Abstract
The Escherichia coli RuvA–RuvB complex promotes branch migration of Holliday junction DNA, which is the central intermediate of homologous recombination. Like many DNA motor proteins, it is suggested that RuvA–RuvB promotes branch migration by driving helical rotation of the DNA. To clarify the RuvA–RuvB-mediated branch migration mechanism in more detail, we observed DNA rotation during Holliday junction branch migration by attaching a bead to one end of cruciform DNA that was fixed to a glass surface at the opposite end. Bead rotation was observed when RuvA, RuvB, and ATP were added to the solution. We measured the rotational rates of the beads caused by RuvA–RuvB-mediated branch migration at various ATP concentrations. The data provided a Km value of 65 μM and a Vmax value of 1.6 revolutions per second, which corresponds to 8.3 bp per second. This real-time observation of the DNA rotation not only allows us to measure the kinetics of the RuvA–RuvB-mediated branch migration, but also opens the possibility of elucidating the branch migration mechanism in detail.
Keywords: AAA+ ATPase family, motor protein, single-molecule analysis
Homologous recombination plays important biological roles in regulating genetic diversity and in repairing damaged chromosomes. One important intermediate of homologous recombination is a DNA structure called the Holliday junction that consists of two homologous duplex DNA molecules linked by a single-stranded crossover. In the late stage of the Escherichia coli recombination reaction, RuvA, RuvB, and RuvC proteins are involved in the processing of Holliday junction DNA into mature recombinant DNA molecules. The RuvA–RuvB complex is an ATP-dependent motor that promotes the branch migration of Holliday junction DNA (1, 2). RuvC is a structure-specific endonuclease that resolves Holliday junctions (2, 3). The nicked ends of the DNA molecules are sealed by DNA ligase to complete the recombination reaction.
Structural, biochemical and mutational analyses show that RuvA is a Holliday junction-specific DNA binding protein. One RuvA tetramer binds to the Holliday junction or two RuvA tetramers sandwich the Holliday junction, which unfolds the junction from the stacked X-structure into a square-planer conformation. RuvA has three consecutive domains (domains I, II, and III). Domains I and II are involved in DNA binding and tetramer formation. Domain III regulates branch migration through direct contact with RuvB (4). RuvB is a motor protein classified as a member of the AAA+ (ATPase associated with various cellular activities) family (5). Crystallographic studies of RuvB from Thermus thermophilus and Thermotoga maritima showed that RuvB has a crescent-like structure consisting of three consecutive domains (domains N, M, and C) (6, 7). The first two domains (domains N and M) compose the AAA+ ATPase domain, which is involved in ATP binding, ATP hydrolysis, and hexamer formation. A unique β-hairpin protruding from domain N, physically interacts with RuvA and is required for RuvA–RuvB complex formation (8, 9). The folding of domain C is similar to the winged helix DNA binding motif and may play a major role in pumping out DNA duplexes by directly interacting with DNA (10). Specific binding of RuvA to the junction is followed by the loading of RuvB hexameric rings and the formation of a tripartite structure, in which the RuvA tetramer is flanked by two RuvB hexameric rings on opposite sides (11). It has been suggested that the RuvB hexameric ring pumps out dsDNA through the RuvA octameric core with helical rotation of DNA to promote branch migration of Holliday junction DNA (12).
Recently, RuvA–RuvB-mediated Holliday junction branch migration was visualized by single-molecule analyses (13–15). The branch migration rates were measured by monitoring the height of tethered beads. Holliday junction DNA is anticipated to rotate through its helix accompanying branch migration. The visualization of DNA rotation is favorable for the measurement of torque produced by a motor protein as well as the high resolution of DNA translocation. However, the DNA rotation mediated by the RuvA–RuvB at Holliday junction during branch migration has never been observed. In this study, we constructed a system to observe the RuvA–RuvB-mediated DNA rotation at a Holliday junction directly through an optical microscope.
Results
Construction of the Observational System.
Structural and biochemical studies on the RuvA–RuvB-Holliday junction ternary complex imply that the RuvB hexameric rings promote branch migration of Holliday junction by driving a helical rotation of DNA that results in pulling out dsDNA passing through the cavities of the tetrameric RuvA sandwich (Fig. 1A) (12). When one end of the cruciform DNA is fixed to a glass surface and the RuvB hexameric rings bind to both sides of the junction along the horizontal DNA arms, the rotation of the lower DNA branch is restrained while this DNA segment is pulled into and expelled from the RuvA octameric core. This extends the horizontal DNA arms as the RuvA–RuvB complex, together with the horizontal DNA arms, rotates in a right-handed orientation (Fig. 1B Left). Because the upper DNA is also pulled into the RuvA octameric core with a right-handed rotation, a bead attached to the upper end is expected to undergo right-handed rotation as it is pulled down (Fig. 1 B Left and C). In contrast, when the RuvB hexameric ring binds to both sides of the junction along the vertical axis, left-handed rotation of the beads attached to the upper DNA branch is expected to accompany its upward movement (Fig. 1B Right).
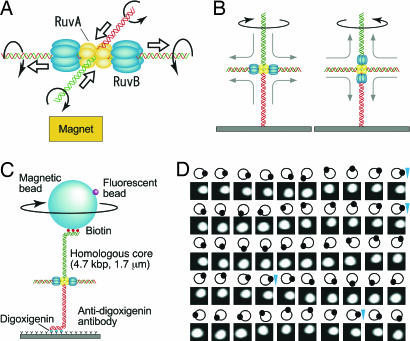
Fig. 1.
Observation of Holliday junction DNA rotation by RuvA–RuvB. (A) Model of branch migration by RuvA–RuvB. Branch migration in solution is depicted. The rotational direction of each DNA strand is indicated by an arrow. Thick arrows indicate the direction of DNA translocation. (B) One end of cruciform DNA is fixed to the glass surface. The RuvB hexameric rings bind to both sides of the junction along the horizontal DNA, the rotation of the lower DNA is restrained, and the right-handed helical rotation of DNA is observed from above (Left). In contrast, when the RuvB hexameric ring binds to both sides of the junction along the vertical axis, left-handed helical rotation of DNA is observed (Right). (C) Observation system (not to scale). The magnetic bead was pulled upwards by a disk-shaped neodymium magnet. The direction of the magnetic field was vertical and did not prevent bead rotation. Daughter fluorescent beads served as markers of rotation. (D) Snapshots of rotating beads at 100-ms intervals at an ATP concentration of 50 μM. The moving white spot is a daughter fluorescent bead. Diagrams show their relative positions. Blue arrowheads indicate completion of a turn.
To examine Holliday junction branch migration by the RuvA–RuvB complex, we used an optical microscopy technique based on the tethered-particle method. The observational system is depicted in Fig. 1C. We constructed Holliday junction DNA with a 4,700-bp (≈1.6 μm) homologous core (see Fig. 6, which is published as supporting information on the PNAS web site). The Holliday junction DNA was constructed with three biotin tags at one end and three digoxigenin tags at the opposite end to tether a magnetic bead to a glass surface. Magnetic beads were decorated with small fluorescent beads to observe their rotation. We pulled the magnetic bead upward with a magnet at ≈0.3 pN to confine the rotation in a horizontal plane. With this system, if DNA rotation induced by branch migration is completely transferred to rotation of the bead tethered to the DNA end, the rotation number of the bead would correspond to the sum of the helical rotation of the two opposed vertical DNAs. Because one turn of a DNA double helix consists of 10.4 bp, one revolution of the bead is estimated to correlate to 5.2 bp of Holliday junction branch migration. Branch migration was initiated by the addition of RuvA, RuvB, magnesium ion, and ATP to the chamber. We observed both clockwise (Fig. 1D and Movie 1, which is published as supporting information on the PNAS web site) and counterclockwise rotation of the beads (Movie 2, which is published as supporting information on the PNAS web site). We also observed that some counterclockwise rotating beads were released, indicating that the horizontal DNA was pulled into the RuvA–RuvB complex to the end (data not shown). We observed 72 rotating beads, and among them, 59% rotated clockwise. We could not observe the maximum number of rotations expected from the length of DNA, which corresponds to ≈900 revolutions, because we had to search over several fields of view before finding a continuously rotating bead and the Holliday junction DNA had no heterologous region to prevent spontaneous branch migration.
Kinetics of RuvA–RuvB-Mediated Branch Migration of Holliday Junction DNA with the Tethered-Particle Method.
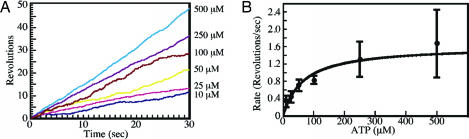
To analyze the kinetics of branch migration rates by RuvA–RuvB, we measured the rotational rates of beads at various ATP concentrations. Time courses of rotation of individual beads are shown in Fig. 2A. As shown in Fig. 2B, the RuvA–RuvB-mediated branch migration activity exhibits Michaelis–Menten kinetics over the ATP concentration range examined (10–500 μM). These data provide a Km value of 65 μM and a Vmax value of 1.6 revolutions per second (rps). To rotate a bead of diameter D = 680 nm in bulk water at this speed, a torque of Γ = 2π[1.6 rps]ξ ≈ 9.9 pN nm is required, where ξ = πηD3 is the rotational frictional drag coefficient and η (= 10−9 pN·nm−2·s) is the viscosity of water.
Fig. 2.
Observation of Holliday junction branch migration. (A) Time courses of bead rotation at various ATP concentrations. Rotation angles were estimated from the images in Fig. 1C by centroid analysis (18). (B) ATP concentration dependence of the rotational rate. The rotational rates at the indicated ATP concentration are an average of 8–22 rotational beads. The error bars indicate the standard deviations.
Under 0.3 pN of tension, the critical torque Γc of DNA for the formation of plectonemes is 11 pN nm (16), which is slightly higher than the Γ described above. If the DNA rotation cannot be faithfully transferred to the rotation of the beads, Holliday junction DNA would be twisted by the DNA rotation, resulting in the formation of plectonemes. Therefore, we could not rule out the possibility that the apparent Vmax value did not reach the maximal rotational rate due to the low critical torque under 0.3 pN of tension. Therefore, we also measured the rotational rate of beads under 2 pN of tension where the critical torque is 26 pN nm (16). If the apparent Vmax value did not reach to the maximal rotational rate due to the low critical torque, the rotational rate under 2 pN of tension is expected to be larger than that under 0.3 pN. However, the rotational rate was ≈1.6 rps, which is comparable to that under 0.3 pN of tension, indicating that the apparent Vmax value is the maximal rotation rate with our observation system (data not shown).
Pause and Resumption of Branch Migration.
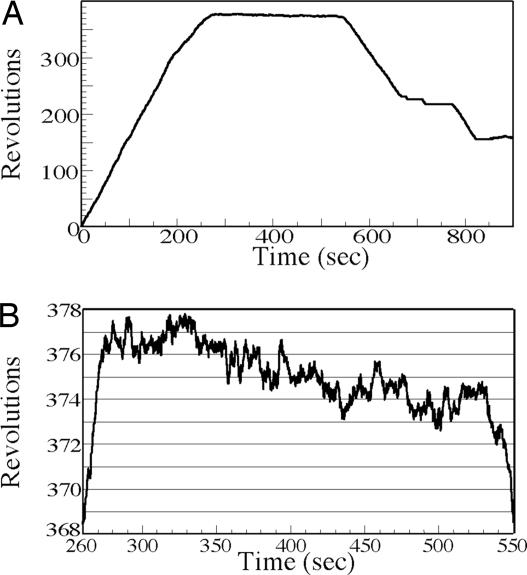
We also observed that some of the beads that rotated changed their direction after a pause (Fig. 3 and Movie 3, which is published as supporting information on the PNAS web site). A similar result was obtained by Stavans and colleagues (15), although their detection system monitored DNA length. Our observation supports the suggestion that pauses are caused by the dissociation of the RuvB motors or the RuvA–RuvB complex from the junction and that resumption of branch migration in either direction corresponds to the equal probabilities of reassembly of RuvB hexamers or the RuvA–RuvB complex on the two alternative sets of branches. In addition, the RuvB motors may simply pause and resume branch migration in the same direction. The paused beads fluctuated as shown in Fig. 3B and Movie 3. We observed 20 events of resumption of branch migration and the time of the pause was ≈100 s at any ATP concentrations (see Table 1, which is published as supporting information on the PNAS web site). Although we observed only 20 events, there was a direction bias upon resumption of branch migration (see Table 2, which is published as supporting information on the PNAS web site). When the clockwise rotating beads paused, the frequency of resumption accompanied by reversal of rotation was three times that of continued rotation in the same direction. Alternately, when the counterclockwise rotating beads paused, the frequency of resumption accompanied by reversal rotation was comparable with that by the same direction. This bias may be explained if pausing of some of the clockwise rotating beads was caused by close approach and capture of the RuvA–RuvB complex by the glass surface. Then, resumption of rotation for those beads would be possible only after reassembly of the complex on the other face of the Holliday junction resulting in reversal of rotation.
Fig. 3.
Branch migration of an individual Holliday junction. (A) The rotational number of a bead indicates clockwise rotation as positive. Branch migration is interrupted by a pause and then restarts in same direction or changes direction. (B) The state of the paused bead. The graph shows the time course of the bead in A from 260 to 550 s.
Biochemical Analysis of Branch Migration Rate by RuvA–RuvB.
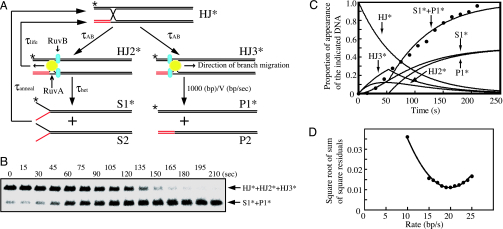
To confirm that the rotation rates of beads are consistent with the branch migration rates in solution, RuvA–RuvB-mediated branch migration was examined biochemically. As reported in ref. 14 and described in Supporting Text, which is published as supporting information on the PNAS web site, the Holliday junction DNA is converted into dsDNA products through a multistage process (Fig. 4A). To determine the branch migration rate, we formulated the difference equations as described in Supporting Text. So that we would not need to take into account the processivity of branch migration (more than 4,400 bp/s) (14), we used a 1-kbp arm of Holliday junction DNA in this study (see Fig. 7, which is published as supporting information on the PNAS web site). Therefore, we were able to use simplified equations compared to the previous report (14).
Fig. 4.
Biochemical analysis of Holliday junction branch migration. (A) Schematic diagram of RuvAB-mediated branch migration. Computer simulations follow the diagram, which is simplified in comparison to the diagram reported in ref. 14. (B) Branch migration assays were carried out as described in Materials and Methods. The result at 500 μM ATP concentration is shown. Aliquots were taken at the indicated times. Deproteinized reaction products were analyzed by 0.8% agarose gel electrophoresis containing 2 mM MgCl2. ∗, 32P labeling. (C) Time course of dissociated-product appearance during branch migration. Filled circles indicate experimental points of dissociated-product appearance during branch migration as shown in B. The lines represent computer-generated simulations of the appearance of the indicated substrates or products. (D) Minimization analysis for the best-fit branch migration rate. Square root of sum of square residuals plot versus branch migration rate was determined by constraining the branch migration rate (bp/s) to have a constant value between 10 and 25 and then optimizing the fit by using Eqs. 1–3, 5, and 6 described in Supporting Text.
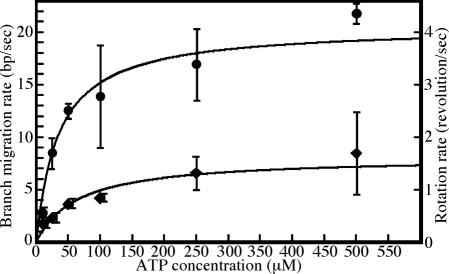
The biochemical time course shows the steep rise in dsDNA appearance (Fig. 4C). These results indicate that a fraction of the Holliday junctions revert to the unpaired branch migration substrates (S1*+S2) before the kinked point and the kinked point corresponds to the beginning of appearance of the duplex products (P1*+P2) (14). To obtain the kinetic parameters shown in Fig. 4A, a nonlinear least-squares (NLLS) fitting of the data to Eqs. 1–3, 5, and 6 in Supporting Text was performed (Fig. 4C). We systematically constrained the branch migration rate to have values from 10 bp/s to 25 bp/s and then optimized the NLLS fit of the time course by allowing the remaining parameter to float for each branch migration rate. The square root of the sum of the square residuals obtained from the analysis for each value of V was plotted as shown in Fig. 4D. The absolute minimum of the square root of the sum of the square residuals occurs at a branch migration rate of 19 bp/s, which is consistent with the result reported previously (14). Using the NLLS analyses, we obtained the branch migration rates at various ATP concentrations (Fig. 5). These results provide a Km value of 35 μM and a Vmax of 20 bp/s. As described above, because the revolution number of a bead corresponds to the sum of the rotation of the opposed vertical DNAs, the rotation rate (rps) multiplied by 5.2 will be the branch migration rate (bp/s). Compared to the results obtained by the tethered particle method (Fig. 5), at an ATP concentration of 10 μM, the branch migration rates by both analyses were comparable to each other, whereas the maximal rate with the tethered particle method was calculated as 8.3 bp/s, which was 40% of that determined by biochemical analysis.
Fig. 5.
Comparison of the rotation rate of beads and branch migration rates. Filled diamonds, the average rotational rates of 8–22 rotational beads at the indicated ATP concentration. Filled circles, branch migration rate obtained by biochemical analysis (Fig. 4). The values are the average of three measurements at each ATP concentration. We consider that the rotational rate (revolutions/s) corresponds to 5.2 bp/s as described in the text.
Discussion
In this study, we constructed a system with which we successfully observed the rotation of Holliday junction DNA branch migration caused by the RuvA–RuvB protein complex using the tethered particle method (Fig. 1 C and D and Movies 1 and 2). We estimated the rates of branch migration from the rotation rates of the tethered beads at various ATP concentrations (Fig. 2). We also estimated the rates of branch migration determined from the time course of product appearance (Fig. 4). Both methods exhibited Michaelis–Menten kinetics, providing a Km of 65 μM and a Vmax of 1.6 rps using single-molecule analysis, and a Km of 35 μM and a Vmax of 20 bp/s using biochemical analysis (Fig. 5). Because the revolution number of a bead corresponds to the sum of the helical rotation of the opposing vertical DNAs, the rotational rate (rps) multiplied by 5.2 will yield the branch migration rate (bp/s) if the DNA rotation is faithfully transferred to the rotation of the beads. At an ATP concentration of 10 μM, the branch migration rates measured by the single-molecule and biochemical analysis were 1.8 ± 0.4 and 2.5 ± 0.5 bp/s, respectively (Fig. 5). These results indicate that the DNA rotation was faithfully transferred to the rotation of the beads. However, the Vmax value of the rotation rate was one-third of that determined by biochemical analysis.
Several possibilities are suggested to explain the reduction of the maximal rate. One of them is the difference of the buffer conditions between the single-molecule and biochemical analyses. To prevent the beads, DNA and protein from binding to glass surface nonspecifically, we used a buffer containing 300 mM KCl, 1 mg/ml α-casein and Hepes·HCl (pH 7.5). Therefore, we also measured the branch migration rate biochemically under various conditions (see Fig. 8 and Supporting Text, which are published as supporting information on the PNAS web site). The addition of α-casein as well as changing pH from 8.0 to 7.5 barely influenced the branch migration rate (Fig. 8 A, C, E, and F). Alternately, under the buffer condition containing 300 mM KCl, the branch migration rate decreased to 70% of the maximal rate determined by bulk measurements at the lower salt concentration (Fig. 8 B and D–F). However, because the Vmax value of the beads rotation is one-third of that by the biochemical analysis, the effect of 300 mM KCl alone does not fully account for the rate reduction.
As shown in Movies 1–3, most of the rotating beads stumble at a particular angle. Imperfect alignment of the magnetic field or mechanical resistance to the swinging motion of the horizontal DNA arms may explain the rotational stumble. Transient interaction of the horizontal DNA with the glass surface could also be a cause of the reduced maximal rate.
The frictional load of the bead rotation is another plausible reason for the slower rate. Further evaluation of the impact of rotational load on the maximum rate would require experiments under different load conditions. As described in Results, incomplete transfer of the DNA rotation to the bead rotation is unlikely to be the cause of the reduced maximum rate observed.
If the branch migration rate is slowed down by the torque load of the rotating bead, long genomic DNA with bound proteins surrounding the Holliday junction inside the cell would also slow down the branch migration by RuvA–RuvB complex. It is currently unknown whether mechanical load slows down the rate of hydrolysis of ATP. If the mechano-chemical coupling of the RuvB motor is tight, the ATPase rate would change in parallel to the branch migration rate. On the other hand, if the coupling is weak, slippage would happen with nonproductive ATP hydrolysis under high load conditions.
As described above, we estimate the torque produced by RuvA–RuvB to be ≈10 pN nm. The torques produced by E. coli RNA polymerase and F1-ATPase are reported as 6 and 40 pN nm, respectively (17, 18). Although the torque produced by the RuvA–RuvB complex is somewhat larger than that by RNA polymerase, it is significantly lower than that of F1-ATPase. The reason for the low torque produced by the RuvA–RuvB complex as well as RNA polymerase is unknown. One possibility is that the torque produced by DNA motor proteins may be limited to prevent overtwisting of the DNA. Further analyses of the DNA and/or RNA related proteins, including RuvA–RuvB and RNA polymerase, are required to elucidate the cause of the low torque.
It has been suggested that the step-wise DNA rotation through the RuvB hexamer from ATP-bound to ADP-bound to nucleotide-free states may couple ATP hydrolysis to DNA translocation (19). Therefore, one might have expected that step-wise rotation of the beads would have been observed. However, we did not observe step-wise rotation. This is understandable considering the relatively long DNA arm length and large rotational friction of the bead used here. To detect the step-wise movement, experiments must be conducted with shorter DNA arm length and with smaller beads with lower rotational friction.
Our system to observe DNA rotation directly by the tethered particle method is appropriate not only for analysis of RuvAB-mediated branch migration, but also for any other helicase. Recently, some of helicases (RecBCD, UvrD, Rep, etc.) have been analyzed by single-molecule techniques (20–23). RecBCD and UvrD are highly processive helicases, and furthermore, RecBCD is a highly processive nuclease, hence their motion could be visualized (20, 21, 23). Rep was analyzed by a refined single-molecule FRET technique (22). However, because most helicases have low processivity to separate double-helical nucleic acids into single strands, it is relatively difficult to resolve changes in the length of the DNA. The unwinding or rewinding of double-stranded nucleic acids accompanies a helical rotation of the nucleic acids, which could be detectable with systems similar to the one reported here. Therefore, our system would also be useful in the elucidation of the detailed mechanism of other helicases.
Materials and Methods
Materials.
RuvA and RuvB proteins were purified as described (8).
Streptavidin-coated fluorescent magnetic beads were constructed as described (17). Fluorescent, carboxylated microbeads (20 nm, excitation 580 nm, emission 605 nm, Molecular Probes) were amino-derivatized with ethylenediamine in the presence of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC). The microbeads and Biotin-X cadaverine (Molecular Probes) were conjugated to 680-nm carboxylated magnetic beads (Seradyn) with EDC, and streptavidin was bound to the biotinylated magnetic beads.
Construction of Holliday Junction.
Details are given in Supporting Text.
Bead Rotation Assays.
A flow chamber was made of two coverslips separated by two spacers of 50 μm thickness. The flow chamber was incubated with anti-digoxigenin antibody (15 μg/ml) in an HBS buffer (20 mM Hepes·HCl, pH 7.5/300 mM KCl/1 mM EDTA) for 10 min at room temperature. The flow chamber was then incubated with α-casein (1 mg/ml) in an HBS buffer for 10 min at room temperature to reduce nonspecific DNA and protein binding to the glass surface. Holliday junction DNA was infused into the flow chamber and incubated for 10 min at room temperature to attach it to the glass surface by its digoxigenin end. The flow chamber was washed with HBS buffer and then incubated with streptavidin-coated magnetic beads for 10 min at room temperature to attach it to Holliday junction DNA by a biotin tag on the DNA. A magnet was positioned above the chamber to pull the magnetic beads upward, and then HBS buffer containing RuvA (100 nM), RuvB (300 nM), MgCl2 (10 mM), and the indicated concentration of ATP were introduced into the chamber to start Holliday junction branch migration.
Microscopy.
Samples were observed at 26 ± 2°C on an Olympus IX71 inverted microscope with a 100× oil-immersion objective. Fluorescent daughter beads were imaged with standard epifluorescence optics. Superimposed bright field and fluorescence images were projected on an electron bombardment CCD camera (C7190–23, Hamamatsu Photonics) and recorded on videotape. A disk-shaped neodymium magnet was placed above the sample to pull the beads (Fig. 1C). The vertical pulling force was calibrated by tethering the magnetic beads with 16-μm-long λ phage DNA and measuring the height of the tethered beads.
Branch Migration Assays.
The ATP-dependent branch migration activity of the RuvA–RuvB complex was assayed by measuring dissociation of the Holliday junctions. The standard reaction mixture (160 μl) contained 20 mM Tris-acetate (pH 8.0), 10 mM Mg(OAc)2, 1 mM DTT, the indicated concentration of ATP, 0.01% (wt/vol) BSA, 0.2 nM 32P-labeled Holliday junction DNA, 100 nM RuvA, and 300 nM RuvB. Reactions were initiated by the addition of RuvA and RuvB and incubated at 26°C. At the indicated time, 10 μl of reaction mixture was sampled and terminated by the addition of 5 μl of stop buffer (20 mM Tris·HCl, pH 7.5/50 mM EDTA/5 mg/ml proteinase K/2% SDS). The products were analyzed by 0.8% agarose gel electrophoresis containing 2 mM Mg(OAc)2 in TAM buffer and visualized with a PhosphorImager (Fuji BAS 1500).
Biochemical Analysis of Branch Migration Kinetics.
Details are given in Supporting Text.
Supplementary Material
Acknowledgments
We thank Drs. K. Yamada, H. Yokota, Y. Sasuga, and T. Miki for helpful discussion. We appreciate Ms. K. Terada for correcting English. This work was supported in part by a research grant from Core Research for Evolutional Science and Technology (CREST) of the Japan Science and Technology Agency (JST); by a Grant-in-Aid for Scientific Research by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to Y.H. and H.S.); and by the Human Frontier Science Program (to Y.H.). Y.H. acknowledges the financial support from the Toray Science Foundation. Y.-W.H. was supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
Abbreviation
- rps
revolutions per second.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Iwasaki H., Takahagi M., Nakata A., Shinagawa H. Genes Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 2.West S. C. Annu. Rev. Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 3.Iwasaki H., Takahagi M., Shiba T., Nakata A., Shinagawa H. EMBO J. 1991;10:4381–4389. doi: 10.1002/j.1460-2075.1991.tb05016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishino T., Iwasaki H., Kataoka M., Ariyoshi M., Fujita T., Shinagawa H., Morikawa K. J. Mol. Biol. 2000;298:407–416. doi: 10.1006/jmbi.2000.3675. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki H., Han Y. W., Okamoto T., Ohnishi T., Yoshikawa M., Yamada K., Toh H., Daiyasu H., Ogura T., Shinagawa H. Mol. Microbiol. 2000;36:528–538. doi: 10.1046/j.1365-2958.2000.01842.x. [DOI] [PubMed] [Google Scholar]
- 6.Yamada K., Kunishima N., Mayanagi K., Ohnishi T., Nishino T., Iwasaki H., Shinagawa H., Morikawa K. Proc. Natl. Acad. Sci. USA. 2001;98:1442–1447. doi: 10.1073/pnas.031470598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putnam C. D., Clancy S. B., Tsuruta H., Gonzalez S., Wetmur J. G., Tainer J. A. J. Mol. Biol. 2001;311:297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
- 8.Han Y. W., Iwasaki H., Miyata T., Mayanagi K., Yamada K., Morikawa K., Shinagawa H. J. Biol. Chem. 2001;276:35024–35028. doi: 10.1074/jbc.M103611200. [DOI] [PubMed] [Google Scholar]
- 9.Yamada K., Miyata T., Tsuchiya D., Oyama T., Fujiwara Y., Ohnishi T., Iwasaki H., Shinagawa H., Ariyoshi M., Mayanagi K., Morikawa K. Mol. Cell. 2002;10:671–681. doi: 10.1016/s1097-2765(02)00641-x. [DOI] [PubMed] [Google Scholar]
- 10.Ohnishi T., Hishida T., Harada Y., Iwasaki H., Shinagawa H. J. Biol. Chem. 2005;280:30504–30510. doi: 10.1074/jbc.M502400200. [DOI] [PubMed] [Google Scholar]
- 11.Shinagawa H., Iwasaki H. Trends Biochem. Sci. 1996;21:107–111. [PubMed] [Google Scholar]
- 12.Hiom K., West S. C. Cell. 1995;80:787–793. doi: 10.1016/0092-8674(95)90357-7. [DOI] [PubMed] [Google Scholar]
- 13.Dawid A., Croquette V., Grigoriev M., Heslot F. Proc. Natl. Acad. Sci. USA. 2004;101:11611–11616. doi: 10.1073/pnas.0404369101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis C., Fedorov A., Kas E., Salome L., Grigoriev M. EMBO J. 2004;23:2413–2422. doi: 10.1038/sj.emboj.7600249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amit R., Gileadi O., Stavans J. Proc. Natl. Acad. Sci. USA. 2004;101:11605–11610. doi: 10.1073/pnas.0404332101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strick T., Allemond J.-F., Bensimon D., Lavery R., Croquette V. Physica A. 1999;263:392–404. [Google Scholar]
- 17.Harada Y., Ohara O., Takatsuki A., Itoh H., Shimamoto N., Kinosita K., Jr Nature. 2001;409:113–115. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 18.Yasuda R., Noji H., Kinosita K., Jr, Yoshida M. Cell. 1998;93:1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 19.Hishida T., Han Y. W., Fujimoto S., Iwasaki H., Shinagawa H. Proc. Natl. Acad. Sci. USA. 2004;101:9573–9577. doi: 10.1073/pnas.0403584101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dohoney K. M., Gelles J. Nature. 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 21.Bianco P. R., Brewer L. R., Corzett M., Balhorn R., Yeh Y., Kowalczykowski S. C., Baskin R. J. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 22.Ha T., Rasnik I., Cheng W., Babcock H. P., Gauss G. H., Lohman T. M., Chu S. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 23.Dessinges M. N., Lionnet T., Xi X. G., Bensimon D., Croquette V. Proc. Natl. Acad. Sci. USA. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.