Abstract
Pulsed-field gel electrophoresis (PFGE) has been used extensively to investigate the epidemiology of Escherichia coli O157:H7, although it has not been evaluated as a tool for establishing genetic relationships. This is a critical issue when molecular genetic data are used to make inferences about pathogen dissemination. To evaluate this further, genomic DNAs from 62 isolates of E. coli O157:H7 from different cattle herds were digested with XbaI and BlnI and subjected to PFGE. The correlation between the similarity coefficients for these two enzymes was only 0.53. Four additional restriction enzymes (NheI, PacI, SfiI, and SpeI) were used with DNAs from a subset of 14 isolates. The average correlations between similarity coefficients using sets of one, two, and three enzymes were 0.405, 0.568, and 0.648, respectively. Probing with lambda DNA demonstrated that some DNA fragments migrated equal distances in the gel but were composed of nonhomologous genetic material. Genome sequence data from EDL933 indicated that 40 PFGE fragments would be expected from complete XbaI digestion, yet only 19 distinguishable fragments were visible. Two reasons that similarity coefficients from single-enzyme PFGE are poor measures of relatedness (and hence are poorly correlated with other enzymes) are evident from this study: (i) matching bands do not always represent homologous genetic material and (ii) there are limitations to the power of PFGE to resolve bands of nearly identical size. The findings of the present study indicate that if genetic relationships must be inferred in the absence of epidemiologic data, six or more restriction enzymes would be needed to provide a reasonable estimate using PFGE.
Escherichia coli O157:H7 (referred to as O157 hereafter) is a food-borne human pathogen that can cause bloody diarrhea, hemolytic uremic syndrome, and death (66). Studies of animal reservoirs have focused on domestic cattle because early epidemiologic evidence indicated that foods of bovine origin were the primary vehicles of infection and O157 was detected in herds associated with outbreaks (25). Although outbreaks associated with foods of nonbovine origin have been detected with increasing frequency, bovine sources have been consistently implicated in the plurality of human infections (35, 46, 52).
Pulsed-field gel electrophoresis (PFGE) has been used effectively as a molecular subtyping tool in outbreak investigations (5, 11-16, 23, 33, 37, 59) and surveillance (6, 65), and PFGE has been used to infer genetic relatedness between isolates of O157 from different cattle herds (41, 64). Although the discriminatory power of PFGE has compared favorably to that of other subtyping methods (21, 29, 30, 38, 48), the similarity of PFGE patterns has not been stringently evaluated as a measure of genetic relatedness. This is most critical for transmission studies, which rely on molecular genetic data for making inferences about routes of pathogen spread (17, 20, 40, 47, 60, 69).
A Dice similarity coefficient is often used to quantify the similarity between PFGE banding patterns (3, 20, 23, 31, 38, 39, 43, 44). The Dice coefficient is calculated as 2h/(a + b), where h is the number of matching bands and a + b is the total number of bands being compared, including matching and nonmatching bands (19). The use of the Dice coefficient to measure genetic similarity assumes that bands of identical size are, with high probability, genetically homologous. But in theory, a restriction enzyme may cleave two nonhomologous genomes in such a way as to yield similar fragment sizes, and thus produce spurious matches. Equally, a minor genetic event (such as a single-nucleotide mutation) could produce the same banding change as a major genetic event (such as a very long insertion or deletion) (67). There are also empirical reasons for uncertainty that XbaI PFGE by itself is an adequate measure of relatedness. Spurious matches have been reported from surveillance data (W. E. Keene, V. K. Balan, and P. R. Cieslak, poster, International Conference on Emerging Infectious Diseases, Atlanta, Georgia, 1998) and in conjunction with outbreak investigations (4, 5, 32, 71). Furthermore, different restriction enzymes may assign conflicting relatedness to isolate pairs. For example, Harsono et al. (29) found that some XbaI subtypes contained multiple SfiI subtypes and some SfiI subtypes contained multiple XbaI subtypes, so that some pairs were 100% similar by SfiI but unrelated by XbaI and the reverse. Rice et al. (57) found marked differences in NotI banding patterns between isolates that were indistinguishable by XbaI. Malorny et al. (45) also found discrepancies using three different restriction enzymes and PFGE for Salmonella enterica serovar Typhimurium DT104. Others have found a poor correlation between similarities derived from PFGE and randomly amplified DNA for O157 (24, 56). Olsen et al. (49) found low correlations between similarity coefficients computed from PFGE and two other genotyping methods for serovar Typhimurium.
To the extent that PFGE patterns are a measure of isolate-specific genetic composition, one would predict that closely related isolates would be more similar and distantly related isolates less similar, regardless of the restriction enzyme used. It would also follow that there should be a good correlation between Dice coefficients generated by two enzymes, and nonhomology between matching bands should be infrequent. Also, the distribution of fragment sizes from a restriction enzyme profile would be expected to reflect the distribution of restriction enzyme recognition sites throughout the genome. The goal of this study was to determine the suitability of PFGE for measuring relatedness by (i) measuring the degree of correlation among similarity coefficients obtained by different enzymes and determining if this correlation was increased by adding information from other enzymes, (ii) determining whether bands that match by PFGE sometimes contain nonhomologous genetic material, and (iii) determining the degree to which theoretical bands from a sequenced isolate match the bands resolved by PFGE.
MATERIALS AND METHODS
Isolates.
Sixty-two O157 isolates banked from previous studies (26-28) were used for the present study. The sources of all isolates used in the present study were bovine fecal samples from herds sampled from 1994 to 1995. One isolate from each herd was used. A subset of 14 of these isolates was used for the multiple-enzyme analysis. Isolate pairs with 100% similarity by XbaI PFGE were used for band comparison.
PFGE.
A standard protocol for PFGE of O157 was used, with some modifications (14a). Plugs were prepared from bacterial suspensions (200 μl) to which proteinase K (10 μl at 20 mg/ml) was added. InCert (FMC Bio Products, Rockland, Maine) agarose was added (1.6% with 1% sodium dodecyl sulfate in 200 μl of TE buffer [100 mM Tris and 100 mM EDTA]), and the mixture was immediately dispensed into plug molds (Bio-Rad, Hercules, Calif.). The plugs were lysed in ES buffer (0.5 M EDTA, pH 9.0, 1% sodium-lauroyl-sarcosine) with proteinase K at 54°C for 1 to 2 h and then washed at 50°C in sterile distilled water (two times for 15 min each time) and TE buffer (four times for 15 min each time). Plug slices (2 mm thick) were digested with a restriction endonuclease (Table 1 shows the times and temperatures). PFGE was performed on a CHEF-DRII PFGE apparatus (Bio-Rad) using the following parameters: separation on a 1% agarose gel (Seakem Gold agarose; FMC Bio Products) in 0.5× Tris-Borate-EDTA at 14°C and 6 V/cm. The run times and pulse times differed according to the enzyme used (Table 1). The gels were stained with ethidium bromide and photographed with UV transillumination. The photographic image was captured digitally using a gel documentation system (AlphaImager 2000; Alpha Innotech Corp., San Leandro, Calif.). After visual evaluation of digital images, the bands were marked by hand and analyzed using Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium).
TABLE 1.
Restriction enzyme digest and electrophoresis conditions for each restriction enzyme used
| Enzyme | Digestion temp (°C) | Digestion time (h) | Units of enzyme per plug | Run time (h) | Initial switch time (s) | Final switch time (s) |
|---|---|---|---|---|---|---|
| BlnI | 37 | 16 | 10 | 22 | 2.2 | 54.2 |
| NheI | 37 | 14 | 15 | 24 | 1 | 15 |
| PacI | 37 | 21 | 24 | 20 | 0.1 | 15 |
| SfiI | 50 | 17 | 29 | 22 | 5 | 25 |
| SpeI | 37 | 17 | 25 | 22 | 5 | 25 |
| XbaI | 37 | 4-16 | 30 | 20 | 2.2 | 54.2 |
Data analysis.
Standard isolate G5244 (courtesy of Mansour Samadpour, Department of Environmental Health, University of Washington, Seattle) was used as the standard for the 62 isolates for which XbaI results were compared to BlnI results. Each gel included 20 lanes with four standards for every gel. Dice similarity coefficients were generated for all possible pairwise comparisons. The unweighted pair group method of analysis was used to generate dendrograms. For each isolate pair, the Dice similarity coefficient derived from the XbaI pattern was compared to the Dice coefficient derived from BlnI digestion. This was done by exporting similarity matrices from Bionumerics into Excel (Microsoft, Redmond, Wash.) and calculating the Pearson product-moment correlation coefficient between the XbaI Dice similarity coefficients and the BlnI Dice similarity coefficients. For a subset of 14 isolates, each of four additional restriction enzymes (NheI, PacI, SfiI, and SpeI) was used to generate a set of similarity indices, so that for each of these 14 isolates a total of six enzyme-specific profiles were generated. The similarity indices for each enzyme were compared to each other, as for BlnI and XbaI. An arithmetic average of the correlations between each enzyme pair was used to estimate the average correlation between two enzymes. To estimate the correlation between pairs of enzymes, the average similarity for one enzyme pair was calculated, and then those average similarities were correlated with the average similarity from a different enzyme pair. Likewise, to compare enzyme triplets, the average similarities for three enzymes were calculated and correlated to the average similarities from a different set of three enzymes.
Band comparison with Southern analysis.
To evaluate whether bands that migrate the same distance on PFGE gels consistently contain homologous genetic material, DNAs from PFGE gels were probed using a standard Southern transfer protocol (62). DNA fragments were transferred to a positively charged nylon membrane (Immobilon-Ny+; Millipore, Bedford, Mass.) under alkaline conditions using capillary transfer. Lambda DNA (2 μg; Promega Corp., Madison, Wis.) was biotinylated by nick translation using the BioNick kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. The biotinylated DNA was cleaned using a QIAquick cleanup kit (Qiagen, Valencia, Calif.), denatured by boiling it for 3 min, and dissolved in 15 ml of hybridization buffer. Hybridization took place overnight at 50°C. A Southern Star detection kit (Applied Biosystems, Foster City, Calif.) was used for probe detection.
Predicted restriction fragments from genome sequence.
A strain of O157 for which the genome sequence is available (EDL933) (53) was acquired from the American Type Culture Collection (ATCC 43895). The predicted fragments from an XbaI digestion of EDL933 were obtained by downloading the entire genome sequence (GenBank number AE005174) and searching for the XbaI recognition site (TCTAGA), using Vector NTI Suite software (InforMax, Bethesda, Md.). PFGE of EDL933 following XbaI digestion was performed under the conditions listed in Table 1. For better fragment resolution, PFGE was also performed following XbaI digestion with a pulse time ramp of 0.1 to 38 s over a run time of 24 h (56).
RESULTS
Enzyme correlation.
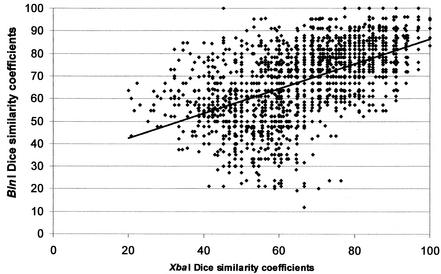
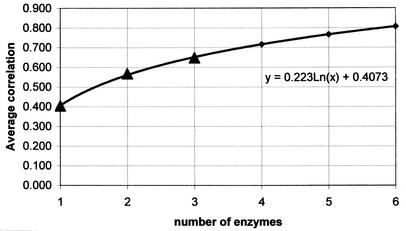
Genomic DNAs from 62 O157 isolates were subjected to PFGE after restriction with XbaI and BlnI, and each PFGE pattern was compared to those of all other isolates, resulting in 1,891 pairwise comparisons of Dice coefficients. The resulting XbaI and BlnI similarity coefficients for each pair of isolates were positively correlated (r = 0.53) (Fig. 1). A subset of 14 of the isolates was then similarly subjected to PFGE following DNA digestion with each of four additional restriction enzymes, resulting in 91 pairwise comparisons for each of six enzymes. The average correlations for Dice coefficients derived from one, two, or three enzymes were 0.405, 0.568, and 0.648, respectively (Tables 2, 3, and 4). A logarithmic function computed from these data projected that six enzymes would be required to reach a correlation coefficient of 0.8 or higher (Fig. 2).
FIG. 1.
Correlation between Dice similarity indices for pairwise analysis of 62 isolates (r = 0.5318).
TABLE 2.
Matrix of correlation coefficients between the Dice similarity coefficients generated by single restriction enzymesa
| Enzyme | Coefficient
|
||||
|---|---|---|---|---|---|
| X | B | N | P | Sf | |
| X | |||||
| B | 0.573158 | ||||
| N | 0.195999 | 0.379299 | |||
| P | 0.521172 | 0.429225 | 0.504657 | ||
| Sf | 0.235544 | 0.259242 | 0.586138 | 0.408706 | |
| Sp | 0.230233 | 0.264564 | 0.505782 | 0.484749 | 0.499297 |
X, XbaI; B, BlnI; N, NheI; P, PacI; Sf, SfiI; Sp, SpeI. Average correlation, 0.405.
TABLE 3.
Correlations between Dice similarity indices averaged from two restriction enzymesa
| Enzyme | Coefficient
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| X-B | X-N | X-P | X-Sf | X-Sp | B-N | B-P | B-Sf | B-Sp | N-Sf | N-Sp | |
| B-N | 0.551 | 0.617 | 0.587 | ||||||||
| B-P | 0.732 | 0.641 | 0.640 | ||||||||
| B-Sf | 0.723 | 0.578 | 0.626 | ||||||||
| B-Sp | 0.670 | 0.569 | 0.614 | ||||||||
| N-P | 0.442 | 0.594 | 0.591 | 0.660 | 0.642 | ||||||
| N-Sf | 0.337 | 0.374 | 0.537 | 0.480 | 0.597 | ||||||
| N-Sp | 0.342 | 0.398 | 0.548 | 0.503 | 0.622 | ||||||
| P-Sf | 0.426 | 0.674 | 0.626 | 0.622 | 0.610 | 0.702 | |||||
| P-Sp | 0.394 | 0.596 | 0.560 | 0.558 | 0.557 | 0.618 | |||||
No correlations were computed if enzyme sets being compared included the same enzyme. Average correlation, 0.568. See Table 2, note a, for abbreviations.
TABLE 4.
Correlations between Dice similarity indices averaged from three restriction enzymesa
| Enzyme | Coefficient
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| X-B-N | X-B-P | X-B-Sf | X-B-Sp | X-N-P | X-N-Sf | X-N-Sp | X-P-Sf | X-P-Sp | X-Sf-Sp | |
| B-N-P | 0.697 | |||||||||
| B-N-Sf | 0.656 | |||||||||
| B-N-Sp | 0.686 | |||||||||
| B-P-Sf | 0.776 | |||||||||
| B-P-Sp | 0.701 | |||||||||
| B-Sf-Sp | 0.727 | |||||||||
| N-P-Sf | 0.614 | |||||||||
| N-P-Sp | 0.596 | |||||||||
| N-Sf-Sp | 0.436 | |||||||||
| P-Sf-Sp | 0.596 | |||||||||
No correlations were computed if enzyme sets being compared included the same enzyme. Average correlation, 0.648. See Table 2, note a, for abbreviations.
FIG. 2.
Effect of increasing numbers of enzymes on the correlation between Dice similarities. Triangles, actual data points; diamonds, extrapolations.
Genetic homology of same-size bands.
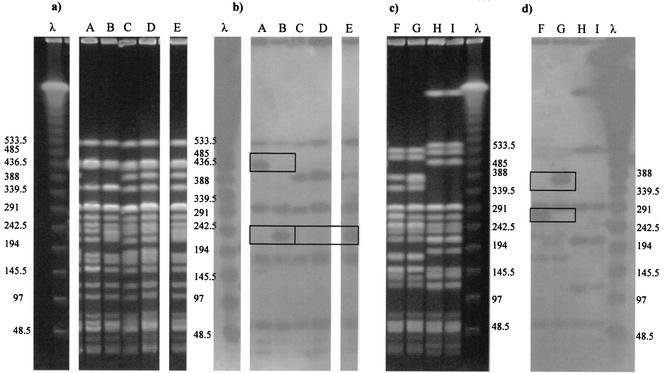
To evaluate whether same-size bands represented homologous genetic sequences, six isolate pairs having 100% similarity by XbaI PFGE (Fig. 1) were subjected to Southern transfer. Probing with lambda DNA demonstrated that isolates that matched by XbaI PFGE (100% similarity) had matching bands that contained nonhomologous genetic material (Fig. 3). For example, lambda probe hybridized with a 436-kbp band from isolate A but not with one from isolate B, and it hybridized with a 200- to 240-kbp band from isolate B but not with one from isolate A. Similarly, isolates C, D, and E were 100% similar by PFGE, yet lambda probe hybridized with a 194- to 242-kbp band of isolate E but not with one from C or D (Fig. 3a and b). Lambda probe hybridized with a 242- to 290-kbp band from isolate F but not with one from G, and it hybridized with a 339- to 388-kbp band from G but not with one from F. Isolate pairs H plus I and C plus D provided an example of matching PFGE profiles consistent with hybridization (Fig. 3c and d).
FIG. 3.
Isolate pairs AB, CD, CE, DE, FG, and HI were 100% similar after digestion with XbaI, PFGE by the standard protocol, and analysis in Bionumerics with a 1% position tolerance. Lanes λ contain lambda DNA concatemers. (a and c) PFGE gel image. (b and d) Nylon membrane with lambda probe hybridized to membrane. See the text for interpretation. The boxes mark occurrences of nonhomologous same-size bands.
Comparison of PFGE banding patterns to sequence data.
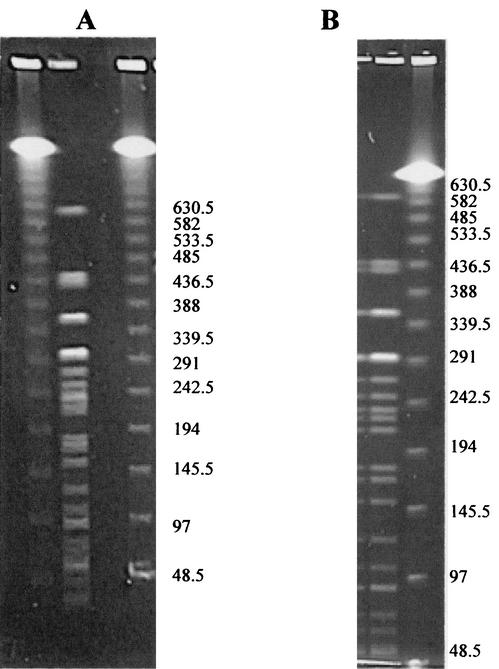
As an empirical test of the accuracy of PFGE in resolving DNA bands, the predicted banding pattern of E. coli O157 EDL933 based on its whole genome sequence was compared with the actual PFGE banding patterns in this study. Genome sequence data indicated that 40 fragments would be expected from complete XbaI digestion and PFGE of EDL933, yet only 19 distinguishable fragments were seen by the standard method (Fig. 4). Eight of the predicted fragments, smaller than 35 kbp, would be expected to migrate off the gel under many PFGE protocols. No predicted fragments were larger than 388 kbp, yet in this study and in that reported by Harsono et al. (29), three or four fragments larger than 388 kbp were observed (Table 5). PFGE of EDL933 using the standard electrophoresis protocol produced some bands that did not resolve well (Fig. 4a). Better resolution was obtained using different ramping times and run times (56) (Fig. 4b).
FIG. 4.
Pulsed-field gels of isolate EDL933 after XbaI digestion. Shown are electrophoresis conditions according to the standard PulseNet protocol with switch times from 2.2 to 54.2 s over a 21-h run time (A) and 0.1 to 38 s over a 24-h run time (B).
TABLE 5.
Results of PFGE of XbaI-digested EDL933
| Size range (kbp) | Expected no. of fragments in range (sequence data) | Observed no. of fragments in range (PFGE)
|
|
|---|---|---|---|
| Present studya | Harsono 1993b | ||
| 48.5 | 12 | NA | 2 |
| 48.5-97 | 9 | 4 | 3 |
| 97-145.5 | 2 | 2 | 3 |
| 145.5-194 | 3 | 3 | 3 |
| 194-242.5 | 5 | 3 | 2 |
| 242.5-291 | 4 | 2 | 3 |
| 291-339.5 | 3 | 1 | 1 |
| 339.5-388 | 2 | 1 | 0 |
| 388-436.5 | 0 | 1 | 1 |
| 436.5-485 | 0 | 1 | 0 |
| 485-533.5 | 0 | 0 | 2 |
| 533.5-582 | 0 | 0 | 0 |
| 582-630.5 | 0 | 1 | 0 |
| >630.5 | 1 | ||
DISCUSSION
The Dice similarity coefficient has often been presented as virtually synonymous with genetic relatedness (3, 7, 18, 42, 50, 51, 54, 58, 63), yet there is a very low correlation between similarity estimates derived from single enzymes (average r, 0.4), and thus, the Dice coefficient is only a poor estimate of genetic relatedness between two isolates. If the Dice similarity coefficient from a single enzyme digest were an accurate estimate, then the correlation between Dice coefficients from two different restriction enzymes should be close to 1. If, however, one enzyme assigns a low similarity to two isolates while the other assigns a high similarity to the same pair, one or both must be an inaccurate measure of relatedness. Nevertheless, there is a positive, albeit low, correlation between all pairs of enzyme results. The most likely basis for this is that any single enzyme provides at least a crude measure of true genetic relatedness. This supposition is supported by our finding that pooling similarity results from multiple enzymes provided better correlation between Dice similarities.
Additional enzymes provide information from a larger proportion of the genome, which could explain improved correlations. Single-enzyme PFGE is likely to correlate poorly with randomly amplified polymorphic DNA (24, 56) or other methods (49), which of necessity only sample a minority of the bacterial genome. Other investigators have found that combining results from additional typing methods and/or restriction enzymes improved both the discriminatory power of the method and the correlation between epidemiology and the molecular data for O157 (1, 4, 24, 32, 55) and Salmonella (2, 40, 49). In the present study, we combined results from multiple enzymes using an unweighted average of similarity coefficients. Using a composite of all banding patterns might have introduced bias by weighting the results in favor of those enzymes that produce a greater number of bands and are therefore more likely to produce spurious matches. This possibility was supported by our finding that enzymes that produced more bands were more likely to have a higher similarity with the lambda concatemer, a molecular size standard unrelated to O157 (data not shown). Computer simulations have also illustrated this point (9).
The results of this study suggest two explanations for the poor correlations between Dice coefficients from single-enzyme PFGE. The first is that fragments that migrate the same distance do not always contain homologous genetic material. The results of our band comparison suggest that this is not a rare occurrence: 62 epidemiologically unrelated isolates yielded six pairs that were 100% similar. Comparing same-size bands by probing with lambda DNA indicated that four of these pairs were spurious matches. When those isolate pairs with 100% similarity were assayed side by side on the same gel, it became apparent that there were some differences in band widths and intensities between some of the “matching” bands. But when large sets of isolates are being analyzed, as in a surveillance program or a large research project, it seems probable that fragments migrating the same distance on a gel will be marked and considered to be a “vote for similarity” whether or not they have the same band intensity or width. Our finding was the result of one probe type, representing only a small proportion of the genome; the use of other probes would likely detect more instances of nonhomology between same-size bands.
Another reason that the similarity coefficient is a poor estimate of relatedness is the failure of PFGE to resolve bands of nearly identical sizes. Among the expected 40 fragments resulting from XbaI digestion of EDL933, 2 differ in size from another fragment by <1%, and 15 differ by <5%. Depending on the resolving power of the technique, as many as 17 fragments could be obscured by bands of similar sizes. Thus, poor resolution of fragment size could account for the discrepancy between the 18 to 25 fragments detected by most published PFGE protocols for O157 (8, 22, 29) and the 40 fragments expected based on sequence data. There are other possible explanations for the discrepancy: (i) the sequence data may be in error or (ii) some of the XbaI recognition sites in the O157 genome may not be available to the enzyme if, for example, they happen to be adjacent to a methylated site. One of these possibilities may explain the large fragment (>600 kb) detected by PFGE of EDL933 (Table 5). The present study demonstrated that, after XbaI digestion, the electrophoresis conditions used by Radu et al. (56) resolved bands that are not resolved by standard electrophoresis conditions. The impact of occult or comigrating bands is potentially great. Homologous but occult bands would not be counted and would bias the Dice index downward. Two comigrating bands in a nonshared position would be counted as only one nonmatching band and would bias the Dice coefficient upward.
Based on the observation that the loss or addition of a single recognition site can result in up to three band differences, The criteria of Tenover et al. for interpretation of PFGE patterns classify two isolates with three or fewer band differences as closely related (67). Under standard PFGE conditions, a significant proportion of fragments comigrate or are too small to visualize, so the Tenover et al. criteria could misclassify unrelated isolates as closely related. There is also potential for the Tenover et al. criteria to misclassify closely related isolates as unrelated if two cells with a recent common ancestor each undergo a minor genetic event and differ from the parent by three bands but differ from each other by six bands. It should be noted that Tenover et al. cautioned against the use of their criteria for the interpretation of PFGE in the absence of epidemiologic data or for extensive sets of isolates (67).
PFGE using one or two enzymes to establish matches between O157 isolates has been used successfully in many investigations of food-borne illness (5, 11-15, 72), where the evidence implicating a food or other exposure has been considerably strengthened by finding indistinguishable (matching) PFGE patterns between the clinical and food (or environmental) isolates. Similarly, the use of PFGE in investigations of farm environments and/or cattle herds associated with human cases have allowed investigators to establish relationships between animal and human isolates (7, 16, 34, 68; W. B. Trevena, R. S. Hooper, C. Wray, G. A. Willshaw, T. Cheasty, and G. Domingue, Letter, Vet. Rec. 138:400, 1996). In this context, where there is a high prior probability that isolates are epidemiologically linked, PFGE of a single enzyme digest is appropriate for identifying genetic-fingerprint matches. Indeed, when an outbreak investigation is under way, time and resources may not accommodate more effort. In the absence of the prior probability conferred by epidemiologic data, however, a valid measure of relatedness, rather than a method that simply identifies matches, is required. Inferences about genetic relationships based solely on one-enzyme PFGE should be interpreted with caution. The availability of computer software programs that make calculation of Dice coefficients and the generation of dendrograms relatively easy, as well as failure to distinguish between the concepts of relatedness and a similarity coefficient, may have contributed to acceptance of dendrograms from single-enzyme PFGE as accurate measures of relatedness. Software such as Bionumerics makes analysis of banding patterns from large sets of isolates possible but cannot be relied on to assess the presence or absence of bands without visual evaluation of the gel image.
While PFGE has been used for over a decade in epidemiologic studies of many eukaryotic and prokaryotic organisms and has proved to be a robust typing method for investigations of food-borne outbreaks and for hospital epidemiology, alternative molecular methods have promise for greater precision, as well as higher efficiency, for molecular epidemiologic analysis of microbial pathogens. These include fluorescent amplified fragment length polymorphisms (73) and DNA microarrays (10, 61). The findings of the present study indicate, however, that if epidemiologic relationships between isolates must be inferred from PFGE data, six or more restriction enzymes would be needed to provide a reasonable estimate of genetic relatedness.
Acknowledgments
Funding was provided in part by the National Alliance for Food Safety, USDA Agricultural Research Service (grant 11D-2530-0236), and in part by the Washington State University Agriculture Research Center (grant 10A30720858).
We thank Dan Rice, Melissa Krug, and Yubei Zhang for technical help and Kevin Bertrand, Rowland Cobbold, Ron Digiacomo, and Mansour Samadpour for their intellectual contributions. We also acknowledge all dairy and beef producers who have been willing to participate in O157 field research.
REFERENCES
- 1.Allison, L. J., P. E. Carter, and E. M. Thomson-Carter. 2000. Characterization of a recurrent clonal type of Escherichia coli O157:H7 causing major outbreaks of infection in Scotland. J. Clin. Microbiol. 38:1632-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amavisit, P., P. F. Markham, D. Lightfoot, K. G. Whithear, and G. F. Browning. 2001. Molecular epidemiology of Salmonella Heidelberg in an equine hospital. Vet. Microbiol. 80:85-98. [DOI] [PubMed] [Google Scholar]
- 3.Barkocy-Gallagher, G. A., T. M. Arthur, G. R. Siragusa, J. E. Keen, R. O. Elder, W. W. Laegreid, and M. Koohmaraie. 2001. Genotypic analysis of Escherichia coli O157:H7 and O157 nonmotile isolates recovered from beef cattle and carcasses at processing plants in the Midwestern states of the United States. Appl. Environ. Microbiol. 67:3810-3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell, B. P., M. Goldoft, P. M. Griffin, M. A. Davis, D. C. Gordon, P. I. Tarr, C. A. Bartleson, J. H. Lewis, T. J. Barrett, J. G. Wells, R. Baron, and J. Kobayashi. 1994. A multistate outbreak of Escherichia coli O157:H7-associated bloody diarrhea and hemolytic uremic syndrome from hamburgers: the Washington experience. JAMA 272:1349-1353. [PubMed] [Google Scholar]
- 6.Bender, J. B., C. W. Hedberg, J. M. Besser, D. J. Boxrud, K. L. MacDonald, and M. T. Osterholm. 1997. Surveillance for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. N. Engl. J. Med. 337:388-394. [DOI] [PubMed] [Google Scholar]
- 7.Bielaszewska, M., H. Schmidt, A. Liesegang, R. Prager, W. Rabsch, H. Tschäpe, A. Cízek, J. Janda, K. Bláhova, and H. Karch. 2000. Cattle can be a reservoir of sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains and a source of human diseases. J. Clin. Microbiol. 38:3470-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böhm, H., and H. Karch. 1992. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Call, D. R.,, J. G. Hallett, S. G. Mech, and M. Evans. 1998. Considerations for measuring genetic variation and population structure with multilocus fingerprinting. Mol. Ecol. 7:1337-1346. [Google Scholar]
- 10.Call, D. R., F. J. Brockman, and D. P. Chandler. 2001. Detecting and genotyping Escherichica coli O157:H7 using multiplexed PCR and nucleic acid microarrays. Int. J. Food Microbiol. 67:71-80. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 1996. Outbreak of Escherichia coli O157:H7 infection—Georgia and Tennessee, June 1995. Morb. Mortal. Wkly. Rep. 45:249-251. [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1996. Outbreak of Escherichia coli O157:H7 infections associated with drinking unpasteurized commercial apple juice—British Columbia, California, Colorado, and Washington, October 1996. Morb. Mortal. Wkly. Rep. 45:975.. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1997. Outbreaks of Escherichia coli O157:H7 infection and cryptosporidiosis associated with drinking unpasteurized apple cider—Connecticut and New York, October 1996. Morb. Mortal. Wkly. Rep. 46:4-8. [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 1997. Escherichia coli O157:H7 infections associated with eating a nationally distributed commercial brand of frozen ground beef patties and burgers—Colorado, 1997. Morb. Mortal. Wkly. Rep. 46:777-778. [PubMed] [Google Scholar]
- 14a.Centers for Disease Control and Prevention. 1998. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. CDC training manual. Centers for Disease Control and Prevention, Atlanta, Ga.
- 15.Centers for Disease Control and Prevention. 2000. Outbreak of Escherichia coli O157:H7 infection associated with eating fresh cheese curds—Wisconsin, June, 1998. Morb. Mortal. Wkly. Rep. 49:911-913. [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. 2001. Outbreaks of Escherichia coli O157:H7 infections among children associated with farm visits—Pennsylvania and Washington, 2000. Morb. Mortal. Wkly. Rep. 50:293-297. [PubMed] [Google Scholar]
- 17.Chasseignaux, E., M. T. Toquin, C. Ragimbeau, G. Salvat, P. Colin, and G. Ermel. 2001. Molecular epidemiology of Listeria monocytogenes isolates collected from the environment, raw meat and raw products in two poultry- and pork-processing plants. J. Appl. Microbiol. 91:888-899. [DOI] [PubMed] [Google Scholar]
- 18.Chinen, I., J. D. Tanaro, E. Milibewsky, L. H. Lound, G. Chillemi, S. Ledri, A. Baschkier, M. Scarpin, E. Manfredi, and M. Rivas. 2001. Isolation and characterization of Escherichia coli O157:H7 from retail meats in Argentina. J. Food Prot. 64:1346-1351. [DOI] [PubMed] [Google Scholar]
- 19.Dice, L. R. 1945. Measures of the amount of ecologic association between species. Ecology 26:297-302. [Google Scholar]
- 20.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M.-S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farber, J. M. 1996. An introduction to the hows and whys of molecular typing. J. Food Prot. 59:1091-1101. [DOI] [PubMed] [Google Scholar]
- 22.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouveia, S., M. E. Proctor, M. S. Lee, J. B. Luchansky, and C. W. Kaspar. 1998. Genomic comparisons and Shiga toxin production among Escherichia coli O157:H7 isolates from a day care center outbreak and sporadic cases in southeastern Wisconsin. J. Clin. Microbiol. 36:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grif, K., H. Karch, C. Schneider, F. D. Daschner, L. Beutin, T. Cheasty, H. Smith, B. Rowe, M. P. Dierich, and F. Allerberger. 1998. Comparative study of five different techniques for epidemiological typing of Escherichia coli O157. Diagn. Microbiol. Infect. Dis. 32:165-176. [DOI] [PubMed] [Google Scholar]
- 25.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 26.Hancock, D. D., T. E. Besser, M. L. Kinsel, P. I. Tarr, D. H. Rice, and M. G. Paros. 1994. The prevalence of Escherichia coli O157.H7 in dairy and beef cattle in Washington State. Epidemiol. Infect. 113:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hancock, D. D., T. E. Besser, D. H. Rice, D. E. Herriott, and P. I. Tarr. 1997. A longitudinal study of Escherichia coli O157:H7 in fourteen cattle herds. Epidemiol. Infect. 118:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock, D. D., T. E. Besser, and D. H. Rice. 1998. Ecology of Escherichia coli O157:H7 in cattle and impact of management practices, p. 85-91. In J. B. Kaper and A. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 29.Harsono, K. D., C. W. Kaspar, and J. B. Luchansky. 1993. Comparison and genomic sizing of Escherichia coli O157:H7 isolates by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 59:3141-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkins, K. L., and A. C. Hilton. 2000. Methods available for the sub-typing of Escherichia coli O157. World J. Microbiol. Biotechnol. 16:741-748. [Google Scholar]
- 31.Houang, E. T. S., Y. W. Chu, and A. F. B. Cheng. 1998. Study of the relatedness of isolates of Shigella flexneri and Shigella sonnei obtained in 1986 and 1987 and in 1994 and 1995 from Hong Kong. J. Clin. Microbiol. 36:2404-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izumiya, H., J. Terjima, A. Wada, Y. Inagaki, K. Itoh, K. Tamura, and H. Watanabe. 1997. Molecular typing of enterohemorrhagic Escherichia coli O157:H7 isolates in Japan by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 35:1675-1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, J. M., S. D. Weagant, K. C. Jinneman, and J. L. Bryant. 1995. Use of pulsed-field gel electrophoresis for epidemiological study of Escherichia coli O157:H7 during a food-borne outbreak. Appl. Environ. Microbiol. 61:2806-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnsson, M. E., A. Aspán, E. Eriksson, and I. Vågsholm. 2001. Persistence of verocytotoxin-producing Escherichia coli O157:H7 in calves kept on pasture and in calves kept indoors during the summer months in a Swedish dairy herd. Int. J. Food Microbiol. 66:55-61. [DOI] [PubMed] [Google Scholar]
- 35.Karch, H., M. Bielaszewska, M. Bitzan, and H. Schmidt. 1999. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34:229-243. [DOI] [PubMed] [Google Scholar]
- 36.Keene, W. E., E. Sazie, J. Kok, D. H. Rice, D. D. Hancock, V. K. Balan, T. Zhao, and M. P. Doyle. 1997. An outbreak of Escherichia coli O157:H7 infections traced to jerky made from deer meat. JAMA 277:1229-1231. [DOI] [PubMed] [Google Scholar]
- 37.Keene, W. E., K. Hedberg, D. E. Herriott, D. D. Hancock, R. W. McKay, T. J. Barrett, and D. W. Fleming. 1997. A prolonged outbreak of Escherichia coli O157:H7 infections caused by commercially distributed raw milk. J. Infect. Dis. 176:815-818. [DOI] [PubMed] [Google Scholar]
- 38.Krause, U., F. M. Thomson-Carter, and T. H. Pennington. 1996. Molecular epidemiology of Escherichia coli O157:H7 by pulsed-field gel electrophoresis and comparison with that by bacteriophage typing. J. Clin. Microbiol. 34:959-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kusiluka, L. J., B. Kokotovic, B. Ojeniyi, N. F. Friis, and P. Ahrens. 2000. Genetic variations among Mycoplasma bovis strains isolated from Danish cattle. FEMS Microbiol. Lett. 192:113-118. [DOI] [PubMed] [Google Scholar]
- 40.Landeras, E., M. A. González-Hevia, and M. C. Mendoza. 1998. Molecular epidemiology of Salmonella serotype Enteritidis. Relationships between food, water, and pathogenic strains. Int. J. Food Microbiol. 43:81-90. [DOI] [PubMed] [Google Scholar]
- 41.Lee, M., C. W. Kaspar, R. Brosch, J. Shere, and J. B. Luchansky. 1996. Genomic analysis using pulsed-field gel electrophoresis of Escherichia coli O157:H7 isolated from dairy calves during the United States national dairy heifer evaluation project (1991-1992). Vet. Microbiol. 48:223-230. [DOI] [PubMed] [Google Scholar]
- 42.Liesegang, A., U. Sachse, R. Prager, H. Claus, H. Steinrück, S. Aleksic, W. Rabsch, W. Voigt, A. Fruth, H. Karch, J. Bockemühl, and H. Tschäpe. 2000. Clonal diversity of Shiga toxin-producing Escherichia coli O157:H7/H− in Germany—a ten-year study. Int. J. Med. Microbiol. 290:269-278. [DOI] [PubMed] [Google Scholar]
- 43.Lortal, S., A. Rouault, S. Guezenec, and M. Gautier. 1997. Lactobacillus helveticus: strain typing and genome size estimation by pulsed-field gel electrophoresis. Curr. Microbiol. 34:180-185. [DOI] [PubMed] [Google Scholar]
- 44.Louie, M., S. Read, L. Louie, K. Ziebell, K. Rahn, A. Borczyk, and H. Lior. 1999. Molecular typing methods to investigate transmission of Escherichia coli O157:H7 from cattle to humans. Epidemiol. Infect. 123:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malorny, B., A. Schroeter, C. Bunge, B. Hoog, A. Steinbeck, and R. Helmuth. 2001. Evaluation of molecular typing methods for Salmonella enterica serovar Typhimurium DT104 isolated in Germany from healthy pigs. Vet. Res. 32:119-129. [DOI] [PubMed] [Google Scholar]
- 46.Mead, P. S., L. Finelli, M. A. Lambert-Fair, D. Champ, J. Townes, L. Hutwagner, T. Barrett, K. Spitalny, and E. Mintz. 1997. Risk factors for sporadic infection with Escherichia coli O157:H7. Arch. Intern. Med. 157:204-208. [PubMed] [Google Scholar]
- 47.Midgley, J., and P. Desmarchelier. 2001. Pre-slaughter handling of cattle and Shiga toxin-producing Escherichia coli (STEC). Lett. Appl. Microbiol. 32:307-311. [DOI] [PubMed] [Google Scholar]
- 48.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen, J. E., M. N. Skov, Ø. Angen, E. J. Threlfall, and M. Bisgaard. 1997. Genomic relationships between selected phage types of Salmonella enterica subsp. enterica serotype typhimurium defined by ribotyping, IS200 typing, and PFGE. Microbiology 143:1471-1479. [DOI] [PubMed] [Google Scholar]
- 50.Osek, J. 2001. Molecular characterisation of Shiga toxin-producing Escherichia coli O157:H7 strains isolated in Poland. Res. Vet. Sci. 70:175-177. [DOI] [PubMed] [Google Scholar]
- 51.Oshibe, T., H. Tsuji, and K. Hamada. 2001. Phylogenetic analysis of genotypic variations of Escherichia coli O157:H7 isolates from sporadic infections by using pulsed-field gel electrophoresis from March 1999 to February 2001 in Hyogo prefecture. Jpn. J. Infect. Dis. 54:162-165. [PubMed] [Google Scholar]
- 52.Park, S., R. W. Worobo, and R. A. Durst. 2001. Escherichia coli O157:H7 as an emerging foodborne pathogen: a literature review. Crit. Rev. Biotechnol. 21:27-48. [DOI] [PubMed] [Google Scholar]
- 53.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 54.Pradel, N., K. Boukors, Y. Bertin, C. Forestier, C. Martin, and V. Livrelli. 2001. Heterogeneity of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome patients, cattle, and food samples in central France. Appl. Environ. Microbiol. 67:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preston, M. A., W. Johnson, R. Khakhria, and A. Borczyk. 2000. Epidemiologic subtyping of Escherichia coli serogroup O157 strains isolated in Ontario by phage typing and pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:2366-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radu, S., O. W. Ling, G. Rusul, M. I. A. Karim, and M. Nishibuchi. 2001. Detection of Escherichia coli O157:H7 by multiplex PCR and their characterization by plasmid profiling, antimicrobial resistance, RAPD, and PFGE analyses. J. Microbiol. Methods 46:131-139. [DOI] [PubMed] [Google Scholar]
- 57.Rice, D. H., K. M. McMenamin, L. C. Pritchett, D. D. Hancock, and T. E. Besser. 1999. Genetic subtyping of Escherichia coli O157 isolates from 41 Pacific Northwest USA cattle farms. Epidemiol. Infect. 122:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rios, M., V. Prado, M. Trucksis, C. Arellano, C. Borie, M. Alexandre, A. Fica, and M. M. Levine. 1999. Clonal diversity of Chilean isolates of enterohemorrhagic Escherichia coli from patients with hemolytic-uremic syndrome, asymptomatic subjects, animal reservoirs, and food products. J. Clin. Microbiol. 37:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts, C. L., P. A. Mschar, M. L. Cartter, J. L. Hadler, D. M. Sosin, P. S. Hayes, and T. J. Barrett. 1995. The role of heightened surveillance in an outbreak of Escherichia coli O157:H7. Epidemiol. Infect. 115:447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodgers, J. D., J. J. McCullagh, P. T. McNamee, J. A. Smyth, and H. J. Ball. 1999. Comparison of Staphylococcus aureus recovered from personnel in a poultry hatchery and in broiler parent farms with those isolated from skeletal disease in broilers. Vet. Microbiol. 69:189-198. [DOI] [PubMed] [Google Scholar]
- 61.Salazar, N. M., and G. Caetano-Anollés. 1996. Nucleic acid scanning-by-hybridization of enterohemorrhagic Escherichia coli isolates using oligodeoxynucleotide arrays. Nucleic Acids Res. 24:5056-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 63.Sharma, M., K. Riederer, L. B. Johnson, and R. Khatib. 2001. Molecular analysis of coagulase-negative Staphylococcus isolates from blood cultures: prevalence of genotypic variation and polyclonal bacteremia. Clin. Infect. Dis. 33:1317-1323. [DOI] [PubMed] [Google Scholar]
- 64.Shere, J. A., K. J. Bartlett, and C. W. Kaspar. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl. Environ. Microbiol. 64:1390-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and the CDC PulseNet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tarr, P. I. 1995. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin. Infect. Dis. 20:1-8. [DOI] [PubMed] [Google Scholar]
- 67.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trevena, W. B., G. A. Willshaw, T. Cheasty, G. Domingue, and C. Wray. 1999. Transmission of Vero cytotoxin producing Escherichia coli O157 infection from farm animals to humans in Cornwall and west Devon. Commun. Dis. Public Health 2:263-268. [PubMed] [Google Scholar]
- 69.Wang, S. M., M. A. Deighton, J. A. Capstick, and N. Gerraty. 1999. Epidemiological typing of bovine streptococci by pulsed-field gel electrophoresis. Epidemiol. Infect. 123:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weigel, R. M., B. Qiao, D. A. Barber, B. Teferedegne, S. Kocherginskaya, B. A. White, and R. E. Isaacson. 2001. Identification of patterns of transmission of Salmonella within swine production systems using pulsed-field gel electrophoresis (PFGE) and repetitive sequence polymerase chain reaction (REP-PCR): a quantitative analysis. Berl. Münch. Tieräztl. Wschr. 114:397-400. [PubMed] [Google Scholar]
- 71.Welinder-Olsson, C., M. Badenfors, E. Kjellin, and B. Kaijser. 2002. Genetic profiling of enterohemorrhagic Escherichia coli strains in relation to clonality and clinical signs of infection. J. Clin. Microbiol. 40:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Willshaw, G. A., H. R. Smith, T. Cheasty, and S. J. O'Brien. 2001. Use of strain typing to provide evidence for specific interventions in the transmission of VTEC O157 infections. Int. J. Food Microbiol. 66:39-46. [DOI] [PubMed] [Google Scholar]
- 73.Zhao, S., S. E. Mitchell, J. Meng, S. Kresovich, M. Doyle, R. E. Dean, A. Casa, and J. Weller. 2000. Genomic typing of Escherichica coli O157:H7 by semi-automated fluorescent AFLP analysis. Microb. Infect. 2:107-113. [DOI] [PubMed] [Google Scholar]