Abstract
The goal of this study was to develop a simple plating medium to allow large-scale screening of water samples for the presence of Helicobacter pylori. Five conventional plating media (brain heart infusion, brucella agar, Columbia blood agar base, campylobacter agar kit Skirrow, and HPSPA medium), each containing a commercial antibiotic supplement, were initially evaluated. Eight strains selected as common waterborne organisms (Acinetobacter, Aeromonas, Bacillus, Escherichia coli, Enterobacter, Enterococcus, Helicobacter pylori, and Pseudomonas strains) were individually plated onto each of these media. Three organisms (Acinetobacter, E. coli, and H. pylori) were able to grow on all five media. This growth was unacceptable since Helicobacter grows very slowly and competing organisms must be inhibited for up to 7 days. Therefore, a more selective medium (HP agar) containing a novel mixture of growth supplements plus amphotericin B and polymyxin B was developed. This medium also included a phenol red color indicator for urease production. Aliquots of nonsterile well water that contained native flora (Flavobacterium, Serratia, Citrobacter, Pasteurella, Ochrobactrum, Rahnella, and unidentified molds) and were further adulterated with the eight strains listed above (106 CFU of each strain per 100 ml) were spiked with H. pylori and were plated. In spite of the heavy mixed microbial load, only H. pylori colonies grew during 7 days of incubation at 37°C. The color indicator system allowed presumptive identification of H. pylori colonies sooner (12 to 20 h) than the conventional media tested allowed. The HP formulation developed in this study provides a medium with superior selectivity for H. pylori from mixed microbial populations in water and reduces the time required to complete the assay.
A scientific breakthrough occurred in 1982 when J. R. Warren and B. Marshall isolated a bacterium and showed that it causes gastritis and stomach ulcers that affect millions of humans worldwide (10, 12). Today this etiology has been proven to the extent that the National Institutes of Health recommends treatment with antibiotics for all patients with peptic ulcers, which are almost exclusively attributed to infection with the bacterium Helicobacter pylori (4). The scope of gastric illnesses around the world is vast, and in the United States alone, over 5,000,000 people are diagnosed annually with ulcers, 1,000,000 people are hospitalized, 40,000 people undergo surgery, and 6,500 people die from ulcer-related complications (11, 21). Estimates suggest that as many as 50% of adult Americans carry the pathogen, most asymptomatically, and in less-developed countries human carriers represent up to 90% of the populations (15).
The source of human infection is not yet known, and until recently, the natural reservoir for H. pylori was thought to be the human gastrointestinal tract (1). However, the association of Helicobacter with nonhuman sources, such as livestock (23), domestic cats, (17), and vegetables (6), prompted researchers to look at environmental sources as vectors to humans. Previous studies suggested that H. pylori is present in groundwater, surface water, and other drinking water (5, 7, 8, 13, 14, 20, 24), implying that there is a waterborne route of transmission to humans. The methods used in these studies (PCR, immunomagnetic separation, autoradiography, enzyme immunoassay) could not differentiate between viable and dead cells and would not be cost-effective for screening a large number of samples representing a substantial geographical area. A low-cost and effective test to isolate viable H. pylori from groundwater and surface water, similar to selective media used for Salmonella or Escherichia coli, would enable the drinking water industry to routinely screen samples and perhaps establish demographics for the propensity of drinking water to contain this pathogen. Therefore, we focused our efforts on development of a plating medium that selects viable H. pylori from water samples containing mixed microbial populations.
MATERIALS AND METHODS
Bacterial strain management.
Five clinical infection H. pylori strains (no environmental isolates are available in United States or European type culture collections) were obtained from the American Type Culture Collection (Manassas, Va.) or the Wisconsin State Laboratory of Hygiene culture collection (Madison, Wis.) (Table 1) and were cultured on brain heart infusion (BHI) (Becton Dickinson, Sparks, Md.) agar supplemented with 7% calf serum in the presence of a microaerobic gas mixture (5% CO2, 10% H2, 85% N2; Praxair, Inc., Danbury, Conn.) at 37°C. Frozen stock cultures were prepared by picking isolated colonies from agar plates and homogenizing them in sodium phosphate buffer to a concentration of about 107 CFU/ml with a McFarland nephelometer. Each of the five isolates (Table 1) was then frozen in BHI broth containing 10% glycerol. Sufficient quantities were prepared to complete the entire study in order to avoid multiple passages of strains, which can sometimes lead to phenotypic variability. Other bacterial strains used to create adulterated water samples (Table 1) were obtained from the Wisconsin State Laboratory of Hygiene culture collection, grown on BHI agar slants at 35°C, and then stored at 4°C for up to 3 weeks before reculturing.
TABLE 1.
Bacterial strains used to prepare water samples containing known levels of contaminants
| Taxon | Straina |
|---|---|
| Helicobacter pylori | ATCC 43504 |
| Helicobacter pylori | ATCC 700392 |
| Helicobacter pylori | ATCC 49503 |
| Helicobacter pylori | WSLH 95-10882 |
| Helicobacter pylori | WSLH 409013 |
| Acinetobacter | WSLH Envir. Iso.1 |
| Aeromonas hydrophila | ATCC 7966 |
| Escherichia coli | ATCC 25922 |
| Pseudomonas aeruginosa | ATCC 10145 |
| Enterobacter cloacae | ATCC 13047 |
| Enterococcus faecalis | ATCC 19433 |
| Bacillus cereus | ATCC 11778 |
ATCC, American Type Culture Collection; WSLH, Wisconsin State Laboratory of Hygiene environmental isolate.
Preparation of conventional media.
Conventional dehydrated medium preparations were chosen for selection of H. pylori from mixed microbial populations based on previously published data. The five media chosen for evaluation were BHI containing 7% calf serum (16), brucella agar (19), campylobacter agar kit Skirrow (3), Columbia blood agar base (2) (Becton Dickinson), and HPSPA medium (9, 22). Table 2 shows the ingredients of these media and the common and unique features of each medium. Inclusion of either H. pylori selective supplement (Oxoid Limited, Hampshire, Basingstoke, England) or Campylobacter selective supplement S (Becton Dickinson) provided antibiotics to prevent background contamination while permitting H. pylori growth. The positive controls consisted of the five conventional media without the selective supplements, while the negative controls consisted of uninoculated plates. All media were prepared according to the manufacturer's or authors' instructions.
TABLE 2.
Formulations of conventional media used to culture and select H. pylori from mixed microbial samples
| Medium | Proteose peptone (g/liter) | Dex- trose (g/liter) | Sodium chloride (g/liter) | Agar (g/liter) | Di- sodium phos- phate (g/liter) | Calf serum with Fe (ml/liter) | Pep- tamin (g/liter) | Tryp- tone (g/liter) | Yeast extract (g/liter) | Sodium bisulfite (g/liter) | Pantone (g/liter) | Bitone (g/liter) | Beef heart digest (g/liter) | Special peptone (g/liter) | Porcine mucin (g/liter) | Beef extract (g/liter) | Ferrous sulfate (g/liter) | Sodium pyruvate (g/liter) | Urea (g/liter) | Liver digest (ml/liter) | H. pylori selective supple- menta | Campylobacter selective supple- ment Sb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BHI | 10 | 2 | 5 | 15 | 2.5 | 70 | Yes | |||||||||||||||
| Brucella agar | 1 | 5 | 15 | 10 | 10 | 2 | 0.1 | Yes | ||||||||||||||
| Colombia blood agar base | 1 | 5 | 15 | 10 | 10 | 3 | Yes | |||||||||||||||
| HPSPA mediumc | 5 | 15 | 70 | 5 | 15 | 2 | 5 | 0.5 | 0.5 | 0.6 | Yes | |||||||||||
| Campylobacter agar kit Skirrow | 15 | 5 | 15 | 5 | 2.5 | Yes |
H. pylori selective supplement contains vancomycin (10 mg/liter), cefsulodin (5 mg/liter), trimethoprim (5 mg/liter), and amphotericin B (5 mg/liter).
Campylobacter selective supplement S contains vancomycin (10 mg/liter), trimethoprim (5 mg/liter), and polymyxin B (2,500 Iu/liter).
Formulation described by Jiang and Doyle (9).
Sample preparation. (i) Growth of pure cultures on conventional media.
Each strain listed in Table 1 was separately cultured on each of the five media listed in Table 2 in order to evaluate the growth and inhibition spectra of individual formulations. First, pure colonies of each strain were picked from solid growth medium (BHI agar plates) and homogenized in sodium phosphate buffer (4%; pH 7.2). Serial dilutions of each pure homogenate were immediately spread (0.1 ml/plate) onto each solid medium with and without the selective supplement added. All plates were incubated under microaerobic conditions at 37°C for up to 7 days and examined to determine the presence of colonies on the media compared to the positive controls. The ideal formulation would permit growth of H. pylori while preventing the growth of all background strains.
(ii) Selectivity of H. pylori in adulterated samples.
Well water containing native flora (see Table 4), which were identified by using the API 20E identification system (Biomerieux Vitek, Inc., Hazelwood, Mo.), was further adulterated with the seven strains of background bacteria and H. pylori to obtain a highly contaminated water sample (10,000 cells of each strain per 100 ml). The adulterated water was then serially diluted, and 0.1 ml of each dilution was spread onto duplicate plates of each of the five conventional media supplemented with either the H. pylori or Campylobacter selective supplement (Table 2). The positive controls were the same as those described above. All plates were incubated in the microaerobic atmosphere at 37°C for up to 7 days to provide the optimal environment for culturing Helicobacter. The media were evaluated for recovery of H. pylori colonies and inhibition of background flora.
TABLE 4.
Growth and inhibition of water-related bacterial strains on conventional media with and without H. pylori selective supplement and on newly formulated HP agar
| Bacterium | Growth on solid media
|
||
|---|---|---|---|
| Conventional media without selective supplementa | Conventional media with selective supplement | HP mediumb | |
| Supplementary | |||
| Acinetobacter | +c | + | − |
| Aeromonas | + | − | − |
| Bacillus | + | − | − |
| Escherichia coli | + | + | − |
| Enterobacter cloacae | + | − | − |
| Enterococcus faecalis | + | − | − |
| Helicobacter | + | + | + |
| Pseudomonas aeruginosa | + | − | − |
| Flora native to well water | |||
| Flavobacterium | + | + | − |
| Serratia | + | − | − |
| Citrobacter | + | − | − |
| Pasteurella | + | + | − |
| Ochrobactrum | + | + | − |
| Rahnella | + | − | − |
Oxoid selective supplement.
HP medium contained (per liter of distilled water) 12 g of special peptone, 5 g of yeast extract, 5 g of beef extract, 5 g of NaCl, 70 ml of calf serum with Fe, 3,500 U of polymyxin B, 7.5 mg of amphotericin B, 10 mg of vancomycin, 5 mg of trimethoprim, 5 mg of cefsulodin, 600 mg of urea, 100 mg of phenol red, 0.8 ml of 1 N HCl, and 15 g of agar. The pH was 6.0 at 22°C. The calf serum with Fe, polymyxin B, amphotericin B, vancomycin, trimethoprim, cefsulodin, and urea were added aseptically after autoclaring and tempering to 50°C. The HCl was added dropwise to cause a color change from red to yellow.
+, growth; −, inhibition.
Formulation of enhanced selectivity medium (HP medium).
Since the selectivity of the five conventional media tested proved to be inadequate for isolating H. pylori from the adulterated water samples, we determined that an enhanced selective medium was required. To develop the formula for this new medium, components were individually evaluated to determine their contributions to the selective and nutritive properties necessary to isolate H. pylori from a mixed population of microbial contaminants. Some nutritive components (yeast extract, beef extract, special peptone [Oxoid], NaCl) were incorporated at conventional concentrations without further evaluation. Table 3 shows the additional components evaluated and the ranges of concentrations tested. The optimum level of each additive was determined based on the shortest incubation time needed to presumptively identify an H. pylori colony, coupled with the ability of the component to prevent colony formation by any extraneous flora present in the adulterated water sample. The novel combination consisting of urea, phenol red, and hydrochloric acid (HCl) was incorporated into the medium to expedite presumptive identification of H. pylori colonies by color enhancement induced by urease activity.
TABLE 3.
Medium components evaluated at a range of concentrations to develop the final HP formulation
| Component | Range tested (liter−1) | Optimum level (liter−1)a |
|---|---|---|
| Porcine mucin | 0-4 g | 0 |
| Ferrous sulfate | 0-500 mg | 0 |
| Sodium pyruvate | 0-500 mg | 0 |
| Polymixin B | 0-4,000 U | 3,500 U |
| Amphotericin B | 0-7.5 mg | 7.5 mg |
| Vancomycin | 0-10 mg | 10 mg |
| Trimethoprim | 0-5 mg | 5 mg |
| Cefsulodin | 0-5 mg | 5 mg |
| Urea | 0-1.0 g | 600 mg |
| Phenol red | 0-200 mg | 100 mg |
| HCl (1 N) | 0-2 ml | 0.8 ml |
Optimum level for recovering H. pylori while inhibiting the background flora.
Preparing HP medium required sequential addition of components. The mixture containing special peptone, beef extract, yeast extract, NaCl, phenol red, agar, and water was autoclaved for 20 min at 121°C and then tempered to 50°C. Then calf serum with iron, antibiotics (vancomycin, trimethoprim, cefsulodin, amphotericin B, and polymixin B), and urea were aseptically added with constant stirring. Finally, 0.8 ml of 1 N HCl was added dropwise to the medium as the color changed from red to yellow-orange (final pH at 45°C, 5.7; final pH at 22°C, 6.0). The medium was then poured into petri plates.
RESULTS
Medium evaluation. (i) Growth of pure cultures on conventional media
Table 2 shows the five conventional medium formulations evaluated. Each of these media was evaluated for its ability to recover H. pylori from a population containing seven spiked strains and at least six indigenous strains in a sample of well water (Table 4). As expected, media without the antibiotic supplement allowed growth of all of the organisms tested. Addition of selective supplements provided some measure of selective pressure; however, some of the organisms (Acinetobacter, E. coli, Flavobacterium, Pasteurella, Ochrobactrum) were not inhibited, and unacceptable levels of overgrowth occurred within 24 h. The selectivity profiles were identical for the five conventional medium formulations, although we noted that H. pylori colonies formed most rapidly (84 h) on HPSPA medium.
Greater selective pressure was needed to isolate H. pylori from environmental samples as the competition of even a single background contaminant rendered the assay unacceptable because the relatively vigorous growth of most background flora outcompeted the slowly growing Helicobacter.
In order to develop a medium with enhanced selectivity for Helicobacter, a number of selective, nutritional, and differential components were evaluated (Table 3). The properties of an improved formulation should include a broader inhibition spectrum that includes E. coli, Acinetobacter, Flavobacterium, Pasteurella, and Ochrobactrum, as well as molds.
The growth of H. pylori was not enhanced by the presence of the various concentrations of ferrous sulfate, sodium pyruvate, or porcine mucin tested. Therefore, these components were omitted from the final formulation. However, more rapid colony formation on HPSPA medium was attributed to the presence of special peptone and calf serum with iron, so these remained as nutritional components. Increasing the level of vancomycin or cefsulodin to more than 10 mg/liter resulted in retarding the growth of Helicobacter, as did increasing the level of trimethoprim to more than 5 mg/liter. Conversely, reducing the concentrations below these levels allowed background contamination. Therefore, these concentrations (present in the selective supplement) were considered optimal. Increasing the level of amphotericin B from 5 to 7.5 mg/liter was adequate to broaden the spectrum of inhibition to include all apparent background bacteria. Addition of polymyxin B at a concentration of 3,500 U/liter appreciably reduced the occurrence of mold contaminants and had few deleterious effects on H. pylori colony development.
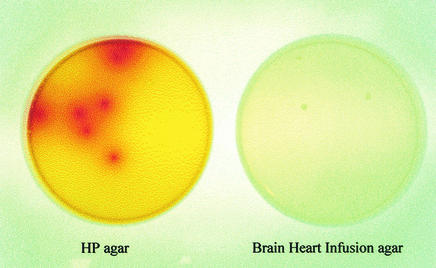
The novel color indicator system that easily detected H. pylori among organisms that do not produce urease greatly enhanced the utility of the medium (Fig. 1). Colony growth and subsequent urease production resulted in hydrolysis of urea to ammonium and bicarbonate, thus neutralizing a discrete area around each colony. This area of neutralization was marked by a zone of red around the colony as the pH of the medium changed from about 6.0 to >7.5. Incorporating the color indicator accelerated presumptive identification of H. pylori colonies by at least 12 h (from 84 to 72 h).
FIG. 1.
Growth of H. pylori on HP and BHI agar plates.
(ii) Selectivity of modified HP medium.
The local well water used in these experiments contained native heterotrophic bacteria at lower levels than the levels of the artificially added background strains (400 cells per 100 ml compared to 10,000 cells per 100 ml). Some of the native organisms were identified as Flavobacterium, Serratia, Citrobacter, Pasteurella, Ochrobactrum, and Rahnella by using the API 20E identification system (Table 4). In addition to the native flora, seven additional strains and an H. pylori cocktail were added at levels of about 10,000 CFU of each organism per 100 ml. Dilutions of the adulterated well water were then plated onto BHI agar with 7% calf serum (positive control) and HP agar. The plates were incubated under microaerobic conditions for up to 7 days and were monitored for colony development. The positive control media without the selective agents became overgrown with bacterial colonies within 24 h of inoculation. The HP agar plates, however, contained only colonies of H. pylori during the 7-day incubation period, and the recovery efficiencies ranged from 20 to 50 cells per ml. Colonies were presumptively recognizable within 72 ± 8 h (ca. 14% shorter incubation period) because of the pH indicator and resultant red halo showing urease production. In addition to shorter incubation times, interference from background bacteria and/or molds was not problematic because of the increased levels of antibiotics.
DISCUSSION
An examination of five conventional medium formulations showed that all of them were comparably nutritious for culturing H. pylori, as well as the native and added background organisms (Table 4). However, developing an acceptable selective medium for H. pylori in water presented the classical microbiological problem of finding a medium that is nutritionally rich enough to resuscitate and grow a fastidious organism while managing to inhibit the growth of all the other organisms found in water samples. H. pylori is a relatively slowly growing bacterium, usually requiring about 4 days to develop discernible colonies. This organism can easily be overgrown on solid media by robust strains that grow readily within 24 h and thereby conceal the presence of the pathogen. The antibiotic resistance spectrum of H. pylori is well defined, but the concentrations in media vary widely depending on the matrix in which the research is done. For example, trimethoprim and polymyxin B were used at concentrations of 5 mg/liter and 3,500 U/liter, respectively, to isolate H. pylori in a water matrix (18), but the concentrations were increased to 40 mg/liter and 62,000 U/liter for selection in cattle mucosa (22). In this study, we focused on antibiotic levels established by commercial vendors (Oxoid, Becton Dickinson) and adjusted these levels in order to achieve acceptable results.
Commercial supplements allowed some background strains (E. coli, Acinetobacter, Flavobacterium, Pasteurella, and Ochrobactrum) to grow during the moist, warm (37°C), and relatively lengthy (up to 7 days) incubation. In spite of extraordinary care to maintain sterility, molds also frequently developed and overwhelmed the plates. Thus, the concentrations of some of the selective components were increased after each antibiotic was evaluated individually. Increasing the amphotericin concentration from 5 to 7.5 mg/liter and adding polymyxin B at a concentration of 3,500 U/liter were adequate to solve the contamination problems and had no apparent deleterious effects on H. pylori growth.
Although Jiang and Doyle (9) reported that porcine mucin, ferrous sulfate, and sodium pyruvate were important in improving the recovery of H. pylori cells, there was no appreciable difference between the results obtained in the presence and in the absence of these components in this study. The idea of using the characteristic urease production and subsequent color change from yellow to red appeared in a clinical assay (CLO rapid urease test; Delta West Pty Ltd., Bentley, Australia) but not previously in solid media designed for culturing Helicobacter. The addition of this color system substantially reduces (by about 14%) the time required to grow colonies to the point of presumptive visual identification. Even on HP medium inoculated with heavy bacterial loads, the occasional background colony that developed was easily differentiated from H. pylori.
In summary, the HP formulation provides a medium with superior selectivity for H. pylori from mixed microbial populations in water and reduces the time required to complete the assay. Future applications for HP medium will include a geographical survey of Wisconsin water to identify discrete pockets of H. pylori in drinking water.
Acknowledgments
This is work supported by the Wisconsin Department Natural Resources Groundwater Coordinating Council.
We acknowledge the valuable contributions of Becky Leidner and Linda Peterson from the Wisconsin State Laboratory of Hygiene.
REFERENCES
- 1.Axon, A. T. R. 1996. The transmission of Helicobacter pylori: which theory fits the facts? Eur. J. Gastroenterol. Hepatol. 8:1-2. [DOI] [PubMed] [Google Scholar]
- 2.Baron, E. J., L. R. Peterson, and S. M. Finegold. 1994. Bailey & Scott's diagnostic microbiology, 9th ed. Mosby-Year Book, Inc., St. Louis, Mo.
- 3.Corry, J. E. L., D. E. Post, P. Colin, and M. J. Laisney. 1995. Culture media for the isolation of campylobacters. Elsevier Science, Amsterdam, The Netherlands. [DOI] [PubMed]
- 4.Graham, D. Y., H. M. Malaty, D. G. Evans, D. J. Evans, P. D. Klein, and E. Adam. 1991. Epidemiology of Helicobacter pylori in an asymptomatic population in the United States. Gastroenterology 100:1494-1501. [DOI] [PubMed] [Google Scholar]
- 5.Hegarty, J. P., M. T. Dowd, and K. T. Baker. 1999. Occurrence of Helicobacter pylori in surface water in the United States. J. Appl. Microbiol. 87:697-701. [DOI] [PubMed] [Google Scholar]
- 6.Hopkins, R. J., P. A. Vial, C. Ferreccio, J. Ovalle, P. Prado, V. Sotomayor, R. G. Russel, S. S. Wasserman, and J. G. Morris. 1993. Seroprevalence of Helicobacter pylori in Chile—vegetables may serve as one route of transmission. J. Infect. Dis. 168:222-226. [DOI] [PubMed] [Google Scholar]
- 7.Hulten, K., H. Enroth, T. Nystrom, and L. Engstrand. 1998. Presence of Helicobacter species DNA in Swedish water. J. Appl. MIcrobiol. 85:282-286. [DOI] [PubMed]
- 8.Hulten, K., S. W. Han, and F. A. K. El-Zaatari. 1995. Detection of Helicobacter pylori in Peruvian water sources by two PCR assays based on independent genes. Gut 37:S1A10.
- 9.Jiang, X. P., and M. P. Doyle. 2000. Growth supplements for Helicobacter pylori. J. Clin. Microbiol. 38:1984-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd, M., and I. M. Modlin. 1998. A century of Helicobacter pylori: paradigms lost-paradigms regained. Digestion 59:1-15. [DOI] [PubMed] [Google Scholar]
- 11.Levin, T. R., J. A. Schmittdiel, J. M. Henning, K. Kunz, C. J. Henke, C. J. Colby, and J. V. Selby. 1998. A cost analysis of a Helicobacter pylori eradication strategy in a large health maintenance organization. Am. J. Gastroenterol. 93:743-747. [DOI] [PubMed] [Google Scholar]
- 12.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed]
- 13.Mazari-Hiriart, M., Y. Lopez-Vidal, and J. J. Calva. 2001. Helicobacter pylori in water systems for human use in Mexico City. Water Sci. Technol. 43:93-98. [PubMed] [Google Scholar]
- 14.McKeown, I., P. Orr, S. MacDonald, A. Kabani, R. Brown, G. Coghlan, M. Dawood, J. Embil, M. Sargent, G. Smart, and C. N. Bernstein. 1999. Helicobacter pylori in the Canadian arctic: seroprevalence and detection in community water samples. Am. J. Gastroenterol. 94:1823-1829. [DOI] [PubMed] [Google Scholar]
- 15.Munnangi, S., and A. Sonnenberg. 1997. Time trends of physician visits and treatment patterns of peptic ulcer disease in the United States. Arch. Intern. Med. 175:1489-1494. [PubMed] [Google Scholar]
- 16.Osaki, T., H. Taguchi, H. Yamaguchi, and K. Shigeru. 1998. Detection of Helicobacter pylori in fecal samples of gnotobiotic mice infected with H. pylori by an immunmomagnetic-bead separation technique. J. Clin. Microbiol. 36:321-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otto, G., S. H. Hazell, J. G. Fox, C. R. Howlett, J. C. Murphy, J. L. O'Rourke, and A. Lee. 1994. Animal and public health implications of gastric colonization of cats by Helicobacter-like organisms. J. Clin. Microbiol. 32:1043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penner, J. L. 1991. Campylobacter, Helicobacter, and related spiral bacteria, p. 406-407. In A. Balows, W. J. Hausler, K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 19.Poms, R. E., and S. R. Tatini. 2001. Survival of Helicobacter pylori in ready-to-eat foods at 4oC. Int. J. Food Microbiol. 63:281-286. [DOI] [PubMed] [Google Scholar]
- 20.Shahamat, M., U. Mai, C. Paszko-kolva, M. Kessel, and R. R. Colwell. 1993. Use of autoradiography to assess viability of Helicobacter pylori in water. Appl. Environ. Microbiol. 59:1231-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonnenberg, A., and J. E. Everhart. 1997. Health impact of peptic ulcer in the United States. Am. J. Gastroenterol. 92:614-620. [PubMed] [Google Scholar]
- 22.Stevenson, T. H., L. M. Lucia, and G. R. Acuff. 2000. Development of a selective medium for isolation of Helicobacter pylori from cattle and beef samples. Appl. Environ. Microbiol. 66:723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaira, D., P. Ferron, R. Negrinin, L. Cavazzini, J. Holton, C. Ainley, M. Londei, M. Vergura, R. Dei, and A. Colecchia. 1992. Detection of Helicobacter pylori-like organisms in the stomach of some food-source animals using a monoclonal antibody. Ital. J. Gastroenterol. 24:181-184. [PubMed] [Google Scholar]
- 24.Yingzhi, L., T. E. Redlinger, R. Avitia, G. Adriana, and K. Goodman. 2002. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl. Environ. Microbiol. 68:1436-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]