Abstract
Campylobacter jejuni is a leading cause of food-borne disease in developed countries. The goal of this study was to develop a plasmid-based reporter system with green fluorescent protein (GFP) to facilitate the study of C. jejuni in a variety of niches. C. jejuni transformants harboring the pMEK91 GFP gene (gfp)-containing vector were readily detectable by both fluorescence microscopy and flow cytometry. Given the ease of detecting these organisms, additional experiments were performed in which BALB/c mice were injected intraperitoneally with C. jejuni harboring the gfp-containing vector. Four hours after injection of the mice, flow cytometry analyses determined that C. jejuni synthesizing GFP were predominantly associated with granulocytes. More specifically, the proportion of CD11b+ Gr-1+ lavage neutrophils with green fluorescence ranged from 99.7 to 100%, while the proportion of CD11b+ Gr-1− lavage macrophages ranged from 77.0 to 80.0%. In contrast, few CD11b− CD45R+ B lymphocytes from the lavage of the C. jejuni-injected mice were associated with green-fluorescent C. jejuni (proportions ranged from 0.75 to 0.77%). Cell-free C. jejuni was recovered from tissue homogenates after intraperitoneal injection. Macrorestriction profiling with pulsed-field gel electrophoresis identified a genotypic variant of the C. jejuni F38011 wild-type isolate. In vivo this variant displayed a phenotype identical to that of the wild-type isolate. In summary, we demonstrate that C. jejuni associates with marker-defined cellular subsets in vivo with a novel gfp reporter system and that C. jejuni genotypic variants can be isolated from both in vitro and in vivo systems.
Campylobacteriosis, gastroenteritis resulting from infection caused by a thermotolerant Campylobacter spp., is one of the leading food-borne diseases in developed countries. The economic impact of this disease is significant; in the United States alone, it is estimated that the total cost due to campylobacteriosis exceeds US$1.2 billion annually (3). Despite the prevalence of Campylobacter jejuni infections, C. jejuni-host cell interactions are less well characterized than the interactions of some gastrointestinal bacterial pathogens with host cells. Development of molecular and genetic tools to study Campylobacter spp. in a variety of niches would facilitate our understanding of the biology of this organism.
The use of a reporter system would allow the detection of C. jejuni in in vivo situations, enabling the examination of C. jejuni persistence and survival in different ecological contexts. Ideally, a C. jejuni reporter system would use a gene whose product does not require an exogenous substrate for its function, and this gene would be expressed with an endogenous C. jejuni promoter. The promoter itself would be constitutively expressed at a high level to facilitate reporter detection. While several reporters have been used for Campylobacter spp. (e.g., lacZ, luxAB, cat, cfp, gfp, and yfp), the cfp, gfp, and yfp genes do not require an exogenous substrate (1, 13, 19, 20, 34). The expression of the gene (gfp) encoding green fluorescent protein (GFP) is stable and resistant to photobleaching.
A major advance was reported by Miller et al., who described a fluorescence-based reporter system for C. jejuni (13). In this system, three distinct Campylobacter vectors that contained either gfp, cfp, or yfp constitutively expressed from the C. jejuni consensus promoter sequence identified by Wösten et al. (34) were generated. While this system effectively allowed the detection of C. jejuni on chicken breast tissue and in Caco-2 cells, it was never tested in a live animal model.
Flow cytometry can simultaneously characterize a cell population for size-dependent laser scattering and fluorescence properties with a broad range of detection sensitivities. Investigators routinely use cytometry to analyze cell surface protein expression in heterogeneous cell suspensions from tissue samples and to detect levels of fluorescent protein expression (6). More recently, investigators have coupled multiparameter cell surface phenotype analyses with the detection of fluorescent microbes. For example, investigators have analyzed the dynamics of the host-microbe interaction in animals inoculated with GFP-synthesizing bacteria, characterizing microbe tropism for specific cells through cell subset-specific markers (35). Similar experiments detected GFP-synthesizing vaccinia virus after infection (5). Other cytometric analyses of GFP-synthesizing microbes assessed fluorescent bacteria associated with cultured mammalian cells (28) and determination of bacterial density in soil samples over time (27). Refinements of applications employing fluorescent protein and cytometric detection continue to be reported. We employed cytometry and confocal microscopy to identify the murine cell subsets that retained C. jejuni after intraperitoneal injection.
Epidemiological studies have been used to assess the dynamics of pathogen populations in animals and humans. More specifically, investigators determined that the colonization of animals by Campylobacter spp. is not restricted to a single isolate within a species or, in fact, a single species (31, 32). After experimental infection of chickens with an apparently homogeneous inoculum, more than one isolate type was detected, as judged by restriction fragment length polymorphisms in the flaA gene (15, 30) and by alterations in macrorestriction profiles as determined by pulsed-field gel electrophoresis (mrp-PFGE) (7, 31). The investigators hypothesized that newly isolated C. jejuni variants, which displayed unique genotypic profiles compared with the C. jejuni wild-type isolates, represented clonal isolates that originated in the animal host. However, whether these C. jejuni variants retain their ability to induce a host cell response similar to that of the C. jejuni wild-type isolate has not been tested. Although the frequency of coinfection is less than in animals, Campylobacter coinfections have been observed in humans (21, 22).
In this study, we describe the construction of a plasmid-based reporter system in which gfp is constitutively expressed at high levels with the promoter for the C. jejuni major outer membrane protein. We demonstrated the usefulness of the gfp reporter system in an in vivo system by intraperitoneally injecting mice with C. jejuni that had been transformed with a gfp-containing vector. While mice do not represent a suitable model for Campylobacter-mediated enteritis, investigators have used intraperitoneal injection of mice to examine the interaction of C. jejuni with various components of the immune system (17, 18, 29). In conjunction with flow cytometry analysis, we demonstrate the application of the new vector containing gfp to show the cellular subset associated with C. jejuni in BALB/c mice. Finally, we describe the isolation of a genotypic variant of the C. jejuni F38011 wild-type isolate and its host response phenotype in the mouse model.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All C. jejuni isolates were cultured on Mueller-Hinton agar plates supplemented with 5% (vol/vol) citrated blood (MHB) at 37°C in a microaerophilic atmosphere and passaged every 24 to 48 h. Plates were supplemented with tetracycline (40 μg/ml) as appropriate. For mrp-PFGE experiments, single colonies of C. jejuni F38011 and the F38011 variants were isolated from freezer stocks and used to make clonal cultures.
Molecular biology and polyacrylamide gel electrophoresis.
Cloning of PCR-purified DNA fragments was done by standard molecular biology techniques. All PCR-amplified DNA fragments were cloned into pCR2.1 (Invitrogen). DNA extractions from agarose gels were performed with a Qiaex kit (Qiagen Inc., Valencia, Calif.) according to the manufacturer's instructions. Oligonucleotide primers were synthesized by Invitrogen (Gaithersburg, Md.). In vitro transcription-translation analyses of purified recombinant plasmids were performed with an Escherichia coli S30-coupled transcription-translation system as described by the supplier (Promega, Madison, Wis.). The translated products were labeled with [35S]methionine (Perkin Elmer, Boston, Mass.). For one-dimensional gel electrophoresis, samples were mixed with an equal volume of double-strength electrophoresis sample buffer, and proteins were denatured by placing the tube in a boiling-water bath for 5 min. Proteins were resolved in sodium dodecyl sulfate-10% polyacrylamide gels with the discontinuous buffer system described by Laemmli (10). Labeled proteins in dried gels were detected by autoradiography.
Intraperitoneal infection of mice, recoverable CFU, and flow cytometric analysis.
Female BALB/c mice, 4 to 12 weeks of age, were maintained in specific-pathogen-free conditions in a biosafety level 2 facility and treated humanely in compliance with institutional guidelines. At the times indicated after intraperitoneal injection with C. jejuni in phosphate-buffered saline (PBS), mice were euthanized. The peritoneum was lavaged with ice-cold, sterile PBS (pH 7.2) supplemented with 5% (vol/vol) heat-inactivated bovine calf serum (HyClone, Logan, Utah) to collect both cells and bacteria. The lavage fluid was used directly for CFU determinations and bound to sterile coverslips for microscopy, while cells from the lavage were centrifuged and washed with PBS for cytometric analyses. Liver and spleen tissues were removed aseptically, rinsed with PBS, weighed, and ground through sterile mesh. The resulting suspensions were used either directly for CFU determinations or further prepared for flow cytometry detection of cell-associated bacteria.
For flow cytometry, samples were counted, and 106 lavage cells or splenocytes were centrifuged (500 × g, 10 min) at 4°C. The cell pellets were suspended in 0.1 ml of cold PBS supplemented with 0.01% (wt/vol) NaN3, 1% (wt/vol) bovine serum albumin (Fraction V; Sigma, St. Louis, Mo.), 10 μg of rat gamma globulin (Jackson ImmunoResearch, West Grove, Pa.), and anti-mouse CD16 antibody (1:100 dilution; Fc Block; BD Biosciences, San Diego, Calif.). Cells were incubated on ice for 10 min before the addition of combinations of anti-mouse CD11b antibody conjugated to Tricolor (1:100 dilution; Caltag, Burlingame, Calif.), anti-mouse CD45R conjugated to phycoerythrin (PE; 1:100 dilution; Caltag) or PE-conjugated anti-mouse Gr-1 (1:200 dilution; BD Biosciences). Isotype-matched control reagents included rat IgG2b conjugated to Tricolor (1:100 dilution; Caltag) and rat IgG2a conjugated to PE (1:100 dilution; Caltag). Following an additional 30 min of incubation with antibody on ice, cells were washed in cold PBS containing 0.01% (wt/vol) NaN3.
Cytometric data were acquired with a FACScalibur cytometer (BD Biosciences) after adjustment of machine settings based on single-color and negative isotype staining controls. For analyses of lavage cells and splenocytes, cytometric data were gated according to size scatter properties and then displayed as two-color dot plots. Plots were analyzed to determine the staining characteristics of the gated populations, and proportions were determined with CellQuest software (BD Biosciences). The data were analyzed statistically for some treatment groups, generating means and standard deviations.
Immunofluorescence microscopy.
Lavage fluid samples were applied to sterile coverslips and incubated at 37°C for 4 h to allow the cells to adhere. After incubation, the coverslips were rinsed to remove unbound cells, and adherent cells were fixed with methanol. Samples were stained with antibody combinations to detect host cell surface markers and C. jejuni. To detect host cell surface markers, samples were stained with a combination of anti-CD11b conjugated to Tricolor (Caltag) and anti-Gr-1 conjugated to PE (BD Biosciences). Control samples were stained with isotype-matched rat IgG2b conjugated to Tricolor and rat IgG2a conjugated to PE (Caltag). To detect C. jejuni, samples were stained with rabbit anti-C. jejuni serum, followed by a goat anti-rabbit IgG-fluorescein F(ab′)2 fragment (1:500 dilution; Roche, Indianapolis, Ind.) as described previously (14). Samples were visualized with a Nikon inverted microscope with a 60× objective, and images were captured with a Bio-Rad 1024 scanning confocal microscope system equipped with a krypton-argon laser (Bio-Rad, Hercules, Calif.). Images were processed with Adobe Photoshop 4.0 (Adobe Systems, Inc., Mountain View, Calif.).
Cell proliferation assays.
To assess the cellular response to the injected C. jejuni, ex vivo splenocyte proliferation assays were performed. Cell densities of splenocyte suspensions were enumerated by counting trypan blue (Sigma)-excluding cells. Cells were diluted in RPMI 1640 medium (MediaTech, Herndon, Va.) supplemented with 10% (vol/vol) bovine calf serum, 10 mM HEPES (Invitrogen), 5 × 10−5 M 2-mercaptoethanol (Sigma), 800 mg of glutamine per ml (Invitrogen), 50 units of penicillin G per ml (Invitrogen), and 50 μg of streptomycin sulfate per ml (Invitrogen). Thereafter, 5 × 105 cells in 0.2 ml were delivered to quadruplicate tissue culture wells. Each well further received 0.025 ml of medium containing 0.5 μCi of [3H]methylthymidine (Perkin Elmer). Plates were incubated for another 24 h, and supernatant fluids and cell lysates were collected. Samples were spotted onto filters and counted in the presence of scintillation fluid (Fisher Scientific, Fair Lawn, N.J.). The data are presented as the geometric mean counts per minute ± standard deviation for each treatment group. Student's t test was used to calculate P values, and a P value of <0.01 was considered significant.
Pulsed-field gel electrophoresis.
C. jejuni was harvested from MHB agar plates in 3 ml of PETT IV buffer (1 M NaCl, 10 mM Tris, 10 mM EDTA, pH 8.0), and cell densities were adjusted to a McFarland standard of approximately 1.0. Each bacterial suspension (1.5 ml) was concentrated by centrifugation (5,000 × g, 10 min) and resuspended in 150 μl of PETT IV buffer. Two hundred and fifty microliters of 1.6% (wt/vol) molten (50°C) pulsed-field grade agarose (Bio-Rad) was added to each sample and mixed gently, and a 100-μl aliquot of the cell suspension was pipetted into agarose plug molds. The agarose plugs were removed from the molds and incubated in 1 ml of ESP buffer (500 mM EDTA, 1% [wt/vol] N-lauroyl sarcosine, 0.5 mg of proteinase K per ml) at 50°C for 48 h. Following cell wall lysis, the agarose blocks were washed three times in sterile water and three times in TE (10 mM Tris [pH 8.0], 1 mM EDTA). Each wash was performed at ambient temperature for 30 min.
Individual agarose plugs were incubated with 100 μl of 1× restriction endonuclease buffer containing 20 U of restriction endonuclease. The reaction mixes were incubated at 25°C for a minimum of 4 h. Following incubation, the agarose plugs were loaded into an agarose gel. Restricted genomic DNA was separated in 1% (wt/vol) pulsed-field grade agarose that had been prepared with 0.5× TBE [0.089 M Tris base, 0.089 M boric acid, 0.002 M EDTA (pH 8.0)]. Run parameters consisted of a reorientation angle of 120° with a constant voltage of 120 V and a constant temperature of 14°C. An electrophoresis run time of 22 h and a ramped pulse time of 10 to 35 s were used. Gels were stained for 20 min in 3 μg of ethidium bromide per ml and destained for 20 min in water. Images were captured with a Bio-Rad FluorS system and processed with Adobe Photoshop.
RESULTS
Generation of Campylobacter shuttle vector harboring gfp.
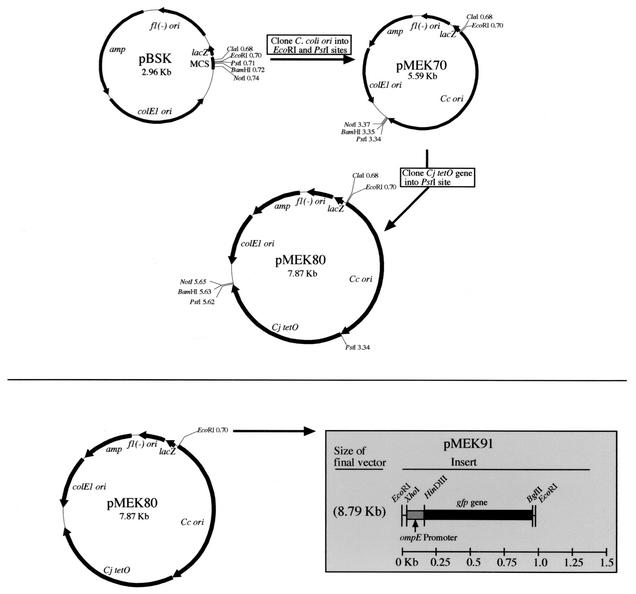
A Campylobacter shuttle vector harboring gfp was generated in order to perform in vivo studies with C. jejuni. Shuttle plasmid replication functions were amplified from the Campylobacter pILL550 shuttle vector (9) by PCR with the pILL550-T3-F4 PstI and pILL550-T7-F2 EcoRI primers (Table 1). The 2.6-kb amplified DNA fragment was subsequently cloned into the pBluescript II(SK+) vector (Fig. 1). The resultant vector was designated pMEK70. The C. jejuni tetO gene (2.28 kb) was PCR amplified from pUOA3 (25) with the primers listed in Table 1, and the resultant fragment was cloned into the pMEK70 shuttle vector following PstI digestion (Fig. 1). The new Campylobacter shuttle vector was designated pMEK80. Consistent with previous work (24), we found it necessary to clone a minimum of 288 nucleotides upstream of the methionine initiation codon to obtain a vector capable of conferring tetracycline resistance on both E. coli and C. jejuni transformants. Following construction of pMEK80, gfp (4) was cloned into the EcoRI site. To ensure that the gfp gene would be transcribed in C. jejuni, its expression was driven from the ompE promoter (Fig. 1). The ompE gene in C. jejuni encodes the major outer membrane protein and is constitutively expressed. The resultant vector was designated pMEK91.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | DNA sequence (5′ to 3′) |
|---|---|
| pILL550- T3-F4-PstI | TAA CTG CAG AGA ACT AGG ACA CGA AAG AGC |
| pILL550-T7- F2-EcoRI | TTT GAA TTC GTC TTA GCA TTA TCG TTG G |
| tetO-F-PstI | TAA CTG CAG AGA TTC AGT ATT ATA ACA AGG |
| tetO-R-PstI | TTA CTG CAG CAT CAT AAT TAT CTC TAA TCC |
| ompE-prom- F-XhoI | TTT CTC GAG CTT TAG ATG TTT TTA TCT TCG |
| ompE-prom- R-HinDIII | CGA AAG CTT TCT CCT TGT CAA AAA TTA ATA AAA C |
| gfp-F-HinDIII | TTT AAG CTT AGT AAA GGA GAA GAA CTT TTC ACT GG |
| gfp-R-BgIII | TTT AGA TCT TTT TGA CAC CAG ACA AGT TG |
FIG. 1.
Construction of pMEK91 Campylobacter shuttle vector used in this study. MCS, multiple cloning site; Cc ori, C. coli origin of replication; Cj, C. jejuni; colE1 ori, plasmid origin of replication; fl(−)ori, filamentous phage origin of replication.
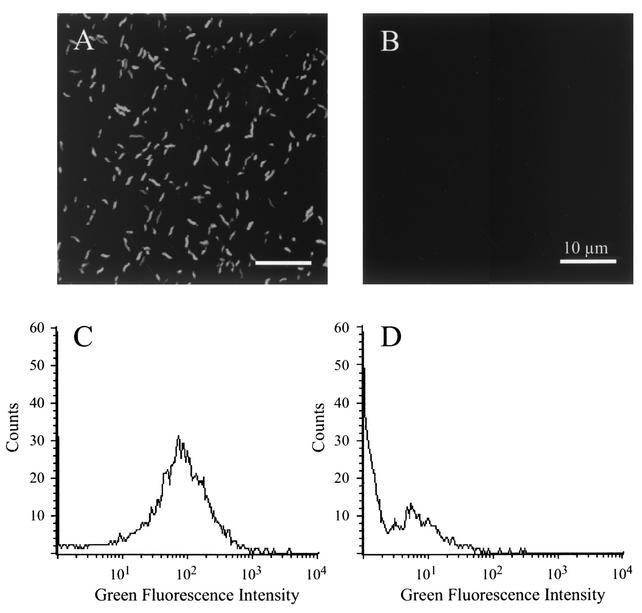
After constructing the Campylobacter pMEK91 shuttle vector and confirming that the genes of interest were expressed, as judged by an in vitro E. coli cell-free transcription translation (S30) system (not shown), the pMEK91 vector was introduced into C. jejuni F38011 by electroporation. Following electroporation, a tetracycline-resistant C. jejuni transformant was isolated and analyzed by fluorescence microscopy and flow cytometry. In contrast to the wild-type C. jejuni isolate, the C. jejuni transformant harboring pMEK91 was readily detectable by both fluorescence microscopy and flow cytometry. Moreover, C. jejuni harboring pMEK91 was brightly fluorescent compared to the C. jejuni wild-type isolate, as judged by fluorescence microscopy (Fig. 2A and B). Similarly, C. jejuni harboring pMEK91 displayed a green fluorescence signal with a geometric mean of 53.04 (Fig. 2C), compared to the C. jejuni wild-type isolate that displayed a green fluorescence signal with a geometric mean of 2.25 (Fig. 2D). Thus, it was clear that the C. jejuni harboring pMEK91 could be used in other applications, as the bacteria fluoresced brightly upon excitation.
FIG. 2.
Green fluorescence of C. jejuni harboring pMEK91. C. jejuni was cultured on MHB agar plates, harvested in PBS, and visualized with a confocal microscope (A and B). In addition, bacterial samples were suspended in PBS and analyzed by flow cytometry (C and D). Data for the C. jejuni F38011 isolate expressing gfp are shown in panels A and C, whereas the data for the nontransformed C. jejuni F38011 wild-type isolate are shown in panels B and D.
In vivo association of C. jejuni with macrophages and neutrophils.
Based on the ease of detecting C. jejuni harboring pMEK91 by fluorescence microscopy and flow cytometry, we sought to assess the utility of C. jejuni synthesizing GFP in an in vivo situation. For this experiment, four mice were injected intraperitoneally with either C. jejuni in PBS or PBS alone (sham injection). Specifically, one mouse was injected with 3.1 × 109 CFU of the C. jejuni F38011 wild-type isolate, two mice (subsequently referred to as GFP-1 and GFP-2) were injected with 3.3 × 109 CFU of C. jejuni synthesizing GFP, and one mouse was injected with PBS. Four hours after injection, the peritoneum was lavaged, and the spleens were aseptically removed from each of the four mice. Cells from the lavage and spleen were washed and stained with antibody combinations to discriminate neutrophils (CD11b+, Gr-1+), macrophages (CD11b+, Gr-1−), and B lymphocytes (CD11b−, CD45R+).
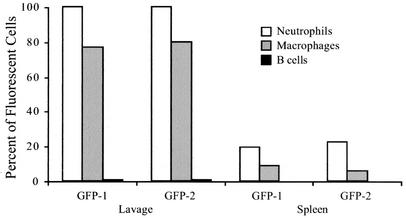
The proportion of cells within each subset with green fluorescence above background levels was determined by flow cytometry (Fig. 3). In these analyses, the GFP-1 and GFP-2 samples displayed similar characteristics. After 4 h, all lavage neutrophils from the mice injected with the C. jejuni pMEK91 transformant had intense green fluorescence (100% of GFP-1 and 99.7% of GFP-2 CD11b+ Gr-1+ lavage neutrophils). A lower but large proportion of lavage macrophages also displayed intense green fluorescence (77.0% of GFP-1 and 80.0% of GFP-2 lavage CD11b+ Gr-1− macrophages). In contrast, few B lymphocytes from the lavage of injected mice were associated with green fluorescence (0.77% of GFP-1 and 0.75% of GFP-2 CD11b− CD45R+ B lymphocytes). Lavage neutrophil, macrophage, and B-cell subsets from the C. jejuni F38011 wild-type-injected mice as well as the sham-injected (PBS-injected) mice displayed levels of green fluorescence below 0.50% (not shown).
FIG. 3.
C. jejuni synthesizing GFP displays a host cell association in the peritoneal lavage and spleen. Four hours after intraperitoneal injection with either PBS or bacteria, mice were euthanized, and cell suspensions were prepared for flow cytometric analysis. The subset of cells expressing the appropriate surface markers was analyzed to determine the proportion of neutrophils (CD11b+ Gr-1+), macrophages (CD11b+ Gr-1−), and B lymphocytes (CD45R+ CD11b−) with green fluorescence above background. The data presented were collected from two mice injected with C. jejuni synthesizing GFP (GFP-1 and GFP-2).
Splenocytes were also examined for C. jejuni synthesizing GFP. Four hours after inoculation, 19.44% and 22.22% of splenic neutrophils from GFP-1 and GFP-2, respectively, displayed green fluorescence. Green fluorescence levels were lower but detectable in splenic macrophages, with 9.06% of splenocytes from GFP-1 and 6.00% from GFP-2 exhibiting green fluorescence intensity above background. In contrast, only 0.01% of splenic B cells from both mice displayed green fluorescence, indicating that the majority of C. jejuni organisms are not associated with splenic B cells. Levels of green fluorescence in splenocytes from the C. jejuni F38011 wild-type-injected mice and sham-injected mice were below 0.50% (not shown). Collectively, these data indicate that neutrophils engulf C. jejuni within the peritoneum and migrate to the spleen.
To determine the CFU of C. jejuni in the various tissue homogenates, samples were plated prior to preparation for cytometric analysis. The number of CFU per homogenate is shown in Table 2. The total CFU recovered from the lavages of the GFP-1-, GFP-2-, and C. jejuni F38011 wild-type-injected mice were similar. The number of bacteria recovered from the lavage homogenates was the greatest, followed by the liver homogenates and then the splenic homogenates. Given the similarity in number of bacteria recovered from the homogenates of the C. jejuni wild-type isolate and C. jejuni pMEK91-injected mice, carriage of the pMEK91 shuttle plasmid and expression of the gfp gene did not appear to cause a selective disadvantage to the organism. At no time was C. jejuni recovered from tissue homogenates of the sham-injected mouse.
TABLE 2.
Number of C. jejuni recovered from the lavage, spleen, and liver 4 h postinjection
| C. jejuni isolate | Inoculum (CFU) | Total CFU per homogenate
|
||
|---|---|---|---|---|
| Lavage | Spleen | Liver | ||
| F38011 | 3.1 × 109 | 5.2 × 108 | 3.4 × 107 | 3.0 × 108 |
| GFP-1 | 3.3 × 109 | 5.2 × 108 | 4.6 × 107 | 2.2 × 108 |
| GFP-2 | 3.3 × 109 | 6.0 × 108 | 7.0 × 107 | 2.2 × 108 |
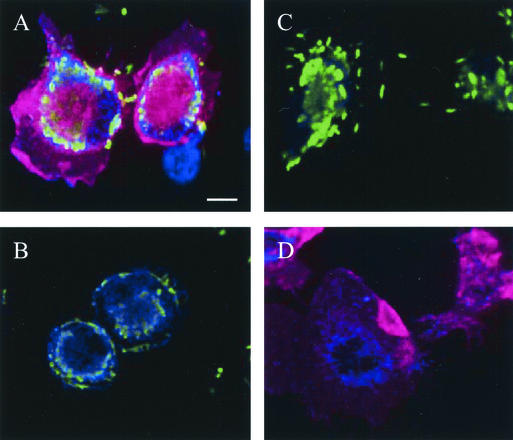
To confirm the association of C. jejuni with granulocytes, lavage fluid samples were applied to sterile coverslips and incubated at 37°C for 4 h to allow the cells to adhere. After incubation, the coverslips were stained to detect host cell surface markers and C. jejuni. Confocal microscopy of cells lavaged from the peritoneum of the injected mice revealed neutrophils (CD11b+ Gr-1+) uniformly associated with C. jejuni (Fig. 4A). The association of C. jejuni with lavage macrophages was also observed (Fig. 4A and B). The specificity of staining reagents was demonstrated simultaneously by running additional samples in parallel (Fig. 4C and D). To demonstrate the specificity of the host cell staining reagents, another lavage sample was incubated with the anti-C. jejuni reagents and isotype-matched rat antibodies conjugated to Tricolor and PE (Fig. 4C); the bacteria were readily detectable, and there was no nonspecific cellular fluorescence. To demonstrate the specificity of the C. jejuni detection, a lavage sample from the sham-injected mouse was stained with the anti-C. jejuni reagents, anti-mouse CD11b antibody, and anti-mouse Gr-1 antibody (Fig. 4D); no green fluorescence was detectable in the sample. Consistent with cytometric data, microscopy revealed that C. jejuni displayed an association for granulocytes.
FIG. 4.
Cellular association of C. jejuni in lavage monocytes. Cells from lavage fluid were bound to sterile coverslips, fixed, and stained with antibody combinations before confocal microscopy images were captured. Panel A shows C. jejuni (green)-infected neutrophils [CD11b+ (blue), Gr-1+ (red)] and an uninfected macrophage (CD11b+ Gr-1−). Panel B shows C. jejuni-infected macrophages. Panel C shows cells stained with nonspecific isotype-matched rat IgGs (rat IgG2b conjugated to Tricolor and rat IgG2a conjugated to PE) in conjunction with antibody specific for C. jejuni. Panel D shows neutrophils from the sham (PBS)-injected mouse stained with Campylobacter-specific, CD11b-specific, and Gr-1-specific antibody combinations. All images were captured at the same magnification. Bar, 10 μm.
Failure of C. jejuni to survive within granulocytes.
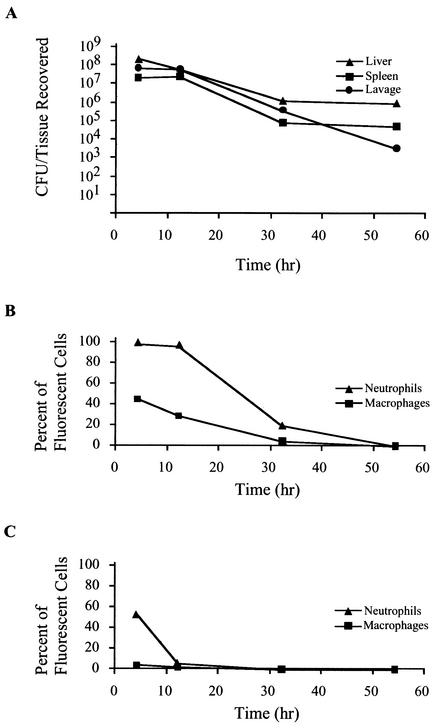
To determine the interaction of C. jejuni with host cells over time, a separate experiment was performed in which mice were injected with 1.3 × 1010 CFU of C. jejuni synthesizing GFP. At various time points following injection, mice were euthanized, and tissues were harvested aseptically, homogenized, and plated for CFU recovery (Fig. 5A). Two mice were injected per time point. A representative of three experiments is displayed. After the first 4 h of incubation, the number of bacteria recovered from lavage was 3.0 × 108 CFU. At 12 h postinjection, the total recoverable CFU from the lavage decreased to 2.4 × 108 CFU, followed by recovery of 1.6 × 106 CFU at 32 h. The levels of recoverable lavage CFU continued to decrease, with 1.4 × 104 CFU being recovered at 54 h postinoculation. Similar reductions were observed in the total CFU recovered from the spleen and liver over time. Levels of CFU recovered from spleen homogenates were nearly identical at 4 and 12 h postinjection but decreased to 1.8 × 105 at 32 h and to 1.0 × 105 total CFU at 54 h postinjection. Liver levels of total CFU also decreased in a nearly linear fashion, with CFU levels of 5.0 × 108 at 4 h, 1.4 × 108 at 12 h, 2.6 × 106 at 32 h, and 1.8 × 106 at 54 h postinoculation. At 54 h after inoculation, the tissue homogenates contained recoverable CFU, while the levels of cell-associated C. jejuni synthesizing GFP were near background levels (see below). These experiments revealed that the total recoverable CFU of C. jejuni in all tissues decreased over time. The cytometry data (Fig. 5) further indicated that in the lavage and spleen, C. jejuni was predominantly associated with neutrophils and macrophages rather than lymphocytes.
FIG. 5.
Clearance of C. jejuni synthesizing GFP associated with neutrophils and macrophages. Mice were injected with C. jejuni synthesizing GFP. At indicated times, tissues from euthanized mice were homogenized and plated for CFU recovery. Panel A shows reduction of recoverable total CFU per tissue over time. Cells from the homogenates were washed, stained for cell surface markers, and analyzed by flow cytometry. The proportion of cells within a marker-defined subset with green fluorescence was determined. Panel B shows the proportion of lavage neutrophils (CD11b+ Gr-1+) and macrophages (CD11b+ Gr-1−) associated with C. jejuni synthesizing GFP over time. Panel C shows the proportion of splenic neutrophils (CD11b+ Gr-1+) and macrophages (CD11b+ Gr-1−) associated with C. jejuni synthesizing GFP over time.
In addition to determining the number of C. jejuni in each homogenate, the cells from tissue homogenates were washed and stained with antibody combinations to determine the percentage of neutrophils (CD11b+, Gr-1+) and macrophages (CD11b+, Gr-1−) associated with C. jejuni. After staining, the samples were analyzed by flow cytometry, and the proportion of cells within each subset with green fluorescence above background levels was determined (Fig. 5). Nearly all lavage neutrophils (99.1%) harbored C. jejuni after 4 h (Fig. 5B). The level of lavage neutrophils harboring C. jejuni decreased only slightly after 12 h (96.0%) but more dramatically after the 32-h time point (19.5%). After 54 h, the number of neutrophils harboring C. jejuni was near background (0.01%). These findings indicate clearance of C. jejuni from neutrophils over time. A reduction in C. jejuni from splenic neutrophils was also observed (Fig. 5C), with a decrease from 54.0% at 4 h to 5.80% at 12 h and then to 0.28% at 32 h. No splenic neutrophils with C. jejuni were detected at 54 h postinjection. At all time points, the association of C. jejuni synthesizing GFP with lavage B lymphocytes (CD45R+ CD11b−) was below 1.0%.
The reduction in C. jejuni within the macrophage population over time was analogous to that observed within the neutrophils (Fig. 5B). At 54 h postinjection, only 0.09% of lavage macrophages were associated with green fluorescence. Splenic macrophages showed low levels of green fluorescence, with 4.97% harboring C. jejuni synthesizing GFP 4 h after inoculation (Fig. 5C). These levels diminished to 2.25% at 12 h and 0.54% at 32 h. No C. jejuni-associated splenic macrophages were detected 54 h after injection. In B lymphocytes (CD45R+, CD11b−), the level of C. jejuni synthesizing GFP was below 0.5% throughout the experiment. Collectively, these data suggest that innate immune components are responsible for the reduction in recoverable bacteria.
C. jejuni F38011 variant induces pathology in mice similar to that with the wild-type isolate.
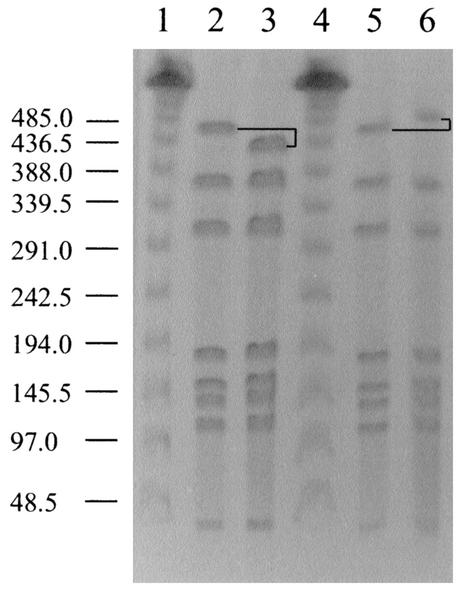
C. jejuni cells recovered from the mouse organs and peritoneal lavages over the course of the survival experiment were analyzed by SmaI mrp-PFGE to confirm the C. jejuni F38011 wild-type genotype. Unexpectedly, a second genotype was isolated from the mice that differed from that of the original C. jejuni F38011 wild-type genotype (Fig. 6). The organism with the altered genotype was observed at low frequency at the first sampling time point (1 of 12 colonies examined by mrp-PFGE at 4 h) and was evident throughout the entire experiment (4, 12, 32, and 54 h). At 54 h, the C. jejuni F38011 variant was recovered at a frequency similar to that of the C. jejuni F38011 wild-type isolate (7 of 12 colonies examined displayed the altered macrorestriction profile). As the profiles of the two isolates differed in only one band, and given the temporal and geographical relationship of the two isolates, we concluded that these isolates were clonal. This is consistent with previously published interpretative criteria which state that isolates that have three band differences or fewer, compared to an outbreak isolate, are closely related (26). Additionally, since no Campylobacter organisms were recovered from sham-injected mice, we conclude that the alteration in macrorestriction profile revealed an F38011 variant rather than a distinct C. jejuni isolate. Thus, the isolate with the altered genotype was designated C. jejuni mouse variant F38011mv.
FIG. 6.
Genotypic differences in C. jejuni F38011 isolates observed by mrp-PFGE. Bacteria were lysed in PFGE-grade agarose, and DNA was digested with the restriction enzyme SmaI. DNA fragments were separated by contour-clamped homogeneous electric field electrophoresis through a 1% (wt/vol) agarose gel. Lanes: 1 and 4, phage lambda size markers (in kilobases) (New England Biolabs); 2 and 5, C. jejuni F38011 wild-type isolate; 3, C. jejuni F38011 mouse-derived variant F38011mv; 6, C. jejuni F38011 plate-derived variant. The altered fragment is noted in black. The molecular sizes are provided to the left of the figure.
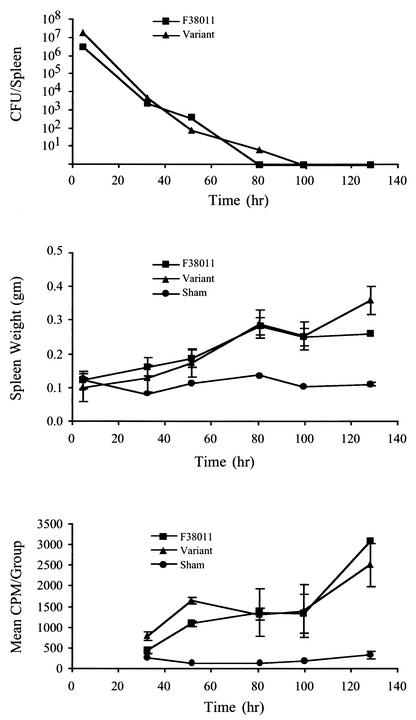
To determine whether C. jejuni F38011mv acted like the C. jejuni F38011 wild-type isolate in mice, one cohort of BALB/c mice was injected with the wild-type C. jejuni and a second with the F38011mv isolate. A minimum of three mice were euthanized at the times indicated after injection for both the C. jejuni F38011 wild-type and F38011mv cohorts, with the exception that only two mice were euthanized at 128 h for the C. jejuni F38011 wild-type isolate. In addition, a minimum of two PBS-injected mice were euthanized and analyzed at each time point. C. jejuni was subsequently recovered from mouse organs and peritoneal lavages. Similar numbers of each bacterial isolate were recovered at all time points, with neither isolate having an obvious growth or survival advantage in vivo (Fig. 7A).
FIG. 7.
C. jejuni F38011 mouse variant exhibits the same survival kinetics and host response phenotype as the C. jejuni F38011 wild-type isolate in mice. The wild-type C. jejuni F38011 and F38011 mouse variant were intraperitoneally injected into separate cohorts of mice. At each time point, mice were euthanized, and samples were subjected to various analyses. Panel A shows the total CFU recovered from the splenic homogenates. Panel B shows the mean weights of spleens from the C. jejuni- and sham-injected mice. Panel C shows the ex vivo proliferation of splenocytes from C. jejuni- and sham-injected mice.
Based on the data, it was evident that both isolates were capable of inducing pathology in mice, as judged by statistically significant increases in spleen weights of the C. jejuni-injected mice versus the sham-injected mice (Fig. 7B). Evidence that the splenocytes of the C. jejuni-injected mice were activated was obtained from ex vivo proliferation assays (Fig. 7C). Both the C. jejuni F38011 wild-type and F38011mv isolates induced a similar level of splenocyte proliferation. Nevertheless, mrp-PFGE analysis of the C. jejuni F38011 wild-type and C. jejuni F38011mv isolates recovered from the liver (12 isolates in each cohort per time point) revealed no genotype switching. While these results demonstrated that the C. jejuni variant was capable of inducing the same pathology in the mice as the C. jejuni F38011 wild-type isolate, it was unclear whether the variant arose prior to or after C. jejuni exposure to the mouse immune system.
To address whether C. jejuni F38011mv arose prior to the injection of the mice, an experiment was performed in which the C. jejuni F38011 wild-type isolate was streaked onto an MHB plate to generate single colonies. Following incubation, a single colony was subsequently passaged to generate more single colonies. Twelve random colonies were picked at each of three subpassages, resulting in a total of 36 individual colonies. SmaI macrorestriction profiles of these 36 colonies exposed one genotypic variant. In this case, the variant (F38011 plate variant) appeared after the initial subpassage and was a result of the acquisition of DNA in the largest SmaI fragment (Fig. 6).
Given that the SmaI macrorestriction profiles of the original inoculum injected into mice did not reveal the presence of the C. jejuni F38011mv isolate, this isolate either arose after injection of the mice or was a minor constituent of the original inoculum. Based on the observation that the C. jejuni F38011mv isolate was recovered from every mouse injected with C. jejuni F38011 in the initial experiment and the fact that no genotypic switching was evident in the F38011-injected mice during the follow-up experiment, the C. jejuni F38011mv isolate may have been a minor constituent of the original inoculum. However, it was not possible to definitely conclude whether C. jejuni F38011mv arose in vitro or in vivo.
DISCUSSION
The construction of a new gfp vector allowed detection of C. jejuni in vitro and in vivo. Intraperitoneal injection of mice proved to be a useful in vivo model to demonstrate the ease of detecting C. jejuni synthesizing GFP. Although other investigators previously used intraperitoneal injection of BALB/c mice with Campylobacter strains and focused on bacterial recovery and gross pathology (29), the use of C. jejuni synthesizing GFP as described here further allowed identification of host cell subsets through staining of cell surface markers. The inoculation of mice with C. jejuni synthesizing GFP revealed the association of C. jejuni with granulocytes and their decrease over time in this cell population. During the analyses of CFU recovered from the BALB/c mice, a C. jejuni variant was identified that displayed an altered genotype. Based on this finding, additional experiments revealed that variation occurred within the C. jejuni genome in the absence of immunological pressure. In summary, flow cytometric analysis allowed us to rapidly identify the cellular subsets associated with C. jejuni transformed with a new gfp plasmid-based reporter system. This technique proved to be simple and reproducible and allowed the analysis of viable bacterial and host cells.
The use of GFP as a means of detecting C. jejuni has been described previously. In fact, we first reported that a C. jejuni isolate harboring a Campylobacter shuttle vector could be used to detect C. jejuni binding to cultured epithelial cells (8). In this study, we generated the vector pMEK91, which contains gfp driven by the ompE promoter. Miller et al. (13) reported the construction of two sets of Campylobacter shuttle vectors containing various reporter genes, including gfp. In their vectors, a constitutive yet artificial promoter based on the C. jejuni consensus promoter (34) drove gfp expression. To determine the fluorescence intensity of C. jejuni harboring pMEK91 versus the previously described gfp vector pMW1007, pMW1007 was introduced into wild-type C. jejuni F38011, and both organisms synthesizing GFP were subjected to flow cytometric analysis. The intensity of the green fluorescence signal was 10-fold greater from C. jejuni F38011 harboring pMEK91 compared to C. jejuni F38011 harboring pMW1007 (not shown). The copy number of pMEK91 and pMW1007 in C. jejuni F38011 is most likely identical, because the repB genes in both plasmids originated from the same Campylobacter coli cryptic plasmid, pIP1455. The simplest explanation for the difference in fluorescence between C. jejuni F38011 harboring pMEK91 versus pMW1007 is the relative strength of the promoter driving the gfp gene. Thus, when a high level of transcription is important, a strong Campylobacter promoter should be used to drive gene transcription.
Intraperitoneal injection of mice with C. jejuni synthesizing GFP coupled with flow cytometric analysis demonstrated selective bacterial association with granulocytes. In comparing the splenocytes to the cells harvested from the lavage 4 h after infection, a lower proportion of splenic neutrophils (19.4 to 22.2%) harbored C. jejuni than lavage neutrophils (99.7 to 100%). Similarly, a lower proportion of splenic macrophages harbored C. jejuni than macrophages in the lavage (6.0 to 9.1% splenic macrophages, as opposed to 77.0 to 80.0% lavage macrophages). The reason for the lower percentage of granulocytes associated with C. jejuni in the spleen compared to the lavage is presumably that the C. jejuni-infected cells in the lavage are trafficked to other sites, including the spleen (11). Because the proportion of cells in the lavage and spleen that were associated with C. jejuni decreased over time, the data support a model in which C. jejuni cannot persist intracellularly within granulocytes. We hypothesize that the observed decrease in C. jejuni CFU in the various cells and tissues following injection is due to the combined action of components of the innate immune system.
As judged by flow cytometry, the green fluorescence signal associated with the splenic granulocytes decreased dramatically following a 4-h time period, as did the fluorescence signal associated with the lavage granulocytes after a 12-h incubation (Fig. 5). However, C. jejuni CFU in the spleen and lavage homogenates at the corresponding time points were clearly still present. Given this observation, an additional experiment was performed in which the number of CFU present in the lavage and spleen homogenates was compared to that from the cells washed in preparation for flow cytometric analysis. While the number of CFU recovered from the tissue homogenates was consistent with the data from the earlier experiments, the cytometry samples with little green fluorescence yielded few recoverable CFU (not shown). Thus, the detectable green fluorescence correlated well with the cell-associated bacterial load. This finding indicated that the bacteria recovered from the spleen homogenates at the later time points were not likely cell associated, even though they arrived at that site within granulocytes. Hence, these results suggest that a subpopulation of the bacteria being trafficked from the lavage to the spleen possess the ability to escape from granulocytes.
In a previous report, Vuckovic et al. (29) noted reductions in bacterial load in BALB/c mice over time but were able to recover C. jejuni from the spleen up to 6 days following injection and from the liver up to 17 days following injection. While differences are apparent in the persistence of C. jejuni in mice between their study and ours, the data generated in both studies are in agreement with respect to the eventual decline in C. jejuni CFU and the pathology observed in the injected mice (e.g., enlarged spleen). Possible explanations for the difference in C. jejuni persistence in mice in the previous report versus our work include strain differences and variations in experimental protocols. We also observed differences in the persistence of C. jejuni in mice that correlated with the number of injected organisms.
The lack of association of C. jejuni with lavage B lymphocytes may be attributed to the paucity of high affinity ligands for bacteria on prevalent B-1 lymphocytes in the lavage or lack of components for signaling by B-1 lymphocytes (12). However, other B lymphocyte subsets respond robustly to C. jejuni products such as lipopolysaccharide, rapidly producing antibody, and promoting adaptive immunity (2). In humans, 0.5 to 1.0% of patients infected with C. jejuni subsequently develop autoimmune neuropathies, such as Guillain-Barré syndrome, due to the production of anti-C. jejuni antibodies that bind ganglioside residues on peripheral nerve cells, inciting damage (33). Our data support a model in which indirect effects of immune regulation allow production of self-reactive antibody rather than direct C. jejuni-B-lymphocyte association.
The lack of stability of the C. jejuni genome after passage of organisms through an animal, or from human infections, has been previously demonstrated (7, 22, 31). Wassenaar et al. (31) observed C. jejuni clonal variants in samples of processed poultry from the same producer. To assess the stability of one of these variants, a C. jejuni variant was subsequently fed to one-day-old chicks and reisolated after five days. As judged by the SmaI-mrp, this isolate's genotype was indistinguishable from that of the initial inoculum. Genotypic variants were also noted by Hänninen et al. (7), who inoculated newly hatched chicks with twelve C. jejuni isolates and observed mrp genotypic alterations in two of twelve isolates. With respect to humans, Steinbrueckner et al. (22) observed variations in mrp genotypes of C. jejuni isolates cultured from human stool samples from several patients over time. In our study, a clonal SmaI-mrp variant of the C. jejuni isolate F38011 was observed after intraperitoneal injection in mice. While variation at the individual gene level has been observed in C. jejuni after in vitro passage, notably in the flagellin filament genes (15, 30), no work has been done to investigate changes to the mrp genotype. To our knowledge, this is the first time a mrp genotypic variant has been recovered from mice.
To determine whether the C. jejuni F38011mv isolate differed from the C. jejuni F38011 wild-type isolate with respect to the induced host cell phenotype, we injected both isolates into separate cohorts of mice. The mouse-derived F38011mv isolate displayed the same survival kinetics and induced the same pathology as the C. jejuni F38011 wild-type isolate. To our knowledge, this is the first time a C. jejuni genotypic variant has been tested for whether it induces the same host response phenotype as the wild-type isolate in an animal model system.
While the genotype of the C. jejuni F38011mv isolate was stable as judged by a second passage of the isolate in mice, we did observe an SmaI variant of the C. jejuni F38011 wild-type isolate after in vitro passage. The macrorestriction profile of the C. jejuni plate variant (F38011 plate variant) differed significantly from the genotypic variant isolated from mice. Similar to the findings of other investigators, an increase was noted in the total genomic content of the F38011 plate variant (22, 23). Variant stability, as determined by mrp-PFGE, has been tested previously by in vitro passage; however, other investigators have not observed in vitro macrorestriction profile genotypic instability with regard to C. jejuni (22, 31). One possible reason for the difference in our findings versus others may be variations in experimental protocols. More specifically, Wassenaar et al. (31) continually passaged an isolate on solid medium ten times, with all but the last passage consisting of a mixture of cells from the most densely populated portion of the plate. In contrast, our method involved streaking the C. jejuni F38011 wild-type isolate onto a solid medium to generate single colonies. Following incubation, a single colony was subsequently passaged to generate more single colonies. Twelve random colonies were picked at each of three subpassages, resulting in a total of 36 individual colonies. The C. jejuni F38011 plate variant isolate was observed in one of these 36 colonies. Of interest, in vitro macrorestriction profile genotypic variation has been observed previously in C. coli (16).
Given the identification of a C. jejuni SmaI macrorestriction profile variant in vitro, it seems unlikely that the molecular mechanism responsible for the observed genotypic instability is either natural transformation or horizontal gene transfer. It is more plausible that the genomic instability occurred by intramolecular recombination. Because the genotypic variation was observed in the same SmaI-restricted band during both the in vitro subculturing and in vivo mouse survival experiments, characterization of these genomic alterations may identify a hot spot for recombination.
In summary, we generated a new Campylobacter shuttle vector and demonstrated the usefulness of gfp in the detection of C. jejuni following intraperitoneal injection of mice by flow cytometric analyses. Given the intensity of the fluorescence signal emitted by C. jejuni harboring pMEK91, this vector may prove useful to study the biology of C. jejuni in various niches. During characterization of bacteria recovered from various murine tissues, a clonal variant of the C. jejuni F38011 wild-type isolate was identified by mrp-PFGE analysis. Because C. jejuni macrorestriction profile variants were identified after in vivo and in vitro passage and these genotypic alterations occurred in the same fragment, we will characterize the genotypic variants to elucidate the nature of the molecular events leading to recombination.
Acknowledgments
We thank Diane Taylor for plasmid pUOA3, Bill Miller for providing pMW1007, and Marshall Monteville for assistance in preparing samples for immunofluorescence microscopy. We also thank Carolyn Hovde (Department of Microbiology, Molecular Biology and Biochemistry, University of Idaho) and Brent Gilpin (Environmental Science and Research, Ltd., Christchurch, New Zealand) for critical review of the manuscript.
This work was supported by grants from the NIH (DK58911) and the USDA National Research Initiative Competitive Grants Program (USDA/NRICGP, 99-35201-8579) awarded to M.E.K. and by funds from the School of Molecular Biosciences to P.F.M.
REFERENCES
- 1.Alm, R. A., P. Guerry, and T. J. Trust. 1993. The Campylobacter σ54 flaB flagellin promoter is subject to environmental regulation. J. Bacteriol. 175:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser, M. J. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl. 2):S103-S105. [DOI] [PubMed] [Google Scholar]
- 3.Buzby, J. C., T. Roberts, C.-T. J. Lin, and J. M. MacDonald. 1996. Bacterial foodborne diseases: medical costs and productivity losses. Agricultural Economics Report 741. Economic Research Service/USDA. U.S. Department of Agriculture, Washington, D.C.
- 4.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 5.Domínguez, J., M. del Mar Lorenzo, and R. Blasco. 1998. Green fluorescent protein expressed by a recombinant vaccinia virus permits early detection of infected cells by flow cytometry. J. Immunol. Methods 220:115-121. [DOI] [PubMed] [Google Scholar]
- 6.Givan, A. L. 2001. Cells from without: leukocytes, surface proteins, and the strategy of gating, p. 81-113. In A. L. Givan (ed.), Flow cytometry: first principles, 2nd ed. Wiley-Liss, New York, N.Y.
- 7.Hänninen, M.-L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konkel, M. E., L. Joens, and P. F. Mixter. 2000. Molecular characterization of Campylobacter jejuni virulence determinants, p. 217-240. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 9.Labigne-Roussel, A., J. Harel, and L. Tompkins. 1987. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J. Bacteriol. 169:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 11.Lagasse, E., and I. L. Weissman. 1996. Flow cytometric identification of murine neutrophils and monocytes. J. Immunol. Methods 197:139-150. [DOI] [PubMed] [Google Scholar]
- 12.Lajaunias, F., L. Nitschke, T. Moll, E. Martinez-Soria, I. Semac, Y. Chichepotiche, R. M. E. Parkhouse, and S. Izui. 2002. Differentially regulated expression and function of CD22 in activated B-1 and B-2 lymphocytes. J. Immunol. 168:6078-6083. [DOI] [PubMed] [Google Scholar]
- 13.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monteville, M. R., J. E. Yoon, and M. E. Konkel. 2002. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer membrane protein and microfilament reorganization. Microbiology 149:153-165. [DOI] [PubMed] [Google Scholar]
- 15.Nuijten, P. J. M., A. J. G. van den Berg, I. Formentini, B. A. M. van der Zeijst, and A. A. C. Jacobs. 2000. DNA rearrangements in the flagellin locus of an flaA mutant of Campylobacter jejuni during the colonization of chicken ceca. Infect. Immun. 68:7137-7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.On, S. L. W. 1998. In vitro genotypic variation of Campylobacter coli documented by pulsed-field gel electrophoretic DNA profiling: implications for epidemiological studies. FEMS Microbiol. Lett. 165:341-346. [DOI] [PubMed] [Google Scholar]
- 17.Pancorbo, P. L., M. A. de Pablo, E. Ortega, A. M. Gallego, C. Alvarez, and G. Alvarez de Cienfuegos. 1999. Evaluation of cytokine production and phagocytic activity in mice infected with Campylobacter jejuni. Curr. Microbiol. 39:129-133. [DOI] [PubMed] [Google Scholar]
- 18.Pancorbo, P. L., A. M. Gallego, M. de Pablo, C. Alvarez, E. Ortega, and G. Alvarez de Cienfuegos. 1994. Inflammatory and phagocytic response to experimental Campylobacter jejuni infection in mice. Microbiol. Immunol. 38:89-95. [DOI] [PubMed] [Google Scholar]
- 19.Park, S. F. 1999. The use of hipO, encoding benzoylglycine amidohydrolase (hippuricase), as a reporter of gene expression in Campylobacter coli. Lett. Appl. Microbiol. 28:285-290. [DOI] [PubMed] [Google Scholar]
- 20.Purdy, D., and S. F. Park. 1993. Heterologous gene expression in Campylobacter coli: The use of bacterial luciferase in a promoter probe vector. FEMS Microbiol. Lett. 111:233-237. [DOI] [PubMed] [Google Scholar]
- 21.Richardson, J. F., J. A. Frost, J. M. Kramer, R. T. Thwaites, F. J. Bolton, D. R. A. Wareing, and J. A. Gordon. 2001. Coinfection with Campylobacter species: an epidemiological problem? J. Appl. Microbiol. 91:206-211. [DOI] [PubMed] [Google Scholar]
- 22.Steinbrueckner, B., F. Ruberg, and M. Kist. 2001. Bacterial genetic fingerprint: a reliable factor in the study of the epidemiology of human Campylobacter enteritis? J. Clin. Microbiol. 39:4155-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens, C. P., S. L. W. On, and J. A. Gibson. 1998. An outbreak of infectious hepatitis in commercially reared ostriches associated with Campylobacter coli and Campylobacter jejuni. Vet. Microbiol. 61:183-190. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, D. E. 1992. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli to tetracycline, chloramphenicol, and erythromycin, p. 74-86. In I. Nachamkin, M. J. Blaser, and L. S. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, D.C.
- 25.Taylor, D. E., K. Hiratsuka, H. Ray, and E. K. Manavathu. 1987. Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J. Bacteriol. 169:2984-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unge, A., R. Tombolini, L. Mølbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 29.Vuckovic, D., M. Abram, and M. Doric. 1998. Primary Campylobacter jejuni infection in different mice strains. Microb. Pathog. 24:263-268. [DOI] [PubMed] [Google Scholar]
- 30.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1995. Variation of flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141:95-101. [DOI] [PubMed] [Google Scholar]
- 31.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weijtens, M. J. B. M., J. van der Plas, P. G. H. Bijker, H. A. P. Urlings, D. Koster, J. G. van Logtestijn, and J. H. J. Huis in't Veld. 1997. The transmission of campylobacter in piggeries: an epidemiological study. J. Appl. Microbiol. 83:693-698. [DOI] [PubMed] [Google Scholar]
- 33.Willison, H. J., and G. M. O'Hanlon. 2000. Antiglycosphingolipid antibodies and Guillain-Barré syndrome, p. 259-285. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. American Society for Microbiology, Washington, D.C.
- 34.Wösten, M. M. S. M., M. Boeve, M. G. A. Koot, A. C. van Nuene, and B. A. M. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yrlid, U., M. Svensson, A. Håkansson, B. J. Chambers, H.-G. Ljunggren, and M. J. Wick. 2001. In vivo activation of dendritic cells and T cells during Salmonella enterica serovar Typhimurium infection. Infect. Immun. 69:5726-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]