Response to Cytokinin.
To explore the subcellular localization of the CRF proteins, we examined protoplasts transformed with GFP:CRF fusions expressed from the cauliflower mosaic (CaMV) 35S promoter. In the absence of cytokinin, we observed relatively uniform fluorescence throughout the cell. After treatment with cytokinin, all six GFP:CRF fusion proteins accumulated in the nucleus (Fig. 2A; see Fig. 7, which is published as supporting information on the PNAS web site). This nuclear accumulation was never observed in protoplasts treated with a DMSO vehicle control. Nuclear accumulation in response to cytokinin was observed in >50% of the transformed protoplasts and was very was rapid, occurring within 5–10 min, which is consistent with a relocation of preexisting protein. Additionally, this characteristic is independent of transcriptional induction, as these CRF fusions were driven by the constitutive CaMV 35S promoter. Cytokinin-induced nuclear accumulation is not a general property of ERF transcription factors, as two other ERFs that cluster outside of the CRF subgroup display constitutive nuclear localization (25). The rapid cytokinin-induced intracellular relocalization of CRFs was also observed in wild-type Arabidopsis stably transformed with CRF2:GFP, CRF5:GFP, or CRF6:GFP translation fusions expressed from their native promoters (Fig. 2C; data not shown). This relocalization was observed in the majority of the root hair cells that were examined, and occurred in multiple other cell types as well (data not shown). Consistent with the rapid kinetics, treatment with cycloheximide (an inhibitor of protein synthesis) did not inhibit nuclear accumulation of CRF5:GFP in seedlings (Fig. 2C) or in protoplasts (data not shown), indicating that preexisting CRF protein moves into the nucleus.
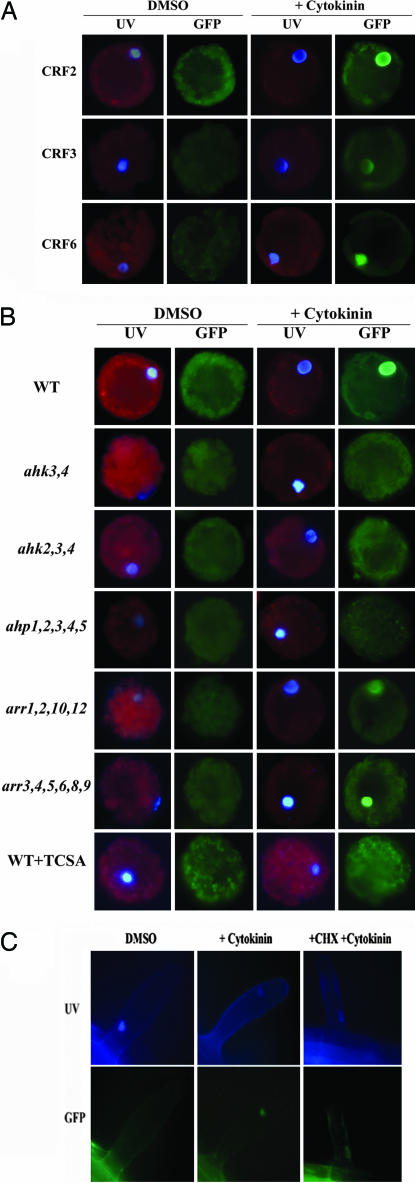
Fig. 2.
CRF proteins accumulate in the nucleus in response to cytokinin. (A) CaMV 35S:GFP:CRF constructs were transformed into wild-type protoplasts and examined by using epifluorescent microscopy after treatment with cytokinin or DMSO control as indicated for 5–10 min. (B) CRF protein nuclear localization requires the AHKs and AHPs. A 35S:GFP:CRF2 construct was transformed and examined following cytokinin treatment in protoplasts from wild type, ahk3,4, ahk2,3,4, ahp1,2,3,4,5, arr1,2,10,12, or arr3,4,5,6,8,9 leaves. Additionally, wild-type protoplasts were treated with 10 mM of the histidine kinase inhibitor TCSA for 30 min before treatment. (C) Visualization of CRF5:GFP in root hair cell of a transgenic plant harboring a genomic CRF5 fragment fused to GFP. Seedlings were examined after cytokinin treatment as in A.
CRF Protein Relocalization Defines a Branch Point in the Cytokinin Phosphorelay Response Pathway.
We examined the role of two-component elements in the cytokinin-induced nuclear accumulation of the CRFs. To this end, CaMV 35S:GFP:CRF2 constructs were transformed into protoplasts derived from cytokinin signaling mutants. In homozygous double ahk3,4 and triple ahk2,3,4 receptor mutant protoplasts, the distribution of the fluorescence signal from the GFP:CRF2 fusion protein was not altered by the addition of cytokinin (Fig. 2B), indicating that cytokinin-mediated nuclear accumulation of the CRFs requires the cytokinin AHK receptors. The histidine kinase inhibitor 3,3′,4′,5-tetrachlorosalicylanilide (TCSA) also blocked the movement of 35S:GFP:CRF2 in response to cytokinin (Fig. 2B), indicating that the histidine kinase activity of the cytokinin receptors is required for relocalization.
Functioning immediately downstream of the cytokinin receptors are the AHPs, which like the CRFs, also exhibit cytokinin-dependent nuclear localization (6). As with the receptor triple mutants, no nuclear accumulation of GFP:CRF2 was observed in response to cytokinin in transformed ahp1,2,3,4,5 protoplasts (C.E.H. and J.J.K., unpublished data) (Fig. 2B). In contrast, disruption of multiple type-B ARRs (i.e., the arr1,2,10,12 mutant) did not affect CRF nuclear movement in response to cytokinin (Fig. 2B). This is unlikely a result of residual cytokinin responsiveness as the arr1,2,10,12 mutant is more insensitive to cytokinin in a variety of cytokinin response assays (data not shown) than is the ahk3,4 double mutant, which blocks the CRF nuclear accumulation. Likewise, disruption of multiple type-A ARRs (arr3,4,5,6,8,9) did not alter the distribution of the CRF proteins (Fig. 2B). Thus, both the AHPs and cytokinin receptors are required for the cytokinin-regulated movement of CRFs into the nucleus, but the downstream ARRs are not.
The CRFs Play a Role in the Development of Cotyledons, Leaves, and Embryos.
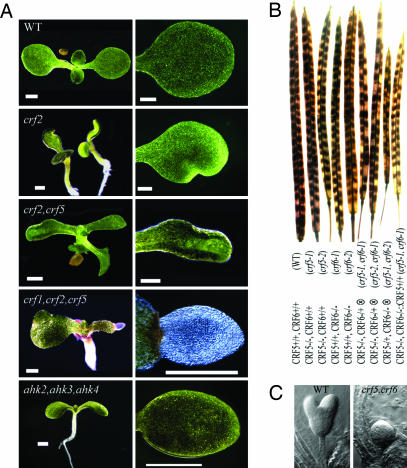
To determine the physiological role of the CRF genes, we characterized plants containing single and multiple loss-of-function CRF mutations (see Fig. 8, which is published as supporting information on the PNAS web site). Single crf mutants showed minor, poorly penetrant defects in cotyledon development (3–5%; Fig. 3A; see Table 3, which is published as supporting information on the PNAS web site). In all single CRF mutant alleles identified, small notches appear in the cotyledons and occasionally the first true leaves, suggesting localized areas of restricted cell expansion or division. The penetrance of the phenotype increased when seedlings were grown at elevated temperatures (data not shown), as has been observed in the asymmetric leaves (as1 and as2) and cup-shaped cotyledons (cuc1 and cuc2) mutants (26, 27). In multiple crf mutants, the penetrance of the phenotype increases, reaching up to ≈50% in the crf1,2,5 triple mutant. These results indicate genetic redundancy among the CRFs. The severity of the phenotype also increases as more genes are disrupted (Fig. 3A). Cotyledon development in the triple crf1,2,5 mutant is severely affected, forming cotyledons that are greatly reduced in size and translucent or white in color. This reduced cotyledon size is primarily the result of reduced cell expansion, as the area of severely affected cotyledons was reduced by almost 96%, but epidermal cell number was reduced only ≈30% (wild type 27.7 ± 1.9 mm2 and 996 ± 103 cells vs. crf1,2,5 1.0 ± 0.1 mm2 and 705 ± 96 cells in 10-day-old seedlings). A triple cytokinin receptor mutant (ahk2,3,4) also displays reduced cotyledon size that results primarily from reduced cell expansion (ahk2,3,4 cotyledon size is 1.9 ± 0.2 mm2 and are comprised of 933 ± 21 cells in 10-day-old seedlings). The lack of cell expansion in the triple receptor mutant cotyledons is distinct from previous studies that found that the reduced size of the seventh leaf of the ahk2,3,4 mutants was the result of reduced cell number (28), which likely reflects distinct effects of the ahk mutations on these two different organs. However, the severely affected crf triple mutant cotyledons have additional defects not seen in the cytokinin receptor knockouts, such as the lack of normal pigmentation, suggesting that there are aspects of this phenotype that may be cytokinin independent.
Fig. 3.
Phenotypes of crf loss-of-function mutations. (A) Representative whole seedlings and a close-up of cotyledons. CRF mutant cotyledon defects increase in severity from a notch in the single mutants to an extreme lack of cell expansion of the triple mutant. The cytokinin triple receptor knockout mutant ahk2,ahk3,ahk4 also has small cotyledons, but no shape abnormality. (Scale bar, 1 mm.) (B) The double crf5,crf6 mutant is embryo lethal. Wild-type and single crf5 and crf6 mutant allele siliques display complete seed set, whereas the self-set (⊗) siliques of crf5/crf5,CRF6/crf6 or CRF5/crf5,crf6/crf6 individuals result in ≈25% aborted seeds. The transgenic addition of CRF5 cDNA complements the embryo lethal phenotype in a crf5/crf5,crf6/crf6 double mutant background. (C) Developing embryos, shown from a self-fertilized (⊗) crf5/crf5,CRF6/crf6 plant, reveals ≈25% of the embryos fail to develop past the early heart stage.
Interestingly, cytokinins have been closely linked to cotyledon expansion, and this expansion was used as a bioassay to quantify cytokinin until the advent of MS-based methods. The CRFs may mediate the effect of cytokinin on cotyledon cell expansion. In addition to cotyledons, the crf mutants also occasionally affect juvenile leaves, but otherwise do not have any other obvious effect on morphology or development, except for the embryo-lethal phenotype of the crf5, crf6 double mutant discussed below.
In contrast to other crf mutant combinations tested, all combinations of independent alleles of both crf5 and crf6 resulted in embryo lethality that was fully penetrant (Fig. 3B; data not shown). In self-fertilized crf5–1/crf5–1 crf6–1/CRF6, crf5–2/crf5–2 crf6–1/CRF6, or crf5–1/CRF5 crf6–2/crf6–2 plants, ≈25% of the seeds are absent from the silique (Fig. 3B). Approximately 25% of the embryos in these siliques never progress beyond the late globular to early heart stage of development (Fig. 3C). However, none of the single crf5 and crf6 mutant alleles display any detectable embryo defects. In addition, the crf5–1/crf5–1 crf6–1/crf6–1 embryo lethal phenotype can be fully complemented by transformation with a genomic CRF5 fragment (Fig. 3B). These results suggest that the CRF5 and CRF6 genes are redundant and necessary for embryo development.
Unlike mutants in the Arabidopsis two-component signaling pathway, we found little effect of crf single, double and triple mutants in other cytokinin response assays. For example, there is little or no significant effect of these mutants on the response of seedlings to cytokinin in root elongation (see Fig. 9, which is published as supporting information on the PNAS web site), and only very minor effects on in vitro shoot initiation assays (see Fig. 10, which is published as supporting information on the PNAS web site). Furthermore, the morphology of crf mutant seedlings grown on cytokinin did not differ substantial from the wild type (data not shown).
The CRFs Mediate Gene Expression in Response to Cytokinin Together with the Type-B ARRs.
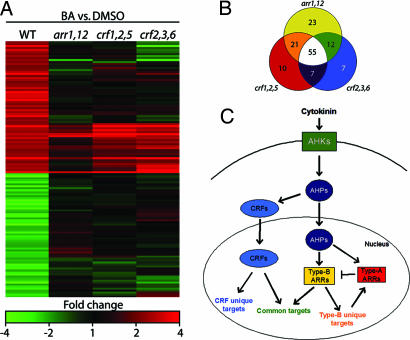
Because the CRFs are predicted to function in the regulation of gene expression, we examined the response of the transcriptome to cytokinin in the crf mutants. Wild-type and crf mutant seedlings were treated with cytokinin for 1 h and gene expression examined in triplicate by using a 29,000 element Arabidopsis oligonucleotide microarray (Fig. 4A; see Table 4 and Fig. 11, which are published as supporting information on the PNAS web site). Strikingly, of 169 genes whose transcript level was altered by cytokinin in wild-type seedlings, 93 (55%) exhibited a 2-fold or greater decrease in their responsiveness in the crf1,2,5 mutant, and 81 (48%) in the crf2,3,6 mutant (Fig. 4B). This indicates that CRF genes regulate a significant portion of the early transcriptional response to cytokinin. The effect of the crf triple mutants on several of these genes was verified by using quantitative PCR (Table 1).
Fig. 4.
CRFs act in parallel with type-B ARRs to mediate cytokinin regulated gene expression. (A) Wild-type, arr1,12, crf1,2,5, and crf2,3,6 seedlings were treated with either 10 μM BA or a DMSO control for 1 h and gene expression analyzed by using a microarray. Genes that displayed a ≥2-fold change in response to cytokinin in the wild type are shown. (B) Venn diagram of the 135 cytokinin-regulated genes affected by the arr1,12, crf1,2,5, and/or crf2,3,6 mutations. (C) Model of cytokinin signaling. Both AHPs and CRFs move into the nucleus in response to cytokinin. Once there, the AHPs phosphorylate the type-B ARRs, which, together with CRFs, mediate cytokinin-regulated gene expression. See text for further details.
Table 1.
Real-time PCR confirmation of cytokinin-regulated microarray results
| Gene name | At number | Wild type + vs. − BA |
crf1,2,5 + vs. − BA |
crf2,3,6 + vs. − BA |
|||
|---|---|---|---|---|---|---|---|
| Microarray | RT-PCR | Microarray | RT-PCR | Microarray | RT-PCR | ||
| Zinc Finger B-box | At1g68520 | 3.1 | 2.8 ± 0.3 | 1.0 | 1.3 ± 0.1 | 1.1 | 1.1 ± 0.1 |
| NAM AtNAC6 | At4g27410 | 2.7 | 2.7 ± 0.2 | 1.3 | 0.7 ± 0.1 | 1.4 | 1.0 ± 0.1 |
| Speckled POZ | At3g48360 | 3.5 | 2.7 ± 0.3 | 1.1 | 0.8 ± 0.1 | 0.9 | 1.0 ± 0.5 |
| Senescene Assoc. P. | At1g53885 | 3.9 | 2.7 ± 0.3 | 1.3 | 1.7 ± 0.1 | 2.0 | 1.5 ± 0.3 |
| ARR5 | At3g48100 | 7.3 | 14.3 ± 0.4 | 3.2 | 26.6 ± 1.1 | 5.0 | 25.4 ± 0.9 |
| ARR7 | At1g19050 | 3.6 | 9.5 ± 0.3 | 5.7 | 15.1 ± 0.6 | 5.5 | 12.1 ± 0.4 |
Four genes regulated by cytokinin in wild type, but not in CRF mutants, were examined by using real-time PCR, as well as two type-A ARR genes and show similar trends to the microarrays in Fig. 4A.
Comparison of the cytokinin response in a type-B ARR double mutant and the crf mutants revealed substantial overlap among the genes regulated by these two divergent families of transcription factors. In the arr1,12, mutant, 111 genes (66%) exhibited a 2-fold or greater decrease in their cytokinin responsiveness as compared to wild type, with 68% of those affected in arr1,12 similarly affected in the crf1,2,5 mutant and 60% in the crf2,3,6 mutant (Fig. 4B). Thus, the CRF and type-B ARR genes regulate an overlapping set of cytokinin-response genes. However, not all of these genes are similarly regulated, most notably the type-A ARRs, which exhibit reduced responsiveness to cytokinin in the arr1,12 mutant, still exhibit a wild type or greater level of induction in the crf1,2,5 and crf2,3,6 mutants (Table 1; see Table 4). The differential regulation of the type-A ARRs may contribute to the difference in cytokinin-dependent phenotypes observed in the crf and type-B arr mutants.
Model for Cytokinin Response Pathway.
These data are consistent with a model for cytokinin function illustrated in Fig. 4C. Cytokinin binding to the AHKs initiates a phosphorelay that results in the phosphorylation of the AHPs. The phosphorylated AHPs move into the nucleus to activate the type-B ARRs, which then increase the transcription of a subset of the CRFs. In parallel, the phosphorylated AHPs induce CRF proteins to accumulate in the nucleus. Thus, the relocalization of the CRF proteins defines a branch point in the cytokinin two-component signal transduction pathway. The activated CRFs, together with the activated type-B ARRs, mediate cytokinin-regulated gene expression, affecting an overlapping set of gene targets. In addition to mediating a portion of the cytokinin response initiated at the AHK receptors, the CRFs may also play a role in other signaling or developmental pathways, based on the difference in phenotypes associated with the crf loss-of-function mutations compared to those of the cytokinin receptor mutants (29, 30). In particular, the cytokinin receptor triple mutant is a viable plant that is distinct from the embryo lethal phenotype of the crf5,crf6 double mutant. Thus, CRFs are likely to receive input either from other cytokinin receptors, or from non-cytokinin-dependent signaling sources.
Materials and Methods
Plant Materials and Treatments.
Seedlings were grown under standard growth conditions as described (13), except for microarray experiments which were grown as described (23). Mutant lines are described in detail (see Supporting Text, which is published as supporting information on the PNAS web site). Cytokinin treatments were 1 μM N6-benzyladenine (BA) for various times for Northern analysis, 2 μM BA for 10 min for protoplast experiments, and 10 μM for 1 h in microarray and real-time PCR experiments (14-day-old plants). Cycloheximide was used at 50 μM as 1h pretreatments. Stable transformation of Arabidopsis was performed by floral dip method (31). Arabidopsis leaf mesophyll protoplasts were isolated and transformed by electroporation as previously described (32) with minor modifications as detailed (see Supporting Text). Plasmids used for transformations are described (see Supporting Text).
Microscopy.
Epiflorescent microscopy with Hoechst dye 33342 (1 ng·ml−1) and an UV source were used to observe cells and identify nuclei. A GFP filter that blocks chlorophyll fluorescence and Hoechst dye 33342 fluorescence was used to examine localization of GFP fusion proteins. Further details of the microscopy are presented in Supporting Text.
Microarray Analysis.
Microarray analyses were conducted by using the 29,000-element Arabidopsis oligonucleotide microarrays (http://ag.arizona.edu/microarray). Genes in these analyses were considered cytokinin regulated if they were found to be significant by a Welch’s t test with an adjusted Bonferroni correction after sample/column normalization (TIGR MeV v3.1 available at www.tm4.org) and had a mean fold change ≥2 or ≤ −2. Further details of the microarray analysis are given in Supporting Text.
Real-Time PCR.
Real-time PCR was carried out (see Supporting Text). At least two biological and two sample replicates were performed for each treatment. Ct values were generated by subtracting blanks and the baseline average over cycles 1–10 with a 10× SD over the cycle range for each sample. Samples with melt curves that did not have a single distinct peak were excluded from further analysis. Fold-change of samples was calculated after normalization to β-tubulin Ct values from each sample.
Supplementary Material
Acknowledgments
We thank members of the J.J.K. laboratory for helpful suggestions. Financial support was provided by a Department of Energy grant (to J.J.K.), a National Institutes of Health grant (to J.J.K. and A.M.R.), a National Science Foundation grant (to J.J.K. and G.E.S.), and a U.S. Department of Agriculture/National Research Initiative Competitive Grants Program grant (to G.E.S.).
Abbreviations
- AHK
Arabidopsis histidine kinase
- AHP
Arabidopsis histidine-containing phosphotransfer protein
- ARR
Arabidopsis response regulator
- ERF
ethylene response factor
- CRF
cytokinin response factor
- CaMV
cauliflower mosaic virus.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Miller C. O., Skoog F., Von Saltza M. H., Strong F. J. Am. Chem. Soc. 1955;77:1392–1393. [Google Scholar]
- 2.Mok D. W., Mok M. C. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;89:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- 3.Davies P. J. Plant Hormones: Biosynthesis, Signal Transduction, Action! Dordrecht, The Netherlands: Kluwer Academic; 2004. [Google Scholar]
- 4.Kakimoto T. J. Plant Res. 2003;116:233–239. doi: 10.1007/s10265-003-0095-5. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira F. J., Kieber J. J. Curr. Opin. Plant Biol. 2005;8:518–525. doi: 10.1016/j.pbi.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 6.Hwang I., Sheen J. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 7.Ueguchi C., Sato S., Kato T., Tabata S. Plant Cell Physiol. 2001;42:751–755. doi: 10.1093/pcp/pce094. [DOI] [PubMed] [Google Scholar]
- 8.Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 9.Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., Yamashino T., Mizuno T. Plant Cell Physiol. 2001;41:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki T., Miwa K., Ishikawa K., Yamada H., Aiba H., Mizuno T. Plant Cell Physiol. 2001;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- 11.Sakai H., Honma T., Aoyama T., Sato S., Kato T., Tabata S., Oka A. Science. 2001;294:1519–1521. doi: 10.1126/science.1065201. [DOI] [PubMed] [Google Scholar]
- 12.Imamura A., Hanaki N., Nakamura A., Suzuki T., Taniguchi M., Kiba T., Ueguchi C., Sugiyama T., Mizuno T. Plant Cell Physiol. 1999;40:733–742. doi: 10.1093/oxfordjournals.pcp.a029600. [DOI] [PubMed] [Google Scholar]
- 13.To J. P. C., Haberer G., Ferreira F. J., Deruère J., Mason M. G., Schaller G. E., Alonso J. M., Ecker J. R., Kieber J. J. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandstatter I., Kieber J. J. Plant Cell. 1998;10:1009–1020. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mähönen A. P., Bonke M., Kauppinen L., Riikonon M., Benfey P., Helariutta Y. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiba T., Naitou T., Koizumi N., Yamashino T., Sakakibara H., Mizuno T. Plant Cell Physiol. 2005;46:339–355. doi: 10.1093/pcp/pci033. [DOI] [PubMed] [Google Scholar]
- 17.Rashotte A. M., Carson S. D. B., To J. P. C., Kieber J. J. Plant Physiol. 2003;123:184–194. doi: 10.1104/pp.103.021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenner W. G., Romanov G. A., Kollmer I., Burkle L., Schmülling T. Plant J. 2005;44:314–333. doi: 10.1111/j.1365-313X.2005.02530.x. [DOI] [PubMed] [Google Scholar]
- 19.Nakano T., Suzuki K., Fujimura T., Shinshi H. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohme-Takagi M., Shinshi H. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGrath K. C., Dombrecht B., Manners J. M., Schenk P. M., Edgar C. I., Maclean D. J., Scheible W.-R., Udvardi M. K., Kazan K. Plant Physiol. 2005;139:949–959. doi: 10.1104/pp.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Fits L., Memelink J. Plant J. 2001;25:43–53. doi: 10.1046/j.1365-313x.2001.00932.x. [DOI] [PubMed] [Google Scholar]
- 23.Mason M. G., Mathews D. E., Argyros D. A., Maxwell B. B., Kieber J. J., Alonso J. M., Ecker J. R., Schaller G. E. Plant Cell. 2005;17:3007–3018. doi: 10.1105/tpc.105.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakai H., Aoyama T., Oka A. Plant J. 2000;24:703–711. doi: 10.1046/j.1365-313x.2000.00909.x. [DOI] [PubMed] [Google Scholar]
- 25.Koroleva O. A., Tomlinson M. L., Leader D., Shaw P., Doonan J. H. Plant J. 2005;41:162–174. doi: 10.1111/j.1365-313X.2004.02281.x. [DOI] [PubMed] [Google Scholar]
- 26.Qi Y., Sun Y., Xu L., Xu Y., Huang H. Planta. 2004;219:270–276. doi: 10.1007/s00425-004-1248-z. [DOI] [PubMed] [Google Scholar]
- 27.Vroemen C. W., Mordhorst A. P., Albrecht C., Kwaaitaal M. A. C. J., de Vries S. C. Plant Cell. 2003;15:1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riefler M., Novak O., Strnad M., Schmülling T. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi M., Pischke M. S., Mahonen A. P., Miyawaki K., Hashimoto Y., Seki M., Kobayashi M., Shinozaki K., Kato T., Tabata S., et al. Proc. Natl. Acad. Sci. USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clough S. J., Bent A. F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 32.Sheen J. Plant Cell. 1991;3:225–245. doi: 10.1105/tpc.3.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.