Abstract
The members of the RecQ family of DNA helicases play conserved roles in the preservation of genome integrity. RecQ helicases are implicated in Bloom and Werner syndromes, which are associated with genomic instability and predisposition to cancers. The human BLM and WRN helicases are required for normal S phase progression. In contrast, Saccharomyces cerevisiae cells deleted for SGS1 grow with wild-type kinetics. To investigate the role of Sgs1p in DNA replication, we have monitored S phase progression in sgs1Δ cells. Unexpectedly, we find that these cells progress faster through S phase than their wild-type counterparts. Using bromodeoxyuridine incorporation and DNA combing, we show that replication forks are moving more rapidly in the absence of the Sgs1 helicase. However, completion of DNA replication is strongly retarded at the rDNA array of sgs1Δ cells, presumably because of their inability to prevent recombination at stalled forks, which are very abundant at this locus. These data suggest that Sgs1p is not required for processive DNA synthesis but prevents genomic instability by coordinating replication and recombination events during S phase.
Keywords: DNA replication/genomic instability/rDNA/RecQ helicases
Introduction
The SGS1 gene of Saccharomyces cerevisiae encodes a DNA helicase of the RecQ family, which is involved in the maintenance of genome integrity in all organisms analyzed to date, from bacteria to human (Karow et al., 2000). Sgs1p is closely related to the human helicases BLM, WRN and RECQL4, defective in Bloom syndrome (BS), Werner syndrome (WS) and Rothmund–Thomson syndrome (RTS), respectively. These diseases are characterized by chromosome instability, hyper-recombination and an increased incidence of cancers (Mohaghegh and Hickson, 2001). However, the molecular events underlying these phenotypes have remained obscure.
New insights into the molecular basis of these disorders came with the finding of functional interactions between RecQ helicases and multiple components of the replication machinery, including proliferating cell nuclear antigen (PCNA), RP-A, topoisomerases and DNA polymerase δ (Gangloff et al., 1994; Watt et al., 1995; Lebel et al., 1999; Brosh et al., 2000; Wu et al., 2000). In yeast and human cells, levels of RecQ helicases peak in S phase (Dutertre et al., 2000; Frei and Gasser, 2000), and these enzymes were shown to co-localize with sites of ongoing DNA synthesis in yeast and in Xenopus (Frei and Gasser, 2000; Chen et al., 2001). Moreover, cells derived from BS and WS patients accumulate abnormal replication intermediates (Lonn et al., 1990; Poot et al., 1992). Taken together, these data argue for a role for RecQ helicases at the replication fork.
Unlike components of the MCM complex—the presumed helicase of the growing fork (Labib and Diffley, 2001)—RecQ proteins are not essential for cell growth in the budding and fission yeasts and are therefore not likely to be involved in processive DNA synthesis. The simultaneous inactivation of Sgs1p and Srs2p, another DNA helicase, induces major growth defects in S.cerevisiae (Gangloff et al., 2000; McVey et al., 2001), and it had been proposed initially that Sgs1p and Srs2p are required for replication fork progression (Lee et al., 1999). However, this low viability of sgs1 srs2 cells is suppressed by the inactivation of the homologous recombination pathway, demonstrating that Sgs1p is not required for processive DNA synthesis, but rather is involved in the suppression of inappropriate recombination in S phase (Gangloff et al., 2000).
Interactions between RecQ helicases and components of the DNA recombinational repair machinery further support a role for these enzymes in the coordination of replication and recombination. In mammals, BLM is a component of BASC, a genome surveillance complex coordinating multiple recombination and repair activities (Wang et al., 2000). Like BLM, the yeast Sgs1 helicase interacts with Rad51p and Top3p (Bennett et al., 2000; Wu et al., 2000, 2001). Moreover, Sgs1p functions as a sensor in the intra-S phase checkpoint, potentially linking DNA recombination to cellular surveillance mechanisms (Cobb et al., 2002; Kolodner et al., 2002).
The partial complementation of the hyper-recombination phenotype of the yeast sgs1Δ mutant by the expression of either BLM or WRN suggests that their function has been conserved from yeast to human (Yamagata et al., 1998). However, RecQ activity seems to be dispensable for S phase progression in yeast, and sgs1Δ cells grow with wild-type kinetics. In contrast, cells derived from BS and WS patients display a prolonged S phase, with slow fork progression and accumulation of abnormal replication intermediates (Hand and German, 1975; Lonn et al., 1990; Poot et al., 1992). Whether this difference reflects a specialization of RecQ function in higher eukaryotes or whether it indicates that the same function is differentially required for S phase progression in yeast and mammals remains to be addressed.
Here, we have monitored S phase progression in wild-type and sgs1Δ yeast cells using flow cytometry, pulsed-field gel electrophoresis (PFGE), two-dimensional gel analysis and a new technique called DNA combing, which allows the analysis of replication on single DNA molecules. We show that sgs1Δ cells traverse S phase significantly faster than wild-type cells because forks are moving more rapidly. In contrast, we find that completion of DNA replication is impaired at ribosomal DNA (rDNA), which contains a high density of replication fork barriers. These data suggest that Sgs1p is required neither for elongation nor for the decatenation of newly replicated chromosomes, but plays an important role in the suppression of illegitimate recombination occurring at stalled replication forks.
Results
The Sgs1 helicase slows down progression through S phase
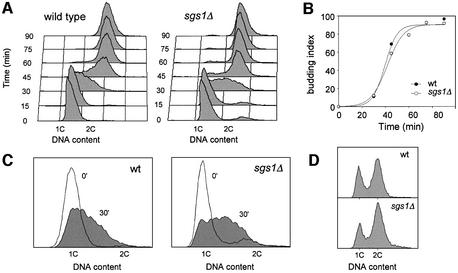
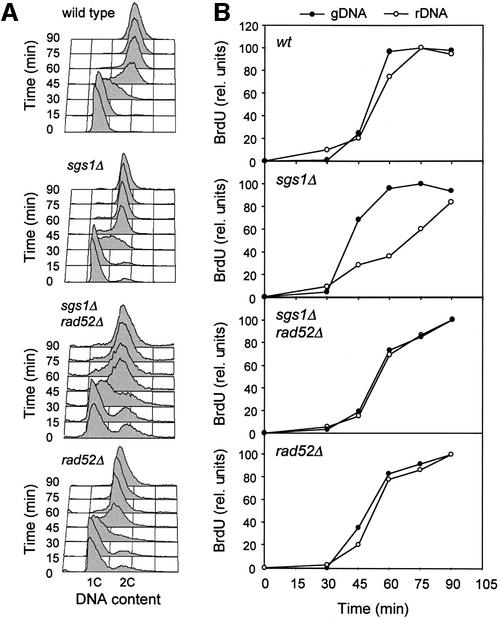
To examine the effect of SGS1 inactivation on DNA replication, congenic wild-type (E1000) and sgs1Δ (E1245) cells were arrested in G1 with α-factor and released synchronously into the cell cycle. Cells were collected every 15 min from release, and DNA content was monitored by flow cytometry (Figure 1A). The appearance of buds, indicative of entry into the cell cycle, was scored (Figure 1B). Surprisingly, the DNA content appeared to increase faster in the sgs1Δ mutant than in wild-type cells (Figure 1A; compare time points 30 and 45 min), although both strains passed START synchronously (Figure 1B). In order to quantify this difference, the area of S phase peaks at t = 30 min was integrated in four independent experiments (example in Figure 1C). The DNA content per haploid cell was 36% (± 8%) greater at this time in the absence of Sgs1 helicase. Interestingly, flow cytometry analysis of log phase cultures grown on synthetic complete medium at 25°C revealed that the proportion of cells with a 2C DNA content is ∼40% higher in the sgs1Δ mutant (Figure 1D), indicating that although sgs1Δ cells progress faster through S phase, they accumulate transiently in G2 or M phase.
Fig. 1. S phase progression is accelerated in sgs1Δ cells. (A–C) Wild-type (E1000) and sgs1Δ (E1245) cells were arrested in G1 with α-mating factor and released synchronously into S phase. (A) Samples were collected at the indicated times after release and DNA content was analyzed by flow cytometry. (B) Budding index. (C) Flow cytometry profiles of wild-type and sgs1Δ cells collected at t0 (G1) and t30 min (early S). The proportion of the area of the t30 min profile extending beyond the G1 peak is 42% larger in sgs1Δ mutants than in wild-type cells. (D) Flow cytometry analysis of exponentially growing wild-type and sgs1Δ cells. The proportion of G2/M cells is 40% (± 6%) higher in the sgs1Δ mutant.
Sgs1 is not required for the termination of DNA replication
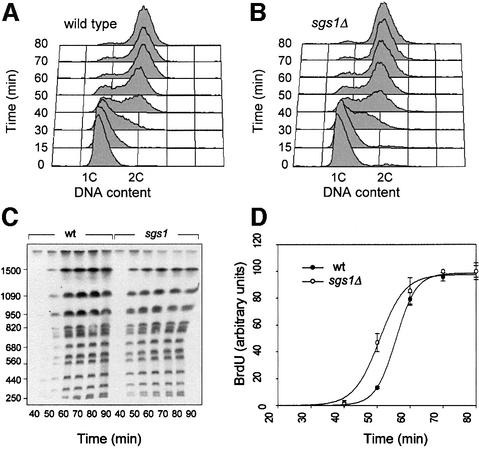
Sgs1p binds topoisomerases II and III, which are required for the decatenation of late replication intermediates (Gangloff et al., 1994; Watt et al., 1995). The elevated rate of missegregation in sgs1Δ cells could therefore reflect their inability to resolve newly replicated chromosomes. To test this hypothesis, we have monitored the completion of DNA replication in wild-type and sgs1Δ cells by PFGE. This approach is based on the fact that incompletely replicated chromosomes cannot be separated by PFGE (Hennessy et al., 1991). Thymidine kinase-expressing (TK+) cells, which are capable of incorporating bromodeoxyuridine (BrdU) (Lengronne et al., 2001), were released synchronously from a G1 arrest in complete medium supplemented with BrdU and were collected at regular intervals throughout the cell cycle (Figure 2A and B). Chromosomal DNA from wild-type and sgs1Δ cells was subsequently purified in agarose plugs and was separated by PFGE. As expected, BrdU-labeled DNA did not enter the gel until the end of S phase (Supplementary figure 1 available at The EMBO Journal Online). The re-emergence of fully replicated chromosomes was quantitated after transfer to a nitrocellulose membrane and detection of BrdU (Figure 2C and D). Interestingly, the bulk of chromosomal DNA re-entered the gel 5–10 min earlier in sgs1Δ cells, which represents approximately a quarter of the length of S phase in wild-type cells. This faster completion of DNA replication is reminiscent of the accelerated S phase progression observed by flow cytometry. Moreover, it shows that Sgs1p is not required for the timely resolution of late replication intermediates. However, the transient accumulation of cells with a 2C DNA content (Figure 1D) suggests that, although the bulk of genomic DNA is rapidly replicated, a significant proportion of the cells may experience problems in separating their chromosomes.
Fig. 2. The Sgs1p helicase is not required for the completion of DNA replication. Wild-type (E1000) and sgs1Δ (E1245) cells were released synchronously from an α-factor block in medium supplemented with 400 µg/ml BrdU and harvested at the indicated times. (A and B) Analysis of DNA content by flow cytometry. (C) Genomic DNA prepared from cells embedded in low-melting agarose plugs was separated by PFGE. The amount of BrdU incorporated in fully replicated chromosomes was quantitated as described in Materials and methods. (D) Kinetics of the re-emergence of fully replicated chromosomes in wild-type and sgs1Δ cells. Relative BrdU incorporation was quantitated for six representative chromosomes. Error bars indicate standard deviation.
Faster S phase progression in sgs1Δ cells is not due to a premature G1/S transition
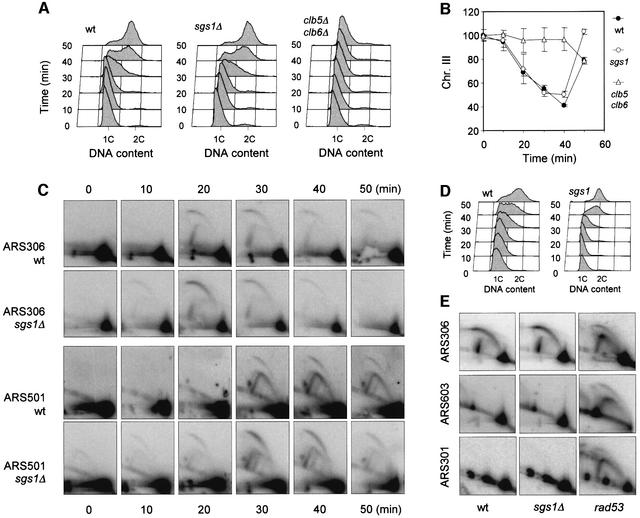
To check whether the faster completion of DNA replication in sgs1Δ cells is due to a premature entry into S phase, we have analyzed the kinetics of origin activation on a specific chromosome (chromosome III) in wild-type and sgs1Δ cells by PFGE (Figure 3A and B). In both strains, chromosome III is excluded progressively from the gel 10 min after release, indicating that cells enter synchronously into S phase. In contrast, chromosome III mobility decreased only after 40 min in clb5Δ clb6Δ cells (Figure 3B), which is consistent with the fact that S phase is delayed by 30 min in this mutant (Schwob and Nasmyth, 1993). Analysis of other yeast chromosomes gave the same result (data not shown). To confirm this observation using an independent approach, we have examined the time of ARS306 activation in wild-type and sgs1Δ cells by two-dimensional gel electrophoresis (Brewer and Fangman, 1987). ARS306 is the first origin to fire on chromosome III. As shown in Figure 3C, it is activated ∼20 min after release from the α-factor arrest in both strains. However, we noticed that replication intermediates disappeared more rapidly in the sgs1Δ mutant, suggesting either that initiation is more synchronous in this population of cells or that forks progress faster in the absence of Sgs1p. Interestingly, the time of ARS306 activation corresponds exactly to the time of the exclusion of chromosome III from the gel. However, this chromosome completed replication faster in sgs1Δ cells (Figure 3B), which is consistent with the data shown in Figure 2D. Taken together, these results indicate that the faster replication in sgs1Δ cells is not due to premature entry into S phase.
Fig. 3. Time of origin firing in wild-type and sgs1Δ cells. (A) Wild-type (E1000), sgs1Δ (E1245) and clb5Δ clb6Δ (E742) cells were arrested in G1 with α-factor. Cells were collected every 10 min after degradation of α-factor, and DNA content was analyzed by flow cytometry. (B) The electrophoretic mobility of chromosome III was analyzed by PFGE, and DNA content was quantitated with a phosphoimager. (C) Analysis of the timing of origin activation in wild-type (E1000) and sgs1Δ (E1245) cells. Cells were released synchronously into S phase from an α-factor block, and samples were taken at the indicated times after release. Neutral/neutral two-dimensional gel analysis (Brewer and Fangman, 1987) was used to monitor initiation at the early replication origin ARS306 and at the late origin ARS501. (D) Flow cytometry profiles of the samples analyzed in (C). (E) Two- dimensional gel analysis of initiation at the early origin ARS306, the late origin ARS603 and the dormant origin ARS301 in wild-type (E1000), sgs1Δ (E1245) and rad53-11 (E1019) cells released into S phase for 90 min in the presence of 200 mM HU.
The mechanisms delaying initiation at late origins are functional in sgs1Δ cells
We next analyzed the time of initiation at the subtelomeric late origin ARS501 and the internal late origin ARS603 using two-dimensional gel electrophoresis. We found that these origins are activated ∼10 min after the early origin ARS306 in both strains (Figure 3C, and data not shown). Although late origins appeared to fire slightly earlier in a fraction of sgs1Δ cells, this suggests that the timing of origin activation is not significantly affected by the absence of Sgs1p. Late origins are normally repressed by the replication checkpoint in the presence of hydroxyurea (HU) (Santocanale and Diffley, 1998), but fire in rpd3Δ cells exposed to HU, presumably because late origin initiation is advanced in these cells (O.Aparicio, unpublished results). To check whether this is also the case for sgs1Δ cells, we analyzed the activity of the late origin ARS603 and of the dormant origin ARS301 by two-dimensional gel electrophoresis. As shown in Figure 3E, these origins are repressed in wild-type and in sgs1Δ cells, but fire in the rad53-11 mutant (Santocanale and Diffley, 1998; Santocanale et al., 1999). Similarly, ARS501 is inactive in sgs1Δ cells exposed to HU (J.Cobb and S.Gasser, submitted). Taken together, these results indicate that the mechanisms controlling the activation of late replication origins are active in sgs1Δ cells.
The accelerated S phase in sgs1Δ cells is not due to a higher frequency of initiation
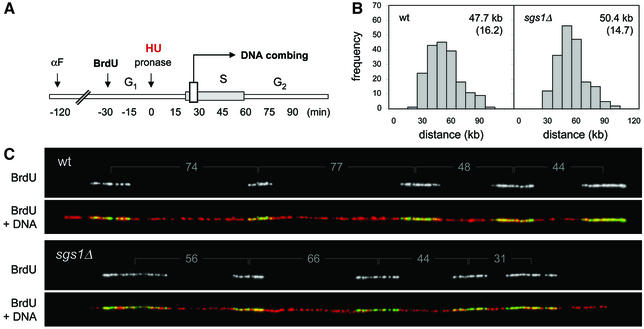
Several lines of evidence indicate that most of the yeast replication origins are active in only a fraction of the cell cycles. In sgs1Δ cells, an overall increase of this initiation rate could explain their faster progression through S phase. To address this possibility, we have compared the density in active origins in wild-type and sgs1Δ cells using BrdU incorporation and dynamic molecular combing (Michalet et al., 1997). Cells were released synchronously into S phase in the presence of BrdU to label initiation sites, and elongation was blocked with the addition of HU (Figure 4A). Chromosomal DNA was prepared in agarose plugs to avoid shearing and was combed on silanized coverslips. The sites of BrdU incorporation were subsequently detected along individual molecules with fluorescent antibodies, and DNA fibers were counterstained with an anti-guanosine antibody (Figure 4C). Since combed DNA fibers are stretched uniformly (Michalet et al., 1997), the statistical analysis of center to center distances between adjacent BrdU tracks provides an accurate indication of origin density (Lengronne and Schwob, 2002; Pasero et al., 2002). Here, the analysis of ∼10 Mb of DNA per strain revealed that active origins are not more abundant in sgs1Δ cells than in wild-type cells (Figure 4B). It is worth noting that initiation at late and dormant origins is not detected in this assay. However, we confirmed by two-dimensional gel electrophoresis that there is no promiscuous activation of dormant origins such as ARS301 in untreated sgs1Δ cells (data not shown). We therefore believe that the faster S phase of sgs1Δ cells is not due to an overall increase of origin activity.
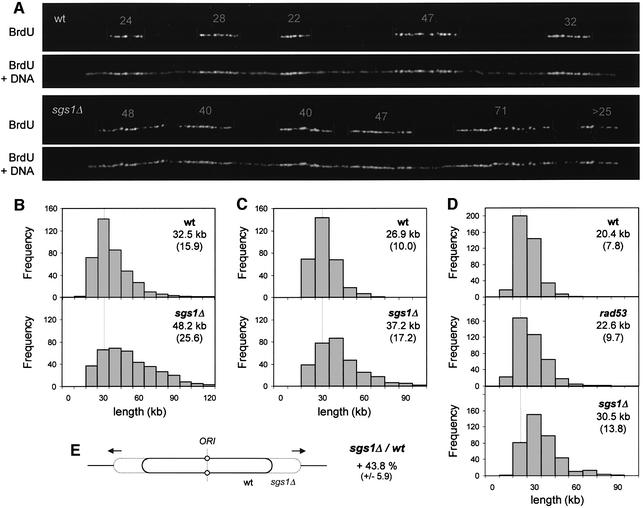
Fig. 4. Single molecule analysis of the initiation rate in wild-type and sgs1Δ cells. (A) Wild-type (E1000) and sgs1Δ (E1245) cells were synchronized in G1 with α-factor and were released for 90 min into S phase in the presence of BrdU and 200 mM HU, to block elongation. (B) DNA fibers were combed on silanized coverslips and the center to center distances between adjacent BrdU tracks were measured as described in Materials and methods. For each strain, a total of 9.4 Mb of DNA was analyzed. Means and standard deviations are indicated. (C) Examples of DNA fibers. The center to center distance between BrdU tracks is indicated in kbp. Green, BrdU; red, DNA.
Progression of replication forks is accelerated in sgs1Δ cells
An alternative explanation for the faster S phase in sgs1Δ cells could be that forks are moving more rapidly in the absence of the Sgs1p helicase. In the experiment described above, we found that BrdU tracks were on average 22% longer in HU-arrested sgs1Δ cells (wild type, 18.4 ± 9.2 kb; sgs1Δ, 22.5 ± 12.2 kb). To examine whether BrdU tracks are also longer in sgs1Δ cells in the absence of drug, wild-type and sgs1Δ cells were released synchronously from G1 and genomic DNA was isolated from cells harvested in mid-S phase (t = 30 min). Individual chromosomes were combed, and newly replicated regions were visualized using anti-BrdU antibodies (Figure 5A). The analysis of a large number of BrdU tracks (∼400 per strain) revealed that newly replicated stretches are 48% longer in sgs1Δ cells (Figure 5B). These data suggest that the rate of elongation is faster in sgs1Δ cells.
Fig. 5. Replication forks progress faster in the absence of Sgs1p. (A) Wild-type (E1000) and sgs1Δ (E1245) cells were released synchronously from an α-factor block in BrdU-supplemented medium as described in Figure 4, but this time HU was omitted. Samples were collected in mid-S phase (30 min after release) and BrdU tracks were detected after DNA combing. The length of BrdU tracks (kb) is indicated for two representative molecules. (B) Size distribution of BrdU tracks in wild-type and sgs1Δ cells. For each strain, ∼400 signals, encompassing >15 Mb in total, were measured. (C) Early log phase cultures of wild-type and sgs1Δ cells were labeled for 25 min with 400 µg/ml BrdU. The distribution of BrdU track length was determined as described above. For each strain, ∼300 BrdU signals were measured (>10 Mb in total). (D) Early log phase cultures of wild-type, sgs1Δ and rad53-11 (E1019) cells were labeled for 22 min and the distribution of BrdU tracks length was determined (400 BrdU signals for each strain). (E) The analysis of four independent sets of experiments indicates that BrdU tracks are reproducibly longer (43.8 ± 5.9%) in the absence of the Sgs1 helicase.
To ensure that longer BrdU tracks in sgs1Δ cells are not due to an artifact of synchronization, exponentially growing cultures of wild-type and sgs1Δ cells were pulse labeled for 25 min with BrdU and the length of newly replicated segments was measured by DNA combing. Again, we found a 40–50% increase of the mean BrdU track length in sgs1Δ cells (Figure 5C and D). The combination of four independent experiments confirmed that BrdU tracks are 44% (± 6%) longer in sgs1Δ cells (Figure 5E). Taken together, these results indicate that the Sgs1p helicase is required neither for the progression of the replication fork nor for the timely resolution of replicated chromosomes. Instead, we find that Sgs1p slows down the global rate of elongation.
The checkpoint kinase Rad53 does not slow down fork progression
In response to fork arrest induced by genotoxic drugs, yeast cells activate the checkpoint kinases Mec1p and Rad53p, which in turn block late origin firing and stabilize stalled forks (Lopes et al., 2001; Tercero and Diffley, 2001; Sogo et al., 2002). Since Sgs1p is required for the activation of Rad53p in HU-arrested cells, at least in Rad24-deficient cells (Frei and Gasser, 2000), we asked whether Sgs1p acts through Rad53p to slow down fork progression in a normal S phase. If this were the case, a rad53 mutant should also display an accelerated fork progression in our assay. To test this possibility, we measured the length of BrdU tracks in congenic wild-type (E1000), sgs1Δ (E1245) and rad53-11 (E1019) cells pulse labeled in mid-log phase as described above. We found that BrdU tracks were roughly identical in wild-type and rad53 cells, while they were again 50% longer in sgs1Δ cells (Figure 5D). These data indicate therefore that Rad53p does not regulate the rate of elongation in unchallenged growth conditions.
Completion of DNA replication is delayed at the rDNA locus of sgs1Δ cells
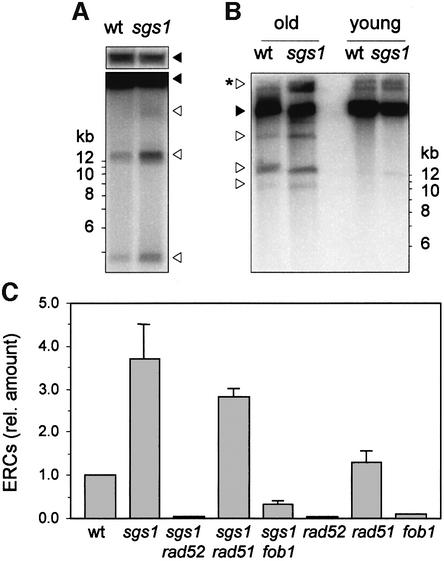
Sgs1-deficient cells are sensitive to sublethal doses of HU, presumably because they fail to stabilize stalled forks (J.Cobb and S.Gasser, submitted). We therefore reasoned that a locus naturally containing a high density of replication fork barriers, namely the rDNA array (Brewer and Fangman, 1988; Linskens and Huberman, 1988), should be more prone to replication fork collapse than bulk genomic DNA. To test this possibility, the rDNA array was excised by restriction digestion (Supplementary figure 2A), and completion of rDNA replication was monitored by PFGE. In wild-type cells, we found that the rDNA array re-entered the gel slightly after genomic DNA, suggesting that branched structures persist for a longer period of time at this locus (Figure 6B). Interestingly, this delay was exacerbated in sgs1Δ cells, the rDNA array re-entering the gel 20–30 min after the other chromosomes (Figure 6B). The same delay was observed when the mobility of rDNA arrays from wild-type and sgs1Δ cells were compared with each other (Supplementary figure 2B and C), or when chromosome XII, which bears the rDNA array, was compared with other chromosomes (Supplementary figure 2F). Interestingly, a similar alteration of chromosome XII mobility, which bears the rDNA array, has been reported recently in sgs1-ts slx4Δ double mutants (Kaliraman and Brill, 2002).
Fig. 6. Altered electrophoretic mobility of the rDNA array in sgs1Δ cells. Wild-type (E1000), sgs1Δ (E1245), rad52Δ (E1384) and sgs1Δ rad52Δ (E1382) cells were released synchronously into S phase in the presence of 400 µg/ml BrdU, and samples were collected at the indicated times. (A) Analysis of DNA content by flow cytometry. (B) Comparative analysis of the electrophoretic mobility of genomic DNA (gDNA) and rDNA in the four different strains. The re-emergence of fully replicated DNA molecules was quantitated as described in Figure 2.
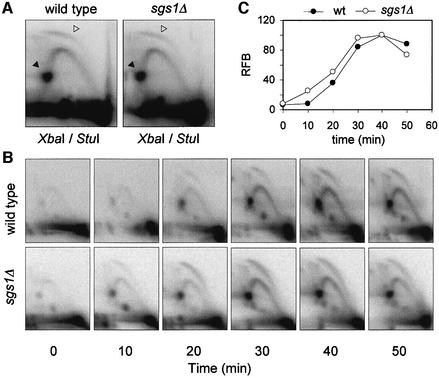
Wild-type and sgs1Δ cells display similar RFB and ARS activities
The rDNA of S.cerevisiae contains ∼150 ARS elements, but only 20% of these potential replication origins are used every cell cycle. Forks moving leftward from the rDNA origin are arrested by a structure called a replication fork barrier (RFB), which persists until it merges with rightward-moving forks (Brewer and Fangman, 1988; Linskens and Huberman, 1988). To check if the altered mobility of the rDNA in sgs1Δ cells was due to a delayed initiation at this locus or to a persistence of stalled forks in late S phase, we have monitored the activity of the rDNA ARS and of the RFB by two-dimensional gel electrophoresis. The quantitation of replication intermediates in three independent experiments showed that origin activity and RFB signals were virtually identical in exponentially growing wild-type and sgs1Δ cells (Figure 7A). To confirm this result, we have analyzed rDNA replication in cells released synchronously in S phase. Bubble arcs were detected throughout the S phase at the rDNA (Figure 7B). Quantitation of RFB signals with a phosphoimager revealed a similar pattern in both strains, although the rDNA replicated slightly earlier in the sgs1Δ mutant (Figure 7C; Supplementary figure 3A). Moreover, the amount of stalled forks decreased after 50 min in both strains, and RFB signals were almost undetectable in sgs1Δ cells 20 min later (Supplementary figure 3B). We therefore assume that the altered electrophoretic mobility of the rDNA in sgs1Δ cells is not due to a replication initiation defect nor to the persistence of stalled replication forks.
Fig. 7. Analysis of rDNA replication and RFB activity by two-dimensional gel electrophoresis. (A) Genomic DNA was extracted from exponentially growing wild-type (E1000) and sgs1Δ (E1245) cells, and a StuI–XbaI rDNA fragment (3.5 kb) centered around the origin was analyzed by two- dimensional gel electrophoresis. Quantitation of bubble arc (open arrowhead) and RFB signals (replication fork barrier, filled arrowhead) with a phosphoimager did not reveal any significant difference between the two strains in three independent experiments. (B) Two-dimensional gel analysis of rDNA ARS and RFB activity in wild-type and sgs1Δ cells released synchronously into S phase (see Figure 3D for the corresponding flow cytometry profiles). (C) Quantitation of RFB signals in wild-type and sgs1Δ cells.
The altered mobility of the rDNA array is due to homologous recombination
Deletion of the SGS1 gene increases homologous recombination at rDNA (Gangloff et al., 1994). To check whether the altered electrophoretic mobility of this locus in sgs1Δ cells is due to the presence of unresolved recombination intermediates, we have inactivated the homologous recombination pathway in these cells. Interestingly, the migration of the rDNA array was restored in rad52Δ sgs1Δ cells (Figure 6B). In the absence of Sgs1p, the abnormal mobility of the rDNA array in late S phase is therefore likely to be attributable to the persistence of unresolved recombination intermediates. It is worth noting that we were unable to detect these intermediates on two-dimensional gels (Figure 7B). In contrast, a strong X-spike, corresponding to Holliday junctions, has been reported at the rDNA of polymerase α and δ mutants (Zou and Rothstein, 1997). We assume that these intermediates are less abundant in sgs1Δ cells, or correspond to different recombination intermediates.
ERCs accumulate faster in young sgs1Δ cells, but not in aging cells
Intra-chromatid recombination at rDNA generates extrachromosomal rDNA circles (ERCs), which have been implicated in replicative senescence (Sinclair and Guarente, 1997). ERCs are present at one or two copies per cell in log phase cultures, but accumulate to very high levels in aging cells (Sinclair and Guarente, 1997; Ivessa et al., 2000). Here, we have examined ERC levels in sgs1Δ cells, as a marker of rDNA recombination, regardless of their role in replicative senescence. We measured a 3.7-fold (± 0.8) increase of ERC levels in young sgs1Δ cells (Figure 8A), which is consistent with the 5-fold increase of rDNA recombination measured in these cells with other techniques (Gangloff et al., 1994; Defossez et al., 1999; Heo et al., 1999; Hoopes et al., 2002; Merker and Klein, 2002).
Fig. 8. Quantitation of extrachromosomal rDNA circles in sgs1Δ cells. (A) Genomic DNA prepared from log phase wild-type (E1000) and sgs1Δ (E1245) cultures was separated by conventional gel electrophoresis. ERCs (empty arrowheads) were detected by Southern blotting and hybridization with a radioactive rDNA probe. A shorter exposure of the signal corresponding to the rDNA array is shown (filled arrowheads). (B) Detection of ERC species in young and old sgs1Δ cells. Cultures of wild-type (E1000) and sgs1Δ (E1245) cells were enriched in old mother cells (seven generations) as described (Sinclair and Guarente, 1997). (C) ERC levels in sgs1Δ (E1245), sgs1Δ rad52Δ (E1382), sgs1Δ rad51Δ (E1450), sgs1Δ fob1Δ (E1457), rad52Δ (E1384), rad51Δ (E1059) and fob1Δ (E1740) were quantitated with a phosphoimager and are expressed relative to the amount of ERCs in wild-type cells.
The faster accumulation of ERCs in old sgs1Δ cells was evoked to explain their premature aging phenotype (Sinclair and Guarente, 1997). However, this model has been challenged in recent studies showing that ERCs are not more abundant in old sgs1Δ cells than in old wild-type cells (Heo et al., 1999; McVey et al., 2001; Kaliraman and Brill, 2002). To reconcile our observations with these reports, we have examined ERC levels in wild-type and sgs1Δ cultures enriched in old mother cells (seven generations). As expected, we observed a dramatic increase of ERC levels in old wild-type and sgs1Δ cells compared with young cells (Figure 8B). However, with the exception of a slow migrating form detected above the rDNA array, the amount of ERC species appeared to be similar in old wild-type and sgs1Δ cells (Figure 8B, asterisked arrowhead). This observation is consistent with earlier reports indicating that ERCs are not responsible for the accelerated aging phenotype of sgs1Δ cells (McVey et al., 2001).
Accumulation of ERCs in sgs1Δ cells is Rad52 and Fob1 dependent
Since deletion of the RAD52 gene relieves the altered electrophoretic mobility of the rDNA array in sgs1Δ cells (Figure 6), we checked whether this would also suppress the accumulation of ERCs. We found that the inactivation of RAD52, but not of RAD51, completely suppressed the formation of ERCs in sgs1Δ cells (Figure 8C; Supplementary figure 4A), which is consistent with the fact that Rad51p is not required for homologous recombination at the rDNA (Zou and Rothstein, 1997). Moreover, deletion of the FOB1 gene, which is essential for RFB activity (Kobayashi and Horiuchi, 1996; Defossez et al., 1999), almost completely suppressed the formation of ERCs in sgs1Δ cells (Figure 8C) and relieved the altered electrophoretic mobility of the rDNA (Supplementary figure 4B). Taken together, these data indicate that rDNA replication is delayed in sgs1Δ cells because of hyper-recombination occurring at replication forks arrested at RFBs.
Discussion
RecQ DNA helicases are important for the maintenance of genome integrity and have been implicated in several aspects of DNA replication. However, the nature of their contribution to DNA synthesis has remained unclear. Here, we report that yeast cells progress faster through S phase in the absence of Sgs1p. We have used two-dimensional gels and PFGE mobility assays to show that this is not due to a premature entry into S phase, nor to the promiscuous activation of dormant replication origins. To investigate the potential causes of this accelerated S phase, we have analyzed the replication of single DNA molecules by dynamic molecular combing. We found that this faster replication is not due to changes in origin usage but rather to a faster progression of replication forks, which was best demonstrated in experiments with unsynchronized cells. It should be noticed that late origins appeared to fire slightly earlier in sgs1Δ cells. Although this effect is much stronger in rad53 mutants, it does not affect the length of the S phase (Shirahige et al., 1998) nor the elongation rate in rad53 cells (this work). We therefore assume that late origins fire slightly earlier in sgs1Δ cells because S phase is shorter, the primary cause for this accelerated S phase being the faster progression of replication forks.
Why DNA replication forks are moving more rapidly in the absence of Sgs1p is an intriguing issue. Since Sgs1p interacts with several components of the replication machinery and co-localizes with sites of ongoing DNA synthesis (Cobb et al., 2002), it could be permanently associated with the replication fork, hindering its progression via an unknown mechanism. Alternatively, Sgs1p could only act at the fork when it encounters an obstacle, such as a lesion of DNA or a replication pause site. Programmed pause sites have been found throughout the yeast genome (Deshpande and Newlon, 1996; Wang et al., 2001; Cha and Kleckner, 2002), and Sgs1p could stabilize forks traversing these sites in order to prevent illegitimate recombination. In sgs1Δ mutants, replication forks may progress faster through these sites, leading to a concomitant increase of the elongation rate and of genomic instability. We currently favor this second model, and experiments are under way to test it.
Genetic evidence indicates that Sgs1p acts together with Rad24p as sensor proteins to activate S phase checkpoints (Frei and Gasser, 2000; Myung and Kolodner, 2002). To test whether Sgs1p acts through Rad53p to regulate fork progression in normal growth conditions, we have measured the rate of elongation in rad53-11 cells by DNA combing. We found that Rad53p alone does not slow down fork progression in a normal S phase, as is the case for cells exposed to DNA damage (Tercero and Diffley, 2001). We therefore propose that Sgs1p modulates fork progression directly, independently of its checkpoint function.
The faster progression of replication forks in RecQ-deficient yeast cells contrasts strikingly with the slower rate of elongation reported previously in human BS and WS cells (Lonn et al., 1990; Poot et al., 1992). This discrepancy suggests that the function of RecQ helicases may have diverged during evolution. Alternatively, it could reflect structural differences between yeast and mammalian chromosomes. Indeed, the genome of S.cerevisiae is rather AT rich and is almost devoid of heterochromatin. In contrast, chromosomes of higher eukaryotes contain complex chromatin structures and a high frequency of repeated sequences, which are prone to homologous recombination. Moreover, mammalian genomes contain GC-rich repetitive elements, which can adopt alternative DNA structures such as hairpins or G-quartets. These structures are resolved efficiently by BLM, WRN and Sgs1p helicases in vitro (Sun et al., 1998, 1999; Fry and Loeb, 1999). In RecQ mutants, they would impede the progression of replication forks and generate genomic instability (Hyrien, 2000; Kamath-Loeb et al., 2001).
In the yeast genome, most of the heterochromatin and the G-rich sequences are found at telomeres and at the ribosomal gene array. Sgs1p is required for telomere maintenance in telomerase-deficient cells (Huang et al., 2001; Johnson et al., 2001). Here, we show that the structure of the rDNA array of sgs1Δ cells is altered uniquely in a replication-dependent manner. This is consistent with a recent report showing a similar alteration in an sgs1 slx4 double mutant (Kaliraman and Brill, 2002). Furthermore, we observed an accumulation of ERCs in young sgs1Δ cells, recapitulating their increased rate of rDNA recombination (Gangloff et al., 1994). Both the altered mobility of the rDNA array and the accumulation of ERCs were totally suppressed by the inactivation of the homologous recombination pathway in a rad52Δ mutant. We therefore propose that in the absence of Sgs1p helicase, unprotected replication increases genomic instability at the rDNA.
The rDNA array contains a high density of replication fork barriers (Brewer and Fangman, 1988; Linskens and Huberman, 1988), which represent a potential source of homologous recombination. We have used two-dimensional gel analysis to compare the abundance of forks arrested at RFBs in wild-type and sgs1Δ cells. No significant differences were detected between the two strains. Moreover, we found that RFB signals disappear almost 20 min before the resolution of intermediates, altering the electrophoretic mobility of the rDNA in sgs1Δ cells. This indicates that RFBs per se are not responsible for the abnormal electrophoretic mobility of the rDNA array in sgs1Δ cells. To check whether RFBs alter the mobility of rDNA via the formation of recombination intermediates, we have deleted the FOB1 gene in sgs1Δ cells. Suppressing RFB activity in these cells relieved this altered PFGE mobility and strongly reduced the formation of ERCs. However, unlike sgs1Δ rad52Δ mutants, low levels of ERCs were detected in RFB-deficient sgs1Δ cells. Fork arrest must therefore occur at other sites in fob1Δ sgs1Δ cells, as is the case for rrm3 mutants (Ivessa et al., 2000).
What is the primary source of homologous recombination in sgs1Δ cells? A branched structure, sometimes referred to as a ‘chicken foot’, has been proposed to form by reversion of stalled forks and to induce a DSB (Klein and Kreuzer, 2002). Such a structure is found frequently in rad53 cells exposed to HU (Sogo et al., 2002) and is resolved efficiently in vitro by Sgs1p (Bennett et al., 1999). However, recent genetic evidence argues against a role for DSBs in the genomic instability of sgs1Δ cells (Fabre et al., 2002). According to this report, Sgs1p would instead prevent the formation of single-stranded DNA (ssDNA) at stalled forks, on which recombination is initiated. ssDNA is not normally found at RFB-arrested forks in wild-type cells (Gruber et al., 2000), and its presence in sgs1Δ cells remains to be demonstrated. Interestingly, a loss of coordination of DNA polymerases during fork arrest has been observed recently in these cells (J.Cobb and S.Gasser, submitted). This provides a plausible mechanism for the formation of ssDNA in sgs1Δ mutants.
In conclusion, our data suggest that forks progress faster in the absence of Sgs1p helicase but are less stable, particularly at regions of the yeast genome containing a high density of replication fork barriers such as the rDNA array. It is worth noting that the replication defects observed specifically at the yeast rDNA are reminiscent of the replication defects observed in human BS and WS cells (Lonn et al., 1990; Poot et al., 1992). We therefore propose that yeast and human RecQ orthologs are equally involved in the protection of genome integrity during DNA replication, but that the complexity of vertebrate genomes makes them more dependent on RecQ helicases to replicate their chromosomes safely.
Materials and methods
Strains and synchronization procedure
Congenic E1000 (MATa, ade2-1 trp1-1 can1-100 leu2-3 112 his3-11,15 ura3-1 GAL, ura3::URA3/GPD-TK7x), E1245 (MATa, sgs1::LEU2, TK+), E742 (MATa, clb5::HIS3, clb6::LEU2, TK+), E1019 (MATa, rad53-11, TK+), E1382 (MATa, sgs1::LEU2, rad52::TRP1, TK+), E1384 (MATa, rad52::TRP1, TK+), E1059 (MATα, rad51::LEU2), E1450 (MATa, rad51::LEU2, sgs1::LEU2, TK+), E1740 (MATa, fob1::URA3, TK+) and E1457 (MATa, sgs1::HIS3, fob1::URA3, TK+) cells were grown at 25°C in complete synthetic medium. Mid-log phase cultures (5 × 106 cells/ml) were arrested for 2.5 h in G1 with 2 µg/ml α-factor and released into S phase with the addition of 50 µg/ml pronase (Calbiochem) in complete synthetic medium. To label newly replicated DNA, 400 µg/ml BrdU (Sigma) was added to the medium 30 min before release. To block elongation, 200 mM HU (Sigma) was added simultaneously to BrdU and cells were collected 90 min after release from the G1 arrest. Cell cycle progression was monitored by flow cytometry (FACScan) as described (Epstein and Cross, 1992).
Pulsed-field gel electrophoresis and quantitation of BrdU incorporation
Yeast cells were embedded in low-melting agarose plugs (5 × 107 cells/ml) and genomic DNA was extracted as described (Lengronne et al., 2001). Yeast chromosomes were separated by PFGE (Gene Navigator, AP Biotech) and transferred to a nitrocellulose membrane (Protran, Schleicher & Schüll). BrdU was detected with a mouse monoclonal antibody (Dako) and two layers of IgG coupled to Alexa 488 (Molecular Probes) as described (Lengronne et al., 2001). The membrane was scanned with a FluorImager and signals were quantitated with ImageQuant (Amersham Biosciences).
Dynamic molecular combing
DNA combing was performed as described (Michalet et al., 1997), with the following modifications. Genomic DNA prepared in agarose plugs (800 ng DNA/plug) was stained with YOYO-1 (Molecular Probes) and resuspended at 150 ng/ml in 50 mM MES pH 5.7 after digestion of the plugs with agarase (Roche). Combed DNA fibers were denatured with 1 M NaOH, and BrdU was detected with a rat monoclonal antibody (Sera Lab) and a secondary antibody coupled to Alexa 488 (Molecular Probes). Since denaturation eliminates YOYO-1 staining, DNA molecules were counterstained with an anti-guanosine antibody (Argene) and an anti-mouse IgG coupled to Alexa 546 (Molecular Probes). Images were recorded with a Leica DMRA microscope coupled to a CCD camera, and signals were measured with MetaMorph (Universal Imaging Corp.). Physical distances between signals were converted into base pairs (1 pixel = 340 bp) using adenovirus DNA molecules as size standard.
Two-dimensional gel analysis and quantitation of ERC species
Neutral/neutral two-dimensional gel analyses were performed as described (Brewer and Fangman, 1987). To monitor the amount of ERC species, genomic DNA was prepared in agarose plugs to avoid shearing, and was separated on a 0.7% agarose gel in 1× TAE without ethidium bromide for 22 h at 1 V/cm. rDNA species were detected after Southern blotting and hybridization with a 3 kb rDNA probe (Pasero et al., 2002). Quantification of autoradiograms was performed by storage phosphoimaging (Amersham). ERC levels were normalized against the chromosomal rDNA.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to S.Gasser, S.Gangloff and P.A.Defossez for critical comments on the manuscript, A.Bensimon for the gift of silanized coverslips for DNA combing, O.Aparicio and S.Gasser for sharing unpublished results, and P.A.Defossez for strains. This work was supported by grants from the CNRS (Physique et Chimie du Vivant) to E.S., NSF MCB RUI 113937 to L.H. and ARC (Association pour la Recherche contre le Cancer) to P.P.
References
- Bennett R.J., Keck,J.L. and Wang,J.C. (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S.cerevisiae. J. Mol. Biol., 289, 235–248. [DOI] [PubMed] [Google Scholar]
- Bennett R.J., Noirot-Gros,M.F. and Wang,J.C. (2000) Interaction between yeast sgs1 helicase and DNA topoisomerase III. J. Biol. Chem., 275, 26898–26905. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S.cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell, 55, 637–643. [DOI] [PubMed] [Google Scholar]
- Brosh R.M. Jr, Li,J.L., Kenny,M.K., Karow,J.K., Cooper,M.P., Kureekattil,R.P., Hickson,I.D. and Bohr,V.A. (2000) Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem., 275, 23500–23508. [DOI] [PubMed] [Google Scholar]
- Cha R.S. and Kleckner,N. (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science, 297, 602–606. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Graham,J. and Yan,H. (2001) Evidence for a replication function of FFA-1, the Xenopus orthologue of Werner syndrome protein. J. Cell Biol., 152, 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J.A., Bjergbaek,L. and Gasser,S.M. (2002) RecQ helicases: at the heart of genetic stability. FEBS Lett., 529, 43–48. [DOI] [PubMed] [Google Scholar]
- Defossez P.A., Prusty,R., Kaeberlein,M., Lin,S.J., Ferrigno,P., Silver,P.A., Keil,R.L. and Guarente,L. (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell, 3, 447–455. [DOI] [PubMed] [Google Scholar]
- Deshpande A.M. and Newlon,C.S. (1996) DNA replication fork pause sites dependent on transcription. Science, 272, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Dutertre S., Ababou,M., Onclercq,R., Delic,J., Chatton,B., Jaulin,C. and Amor-Gueret,M. (2000) Cell cycle regulation of the endogenous wild type Bloom’s syndrome DNA helicase. Oncogene, 19, 2731–2738. [DOI] [PubMed] [Google Scholar]
- Epstein C.B. and Cross,F.R. (1992) CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev., 6, 1695–1706. [DOI] [PubMed] [Google Scholar]
- Fabre F., Chan,A., Heyer,W.-D. and Gangloff,S. (2002) Alternate pathways involving Sgs1/Top3, Mus81/Mms4 and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA, 99, 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C. and Gasser,S.M. (2000) The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev., 14, 81–96. [PMC free article] [PubMed] [Google Scholar]
- Fry M. and Loeb,L.A. (1999) Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- Gangloff S., McDonald,J.P., Bendixen,C., Arthur,L. and Rothstein,R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol., 14, 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S., Soustelle,C. and Fabre,F. (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet., 25, 192–194. [DOI] [PubMed] [Google Scholar]
- Gruber M., Wellinger,R.E. and Sogo,J.M. (2000) Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol. Cell. Biol., 20, 5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R. and German,J. (1975) A retarded rate of DNA chain growth in Bloom’s syndrome. Proc. Natl Acad. Sci. USA, 72, 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy K.M., Lee,A., Chen,E. and Botstein,D. (1991) A group of interacting yeast DNA replication genes. Genes Dev., 5, 958–969. [DOI] [PubMed] [Google Scholar]
- Heo S.J., Tatebayashi,K., Ohsugi,I., Shimamoto,A., Furuichi,Y. and Ikeda,H. (1999) Bloom’s syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells, 4, 619–625. [DOI] [PubMed] [Google Scholar]
- Hoopes L.L.M., Budd,M., Choe,W., Weitao,T. and Campbell,J.L. (2002) Mutations in DNA replication genes reduce yeast life span. Mol. Cell. Biol., 22, 4136–4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Pryde,F.E., Lester,D., Maddison,R.L., Borts,R.H., Hickson,I.D. and Louis,E.J. (2001) SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol., 11, 125–129. [DOI] [PubMed] [Google Scholar]
- Hyrien O. (2000) Mechanisms and consequences of replication fork arrest. Biochimie, 82, 5–17. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou,J.Q. and Zakian,V.A. (2000) The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell, 100, 479–489. [DOI] [PubMed] [Google Scholar]
- Johnson F.B., Marciniak,R.A., McVey,M., Stewart,S.A., Hahn,W.C. and Guarente,L. (2001) The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J., 20, 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V. and Brill,S.J. (2002) Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet., 41, 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath-Loeb A.S., Loeb,L.A., Johansson,E., Burgers,P.M. and Fry,M. (2001) Interactions between the Werner syndrome helicase and DNA polymerase δ specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem., 276, 16439–16446. [DOI] [PubMed] [Google Scholar]
- Karow J.K., Wu,L. and Hickson,I.D. (2000) RecQ family helicases: roles in cancer and aging. Curr. Opin. Genet. Dev., 10, 32–38. [DOI] [PubMed] [Google Scholar]
- Klein H.L. and Kreuzer,K.N. (2002) Replication, recombination and repair: going for the gold. Mol. Cell, 9, 471–480. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Horiuchi,T. (1996) A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells, 1, 465–474. [DOI] [PubMed] [Google Scholar]
- Kolodner R.D., Putnam,C.D. and Myung,K. (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science, 297, 552–557. [DOI] [PubMed] [Google Scholar]
- Labib K. and Diffley,J.F. (2001) Is the MCM2–7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev., 11, 64–70. [DOI] [PubMed] [Google Scholar]
- Lebel M., Spillare,E.A., Harris,C.C. and Leder,P. (1999) The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem., 274, 37795–37799. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Johnson,R.E., Yu,S.L., Prakash,L. and Prakash,S. (1999) Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science, 286, 2339–2342. [DOI] [PubMed] [Google Scholar]
- Lengronne A. and Schwob,E. (2002) The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G1. Mol. Cell, 9, 1067–1078. [DOI] [PubMed] [Google Scholar]
- Lengronne A., Pasero,P., Bensimon,A. and Schwob,E. (2001) Monitoring S phase progression globally and locally using BrdU incorporation in TK+ yeast strains. Nucleic Acids Res., 29, 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M.H. and Huberman,J.A. (1988) Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 8, 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonn U., Lonn,S., Nylen,U., Winblad,G. and German,J. (1990) An abnormal profile of DNA replication intermediates in Bloom’s syndrome. Cancer Res., 50, 3141–3145. [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino,C., Pellicioli,A., Liberi,G., Plevani,P., Muzi-Falconi,M., Newlon,C.S. and Foiani,M. (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature, 412, 557–561. [DOI] [PubMed] [Google Scholar]
- McVey M., Kaeberlein,M., Tissenbaum,H.A. and Guarente,L. (2001) The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics, 157, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker R.J. and Klein,H.L. (2002) hpr1{Delta} affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae. Mol. Cell. Biol., 22, 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X. et al. (1997) Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science, 277, 1518–1523. [DOI] [PubMed] [Google Scholar]
- Mohaghegh P. and Hickson,I.D. (2001) DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet., 10, 741–746. [DOI] [PubMed] [Google Scholar]
- Myung K. and Kolodner,R.D. (2002) Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 99, 4500–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P., Bensimon,A. and Schwob,E. (2002) Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev., 16, 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M., Hoehn,H., Runger,T.M. and Martin,G.M. (1992) Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp. Cell Res., 202, 267–273. [DOI] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J.F. (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature, 395, 615–618. [DOI] [PubMed] [Google Scholar]
- Santocanale C., Sharma,K. and Diffley,J.F. (1999) Activation of dormant origins of DNA replication in budding yeast. Genes Dev., 13, 2360–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E. and Nasmyth,K. (1993) CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev., 7, 1160–1175. [DOI] [PubMed] [Google Scholar]
- Shirahige K., Hori,Y., Shiraishi,K., Yamashita,M., Takahashi,K., Obuse,C., Tsurimoto,T. and Yoshikawa,H. (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature, 395, 618–621. [DOI] [PubMed] [Google Scholar]
- Sinclair D.A. and Guarente,L. (1997) Extrachromosomal rDNA circles—a cause of aging in yeast. Cell, 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sogo J.M., Lopes,M. and Foiani,M. (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science, 297, 599–602. [DOI] [PubMed] [Google Scholar]
- Sun H., Karow,J.K., Hickson,I.D. and Maizels,N. (1998) The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem., 273, 27587–27592. [DOI] [PubMed] [Google Scholar]
- Sun H., Bennett,R.J. and Maizels,N. (1999) The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G–G paired DNAs. Nucleic Acids Res., 27, 1978–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero J.A. and Diffley,J.F. (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature, 412, 553–557. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cortez,D., Yazdi,P., Neff,N., Elledge,S.J. and Qin,J. (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev., 14, 927–939. [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Vujcic,M. and Kowalski,D. (2001) DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol. Cell. Biol., 21, 4938–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt P.M., Louis,E.J., Borts,R.H. and Hickson,I.D. (1995) Sgs1: a eukaryotic homolog of E.coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell, 81, 253–260. [DOI] [PubMed] [Google Scholar]
- Wu L., Davies,S.L., North,P.S., Goulaouic,H., Riou,J.F., Turley,H., Gatter,K.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product interacts with topoisomerase III. J. Biol. Chem., 275, 9636–9644. [DOI] [PubMed] [Google Scholar]
- Wu L., Davies,S.L., Levitt,N.C. and Hickson,I.D. (2001) Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem., 276, 19375–19381. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Kato,J., Shimamoto,A., Goto,M., Furuichi,Y. and Ikeda,H. (1998) Bloom’s and Werner’s syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl Acad. Sci. USA, 95, 8733–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H. and Rothstein,R. (1997) Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell, 90, 87–96. [DOI] [PubMed] [Google Scholar]