Abstract
Widespread losses of heterozygosity (LOH) in human cancer have been thought to result from chromosomal instability caused by mutations affecting DNA repair/genome maintenance. However, the origin of LOH in most tumors is unknown. The present study examined the ability of carcinogenic agents to induce LOH at 53 sites throughout the genome of normal diploid mouse ES cells. Brief exposures to nontoxic levels of methylnitrosourea, diepoxybutane, mitomycin C, hydroxyurea, doxorubicin, and UV light stimulated LOH at all loci at frequencies ranging from 1–8 × 10−3 per cell (10–123 times higher than in untreated cells). This greatly exceeds the frequencies at which these agents have been reported to induce point mutations and is comparable to the rates of LOH observed in ES cells lacking the gene responsible for Bloom syndrome, an inherited DNA repair defect that results in greatly increased risk of cancer. These results suggest that LOH contributes significantly to the carcinogenicity of a variety of mutagens and raises the possibility that genome-wide LOH observed in some human cancers may reflect prior exposure to genotoxic agents rather than a state of chromosomal instability during the carcinogenic process. Finally, as a practical matter, chemically induced LOH is expected to enhance the recovery of homozygous recessive mutants from phenotype-based genetic screens in mammalian cells.
Keywords: cancer etiology, cancer genetics, carcinogenesis, genome stability
Cancer is thought to arise from the accumulation of somatic mutations in oncogenes and tumor suppressor genes that, when coupled with the selection of clones with increasing capacity for autonomous growth, results in the multistep conversion of normal cells to a malignant state (1, 2). Most cancers are caused by exposure to carcinogens present in the environment or produced by cellular metabolism, often influenced by specific lifestyles (3–5). However, it has become increasingly clear that cancer cells contain extensively altered genomes, widely attributed to an intrinsic state of genomic instability (6–8). Specific genes required to maintain genome integrity and that also function to prevent cancer have been identified in humans with familial cancer syndromes and in mouse knockout models. These include genes involved in recombination, DNA repair, mitotic spindle checkpoint control, and cell-cycle regulation (9–13). Because genomic instability can clearly drive carcinogenesis, presumably by enhancing the likelihood of mutations in oncogenes and tumor suppressor genes, and because chromosome alterations appear to have greater genetic impact than the accumulation of point mutations, genomic instability has been proposed to play a greater role in carcinogenesis than somatic mutations (7, 14–16). However, the origin of most genetic alterations in human cancer cells has not been established; hence, the relative importance of somatic mutations and genomic instability in carcinogenesis remains an active area of controversy (7, 15–17).
Allelic imbalance and losses of heterozygosity (LOH) are the most common genetic alterations in human cancers, which may harbor >10,000 regions of LOH per cell (18–21). LOH contributes to carcinogenesis by altering the dosage of genetically and epigenetically modified genes (22). These include >60 characterized recessive cancer genes (tumor suppressors; ref. 23) and other alleles that may enhance cell fitness. Although mutations in genes required for genome maintenance can produce high levels of LOH, except for a subset of tumors with microsatellite instabilities or those associated with inherited cancer susceptibility syndromes, most tumors appear to lack caretaker gene mutations (6, 14–16). Extensive LOH has been observed in nonmalignant lesions, in some cases at levels comparable to those of invasive tumors (18–21). Thus, genomic instability could be an early event in carcinogenesis. Alternatively, stem cells in the surrounding normal tissues could have equally high levels of LOH that escape detection because, in the absence of clonal growth, sufficiently pure cell populations are not available for analysis (24).
It has been argued that normal mutation rates are not sufficient to account for the levels of genetic alteration found in cancers (7), and, alternatively, that the prevalence of mutations is no higher than would be expected to accumulate in the stem cells, assuming many rounds of cell division (24). The issue is complicated by the possibility that stem cells may possess specialized mechanisms to suppress mutations, possibly as a defense against oncogenic transformation (25–29). Clearly, a better understanding of the origins of LOH will influence opinion about the relative roles of somatic mutations and genomic instability in carcinogenesis. We therefore addressed the question: to what extent are carcinogens, including agents commonly known to induce point mutations, capable of inducing genome-wide LOH in normal diploid stem cells?
To answer this question, genes tagged by a gene trap retrovirus were used to quantify carcinogen-induced LOH at 53 sites in the genome of normal ES cells. The entrapment clones were biologically normal, as assessed by their ability to produce germ-line chimeras and normal offspring, and thus lacked coincidental mutations affecting genome stability. By quantifying the frequencies of LOH at many sites in the genome, this study provides a genome-wide analysis of carcinogen-induced LOH. Finally, the use of ES cells permitted direct comparisons between the effects of chemical carcinogens and the Bloom’s syndrome mutation, a well characterized mutator phenotype that has also been analyzed in genetically deficient ES cells (30–32). We report that limited exposure to a variety of carcinogens induces genome-wide LOH at per-gene frequencies approaching one percent. In short, the carcinogens produced the appearance of chromosomal instability in normal stem cells in the absence of a genetically activated genomic instability phenotype.
Results and Discussion
Carcinogen-Induced LOH.
Carcinogen-induced LOH was measured in a panel of 53 mouse ES cell clones, each containing a neomycin resistance gene (Neo) inserted into a different cellular gene (Fig. 1) by the GTR1.3 gene trap retrovirus. Gene entrapment by GTR1.3 involves selection for inserted Neo sequences that can splice to the 3′ ends of cellular genes (Q.L., S.L.D., and H.E.R., unpublished work). The disrupted genes were identified by sequencing Neo-gene fusion transcripts and were localized on the mouse genome. Previous studies have shown that cells homozygous for GTR1.3-induced mutations can be selected from heterozygous cells simply by selecting for resistance to higher concentrations of G418, a method first shown to select for homozygous mutations induced by gene targeting (33). Mitotic recombination appears to be the preferred mechanism of spontaneous LOH involving Neo genes inserted in ES cells (34) and LOH involving other genes and cell types in vivo (35, 36). LOH doubles the number of Neo genes per cell and thus allows moderately resistant cells to acquire resistance to higher concentrations of G418. The frequencies of spontaneous LOH measured at the 53 different sites ranged from 1.3 × 10−5 to 1.2 × 10−4 (Table 2, which is published as supporting information on the PNAS web site), similar to those reported for other inserted neomycin resistance genes (33, 34) in ES cells, and for loci such as TK and APRT in other cell types (35, 36).
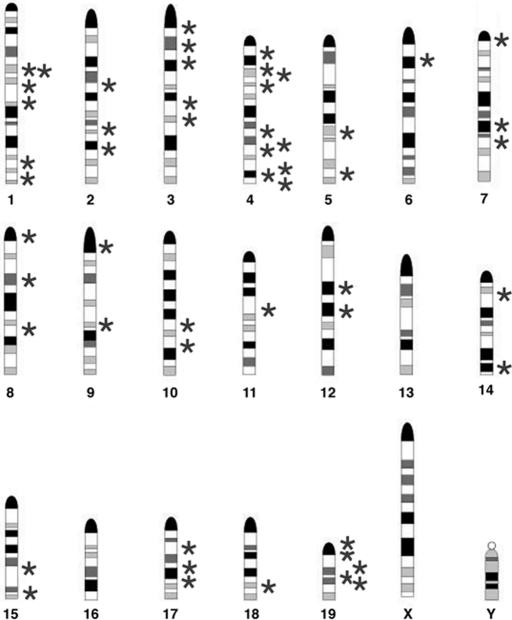
Fig. 1.
Distribution of entrapment mutations in the murine genome. Stars represent the locations of GTR1.3 retroviral vector inserts in 53 clones on murine chromosomes 1–15 and 17–19. The centromere for each chromosome is positioned at the top of the ideogram.
A variety of chemical agents were tested for their ability to enhance the frequencies at which mutant cells survive in 2.0 mg/ml G418. Methylnitrosurea (MNU) is an alkylating agent that produces a variety of monomethylated DNA adducts, hydroxyurea (HU) stalls DNA replication complexes, doxorubicin interferes with DNA synthesis, methotrexate is a competitive inhibitor of dihydrofolate reductase but is not genotoxic, diepoxybutane and mitomycin C induce interstrand DNA crosslinks, UV irradiation causes intra- and interstrand pyrimidine dimmers, and ethidium bromide intercalates between DNA strands to damage DNA (for additional information about these agents, see http://toxnet.nlm.nih.gov and http://lisntweb.swan.ac.uk/cmgt/index.htm). Treatment with 0.5 mM MNU or 0.25 mM HU dramatically increased the number of colonies surviving in high G418 (Fig. 2). The fold increase for MNU and HU ranged from 39–123 and 18–68, respectively (Table 2). The optimal concentrations of each agent to stimulate colony formation with minimal toxicity (<5% loss of cell viability) were determined in advance (Fig. 5, which is published as supporting information on the PNAS web site). Other genotoxic agents that have been reported to promote recombination doxorubicin (0.1 μM), diepoxybutane (100 ng/ml), mitomycin C (50 ng/ml), and UV light (5 J/m2) also enhanced colony formation by an average of 15-, 16-, 14-, and 10-fold. However, ethidium bromide (25 μg/ml) and methotrexate (50 μM) had no significant effect (Table 2 and data not shown). Each of these agents was tested two or more times on at least 25 entrapment lines.
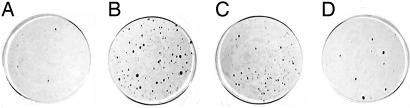
Fig. 2.
Limited carcinogen exposure enhances the survival of mutant ES cells in media containing 2.0 mg/ml G418. ES cells heterozygous for an entrapment mutation in Xrcc5 were selected in high G418 directly (A) or after treatment for 4 h with 0.5 mM MNU (B), 0.25 mM HU (C), or 100 ng/ml diepoxybutane (D). After 12 days in selection, colonies were washed with PBS and stained with crystal violet.
Genotypic analysis of five different mutants confirmed that 100% of colonies that survived high G418 selection after carcinogen treatment had undergone LOH (Fig. 3) compared to 85% of spontaneously resistant colonies. Thus, colony formation in 2.0 mg/ml G418 provided a direct measure of carcinogen-induced LOH at each entrapment locus. The overall extent of LOH induced by a single exposure to nontoxic levels of either MNU or HU was remarkably high (Table 2), in some cases exceeding 1% of the genome.
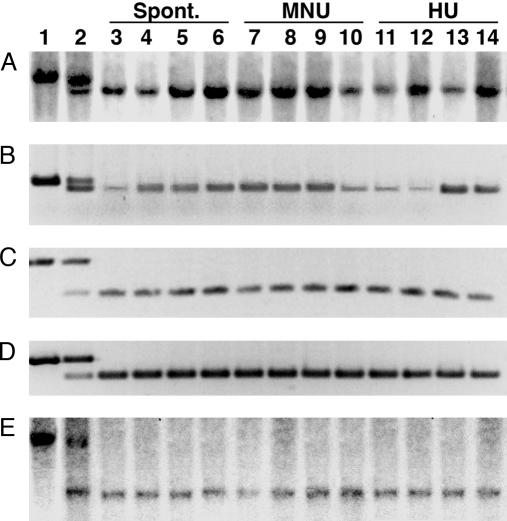
Fig. 3.
LOH at entrapment loci in clones selected in high G418. DNAs from the parental (AC1) ES cells (lane 1), heterozygous mutant entrapment clones (lane 2), and clones isolated in high G418 without treatment (lanes 3–6) and after treatment with MNU (lanes 7–10) or HU (lanes 11–14) were genotyped either by Southern blot hybridization (A and D) or by PCR (B–D) by using gene-specific probes and primers. The mutant clones contained entrapment vectors inserted in the 1810030N24Rik (A), Hesx1 (B), IL8Ra (C), Cradd (D), and Xrcc5 (E) genes. After carcinogen treatment and selection in 2.0 mg/ml G418, LOH was observed in 72, 24, 36, 24, and 232 independent clones, respectively.
LOH Is a Transient Response to Carcinogens.
Two types of experiments were performed to assess whether frequencies of LOH were transiently or stably elevated after carcinogen exposure. First, cells were treated with MNU and HU as before, and the percentages of cells having undergone LOH were determined by selection in 2.0 mg/ml G418 at various times thereafter. Over 90% of the total LOH was induced within 24 h of MNU and HU exposure, and only minimal additional LOH occurred subsequently (Fig. 6, which is published as supporting information on the PNAS web site). Second, we asked whether LOH frequencies at a second locus were elevated in cells having undergone LOH at the entrapment locus. For this, a herpes simplex virus thymidine kinase (TK) gene was introduced into cells containing an entrapment allele of the Hesx1 gene, and frequencies of TK gene loss were measured by selection in gancyclovir. These studies used the cell line containing the TK gene (C8TK1) and derivatives of C8TK1 that had undergone LOH at the entrapment locus induced spontaneously (C8TK1sN) or after treatment with either MNU (C8TKmN) or HU (C8TK1hN). As shown in Table 1, the frequencies of spontaneous TK gene loss were similar in all cells regardless of whether carcinogens had been used previously to induce LOH at the entrapment locus. Moreover, the stability of the TK gene after carcinogen treatment was largely unaffected by prior selection for LOH involving the entrapment locus. Similar results were also obtained with a second TK-containing line (C8TK2; Table 3, which is published as supporting information on the PNAS web site). Together, these experiments indicate that carcinogen-induced LOH results from an acute response rather than from a stably altered cellular phenotype.
Table 1.
Carcinogen-induced LOH does not result from a stably altered cellular phenotype
| Clone | Inducer EL LOH | Genotype | Treatment | Frequency of TK loss | Ratio |
|---|---|---|---|---|---|
| C8TK1 | None | TK+ EL+/− | None | 1.9 × 10−5 | }2.8 |
| C8TK1sN | Spontaneous | TK+ EL−/− | None | 5.3 × 10−5 | |
| C8TK1mN | MNU | TK+ EL−/− | None | 1.4 × 10−5 | }2.6 |
| C8TK1sN | Spontaneous | TK+ EL−/− | None | 5.3 × 10−5 | }0.6 |
| C8TK1hN | HU | TK+ EL−/− | None | 8.4 × 10−5 | |
| C8TK1sN | Spontaneous | TK+ EL−/− | HU | 2.3 × 10−4 | }1.7 |
| C8TK1hN | HU | TK+ EL−/− | HU | 3.9 × 10−4 | |
| C8TK1sN | Spontaneous | TK+ EL−/− | MNU | 2.6 × 10−4 | }1.8 |
| C8TK1mN | MNU | TK+ EL−/− | MNU | 4.7 × 10−4 |
The HSV thymidine kinase (TK) gene was introduced into cells containing an entrapment mutation in the Hesx1 gene. A TK-expressing (TK+) clone (C8TK1) was used to select for cells that had undergone LOH at the entrapment locus (EL) spontaneously (C8TK1sN) or after treatment with HU (C8TK1hN) or MNU (C8TK1mN). The frequencies of TK gene loss were compared in cells with and without prior selection for LOH at the entrapment locus, and the differences were expressed as the indicated ratios.
EL, entrapment locus.
Effect of Chromosome Position on Carcinogen-Induced LOH.
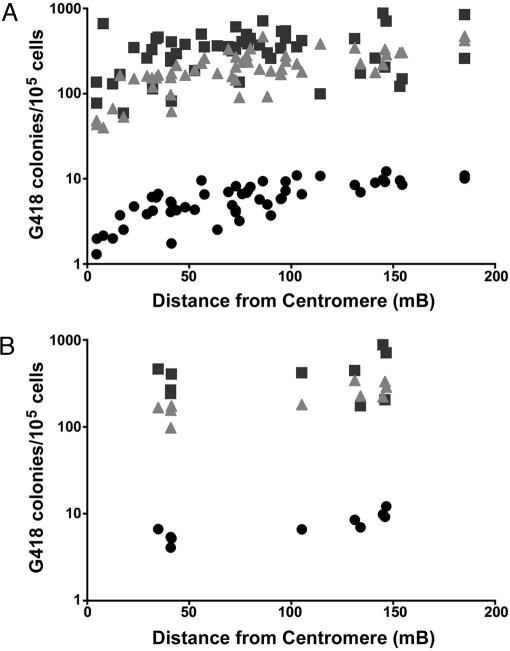
The frequency of spontaneous colony formation in high G418 was previously reported to increase with increasing distance from the centromere (Q.L., S.L.D., and H.E.R., unpublished work), consistent with previous studies suggesting that mitotic recombination plays a significant role in spontaneous LOH (34). Similar chromosome position effects were also observed (Fig. 4) after treatment with HU but not MNU (for example, R2 values for loci on chromosome 4 were 0.65 and 0.11 after HU and MNU treatment, respectively), suggesting that mechanisms other than mitotic recombination (e.g., gene conversion) were responsible for most of the MNU-induced LOH, consistent with studies in mouse lymphocytes (37). However, the ES cells used in the present study were derived from inbred mice and are naturally homozygous at all loci and thus cannot be used to distinguish among the possible mechanisms for generating LOH.
Fig. 4.
Effect of chromosome position on chemically induced survival in high G418. Fifty-three ES cell clones, each containing a single gene trap vector, were treated for 4 h with 0.5 mM MNU (squares) or 0.25 mM HU (triangles) or were untreated (circles) and then placed in media containing 2.0 mg/ml G418. The frequency of colony formation (presumptive LOH) for each clone is plotted against the location of each entrapment mutation (distance from the centromere). Linear regression analysis of all clones in aggregate (A) produced R2 values for untreated and HU- and MNU-treated cells of 0.62, 0.53, and 0.08, respectively. R2 values for all clones with mutations on chromosome 4 (B) were 0.68, 0.65, and 0.11 for untreated or HU- and MNU-treated cells, respectively.
Gene Entrapment in Studies of Genome-Wide LOH.
Entrapment ES cell clones provide an important in vitro model to study spontaneous and chemically induced LOH. ES cells are representative of self-renewing stem cells that serve as the precursors to cancer (38), and their use in mutagenesis studies is potentially important, because stem cells may possess specialized mechanisms to suppress mutations as a defense against oncogenic transformation (25–29). The clones are biologically normal, as assessed by their ability to produce germline chimeras (10 of 10 clones tested) and normal offspring and thus lack coincidental mutations that might affect genome maintenance. Libraries of entrapment clones characterized for mouse genome mutagenesis provide large numbers of genetic markers that allow LOH frequencies to be measured at many sites in the genome. Rates of spontaneous LOH observed in ES cells are similar to those reported in a variety of other mammalian cell types (39, 40). Moreover, the influence of chromosome position indicates that the rates of spontaneous and HU-induced LOH do not primarily reflect localized effects of the integrated gene trap vector.
The use of entrapment ES cells also permits direct comparisons between the effects of chemical carcinogens and specific DNA repair defects such as the Bloom’s syndrome mutation. Given the ease of creating defined mutations that can be transferred back and forth between ES cells and mice, ES cells provide an ideal system to compare the effects of different mutations on spontaneous and carcinogen-induced LOH in a normal and potentially isogenic cellular background. It will be important to test whether endogenous or exogenous carcinogens contribute to genome-wide changes associated with defects in genome maintenance. For example, mice expressing reduced levels of Bub1B, a protein involved in mitotic spindle checkpoint control, form tumors only after carcinogen exposure (41). It should also be possible to assess how specific DNA repair/genome maintenance pathways influence the types of recombination events induced by different genotoxic agents (37).
The GTR1.3 vector has features that allow the selection of homozygous mutant cells except in cases where gene entrapment disrupts genes required for cell growth or viability. The present study suggests that MNU and HU can be used to enhance the recovery of clones homozygous for recessive mutations during phenotype-based genetic screens in mammalian cells (31, 32, 42). The vector may also allow mutagenesis screens to be carried out in a greater variety of cell backgrounds.
LOH as a Somatic Mutation: Implications for the Carcinogenicity of Mutagens Like MNU.
Our results extend previous studies in which mutagens such as MNU have been reported to induce LOH (39, 43–48). However, these studies used nonmammalian systems or tumor-derived cell lines or were limited to only one or two loci. The present study provides a genome-wide analysis of carcinogen-induced LOH and an analysis involving normal diploid stem cells. As with most laboratory assessments of carcinogen risk, it is not possible to extrapolate from the concentrations of carcinogen used experimentally to the levels of exposure in human populations that typically occur over several decades. However, carcinogen concentrations were minimally toxic and were similar to those commonly used to induce tumors in animals.
LOH contributes to carcinogenesis by altering the dosage of genetically and epigenetically modified genes (22), including recessive cancer genes (tumor suppressors) of which >60 have been characterized (23). The ability of MNU and other agents used in the present study to induce point mutations is well established. These agents are also clastogens, as assessed by their ability to induce chromosome aberrations and sister-chromatid exchanges (http://toxnet.nlm.nih.gov). Our results indicate that the induction of LOH by a variety of mutagens occurs in normal stem cells at frequencies 2–4 orders of magnitude higher on a per-gene basis than the reported induction of point mutations. This could contribute to the notion that chromosome alterations such as LOH appear to have a greater impact on tumor cell genomes than the accumulation of point mutations (7, 14–16).
Frequencies of carcinogen-induced LOH were higher in some cases than the reported rates of LOH observed in ES cells homozygous for a mutation in the Bloom syndrome gene (Blm; refs. 30 and 31), an inherited DNA repair defect that results in greatly increased risk of cancer. Higher LOH frequencies were observed, even allowing for differences in plating efficiencies of entrapment clones (10–50%; data not shown), as compared to the Blm-deficient ES cells (30%; ref. 31). In short, the carcinogens tested produced the appearance of chromosomal instability in normal stem cells in the absence of a genetically determined mutator phenotype. Of course, the Blm mutation causes a persistent state of chromosome instability, whereas the rates of carcinogen-induced LOH are elevated only transiently following carcinogen exposure. Although it is not clear how certain mutations affecting DNA repair induce LOH, it would appear that certain types of adducted DNA and/or stalled replication complexes can promote LOH regardless of whether they are caused directly by genotoxic agents or indirectly by genetic attenuation of DNA repair pathways. Just as the carcinogenicity of the Blm mutation has been attributed to high rates of LOH, the carcinogenicity of a variety of mutagens may result as much from their ability to induce LOH as from their ability to induce point mutations.
LOH as Somatic Mutation: Implications Regarding the Origins of LOH in Human Cancer.
Extensive LOH in cancer cells is widely assumed to result from chromosomal instability; however, this conclusion is almost always based on the prevalence of LOH rather than on actual rate measurements (6). The present study showed that extensive LOH is induced in normal stem cells as an acute response to nontoxic levels of various carcinogens. We hypothesize that much of the LOH observed in nonhereditary cancers could result from prior exposure to genotoxic agents rather than from a state of genomic instability during the carcinogenic process. This hypothesis is consistent with the fact that >80% of cancers are caused by carcinogens present in the environment or produced by cellular metabolism (3–5), explains the apparent absence of mutations in genes required for DNA repair/genome maintenance in most cancers (6, 15, 16), and may account for the high levels LOH reported in several types of noncancerous lesions (18–21).
In summary, the present study describes a mechanism capable of generating high levels of LOH in the absence of a genetically activated genomic instability phenotype. Intrinsically low mutation rates and apoptosis in self-renewing stem cells have been proposed as mechanisms to suppress carcinogenesis (25–29). Similarly, the efficient use of sequences from homologous chromosomes to repair DNA damage and/or resolve stalled replication complexes could function to prevent coding sequence mutations. However, the process causes extensive LOH, with the likely consequence of unmasking recessive mutations in tumor suppressor genes.
Materials and Methods
Cell Culture.
The AC1 ES cell line was derived from 3.5-day blastocysts from 129svJ mice. AC1 cells were infected with the GTR1.3 poly(A) gene trap vector, and entrapment clones were isolated in 300 μg/ml G418. GTR1.3 inserts a neomycin phosphotransferase gene (Neo) expressed from the constitutive Pol2 gene promoter. Selection for neomycin (G418) resistance generates cell clones in which the Neo gene splices to 3′ exons of cellular genes (Q.L., S.L.D., and H.E.R., unpublished work). Genes disrupted in the entrapment clones were identified by sequencing cellular sequences appended to Neo fusion transcripts. ES cells were maintained at 37°C in DMEM supplemented with 15% FBS, nonessential amino acids, l-glutamine, 2-mercaptoethanol, and lymphocyte inhibitory factor.
Colony Selection and Chemical Treatment of Cells.
Serially diluted cells were plated onto 150-mm plates containing drug-free media and allowed to attach overnight. Unattached cells were removed, and media containing the indicated concentrations of MNU, HU, ethidium bromide, doxorubicin, methotrexate, diepoxybutane, or mitomycin C were put onto cells for 4 h (or cells were exposed to UV light in the absence of media and allowed to recover in drug-free media for 4 h). Cells were then rinsed twice with drug-free media, and selection media containing 0.0, 0.3, or 2.0 mg/ml G418 were put onto cells. After 12 days of selection, the number of colonies surviving was counted, and the frequency of colony formation was determined by dividing the number of colonies obtained from 2.0 mg/ml G418 selection to that obtained from parallel experiments with 0.3 mg/ml G418 selection. TK gene loss was assessed after selection in media containing 2 μg/ml gancyclovir.
Genotypic Analysis of LOH.
Genotypic analysis was performed by Southern blotting and PCR. Southern blot analysis was performed on 5 μg of genomic DNA that had been digested with a restriction enzyme and resolved on 0.9% agarose gels. Southern blot hybridization was performed by using DNA probes obtained by PCR amplification of genomic DNA adjacent to the site of retroviral vector insertion. PCR analysis was performed on 200 ng of genomic DNA with three primers. The first primer was in the sense orientation and was specific for genomic DNA 5′ to the site of retroviral vector insertion. Two additional primers were added that were in the antisense orientation; one was specific for sequence 3′ of the retroviral vector insertion and the other specific for the LTR portion of the retroviral vector insertion. Using these three primers, PCR amplification of genomic DNA yielded a smaller DNA fragment when the entrapment vector was present and a larger DNA fragment when the entrapment vector was absent.
Supplementary Material
Acknowledgments
We thank Abudi Nashabi and Tracy Moore-Jarrett for technical assistance and Dr. David Adler for use of the murine chromosome idiograms. This work was supported by U.S. Public Health Service Grants R01NS43952, R01RR13166, and P01HL68744 (to H.E.R.). Additional support was provided by Breast Cancer Specialized Program of Research Excellence (SPORE) Grant P50CA98131 and Cancer Center Support Grant P30CA68485 (to the Vanderbilt–Ingram Cancer Center). S.L.D. is supported by a postdoctoral fellowship from the Susan G. Komen Breast Cancer Foundation.
Abbreviations
- LOH
loss(es) of heterozygosity
- MNU
methylnitrosurea
- HU
hydroxyurea.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nowell P. C. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Ponder B. A. Nature. 2001;411:336–341. doi: 10.1038/35077207. [DOI] [PubMed] [Google Scholar]
- 3.Ames B. N., Gold L. S., Willett W. C. Proc. Natl. Acad. Sci. USA. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peto J. Nature. 2001;411:390–395. doi: 10.1038/35077256. [DOI] [PubMed] [Google Scholar]
- 5.Wogan G. N., Hecht S. S., Felton J. S., Conney A. H., Loeb L. A. Semin. Cancer Biol. 2004;14:473–486. doi: 10.1016/j.semcancer.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Lengauer C., Kinzler K. W., Vogelstein B. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 7.Loeb L. A., Loeb K. R., Anderson J. P. Proc. Natl. Acad. Sci. USA. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopalan H., Lengauer C. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 9.Hoeijmakers J. H. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 10.Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 11.Barnes D. E., Lindahl T. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 12.Risinger M. A., Groden J. Cancer Cell. 2004;6:539–545. doi: 10.1016/j.ccr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Weaver B. A., Cleveland D. W. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 14.Boland C. R., Ricciardiello L. Proc. Natl. Acad. Sci. USA. 1999;96:14675–14677. doi: 10.1073/pnas.96.26.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider B. L., Kulesz-Martin M. Carcinogenesis. 2004;25:2033–2044. doi: 10.1093/carcin/bgh204. [DOI] [PubMed] [Google Scholar]
- 16.Duesberg P., Fabarius A., Hehlmann R. IUBMB Life. 2004;56:65–81. doi: 10.1080/15216540410001667902. [DOI] [PubMed] [Google Scholar]
- 17.Marx J. Science. 2002;297:544–546. doi: 10.1126/science.297.5581.544. [DOI] [PubMed] [Google Scholar]
- 18.Stoler D. L., Chen N., Basik M., Kahlenberg M. S., Rodriguez-Bigas M. A., Petrelli N. J., Anderson G. R. Proc. Natl. Acad. Sci. USA. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih I. M., Zhou W., Goodman S. N., Lengauer C., Kinzler K. W., Vogelstein B. Cancer Res. 2001;61:818–822. [PubMed] [Google Scholar]
- 20.Luo L., Li B., Pretlow T. P. Cancer Res. 2003;63:6166–6169. [PubMed] [Google Scholar]
- 21.Chen R., Rabinovitch P. S., Crispin D. A., Emond M. J., Koprowicz K. M., Bronner M. P., Brentnall T. A. Am. J. Pathol. 2003;162:665–672. doi: 10.1016/S0002-9440(10)63860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feinberg A. P. Semin. Cancer Biol. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Futreal P. A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M. R. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomlinson I., Sasieni P., Bodmer W. Am. J. Pathol. 2002;160:755–758. doi: 10.1016/S0002-9440(10)64896-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cairns J. Proc. Natl. Acad. Sci. USA. 2002;99:10567–10570. doi: 10.1073/pnas.162369899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cervantes R. B., Stringer J. R., Shao C., Tischfield J. A., Stambrook P. J. Proc. Natl. Acad. Sci. USA. 2002;99:3586–3590. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saretzki G., Armstrong L., Leake A., Lako M., von Zglinicki T. Stem Cells. 2004;22:962–971. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- 28.Hong Y., Stambrook P. J. Proc. Natl. Acad. Sci. USA. 2004;101:14443–14448. doi: 10.1073/pnas.0401346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aladjem M. I., Spike B. T., Rodewald L. W., Hope T. J., Klemm M., Jaenisch R., Wahl G. M. Curr. Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 30.Luo G., Santoro I. M., McDaniel L. D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R. A., Bradley A. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 31.Yusa K., Horie K., Kondoh G., Kouno M., Maeda Y., Kinoshita T., Takeda J. Nature. 2004;429:896–899. doi: 10.1038/nature02646. [DOI] [PubMed] [Google Scholar]
- 32.Guo G., Wang W., Bradley A. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 33.Mortensen R. M., Conner D. A., Chao S., Geisterfer-Lowrance A. A., Seidman J. G. Mol. Cell. Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefebvre L., Dionne N., Karaskova J., Squire J. A., Nagy A. Nat. Genet. 2001;27:257–258. doi: 10.1038/85808. [DOI] [PubMed] [Google Scholar]
- 35.Gupta P. K., Sahota A., Boyadjiev S. A., Bye S., Shao C., O’Neill J. P., Hunter T. C., Albertini R. J., Stambrook P. J., Tischfield J. A. Cancer Res. 1997;57:1188–1193. [PubMed] [Google Scholar]
- 36.Shao C., Deng L., Henegariu O., Liang L., Raikwar N., Sahota A., Stambrook P. J., Tischfield J. A. Proc. Natl. Acad. Sci. USA. 1999;96:9230–9235. doi: 10.1073/pnas.96.16.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wijnhoven S. W., Sonneveld E., Kool H. J., van Teijlingen C. M., Vrieling H. Carcinogenesis. 2003;24:139–144. doi: 10.1093/carcin/24.1.139. [DOI] [PubMed] [Google Scholar]
- 38.Reya T., Morrison S. J., Clarke M. F., Weissman I. L. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 39.Wijnhoven S. W., Kool H. J., van Teijlingen C. M., van Zeeland A. A., Vrieling H. Mutat. Res. 2001;473:23–36. doi: 10.1016/s0027-5107(00)00163-9. [DOI] [PubMed] [Google Scholar]
- 40.Morley A. A. Mutat. Res. 1991;250:345–349. doi: 10.1016/0027-5107(91)90191-p. [DOI] [PubMed] [Google Scholar]
- 41.Hanks S., Coleman K., Reid S., Plaja A., Firth H., Fitzpatrick D., Kidd A., Mehes K., Nash R., Robin N., et al. Nat. Genet. 2004;36:1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 42.Organ E. L., Sheng J., Ruley H. E., Rubin D. H. BMC Cell Biol. 2004;5:41. doi: 10.1186/1471-2121-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazur-Melnyk M., Stuart G. R., Glickman B. W. Mutat. Res. 1996;358:89–96. doi: 10.1016/0027-5107(96)00174-1. [DOI] [PubMed] [Google Scholar]
- 44.Vogel E. W., Nivard M. J. Mutagenesis. 1993;8:57–81. doi: 10.1093/mutage/8.1.57. [DOI] [PubMed] [Google Scholar]
- 45.Wijnhoven S. W., Van Sloun P. P., Kool H. J., Weeda G., Slater R., Lohman P. H., van Zeeland A. A., Vrieling H. Proc. Natl. Acad. Sci. USA. 1998;95:13759–13764. doi: 10.1073/pnas.95.23.13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stettler P. M., Sengstag C. Mol. Carcinog. 2001;31:125–138. doi: 10.1002/mc.1047. [DOI] [PubMed] [Google Scholar]
- 47.Chen T., Harrington-Brock K., Moore M. M. Mutagenesis. 2002;17:105–109. doi: 10.1093/mutage/17.2.105. [DOI] [PubMed] [Google Scholar]
- 48.Turner D. R., Dreimanis M., Holt D., Firgaira F. A., Morley A. A. Mutat. Res. 2003;522:21–26. doi: 10.1016/s0027-5107(02)00194-x. [DOI] [PubMed] [Google Scholar]
- 49.Wang T. L., Rago C., Silliman N., Ptak J., Markowitz S., Willson J. K., Parmigiani G., Kinzler K. W., Vogelstein B., Velculescu V. E. Proc. Natl. Acad. Sci. USA. 2002;99:3076–3080. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.