Abstract
The dynamics of Hemoglobin I (HbI) from the clam Lucina pectinata, from wild-type sperm whale (SW) myoglobin, and from the L29F/H64Q/V68F triple mutant of SW, both unligated and bound to hydrogen sulfide (H2S), have been studied in molecular dynamics simulations. Features that account for differences in H2S affinity among the three have been examined. Our results verify the existence of an unusual heme rocking motion in unligated HbI that can promote the entrance of large ligands such as H2S. The FQF-mutant partially reproduces the amplitude and relative orientation of the motion of HbI's heme group. Therefore, besides introducing favorable electrostatic interactions with H2S, the three mutations in the distal pocket change the dynamic properties of the heme group. The active-site residues Gln-64(E7), Phe-43(CD1), and His-93(F8) are also shown to be more flexible in unligated HbI than in FQF-mutant and SW. Further contributions to H2S affinity come from differences in hydrogen bonding between the heme propionate groups and nearby amino acid residues.
INTRODUCTION
Hemoglobins and myoglobins can bind small molecules, such as O2, H2O, CO, NO, and CN (1), and the apparent simplicity of this function has resulted in their often being used as model systems in studies of interrelations among structure, dynamics, and function in proteins (2–8). Site-directed mutagenesis of the distal amino acid residues of the heme pocket have helped elucidate the structural basis for differences in ligand affinity (9,10). In studies of the contributions of steric factors, electrostatic interactions, and hydrogen bonding to stabilization of ligand-protein complexes, several residues have been identified as playing key roles in regulating the relative affinities of small molecules (11,12).
Proteins are not rigid, and considerable attention has been given to the role of structural fluctuations in the ligand-binding process (13–15). Access of the ligand to the heme iron atom has been shown to be sensitive to the amino acids that comprise the heme pocket, the manner in which they interconnect, and the fluctuations they are heir to (16). Moreover, the coupling between heme orientational dynamics and the protein environment has prompted thinking that the former may be used to indicate structural changes in the surrounding protein due to ligand dissociation (17).
Analysis of structure-function relationships in nonvertebrate myoglobins and hemoglobins indicates that the ability to bind molecules other than O2 can be acquired by evolutionary adaptation (18–21). Such is the case with the monomeric Hemoglobin I (HbI) (22,23) from the clam Lucina pectinata, which functions as a hydrogen sulfide transport protein (22,24–26). HbI has a high affinity for hydrogen sulfide (K = 2.9 × 106 M−1), the HbI-SH2 association rate (kon) is the highest of the known hemoglobins, and its dissociation rate (koff) is the lowest (24). Table 1 displays a summary of the kinetic and equilibrium constants for the reactions of HbI, FQF-mutant, and SW with oxygen and hydrogen sulfide.
TABLE 1.
Rate constants and equilibrium dissociation constants for reaction of HbI, FQF-mutant, and SW with oxygen and hydrogen sulfide
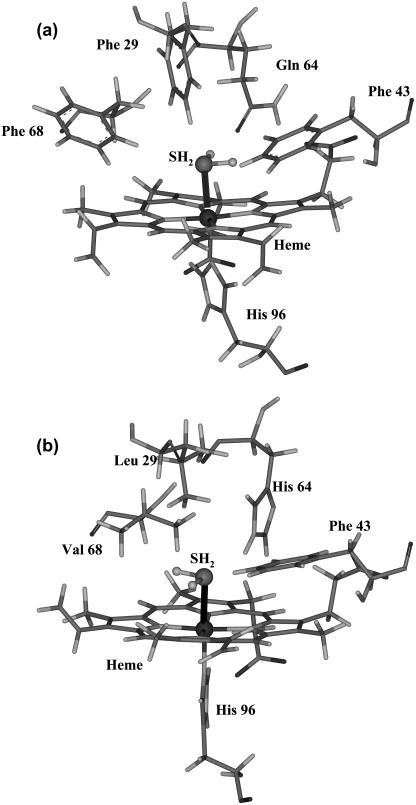
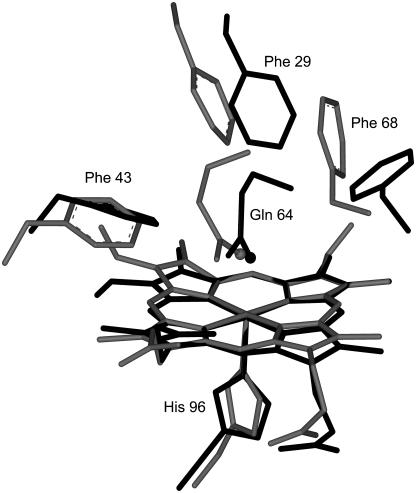
The heme structure of methydrogen sulfide HbI has been analyzed by resonance Raman spectroscopy, and it has been found that H2S ligation of HbI results in a low-spin FeIII complex (27). The affinity of H2S for ferric HbI is ∼5000 times that for wild-type sperm whale (SW) myoglobin (Table 1). The HbI distal pocket has a glutamine at position 64(E7) rather than the histidine found in SW, HbI has Phe-29(B10) and Phe-68(E11) where SW has Leu and Val, respectively, and both have Phe-43(B9). Fig. 1 displays the distal pockets of a), L. pectinata HbI, and b), wild-type SW myoglobin. All else being structurally equal, the FQF triple mutation of SW, viz. His-64(E7) → Gln, Leu-29(B10) → Phe, Val-68(E11) → Phe should mimic the distal pocket of the HbI (28). In fact the FQF mutation results in only ∼700-fold higher sulfide affinity over wild-type SW; HbI still has sevenfold higher sulfide affinity than the FQF-mutant.
FIGURE 1.
Stereo representations of key residues in the heme pocket of (a) HbI from L. pectinata, and (b) wild-type SW myoglobin. The coordinates were taken from optimized structures.
Because the ability of HbI to bind H2S is only partly explained by mimicking the distal heme pocket, further contributions to binding have been sought. Resonance Raman and 1H-NMR spectra have not detected strong hydrogen bonding between the heme propionates and the nearby amino acids (29), suggesting that the heme group is not firmly anchored to the polypeptide chain. A heme rocking freedom that can promote the entrance of large ligands such as H2S has been postulated as a result of this work.
We report here on molecular dynamics (MD) simulations of the three proteins, HbI, FQF-mutant, and SW, both unligated and with H2S bound, to explore the dynamical features of H2S affinity in each. A new set of parameters for the classical AMBER force field (30) was developed to simulate the ferric heme-SH2 complex. Dynamical features examined included rigid body motion and relative orientation of the heme prosthetic group within the protein, hydrogen-bond formation between the heme propionate groups and nearby amino acid residues, and changes in the distal cavity volume. Our analysis reveals differences in behavior among the three systems and confirms the existence of a much greater heme orientational freedom in HbI than in the FQF-mutant or SW.
METHODS
Potential function and molecular dynamics parameters
In all molecular mechanics (MM) and MD calculations the all-hydrogen topology with AMBER94 (30) force field parameters and the TIP3P water model (31) were employed. Following the study of Luthey-Schulten and co-workers (32), minor changes were made to the set of heme force field parameters (33), yielding new AMBER parameters around the iron and in the charge set for heme in its FeIII state.
The model system used to obtain the missing AMBER parameters consisted of an imidazole (Im) ligand bound to a ferric iron-porphyrin (FeP) complex, trans to an H2S ligand. The iron-sulfur (Fe-S) bond parameters were obtained by fitting the stretching frequency from normal mode AMBER force field calculations to the experimental fundamental, νFe-S = 374 cm−1 (27). The Fe-S-H and N(pyrrole)-Fe-S angles and the N(pyrrole)-Fe-S-H torsional parameters were obtained as follows: a), the structure of Im-FeP-SH2 was optimized in density functional theory (DFT) calculations; b), from the minimum energy geometry a set of structures was generated by varying the requisite angle in regular increments, and c), single point DFT energy calculations were carried out on each of these structures; d), MM nonbonding interactions were subtracted from the DFT energy values; and e), the resulting energy profiles were fit to the appropriate AMBER potential functional form (30).
DFT calculations have been extensively applied to metalloproteins in general and to iron-porphyrin systems in particular (34). The DFT calculations reported in this article have employed the BLYP density functional. BLYP consists of Becke's 1988 exchange functional (35) and the correlation functional of Lee, Yang, and Parr (36). The GAUSSIAN03 program (37) was used for single-point energy calculations, and DMol3 (38,39), with polarized double-numerical (DNP) basis sets, was employed for the geometry optimizations. This level of theory has been successfully used to study Im-FeP-XO (X = C, N, O) systems (40,41). In this work the total electronic spin of the Im-FeP-SH2 system has been restricted to S = 1/2, and symmetry has not been imposed.
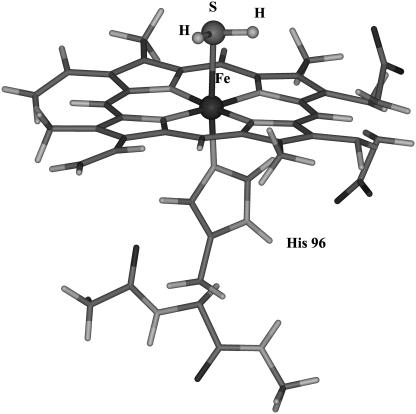
Electrostatic potential (ESP) derived charges for the system shown in Fig. 2 were obtained from single-point BLYP/6-31G* calculations and the Merz-Kollman scheme. Charges were derived for the whole system to ensure that the model would retain its biological relevance (32). A spin state of S = 5/2 was imposed on the system without H2S, and S = 1/2 was used when the ligand was attached. Restricted ESP (RESP) charges (42,43) were obtained by imposing symmetry on equivalent atom types. The charge on the central iron was constrained to its original ESP charge value. Luthey-Schulten and co-workers (32) have pointed out that this is a necessary condition to obtaining a consistent charge distribution on the heme prosthetic group.
FIGURE 2.
Iron porphyrin-hydrogen sulfide-imidazole system used to determine RESP charges.
Molecular dynamics simulations
MD simulations were done for HbI, SW, and the FQF-mutant, both unligated and with H2S bound. These were carried out with the AMBER 7.0 software package (44). Initial coordinates for each protein were taken from x-ray structures. The protein databank identifiers are 1b0b (HbI), 1a6g (SW), and 1obm (FQF-mutant). Each protein was solvated with explicit water molecules in a rectangular periodic box large enough to contain the protein and 10 Å of solvent on all sides. A 1400-step steepest-descent minimization was followed by a 1400-step conjugate gradient minimization, and the systems were then heated for 50 ps to a final temperature of 300 K. During heating a harmonic constraint of 10.0 kcal/Å2-mol was applied to the protein atoms. The time step was 2 fs, and the SHAKE algorithm was employed to constrain bonds involving hydrogen atoms. A cutoff of 10 Å was applied to nonbonded interactions. The system was equilibrated for 80 ps before starting each 1-ns trajectory. Data were collected at 1.0-ps intervals. During the simulations the temperature was held constant at 300 K by the weak-coupling algorithm (45) with a time constant for heat bath coupling of τ = 2.0 ps.
The SURFNET (46) package was used to calculate the volume of the distal pocket. The program defines cavities by filling the empty regions in the interior of the molecule with gap spheres of variable radius. We chose a minimum radius for the gap spheres of 1.0 Å and a maximum of 3.0 Å. The spheres were then used to compute a three-dimensional density map, using a grid separation of 0.8 Å.
RESULTS AND DISCUSSION
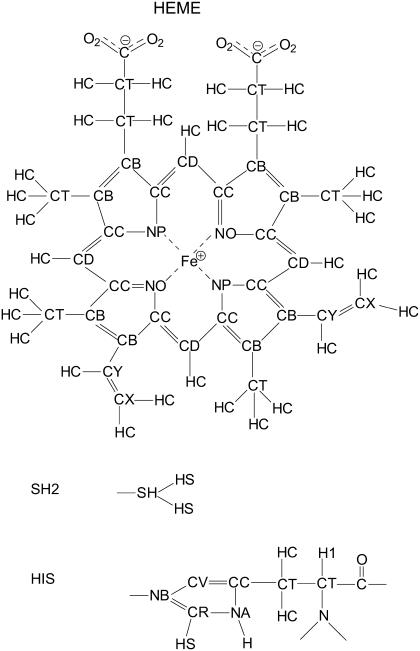
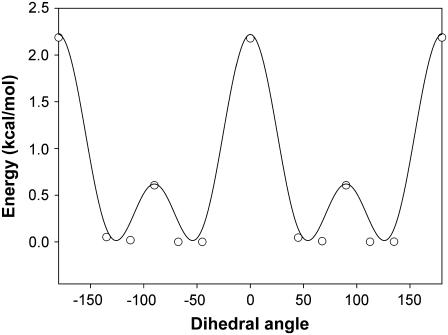
Table 2 summarizes the newly derived force field parameters for the iron-sulfur bond, two bending angles, and one dihedral that define the position of H2S relative to heme. They are denoted in the table as AMBER atom types, following the notation in the original derivation of the AMBER94 force field (30). AMBER notation (30,33) for the atoms of heme, hydrogen sulfide, and the proximal histidine are shown in Fig. 3. The set of Fe-S bond parameters shown in Table 2 results in a harmonic Fe-S stretching frequency of 386 cm−1, which compares well with the experimental fundamental of 374 cm−1 (27). Experimental data were not available for the other parameters. They were determined as described in the previous section by fitting to the results of quantum mechanical (QM) calculations. Fig. 4 shows the energy variation for rotation of H2S around the Fe-S axis. There are two barriers, each with twofold symmetry. Nonbonded interaction energies, calculated using a dihedral force constant of k = 0 kcal/mol, have been subtracted from the total energy. They contribute a miniscule barrier of 0.02 kcal/mol at 90°. The energy profile can be reproduced by a two-component dihedral term with periodicities n = 4 and 2 and a relative phase shift of 90° (Table 2), equally applied over the eight dihedrals that define the Fe-SH2 rotation. The dihedral contributions to the barriers are 2.22 kcal/mol at 0° and 180° and 0.62 kcal/mol at 90°.
TABLE 2.
Bond, angle, and dihedral parameters developed for this study; atoms are labeled as AMBER types
Bond force constants are in kcal/(mol-Å2).
Equilibrium bond distances are in angstroms.
Bending force constants in kcal/(mol-rad2).
Equilibrium angles in degrees.
Magnitude of the torsion in kcal/mol.
Phase offset in degrees.
Periodicity of the torsion. The negative value is not used in calculations; it signifies more than one component around a given bond.
FIGURE 3.
AMBER94 atom types for the heme prosthetic group, hydrogen sulfide, and the proximal histidine, His-F8.
FIGURE 4.
Energy profile for rotation about the Fe-SH bond.
RESP charges for H2S both present and absent are summarized in the Supplementary Material. The charge on the central iron was constrained to its original ESP values of 1.45 and 0.66, respectively.
Table 3 shows selected results of BLYP/DNP optimization of the structure displayed in Fig. 2 and a comparison with the AMBER minimization. The newly derived parameter set reproduces the DFT geometry quite well. Note for example the behavior of the angle Fe-S-H. Although a value (Table 2) of 103.1° was obtained in fitting to the Im-FeP-SH2 model system, for the full system of Fig. 2 an angle of 98° was found, in good agreement with the QM result of 99°.
TABLE 3.
Bond distance (Å), angle (°), and dihedral (°) values for the ferric heme prosthetic group structure after QM and AMBER geometry optimizations; atoms are labeled according to the AMBER types defined in Fig. 3
| Parameters | Optimized QM | AMBER |
|---|---|---|
| FE-NP | 2.04 | 2.02 |
| NP-CC | 1.39 | 1.39 |
| CC-CB | 1.45 | 1.45 |
| CB-CB | 1.40 | 1.36 |
| CB-CD | 1.40 | 1.40 |
| NB-CV | 1.39 | 1.40 |
| NB-CR | 1.34 | 1.35 |
| CV-CC | 1.38 | 1.37 |
| NA-CC | 1.39 | 1.38 |
| NA-CR | 1.36 | 1.44 |
| FE-NB | 2.06 | 2.04 |
| FE-SH | 2.55 | 2.50 |
| CC-CD-CC | 127 | 126 |
| NP-FE-NO | 90 | 90 |
| NP-FE-NP | 177 | 177 |
| CV-NB-CR | 106 | 106 |
| NB-CV-CC | 110 | 108 |
| NB-CR-NA | 110 | 111 |
| NP-FE-SH | 88 | 89 |
| NP-FE-NB | 91 | 91 |
| NB-FE-SH | 175 | 174 |
| FE-SH-HS | 99 | 98 |
| NP-FE-SH-HS | 69 | 62 |
| NP-FE-NB-CV | 3 | 2 |
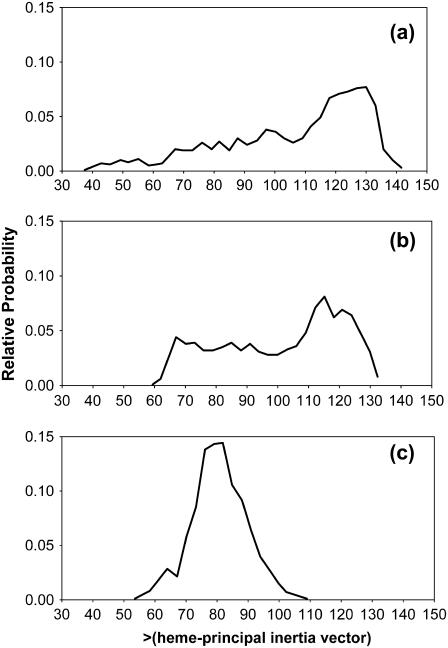
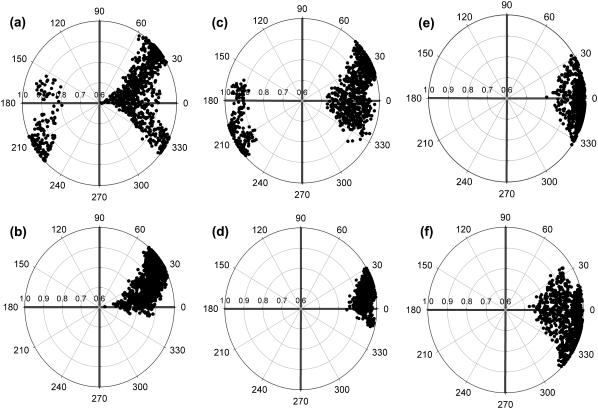
Mobility of the heme prosthetic group in HbI, FQF-mutant, and SW with respect to the rest of the protein was analyzed from the relative orientations of the group and whole-molecule principal axis vectors. The principal axis vectors for heme, Fe, and the 24 atoms of the tetrapyrrole ring were determined by diagonalizing the inertia matrix, which is calculated and stored during the simulations. The eigenvector, vz, with the largest eigenvalue, lying along the axis most difficult to rotate about, is nearly perpendicular to the heme plane. Fig. 5 shows a plot of the polar angle θ between vz and the principal axis of the whole protein, VZ, sampled from MD simulations of the three unligated proteins. The orientational space explored by the heme spans ranges of ∼90°, 65°, and 38°, respectively, for unligated HbI, FQF-mutant, and SW. Fig. 6 presents projections of vz on the surface of a unit sphere. The radial axis is sin(θ), and the circular angle axis displays the corresponding azimuthal angle ϕ in the xy plane (perpendicular to VZ) from the x axis. Results are displayed from snapshots of MD simulations of a), HbI, b), H2S-HbI, c), FQF-mutant, d), H2S-FQF-mutant, e), SW, and f), H2S-SW. Comparison of the three proteins in the absence of H2S (Fig. 6, a, c, and e) reveals that the heme groups of HbI and FQF-mutant are more mobile than that of SW. These results are consistent with the hypothesis drawn from the experiment that, relative to other heme proteins, the heme group of unligated HbI enjoys a rocking freedom that could facilitate the binding of an incoming ligand (29). The distal pocket in the FQF-mutant partially captures this feature. On the other hand comparing unligated to ligated HbI (Fig. 6, a and b, respectively) reveals that the motion of the heme group is markedly reduced when H2S is attached. A similar result is observed with the corresponding states of FQF-mutant (Fig. 6, c and d). This is the behavior to be expected if ligand interactions with the amino acid residues of the distal pocket serve to anchor the heme to the protein. The results are in agreement with the model proposed by Wittenberg and co-workers (24–26,47) in which the carbonyl group of distal glutamine stabilizes H2S-HbI by hydrogen bonding. This argument explains the low dissociation constant and high affinity of HbI for hydrogen sulfide (Table 1) compared to the corresponding values for SW. We have confirmed the model, evaluating the relative orientation and hydrogen-bond interactions between Gln-64(E7) and hydrogen sulfide during the MD simulations. A moderate hydrogen bond was observed with the Oɛ atom of the amide group of Gln-64(E7), pointing toward H2S (Fig. 1 a) in both HbI and FQF-mutant. In contrast Fig. 6, e and f, shows that the mobility of the SW heme group increases upon ligation, an indication that H2S does not interact strongly with apomyoglobin and may in fact act to shield the distal pocket residues from interaction with iron. To explore the capacity of His-64(E7) to stabilize H2S-SW by hydrogen bonding, the Nδ-H and Nɛ-H histidine tautomers were examined. A weak hydrogen bond (15% occupancy) between the nonprotonated Nɛ of the Nδ-H tautomer and hydrogen sulfide was detected, but no significant differences were observed that could represent a contribution to stabilization of the heme-hydrogen sulfide complex in SW.
FIGURE 5.
Histograms for values of the polar angle θ between the principal inertial axis vector of the heme group, vz, and that of the entire protein, VZ, calculated at 1-ps intervals during MD simulations of (a) HbI, (b) FQF-mutant, and (c) SW.
FIGURE 6.
Values of the projections of the heme group principal inertial axis vector, vz, on the surface of a unit sphere during MD simulations. The radial axis indicates the value of sin(θ), where θ is the polar angle of vz with respect to the principal inertial axis, VZ, of the entire protein. The circular angle axis displays the corresponding azimuthal angle ϕ in the XY plane (perpendicular to VZ) from the x axis. The values are from periodic snapshots during the MD simulations of (a) HbI, (b) HbI-H2S, (c) FQF-mutant, (d) FQF-mutant-H2S, (e) SW, and (f) SW-H2S.
Indeed analysis of crystal structures of HbI and FQF-mutant metaquo complexes suggests that differences in relative orientations of Phe-29(B10) and Phe-68(E11) can play an important role in binding and interaction with the heme-hydrogen sulfide complex. Table 4 summarizes the variations in the dihedral angles of these residues in different states of HbI and FQF-mutant. Despite the differences due to the required accommodation, similar values of χ1 and χ2 can be observed for HbI in its unligated state and with either H2S or H2O bound. The same can be said for the values among the different reported states of FQF-mutant. Therefore neither Phe-29(B10) nor Phe-68(E11) change their conformation during the MD simulations. However, both residues in FQF-mutant are significantly rotated about χ2 relative to their positions in HbI (Fig. 7). The calculation of nonbonded interactions of Phe-29(B10) and Phe-68(E11) with the heme-H2S complex reveals that these residues stabilize the complex by ∼1 Kcal/mol and ∼5 Kcal/mol, respectively. Nevertheless no significant differences were observed due to the different orientations of both residues in HbI and FQF-mutant. A more accurate calculation of electrostatic interactions using all-electron DFT calculations may be required to distinguish the differences between the interactions.
TABLE 4.
Comparison of dihedral angles of residues Phe-29(B10), Phe-68(E11), and Gln-64(E7) in crystal structures and in the MD simulations
| Angle | HbI:H2O* | HbI:H2S | HbI | FQF:H2O* | FQF:H2S | FQF |
|---|---|---|---|---|---|---|
| χ1 (Phe-68(E11)) | −172° | −170° ± 10° | −174° ± 9° | 150° | 109° ± 19° | 125° ± 22° |
| χ2 (Phe-68(E11)) | 91° | 103° ± 10° | 103° ± 10° | 66° | 79° ± 12° | 76° ± 15° |
| χ1 (Phe-29(B10)) | −80° | −83° ± 10° | −83° ± 12° | −89° | −88° ± 9° | −90° ± 10° |
| χ2 (Phe-29(B10)) | 87° | 88° ± 21° | 103° ± 14° | 179° | 167° ± 54° | 179° ± 86° |
| χ1 (Gln-64(E7)) | −92° | −71° ± 17° | −65° ± 21°†−175° ± 9°‡ | −180° | −168° ± 9° | −176° ± 9° |
| χ2 (Gln-64(E7)) | −46° | −48° ± 15° | −38° ± 16°†57° ± 10°‡ | 49° | 58° ± 10° | 57° ± 10° |
| χ3 (Gln-64(E7)) | −98° | −67° ± 29° | −77° ± 32°†102° ± 32°‡ | 45° | 62° ± 32° | 37° ± 27° |
FIGURE 7.
Stereo representations of key residues in the heme pocket of HbI from L. pectinata (light lines) and the L29F/H64Q/V68F triple mutant of SW myoglobin (dark lines). The side-chain oxygen atom of the distal glutamine is shown as a ball in each case.
As previously pointed out by Bolognesi et al. (26,47), the fast sulfide association rate constant may be related to the presence of a rather flexible Gln-64(E7) as the distal residue in HbI. Our MD results for unligated HbI confirm this interpretation. Gln-64(E7) interchanged its “open gate” and “closed gate” conformations within 200 ps during MD simulations. The corresponding values for the dihedral angles of these conformations are reported in Table 4. Furthermore, the high flexibility of the Gln-64(E7) in HbI may also be related to the high HbI-O2 association rate (kon), since that would be expected to increase the acceptance of the incoming O2 molecule. On the other hand Gln-64(E7) in unligated FQF does not change its initial “closed gate” conformation throughout the simulation.
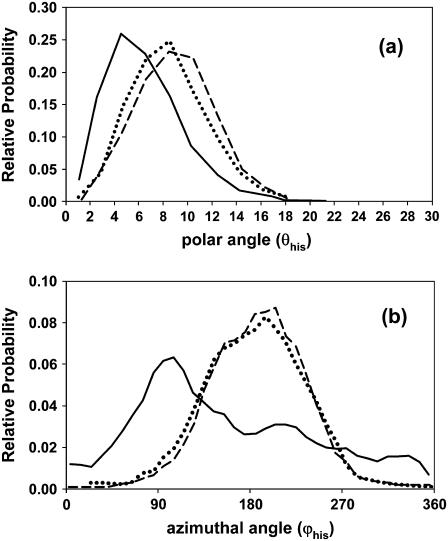
We have also analyzed the relative orientation of heme with respect to the proximal histidine His-F8. For this purpose a coordinate system was defined, the origin on NB (see Fig. 3) of the imidazole ring, and the z axis bisecting angle CV-NB-CR and directed toward heme. The unit vector, vhis, normal to the best least-squares plane through the four nitrogen atoms of the tetrapyrrole ring was calculated at intervals during each 1-ns simulation. Fig. 8 a shows plots of the polar angle, θhis, the angle of vhis with the z axis, for unligated HbI, FQF-mutant, and SW. The distributions for FQF-mutant and SW are similar, whereas HbI is less tilted than these. However, the comparative widths of the distributions indicate that the differences in heme mobility seen in Fig. 6 are not related to changes in the tilt of heme with respect to the His-F8 imidazole ring. On the other hand significant differences are observed between the distributions of the corresponding azimuthal angles, ϕhis, in the xy plane from the x axis (Fig. 8 b). The orientational space explored by the heme of unligated HbI spans a wider range than those of unligated FQF-mutant and SW.
FIGURE 8.
Histograms for (a) the polar angle (θhis) between the unit vector (vhis) normal to the best least-squares plane through the four nitrogen atoms of the tetrapyrrole ring and the z axis, and (b) the corresponding azimuthal angle ϕhis in the xy plane from the x axis. The xyz-coordinate system fixed to the imidazole ring was chosen with its origin on atom NB and the z axis bisecting CV-NB-CR and in the direction of the heme. The values were calculated during MD simulations of the unligated states of HbI (solid line), FQF-mutant (dashed line), and SW (dotted line).
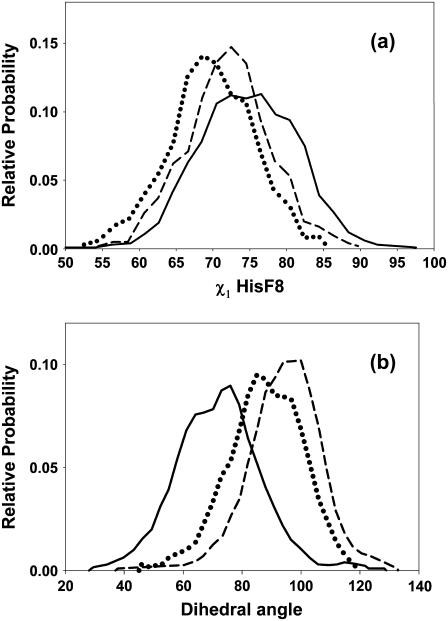
Plots of χ1, the torsion angle of His-F8 (Fig. 9 a), indicate that this residue is more flexible in unligated HbI than in FQF-mutant and SW. The root mean-square deviations of the position of His-F8 imidazole ring are 8.8, 9.2, and 7.7 Å for unligated HbI, FQF-mutant, and SW, respectively. Obviously, differences both in heme mobility with respect to His-F8 and in flexibility of the His-F8 side chain are implicated in the differences in the orientational space explored by the heme group (Fig. 6, a, c, and e). Finally we have analyzed differences in the capacity of the His-F8 imidazole to rotate with respect to the heme plane in the three proteins. Fig. 9 b displays a histogram of the N(pyrrole)-Fe-NB(His-F8)-CR(His-F8) dihedral sampled from MD simulations of the three unligated proteins. The orientational space explored by the His-F8 imidazole ring relative to the heme plane spans ranges of ∼100°, 95°, and 73°, respectively, in unligated HbI, FQF-mutant, and SW. Furthermore, whereas FQF-mutant and SW stabilize an eclipsed orientation of the His-F8 imidazole ring with respect to the nitrogens of the pyrrole rings, HbI displays a distribution with a maximum at ∼75°, corresponding to a more staggered orientation, and in good agreement with the 74° reported in the metaquo HbI crystal structure (26). The average equilibrium values do not change upon ligation. On the contrary, the corresponding orientational spaces are reduced to ∼70° for HbI and FQF-mutant whereas they increase to ∼95° for SW.
FIGURE 9.
Histograms for (a) the χ1 torsion angle of proximal histidine His-F8, and (b) the N(pyrrole)-Fe-NB(His-F8)-CR(His-F8) dihedral from MD simulations of unligated HbI (solid line), FQF-mutant (dashed line), and SW (dotted line).
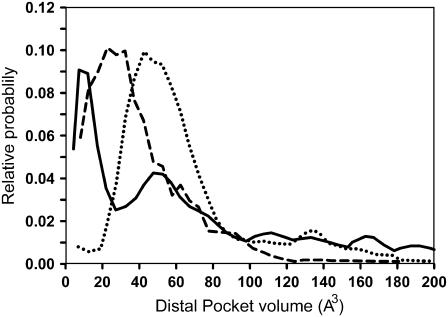
Fig. 10 displays distal pocket volumes during the MD simulations of unligated HbI, FQF-mutant, and SW. The method of calculating the volume has been discussed in the previous section. The most probable volume for SW, 43 Å3, agrees well with previous MD results (16). Cavities were present during substantial fractions of time in each simulation, 60%, 91%, and 75% of the time, respectively, for the simulations of HbI, FQF-mutant, and SW. Substitution of larger residues for smaller, such as the Val → Phe and Leu → Phe that occur in the SW → FQF mutations, results in a smaller FQF-mutant cavity volume, and the smaller distal pocket should be less accommodating for H2S. However, this effect is largely offset by additional hydrogen bond and multi-pole interactions with the ligand (45). Again HbI contrasts with SW and FQF. The distribution of the distal pocket volumes for unligated HbI displays two peaks, at ∼10 and 48 Å3. Analysis of the configurations that contribute to each of the peaks reveals that those corresponding to the maximum at ∼10 Å3 feature short Phe-29-Phe-43 and Phe-29-Leu32 distances compared to those contributing to the maximum at ∼48 Å3. As a consequence the free volume of the distal pocket is partitioned into smaller cavities. Furthermore, in HbI expansion to large volumes is possible due to the opening of the Gln-64 gate, which was not observed in FQF-mutant and SW during the time interval studied.
FIGURE 10.
Histograms of the heme distal pocket volume calculated from MD simulations of unligated HbI (solid line), FQF-mutant (dashed line), and SW (dotted line).
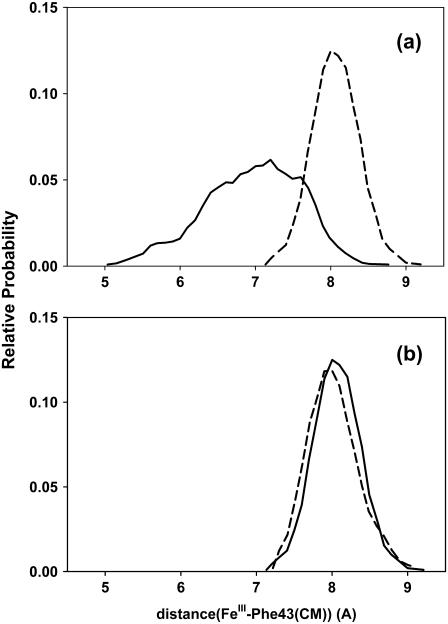
The smaller distal pocket volume seen in unligated HbI with respect to unligated FQF-mutant contrasts with the relatively larger volume of the former suggested by the crystal structures of HbI and FQF-mutant metaquo complexes (28). Table 5 displays a comparison of the distances between FeIII and Cζ of the aromatic residues Phe-29(B10), Phe-68(E11), and Phe-43(CD1) in the crystal structures and those obtained in the MD simulations. In both unligated states and metaquo complexes, distances between FeIII and Phe-29(B10) and Phe-68(E11) in FQF-mutant are shorter than the corresponding distances in HbI. Nevertheless, Phe-43 is ∼1.2 Å closer in unligated HbI than in FQF-mutant. The motion of Phe-43 toward FeIII leads to collapse of the active site in HbI. This feature is not observed in unligated FQF-mutant. Fig. 11 a shows the distribution of the distance between FeIII and the center of mass of Phe-43(CD1), sampled from MD simulations of the unligated states of HbI and FQF-mutant. Despite its closer distance to the FeIII atom, higher flexibility is observed for this residue in unligated HbI than in FQF-mutant. The corresponding distributions for HbI:H2S and FQF-mutant:H2S adducts are displayed in Fig. 11 b. The motion of Phe-43(CD1) is markedly reduced in HbI when H2S is attached, and similar distributions are observed in HbI and FQF-mutant. Thus Fig. 11 b and the values obtained from the crystal structures of HbI:H2O and FQF-mutant:H2O (Table 5) indicate that the high mobility of Phe-43 is a property uniquely associated with the unligated state of HbI.
TABLE 5.
Comparison of distances between FeIII and Cζ of the aromatic residues Phe-29(B10), Phe-68(E11), and Phe-43(CD1) in crystal structures and in the MD simulations
| System | Fe-Cζ(Phe-29(B10)) | Fe-Cζ(Phe-68(E11)) | Fe-Cζ(Phe-43(CD1)) |
|---|---|---|---|
| FQF-mutant | 5.60 ± 0.42 | 7.97 ± 0.51 | 5.46 ± 0.37 |
| HbI | 6.26 ± 0.54 | 8.11 ± 0.58 | 4.21 ± 0.41 |
| FQF-mutant:H2S | 6.31 ± 0.29 | 8.25 ± 0.37 | 5.33 ± 0.31 |
| HbI:H2S | 6.43 ± 0.40 | 7.78 ± 0.58 | 5.11 ± 0.30 |
| FQF-mutant:H2O* | 5.97 | 7.52 | 5.30 |
| HbI:H2O* | 6.36 | 8.06 | 4.92 |
FIGURE 11.
Histograms for distances between FeIII and the center of mass of Phe-43(CD1) from MD simulations of (a) HbI (solid line) and FQF-mutant (dashed line), and (b) HbI-H2S (solid line) and FQF-mutant-H2S (dashed line).
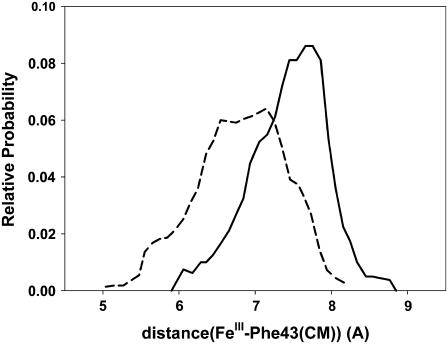
It is important to note that the relative position of Phe-43 is influenced by the opening of the Gln-64 gate. This is evident from Fig. 12, in which the distribution of the distance from FeIII to the center of mass of Phe-43(CD1) is shown separately for the open and closed conformations. The shorter distances observed for the closed conformations points out that the motion of Phe-43 toward FeIII that leads to collapse of the active site takes place mainly when the Gln-64 gate is closed. Since the entrance of the ligand takes place when the Gln-64 gate is open, its accommodation at the sixth coordination site of the iron will not be hindered by the Phe-43 residue. The analysis of CD1 mutants have previously shown the importance of this residue in the orientation of the imidazole ring of the distal histidine (1,48). Moreover, the different possible binding paths have been shown to require the displacement of Phe-43(CD1) for the ligand to enter or leave the globin (49). Analysis of the corresponding distribution of distances for Phe-68(E11) and Phe-29(B10) (not shown) reveals that neither presents significant differences between open and closed conformations.
FIGURE 12.
Histograms for distances between FeIII and the center of mass of Phe-43(CD1) calculated from MD configurations in the open (solid line) and closed (dashed line) conformations of the Gln-64 gate.
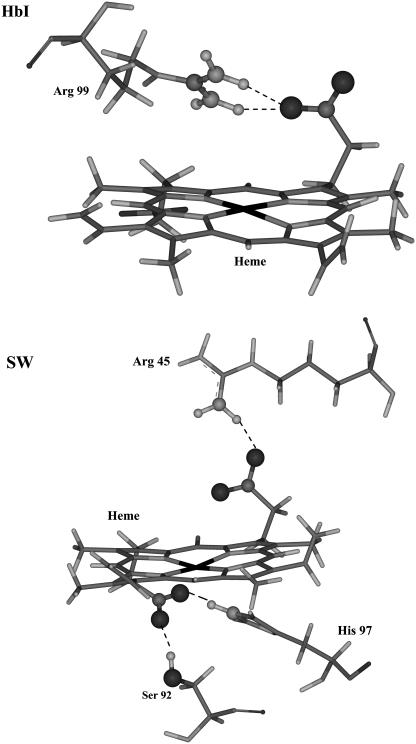
Analyses of resonance Raman spectra of deoxyHbI and metcyano, carbonmonoxy, and oxy Hb derivatives (29) have suggested the presence of moderate hydrogen bonding between Arg-99 and the heme-7-propionate that disappears when the HbI-ligand moiety is formed. 1H-NMR indicates that hydrogen bonding between the heme-6-propionate group and amino acid residues is absent. These studies have given rise to the view that the heme group of HbI is not anchored to the protein chain by its propionates (29). We evaluated the hydrogen-bond interactions of the heme propionates to test that hypothesis. Table 6 lists the percent occupancy >5% and lifetimes of hydrogen bonds between the heme propionates and nearby amino acid residues over the course of the 1-ns simulations. Hydrogen bonding between Arg-99 and heme-7-propionate of HbI becomes less frequent when H2S is attached. It presents a 67.66% occupancy and a moderate lifetime of 1.7 ps in HbI-H2S but only 29.20% with a similar lifetime of 1.8 ps in unligated HbI. The result is in accord with the differences seen in the 370 cm−1 propionate band in HbI and HbI-X (X = CO, O2, and CN) resonance Raman spectra (29). The number and strength of hydrogen bonds increase in going from HbI to either FQF-mutant or SW. In FQF-mutant and SW the heme-6-propionate forms strong hydrogen bonds with Ser-92 and His-93, and heme-7-propionate hydrogen bonds to Arg-45. Occupancies and lifetimes are generally lower in unligated FQF-mutant than in unligated SW, and the heme group of the former can thereby be more flexible (Fig. 6, c and e). Previous work on SW has indicated that the hydrogen-bonding lattice formed by Leu-89, Ser-92, His-93, His-97, and the heme-7-propionate stabilizes the interaction of heme with the apoprotein (50). A detailed comparison of hydrogen bonds between heme propionate groups and the nearby amino acids in HbI and SW is presented in Fig. 13. In addition Arg-45 can hydrogen bond, directly or through a water molecule, with the distal His-64 and with heme-6-propionate (51,52). Previous work has established the relevance of Arg-45 to regulation of myoglobin oxygen affinity and to binding of heme to apomyoglobin (51,53). Replacing residue 64 with Gln (H64Q) disrupts its salt bridge with Arg-45. The percent occupancy of the Arg-45-Gln-64 hydrogen bond in unligated FQF-mutant is only 4%, indicating that Arg-45 is less suitably oriented to form a hydrogen bond with heme-6-propionate. Table 6 indicates a moderate Arg-45-propionate hydrogen bond, with 43.38% occupancy and a 2.1-ps lifetime in unligated FQF-mutant, compared to corresponding values of 76.69% and 4.1 ps in unligated SW. Increased flexibility of the heme group is therefore to be expected (Fig. 6 c) in the mutant. The higher occupancies and lifetimes reported in Table 6 for hydrogen-bond interactions in unligated SW as compared to those seen in SW-SH2 indicates that they may play a role in increased heme mobility upon ligation (Fig. 6, e and f).
TABLE 6.
Values of the percentage occupancy and lifetimes for hydrogen bonds between the heme propionate groups and nearby amino acid residues for the 1-ns simulations; only occupancies higher than 5% are shown
| Donor | % | Lifetime(ps) |
|---|---|---|
| HbI:H2S | ||
| Arg-99 | 29.20 | 1.7 |
| HbI | ||
| Arg-99 | 67.66 | 1.8 |
| FQF-mutant:H2S | ||
| Ser-92 | 65.33 | 3.7 |
| His-97 | 73.97 | 2.1 |
| FQF-mutant | ||
| Ser-92 | 57.21 | 2.8 |
| His-97 | 72.85 | 1.9 |
| Arg-45 | 43.38 | 2.1 |
| SW:H2S | ||
| Ser-92 | 59.80 | 3.3 |
| His-97 | 69.90 | 1.9 |
| Arg-45 | 37.4 | 3.9 |
| SW | ||
| Ser-92 | 61.76 | 3.4 |
| His-97 | 72.31 | 2.2 |
| Arg-45 | 76.69 | 4.1 |
FIGURE 13.
Stereo representations of hydrogen bonds between the heme propionate groups and nearby amino acid residues of (a) HbI, and (b) SW.
Finally, we have also evaluated differences in hydrogen-bond interactions between proximal His-F8 and nearby amino acid residues over the course of the 1-ns simulations. The results are displayed in Table 7. In SW and FQF-mutant, Nδ-H of His-93(F8) forms a bifurcated hydrogen bond with the main-chain >C=O group of Leu-89(F4) and Oγ of Ser-92(F7). These residues form part of a hydrogen-bonding lattice extending from His-F8 to the heme-7-propionate and His-97 (Table 6). The conformation of the His-F8 imidazole appears to be tightly constrained by these interactions. As a result the plane of the imidazole eclipses the equatorial Fe-N(pyrrole) bonds. The absence of this extensive hydrogen-bonding lattice in HbI (Tables 6 and 7) leads to a more staggered conformation of the imidazole ring of His-F8 with respect to the pyrrole nitrogens (Fig. 9 b). Site-directed mutagenesis experiments have shown that replacement of Ser-92 with different nonpolar residues in pig myoglobin leads to increases in the ligand association equilibrium constants (54). Therefore, as has been previously pointed out for leghemoglobins (55,56), the different orientation of His-F8 and the attending disruption of the hydrogen-bonding lattice that connects this residue with the heme-7-propionate can also be related to higher association rates for HbI compared to those for SW (Table 1).
TABLE 7.
Percentage occupancy and lifetimes for hydrogen bonds between Nδ-H of His-F8 and nearby amino acid residues for the 1-ns MD simulations; only occupancies higher than 5% are shown
| Acceptor | % | Lifetime(ps) |
|---|---|---|
| HbI:H2S | ||
| Phe-91 | 55.60 | 2.7 |
| HbI | ||
| Phe-91 | 60.1 | 2.8 |
| FQF-mutant:H2S | ||
| Ser-92 | 24.92 | 1.4 |
| Leu-89 | 5.23 | 1.0 |
| FQF-mutant | ||
| Ser-92 | 31.96 | 1.6 |
| Leu-89 | 4.61 | 1.1 |
| SW:H2S | ||
| Ser-92 | 21.70 | 1.6 |
| Leu-89 | 6.70 | 1.1 |
| SW | ||
| Ser-92 | 18.33 | 1.4 |
| Leu-89 | 14.64 | 1.2 |
CONCLUSIONS
We have presented a comparative study of the dynamical features that can account for the differences in the H2S affinites of L. pectinata HbI, wild-type SW myoglobin, and the L29F/H64Q/V68F mutant of SW (FQF-mutant). The results confirm the hypothesis arising from previous experimental studies that, relative to other heme proteins, the heme group of unligated HbI displays an unusual rocking freedom that can promote the entrance of an incoming ligand. Mimics of HbI's distal pocket in FQF-mutant partially reproduce this feature. The three mutations both stabilize the heme-SH2 adduct and increase the mobility of heme with respect to the rest of the protein in its unligated state.
The MD results also confirm the model proposed by Wittenberg and co-workers (25,26,47) in which the carbonyl of distal glutamine stabilizes H2S-HbI with a hydrogen bond. This argument explained the low-dissociation constant and the high affinity of HbI for hydrogen sulfide (Table 1) as compared to the corresponding values for SW. In addition Phe-29(B10) and Phe-68(E11) were found to stabilize the heme-hydrogen sulfide complex by ∼1 Kcal/mol and ∼5 Kcal/mol, respectively. Nevertheless, no significant differences were observed due to the different orientations of both residues in HbI and FQF-mutant.
A rather flexible Gln-64(E7) was observed in unligated HbI. As previously pointed out by Bolognesi et al. (26,47), this result also contributes to explaining faster sulfide and oxygen association rate constants in HbI. Furthermore, a higher flexibility was also observed for Phe-43(CD1) in unligated HbI with respect to FQF-mutant and SW. The motion of this residue toward FeIII was shown to be influenced by the opening of the Gln-64(E7) gate.
The analysis of the relative orientation of heme with respect to proximal histidine, His-F8, finds that differences in heme mobility relative to His-F8 and in flexibility of the His-F8 side chain are more prominent in unligated HbI than in the FQF-mutant and SW. Moreover, HbI displays a different orientation of His-F8 and disruption of the hydrogen-bonding lattice that connects this residue with the heme-7-propionate. This feature is also associated with higher association rates for HbI compared to those for SW. Further contributions to the high H2S affinity of HbI arise from differences in the degree of hydrogen bonding between the heme propionate groups and nearby amino acid residues. The number and strength of these hydrogen bonds increases in passing from HbI to FQF-mutant and SW. Our results support a model in which the heme propionates of HbI are not tightly anchored to the polypeptide chain by hydrogen bonding.
The study presented here suggests that the mechanism of functional adaptation of HbI as a sulfide-reactive heme protein has taken place both through stabilization of the heme-SH2 adduct and through an increase in active-site flexibility of the unligated protein. The former contributes to a decrease in the HbI-hydrogen sulfide dissociation rate constant (koff), and the latter contributes to an increase in its association rate (kon). Because the HbI-hydrogen sulfide association rate is the highest of the known hemoglobins and its dissociation rate is the lowest, site-directed mutagenesis studies should be able to separate the effects. Sulfide-binding hemoglobins should provide good models for the study of functional adaptation through effects of mutations on dynamical properties.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
This work was supported in part by grants to S.F.-A. from the Universidad de Quilmes and the Agencia Nacional para la Promoción de la Ciencia y Tecnología of the Ministerio de Educación, Ciencia y Tecnología, Argentina. D.E.B. received support from the U.S. National Institutes of Health (NIH) INBRE P20 RR-016470, and J.L.G. received support from National Science Foundation-Molecular and Cellular Biology-0544250 and Minority Biomedical Research Support-Support of Continuous Research Excellence 2S06GM008103-30. S.F.-A., D.E.B., and J.E. wish to acknowledge support from the Consejo Nacional de Investigaciones Científicas y Técnicas de la República Argentina. Support by the Ecole Polytechnique Fédérale de Lausanne is also acknowledged.
References
- 1.Springer, B. A., S. G. Sligar, J. S. Olson, and G. N. Phillips. 1994. Mechanisms of ligand recognition in myoglobin. Chem. Rev. 94:699–714. [Google Scholar]
- 2.Antonini, E., and M. Brunori. 1971. Hemoglobin and Myoglobin and Their Interaction with Ligands. North Holland, Amsterdam.
- 3.Frauenfelder, H., S. G. Sligar, and P. G. Wolynes. 1991. The energy landscapes and motions of proteins. Science. 254:1598–1603. [DOI] [PubMed] [Google Scholar]
- 4.Mcmahon, B. H., B. P. Stojkovic, J. Hay, R. Martin, and A. Garcia. 2000. Microscopic model of carbon monoxide binding to myoglobin. J. Chem. Phys. 113:6831–6850. [Google Scholar]
- 5.Frauenfelder, H., B. H. Mcmahon, R. H. Austin, K. Chu, and J. T. Groves. 2001. The role of structure, energy landscape, dynamics, and allostery in the enzymatic function of myoglobin. Proc. Natl. Acad. Sci. USA. 98:2370–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frauenfelder, H., B. H. Mcmahon, and P. W. Fenimore. 2003. Myoglobin: the hydrogen atom of biology and a paradigm of complexity. Proc. Natl. Acad. Sci. USA. 100:8615–8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourgeois, D., B. Vallone, F. Schotte, A. Arcovito, A. E. Miele, G. Sciara, M. Wulff, P. Anfinrud, and M. Brunori. 2003. Complex landscape of protein structural dynamics unveiled by nanosecond laue crystallography. Proc. Natl. Acad. Sci. USA. 100:8704–8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenimore, P. W., H. Frauenfelder, B. H. Mcmahon, and R. D. Young. 2005. Proteins are paradigms of stochastic complexity. Physica A. 351:1–13. [Google Scholar]
- 9.Olson, J. S., A. J. Mathews, R. J. Rohlfs, B. A. Springer, K. D. Egeberg, S. G. Sligar, J. Tame, J. P. Renaud, and K. Nagai. 1988. The role of the distal histidine in myoglobin and hemoglobin. Nature. 336:265–266. [DOI] [PubMed] [Google Scholar]
- 10.Brunori, M., F. Cutruzzola, and B. Vallone. 1995. Hemoglobin engineering—for fun and money. Curr. Biol. 5:462–465. [DOI] [PubMed] [Google Scholar]
- 11.Carver, T. E., R. J. Rohlfs, J. S. Olson, Q. H. Gibson, R. S. Blackmore, B. A. Springer, and S. G. Sligar. 1990. Analysis of the kinetic barriers for ligand-binding to sperm whale myoglobin using site-directed mutagenesis and laser photolysis techniques. J. Biol. Chem. 265:20007–20020. [PubMed] [Google Scholar]
- 12.Egeberg, K. D., B. A. Springer, S. G. Sligar, T. E. Carver, R. J. Rohlfs, and J. S. Olson. 1990. The role of Val68(E11) in ligand-binding to sperm whale myoglobin—site-directed mutagenesis of a synthetic gene. J. Biol. Chem. 265:11788–11795. [PubMed] [Google Scholar]
- 13.Lee, B., and F. M. Richards. 1971. Interpretation of protein structures—estimation of static accessibility. J. Mol. Biol. 55:379–400. [DOI] [PubMed] [Google Scholar]
- 14.Carlson, M. L., R. M. Regan, and Q. H. Gibson. 1996. Distal cavity fluctuations in myoglobin: protein motion and ligand diffusion. Biochemistry. 35:1125–1136. [DOI] [PubMed] [Google Scholar]
- 15.Mouawad, L., D. Perahia, C. H. Robert, and C. Guilbert. 2002. New insights into the allosteric mechanism of human hemoglobin from molecular dynamics simulations. Biophys. J. 82:3224–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bossa, C., A. Amadei, I. Daidone, M. Anselmi, B. Vallone, M. Brunori, and A. Di Nola. 2005. Molecular dynamics simulation of sperm whale myoglobin: effects of mutations and trapped CO on the structure and dynamics of cavities. Biophys. J. 89:465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry, E. R. 1993. Molecular-dynamics simulations of heme reorientational motions in myoglobin. Biophys. J. 64:869–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolognesi, M., D. Bordo, M. Rizzi, C. Tarricone, and P. Ascenzi. 1997. Nonvertebrate hemoglobins: structural bases for reactivity. Prog. Biophys. Mol. Biol. 68:29–68. [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger, N. B. 1998. Functional adaptations of oxygen-transport proteins. J. Exp. Biol. 201:1085–1098. [DOI] [PubMed] [Google Scholar]
- 20.Vanholde, K. E., and K. I. Miller. 1995. Hemocyanins. In Advances in Protein Chemistry. Academic Press, Orlando, FL. 1–81. [DOI] [PubMed]
- 21.Mangum, C. P., editor. Invertebrate blood oxygen Carriers. In Handbook of Physiology, Section 13. Comparative Physiology, Vol. 2. W. H. Dantzler, editor. Oxford University Press, New York. 1097–1131.
- 22.Bolognesi, M., C. Rosano, R. Losso, A. Borassi, M. Rizzi, J. B. Wittenberg, A. Boffi, and P. Ascenzi. 1999. Cyanide binding to Lucina pectinata hemoglobin I and to sperm whale myoglobin: an x-ray crystallographic study. Biophys. J. 77:1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helbing, J., L. Bonacina, R. Pietri, J. Bredenbeck, P. Hamm, F. Van Mourik, F. Chaussard, A. Gonzalez-Gonzalez, M. Chergui, C. Ramos-Alvarez, C. Ruiz, and J. Lopez-Garriga. 2004. Time-resolved visible and infrared study of the cyano complexes of myoglobin and of hemoglobin I from Lucina pectinata. Biophys. J. 87:1881–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraus, D. W., and J. B. Wittenberg. 1990. Hemoglobins of the Lucina-pectinata bacteria symbiosis. 1. Molecular-properties, kinetics and equilibria of reactions with ligands. J. Biol. Chem. 265:16043–16053. [PubMed] [Google Scholar]
- 25.Kraus, D. W., J. B. Wittenberg, J. F. Lu, and J. Peisach. 1990. Hemoglobins of the Lucina-pectinata bacteria symbiosis. 2. An electron-paramagnetic resonance and optical spectral study of the ferric proteins. J. Biol. Chem. 265:16054–16059. [PubMed] [Google Scholar]
- 26.Rizzi, M., J. B. Wittenberg, A. Coda, M. Fasano, P. Ascenzi, and M. Bolognesi. 1994. Structure of the sulfide-reactive hemoglobin from the clam Lucina-pectinata—crystallographic analysis at 1.5-angstrom resolution. J. Mol. Biol. 244:86–99. [DOI] [PubMed] [Google Scholar]
- 27.Cerda, J., Y. Echevarria, E. Morales, and J. Lopez-Garriga. 1999. Resonance Raman studies of the heme-ligand active site of hemoglobin I from Lucina pectinata. Biospectroscopy. 5:289–301. [Google Scholar]
- 28.Nguyen, B. D., X. F. Zhao, K. Vyas, G. N. La Mar, R. A. Lile, E. A. Brucker, G. N. Phillips, J. S. Olson, and J. B. Wittenberg. 1998. Solution and crystal structures of a sperm whale myoglobin triple mutant that mimics the sulfide-binding hemoglobin from Lucina pectinata. J. Biol. Chem. 273:9517–9526. [DOI] [PubMed] [Google Scholar]
- 29.Cerda-Colon, J. F., E. Silfa, and J. Lopez-Garriga. 1998. Unusual rocking freedom of the heme in the hydrogen sulfide-binding hemoglobin from Lucina pectinata. J. Am. Chem. Soc. 120:9312–9317. [Google Scholar]
- 30.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 117:5179–5197. [Google Scholar]
- 31.Jorgensen, W. L., J. Chandrasekhar, J. D. Madura, R. W. Impey, and M. L. Klein. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935. [Google Scholar]
- 32.Autenrieth, F., E. Tajkhorshid, J. Baudry, and Z. Luthey-Schulten. 2004. Classical force field parameters for the heme prosthetic group of cytochrome C. J. Comput. Chem. 25:1613–1622. [DOI] [PubMed] [Google Scholar]
- 33.Amber Parameter Database. http://pharmacy.man.ac.uk/amber/.
- 34.Siegbahn, E. M. 2003. Mechanisms of metalloenzymes studied by quantum chemical methods. Q. Rev. Biophys. 36:91–145. [DOI] [PubMed] [Google Scholar]
- 35.Becke, A. D. 1988. A multicenter numerical-integration scheme for polyatomic-molecules. J. Chem. Phys. 88:2547–2553. [Google Scholar]
- 36.Lee, C. T., W. T. Yang, and R. G. Parr. 1988. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron-density. Phys. Rev. B. 37:785–789. [DOI] [PubMed] [Google Scholar]
- 37.Frisch, M. J., G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. A. Montgomery Jr., T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Iyengar, J. Tomasi, and others. 2003. Gaussian 2003 Revision B.04. Gaussian, Pittsburgh, PA.
- 38.Delley, B. 1990. An all-electron numerical-method for solving the local density functional for polyatomic-molecules. J. Chem. Phys. 92:508–517. [Google Scholar]
- 39.Delley, B. 2000. From molecules to solids with the Dmol(3) approach. J. Chem. Phys. 113:7756–7764. [Google Scholar]
- 40.Franzen, S. 2002. An electrostatic model for the frequency shifts in the carbonmonoxy stretching band of myoglobin: correlation of hydrogen bonding and the stark tuning rate. J. Am. Chem. Soc. 124:13271–13281. [DOI] [PubMed] [Google Scholar]
- 41.Franzen, S. 2002. Spin-dependent mechanism for diatomic ligand binding to heme. Proc. Natl. Acad. Sci. USA. 99:16754–16759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cieplak, P., W. D. Cornell, C. Bayly, and P. A. Kollman. 1995. Application of the multimolecule and multiconformational RESP methodology to biopolymers—charge derivation for DNA, RNA, and proteins. J. Comput. Chem. 16:1357–1377. [Google Scholar]
- 43.Bayly, C. I., P. Cieplak, W. D. Cornell, and P. A. Kollman. 1993. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the RESP model. J. Phys. Chem. 97:10269–10280. [Google Scholar]
- 44.University of California at San Francisco. http://www.amber.ucsf.edu.
- 45.Berendsen, H. J. C., J. P. M. Postma, W. F. Vangunsteren, A. Dinola, and J. R. Haak. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 46.Laskowski, R. A. 1995. Surfnet—a program for visualizing molecular-surfaces, cavities, and intermolecular interactions. J. Mol. Graph. 13:323–330. [DOI] [PubMed] [Google Scholar]
- 47.Rizzi, M., J. B. Wittenberg, A. Coda, P. Ascenzi, and M. Bolognesi. 1996. Structural bases for sulfide recognition in Lucina pectinata hemoglobin I. J. Mol. Biol. 258:1–5. [DOI] [PubMed] [Google Scholar]
- 48.Li, T., M. L. Quillin, G. N. Phillips Jr., and J. S. Olson. 1994. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 33:1433–1446. [DOI] [PubMed] [Google Scholar]
- 49.Case, D. A., and M. Karplus. 1979. Dynamics of ligand binding to heme proteins. J. Mol. Biol. 132:343–368. [DOI] [PubMed] [Google Scholar]
- 50.Hunter, C. L., E. Lloyd, L. D. Eltis, S. P. Rafferty, H. Lee, M. Smith, and A. G. Mauk. 1997. Role of the heme propionates in the interaction of heme with apomyoglobin and apocytochrome B(5). Biochemistry. 36:1010–1017. [DOI] [PubMed] [Google Scholar]
- 51.Carver, T. E., J. S. Olson, S. J. Smerdon, S. Krzywda, A. J. Wilkinson, Q. H. Gibson, R. S. Blackmore, J. D. Ropp, and S. G. Sligar. 1991. Contributions of residue 45(Cd3) and heme-6-propionate to the bimolecular and geminate recombination reactions of myoglobin. Biochemistry. 30:4697–4705. [DOI] [PubMed] [Google Scholar]
- 52.Takano, T. 1977. Structure of myoglobin refined at 2.0 Å resolution. 1. Crystallographic refinement of metmyoglobin from sperm whale. J. Mol. Biol. 110:537–568. [DOI] [PubMed] [Google Scholar]
- 53.La Mar, G. N., S. D. Emerson, J. T. J. Lecomte, U. Pande, K. M. Smith, G. W. Craig, and L. A. Kehres. 1986. Influence of propionate side-chains on the equilibrium heme orientation in sperm whale myoglobin—heme resonance assignments and structure determination by nuclear Overhauser effect measurements. J. Am. Chem. Soc. 108:5568–5573. [Google Scholar]
- 54.Smerdon, S. J., S. Krzywda, A. J. Wilkinson, R. E. Brantley Jr., T. E. Carver, M. S. Hargrove, and J. S. Olson. 1993. Serine92 (F7) contributes to the control of heme reactivity and stability in myoglobin. Biochemistry. 32:5132–5138. [DOI] [PubMed] [Google Scholar]
- 55.Mims, M. P., A. G. Porras, J. S. Olson, R. W. Noble, and J. A. Peterson. 1983. Ligand binding to heme proteins. An evaluation of distal effects. J. Biol. Chem. 258:14219–14232. [PubMed] [Google Scholar]
- 56.Gibson, Q. H., J. B. Wittenberg, B. A. Wittenberg, D. Bogusz, and C. A. Appleby. 1989. The kinetics of ligand binding to plant hemoglobins. Structural implications. J. Biol. Chem. 264:100–107. [PubMed] [Google Scholar]
- 57.Dou, Y., D. H. Maillett, R. F. Eich, and J. S. Olson. 2002. Myoglobin as a model system for designing heme protein based blood substitutes. Biophys. Chem. 98:127–148. [DOI] [PubMed] [Google Scholar]