Abstract
A binary classification system has been established for group A rotaviruses, with the viral capsid protein VP7 defining G types and VP4 defining P types. At least 15 G types and 21 P types have been isolated globally with various G and P combinations. Most of the currently circulating human rotaviruses belong to G1P[8], G2P[4], G3P[8], and G4P[8]. We report a human rotavirus strain (B1711) with a novel genotypic VP7/VP4 combination of G6P[6]. This unique rotavirus was isolated from a 13-month-old human immunodeficiency virus (HIV)- negative child of an HIV-seropositive Malian mother that was hospitalized with severe diarrhea in Belgium after returning from a trip to Mali. The VP7 and VP4 genes of the rotavirus strain were sequenced, and phylogenetic trees were constructed. Nucleotide and amino acid sequence comparisons with 15 known G genotypes indicated that the VP7 sequence of strain B1711 was most closely related to an American (Se584) and an Italian (PA151) human G6 strain (95 to 96% nucleotide and 98% amino acid identity). Comparison of the VP4 sequence with 21 P types showed the closest similarity to P[6] genotypes, with greatest similarity to a G8P[6] Malawi strain (mw131) (97% nucleotide and 98% amino acid identity). The B1711 strain is the first reported rotavirus isolate with a G6P[6] genotypic combination. The discovery and surveillance of novel human and nonhuman rotavirus G or P types or of novel G/P combinations is essential for the design of future rotavirus vaccines and for our understanding of rotavirus diversity and evolution.
Group A rotaviruses are the single most important etiological agent associated with gastroenteritis in infants and young children (2, 40). Rotavirus-associated diarrhea leads to more than 125 million cases of infantile gastroenteritis and 870,000 deaths each year, primarily in less developed countries (54). In the United States it also results in $274 million in medical care costs and a total of $1 billion in societal costs (including indirect cost of lost parental work time) per year (47). Rotaviruses contain 11 segments of double-stranded RNA within a core shell and are members of the Reoviridae family. Each segment encodes a single viral polypeptide, for a total of five nonstructural and six structural proteins (17). The two outer capsid proteins VP7 and VP4, which independently elicit neutralizing antibodies, are the basis of a binary classification system for rotaviruses: G types (derived from the VP7 glycoprotein) and P types (derived from the protease sensitive VP4 protein). Thus far, 15 different G genotypes and 21 different P genotypes have been reported (2, 26, 31, 48). Because VP4 and VP7 are encoded by different RNA segments, various combinations of G and P types can be observed (40). Most G genotypes were serologically confirmed as serotypes (25). Due to the lack of appropriate antibody reagents, a dual P-typing system (P serotype and P genotype) has been used (18). Strains sharing more than 89% sequence identity at the amino acid level are considered to belong to the same P genotype (17, 27). In the dual typing system, the P serotype is represented by a number immediately after the letter P, and the genotype is denoted by a number in square brackets (17). Thus, a rotavirus strain having P serotype 1A and P genotype 8 is abbreviated as P1A[8]. Nowadays there is a growing trend for naming a rotavirus strain only by genotypes (like G1P[8]) because no serotype has been designated for some of the more recently discovered P genotypes. Whereas only 11 P serotypes have been characterized, to date, at least 21 P genotypes have been discovered (37, 44, 48, 50).
The four major human G types are G1, G2, G3, and G4, and the less common types are G5, G8, G9, and G12 (7, 29, 30, 36, 49). G6 and G10 rotaviruses are the two major types isolated from cattle but have been infrequently encountered in humans. Until now, no rotavirus strains of G7 (chicken and cattle), G11 (pigs), G13 and G14 (horses), and G15 (cattle) have been isolated from humans (9, 10, 48). Only a few epidemiological studies have studied the occurrence of P types. In humans, P[8] is the most common genotype detected worldwide, followed by P[4] and P[6] (20, 40).
Few G6 rotaviruses have been found in human patients. The first characterized human G6 rotavirus strains, PA151 (G6P[9]) and PA169 (G6P[14]), were isolated in 1987 to 1988 from two Italian children hospitalized with severe gastroenteritis (23). Another G6 human rotavirus strain, MG6 (G6P[14]), was isolated in 1993 from a 16-month-old Australian child admitted to the hospital with acute gastroenteritis (46). During 1996 to 1997, two G6P[14] rotavirus strains (MG6.01 and AG6.01) were isolated again in Australia (14). A nucleotide sequence of a G6P[9] human rotavirus strain from the United States (Se584) was recently reported (30).
G6 types are frequently isolated from cattle (12, 24, 41, 51, 55). In a study of Japanese cows, 59.1% of the isolates were G6 (45). Bovine G6 strains were reported in many countries to be combined with P[5], P[2], and P[11] (12, 13, 19). One study on diarrheic piglets from an outbreak in Italy in 1983 and 1984 reported that all strains isolated were G6P[5], a typical bovine G/P combination, and pointed out the high frequency of viral transmission between pigs and cattle (39).
The prevalence of rotaviruses with a P[6] genotype in European countries like Spain (11), the United Kingdom (34), Ireland (43), and France (8) was 1 to 5% during the years 1996 to 1999. In Brazil, 12% of the isolates from children were P[6] in a study during 1997 to 1998 (4). In India (24%) and Bangladesh (24%) a high prevalence of P[6] was found (32, 35, 54). The P[6] genotype was frequently detected in humans in African countries like Nigeria (66.7% during 1999 to 2000), Guinea-Bissau (37.7% during 1996 to 1998), Ghana (25.6% in 1998), and Malawi (32.6% during 1997 to 1999) (1, 5, 6, 15, 16, 21). In South Africa it was the second most (8%) frequently isolated P type (52). All these P[6] types were found to be in a combination with G9, G1, G2, G3, and G8.
We have isolated a unique rotavirus strain from a 13-month-old child who was admitted to the university hospital in Leuven, Belgium, with severe gastroenteritis. The VP7- and VP4- encoding genes were sequenced and characterized as G6P[6]. To our knowledge this is the first reported case of a group A rotavirus infection with this novel G6P[6] combination.
MATERIALS AND METHODS
Rotavirus antigen detection.
Rotavirus antigens were detected in the stool specimen using the Premier Rotaclone solid-phase sandwich-type enzyme immunoassay (Meridian Bioscience, Cincinnati, Ohio). An aliquot of a fecal suspension was added to a plastic microtiter well coated with a monoclonal antibody directed against the rotavirus group-specific antigen VP6 protein. The solution was simultaneously incubated with an antirotavirus monoclonal antibody conjugated to horseradish peroxidase, resulting in the rotavirus antigen being sandwiched between the solid-phase and the enzyme-linked antibodies. After 60 min of incubation at room temperature, the sample well was washed to remove unbound enzyme-labeled antibodies. Urea peroxide and tetramethylbenzidine were added as substrates and incubated for 10 min at room temperature. The enzymatic reaction that converts the colorless substrate to a blue color was stopped with 1 N H2SO4, and the absorbance was determined spectrophotometrically. Specimens with absorbances (A450) greater than 0.150 were considered positive.
RNA extraction.
Viral RNA was extracted from 140 μl of the feces sample using the QIAamp viral RNA minikit (Qiagen/Westburg, Leusden, The Netherlands) according to the manufacturer's instructions.
RT-PCR.
The extracted RNA was denatured on 97°C for 5 min. G and P genotyping was performed as previously described by Gouvea et al. (28) and Gentsch et al. (22), respectively. Reverse transcriptase PCR (RT-PCR) was carried out using the Qiagen OneStep RT-PCR kit (Qiagen/Westburg). For G typing, we amplified a 1,062-bp fragment of the VP7 gene with the forward primer Beg9 (5′-GGCTTTAAAAGAGAGAATTTCCGTCTGG-3′; prototype strain Wa, GenBank accession number M21843, nucleotides [nt] 1 to 28) and the reverse primer End9 (5′-GGTCACATCATACAATTCTAATCTAAG-3′; prototype strain SA11, accession number K02028, nt 1062 to 1036). For P typing, Con3 (5′-TGGCTTCGCCATTTTATAGACA-3′; prototype strain KU, accession number M21014, nt 11 to 32,) and Con2 (5′-ATTTCGGACCATTTATAACC-3′; prototype strain KU, nt 887 to 868) primers were used to amplify an 876-bp fragment of the entire VP8* fragment and the first 40 amino acids of the VP5* fragment of the rotavirus VP4 gene. The reaction was carried out with an initial reverse transcription step at 45°C for 30 min, followed by PCR activation at 95°C for 15 min, 35 cycles of amplification (30 s at 94°C, 45 s at 53°C, 1 min at 72°C), and a final extension of 7 min at 72°C in a GeneAmp PCR system 9700 thermal cycler (Perkin-Elmer, Foster City, Calif.). PCR products were run on a polyacrylamide gel, stained with ethidium bromide, and visualized under UV light.
A multiplex PCR was also performed for different rotavirus non-G6 types (G1, G2, G3, G4, G8, and G9) (28). Type-specific forward primers derived from distinct regions of the VP7 gene for G1 (aBT1; 5′-CAAGTACTCAAATCAATGATGG-3′; prototype strain Wa, nt 314 to 335), G2 (aCT2; 5′-CAATGATATTAACACATTTTCTGTG-3′; prototype strain DS1, accession number M37348, nt 411 to 435), G3 (aET3; 5′-CGTTTGAAGAAGTTGCAACAG-3′; prototype strain P, accession number M37355, nt 689 to 709), G4 (aDT4; 5′-CGTTTCTGGTGAGGAGTTG-3′; prototype strain ST3, accession number M37364, nt 480 to 498), G8 (aAT8; 5′-GTCACACCATTTGTAAATTCG-3′; prototype strain 69 M, nt 178 to 198) (29), and G9 (aFT9; 5′-CTAGATGTAACTACAACTAC-3′; prototype strain WI61, nt 757 to 776) (29) were combined with a reverse primer RVG9 (5′-GGTCACATCATACAATTCT-3′; prototype strain SA11, nt 1062 to 1044). This primer mix allows the amplification of fragments with a G-type-specific segment size on polyacrylamide gel electrophoresis. The reaction was carried out with an initial reverse transcription step at 45°C for 30 min, followed by PCR activation at 95°C for 15 min, 35 cycles of amplification (30 s at 94°C, 45 s at 50°C, 1 min at 72°C), and a final extension of 7 min at 72°.
Nucleotide sequencing.
The PCR amplicons were purified with the QIAquick PCR purification kit (Qiagen/Westburg) and sequenced in both directions using the dideoxy nucleotide chain termination method with the ABI PRISM BigDye terminator cycle sequencing reaction kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.) on an automated sequencer (ABI PRISM 3100) at the Rega Institute core sequencing facility. The Beg9 and End9 RT-PCR primers (for the VP7 gene) and the Con2 and Con3 primers (for the VP4 gene) were also used as sequencing primers.
DNA and protein sequence analysis.
The chromatogram sequencing files were inspected using Chromas 2.2 (Technelysium, Helensvale, Queensland, Australia), and contigs were prepared using SeqMan II (DNASTAR, Madison, Wis.). Nucleotide and protein sequence similarity searches were performed using the National Center for Biotechnology Information (National Institutes of Health, Bethesda, Md.) BLAST (Basic Local Alignment Search Tool) server on GenBank database release 130.0 (3). Pairwise sequence alignments were performed using Lipman and Pearson's ALIGN program on the Southampton BioInformatics Data Server, and multiple sequence alignments were calculated using CLUSTALW (53) on the DDBJ (DNA Data Bank of Japan) server. Sequences were manually edited in the GeneDoc (version 2.6.002) alignment editor (42).
Phylogenetic analysis.
Phylogenetic and molecular evolutionary analyses were conducted using the MEGA version 2.1 software package (38), based on the different G6 and P[6] rotavirus sequences available in GenBank version 130.0. Genetic distances were calculated using the Kimura-2 parameter. The dendrograms were constructed using the neighbor-joining method.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper were deposited in GenBank using the National Center for Biotechnology Information BankIt v3.0 submission tool (http://www3.ncbi.nlm.nih.gov/BankIt/) under accession numbers AF532202 (for the VP7 gene) and AF532203 (for the VP4 fragment).
RESULTS AND DISCUSSION
Case history and rotavirus antigen detection.
Immediately upon returning from a 1-month vacation to Mali in February 2002, a 13-month-old human immunodeficiency virus-negative child from a human immunodeficiency virus-seropositive Malian mother developed gastroenteritis with nausea, vomiting, and severe diarrhea. After 6 days of passing watery stools, progressive dehydration necessitated hospitalization for intravenous rehydration therapy. A stool specimen (B1711) was found positive for rotavirus using a commercial rotavirus antigen enzyme immunoassay (Premier Rotaclone). The further recovery of the child was uneventful, and she was able to leave the hospital after 3 days.
Multiplex rotavirus RT-PCR.
Our routine multiplex RT-PCR strategy (28) that allows detection and discrimination of rotavirus G1, G2, G3, G4, G8, and G9, failed to yield a type-specific rotavirus band with the B1711 sample. Since the general VP7 primers Beg9-End9 amplified a 1,062-bp rotavirus-specific band, this pointed to the presence of a non-G1/G2/G3/G4/G8/G9 rotavirus.
VP7 sequence analysis and determination of G type.
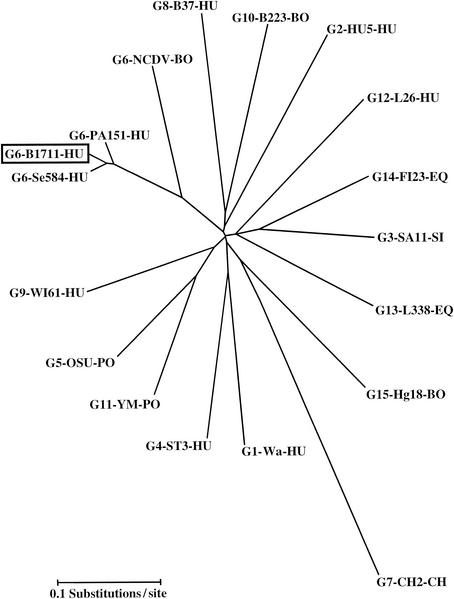
The partial nucleotide (1,007 bp) and deduced amino acid sequences of the VP7-encoding gene of the B1711 strain were determined (GenBank accession number AF532202), and compared with VP7 sequences of prototype strains belonging to G1 to G15 (Table 1 and Fig. 1). Sequence comparison indicated that the VP7 sequence of strain B1711 was most closely related to the VP7 of the G6 prototype strains, such as the American human G6 strain Se584 (96% identity at the nucleotide sequence level and 98% identity at the amino acid sequence level), the Italian human G6 strain PA151 (95% nucleotide and 98% amino acid similarity) and the bovine G6 NCDV strain (82% nucleotide and 90% amino acid similarity). Rotavirus strains representing other G types exhibited much less nucleotide and amino acid similarity (64 to 78% nt and 61 to 85% amino acid similarity) with our B1711 strain.
TABLE 1.
Nucleotide and amino acid sequence similarities of strain B1711 with VP7 sequences with different G genotype specificities
| G type | Strain | Origin | Similarity (%)
|
|
|---|---|---|---|---|
| Nucleotide | Amino acid | |||
| G1 | Wa | Human | 75 | 82 |
| G2 | HU5 | Human | 74 | 76 |
| G3 | SA11 | Simian | 76 | 85 |
| G4 | ST3 | Human | 75 | 79 |
| G5 | OSU | Porcine | 77 | 82 |
| G6 | NCDV | Bovine | 82 | 90 |
| G6 | PA151 | Human | 95 | 98 |
| G6 | Se548 | Human | 96 | 98 |
| G7 | Ch2 | Chicken | 64 | 61 |
| G8 | B37 | Human | 74 | 82 |
| G9 | WI61 | Human | 77 | 83 |
| G10 | B223 | Bovine | 75 | 82 |
| G11 | YM | Porcine | 77 | 83 |
| G12 | L26 | Human | 78 | 79 |
| G13 | L338 | Equine | 74 | 79 |
| G14 | FI23 | Equine | 74 | 80 |
| G15 | Hg18 | Bovine | 75 | 82 |
FIG. 1.
Neighbor-joining phylogenetic tree based on nucleotide sequences of the VP7-encoding genes for B1711 and other established rotavirus G types. The VP7 sequences were obtained from published reports and the GenBank database. The strains sequenced (and their GenBank accession numbers) are as follows: Wa (KO2033), HU5 (A01028), SA11 (K02028), ST3 (X13603), OSU (X04613), NCDV (M12394), PA151 (L20881), Se584 (AJ311740), CH2 (X56784), B37 (J04334), B223 (X57852), YM (M23194), L26 (M58290), L333 (D13549), FI23 (M61876), Hg18 (AF237666). The VP7 sequence of WI61 was obtained from Green et al. (29). Abbreviations: BO, bovine; HU, human; PO, porcine; EQ, equine; CH, chicken.
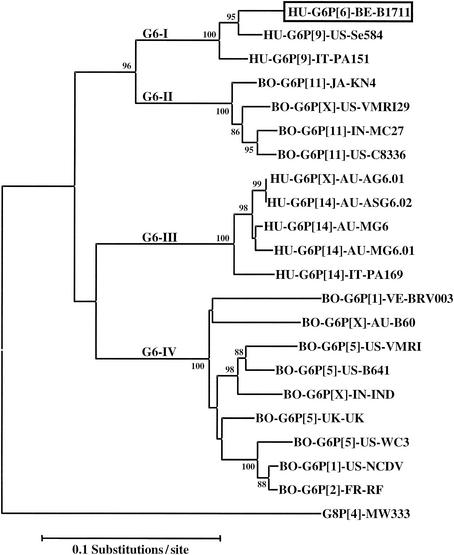
A more detailed phylogenetic analysis (Fig. 2) that included all known G6 VP7 sequences that were available in the GenBank database (release 130.0) confirmed that our B1711 strain clustered with the American human G6P[9] Se584 strain and the Italian human G6P[9] PA151 strain in a cluster of human G6 strains (cluster G6-I) that is related to a bovine cluster (cluster G6-II) of G6P[11] strains. Since the G6 sequence of the B1711 strain is located in a human cluster of the G6 phylogenetic tree, it is likely that the infection was the result of human-to-human transmission, rather than a recent bovine-to-human interspecies transmission event.
FIG. 2.
Phylogenetic tree based on nucleotide sequence of the VP7-encoding gene for B1711 and other G6 rotaviruses in GenBank. The VP7 sequences (and their GenBank accession numbers) are as follows: KN-4 (D12710), VMRI29 (U50332), MC27 (AF162435), C8338 (U14997), AG6.01 (AF207063), ASG6.02 (AF421183), MG6 (U22011), MG6.01 (AF207062), PA169 (L20880), BRV003 (U62154), B60 (M64680), IND (U15000), UK (M22306), WC3 (AY050272), NCDV (M12394), RF (X65940), MW333 (AJ278257). The number adjacent to the nodes represent the percentage of bootstrap support (of 1,000 replicates) for the clusters to the right of the node. Bootstrap values lower than 75% are not shown. Abbreviations: BO, bovine; HU, human.
Since G6 rotaviruses are the most prevalent genotype in cattle worldwide and are only rarely encountered in humans, it is likely that the G6 genotype is originally of bovine origin. Genetic analysis of the human PA151 isolate showed that at least 7 of the 11 RNA segments were of bovine origin (33). Other characterized G6 strains cluster in a separate human cluster G6-III (including G6P[14] PA169) and a bovine cluster G6-IV (including G6P[1] NCDV). It seems likely that there were two major natural bovine-human transmission events that led to the perpetuation of G6 strains in human populations. Since bovine and human G6 strains did not occur interspersed in the same cluster, this also suggested that interspecies transmission between bovines and humans is relatively uncommon. Since the isolates from the same cluster were found in different geographical regions on different continents (e.g., B1711 in Belgium-ex-Mali, Se584 in the United States, PA151 in Italy), it is likely that the two bovine-human transmission events are not of recent origin. Further molecular evolutionary studies might yield insights in the phylogenetic dating of these transmission events.
VP4 sequence analysis and determination of P type.
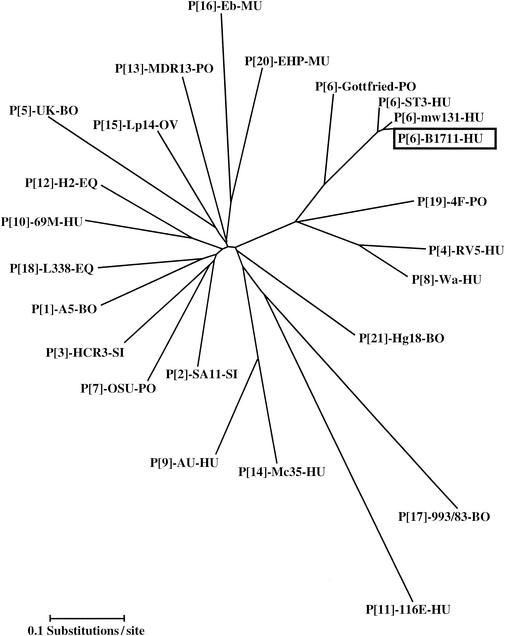
The partial VP4-encoding gene sequence (835 bp; accession number AF532203) of strain B1711 was compared with other established P genotypes (Table 2 and Fig. 3). Strain B1711 showed the highest similarity to rotavirus P[6] types, with the greatest identity to Malawi strain mw131 (97% nucleotide and 98% amino acid identity). Non-P[6] rotavirus strains showed only 58 to 72% nucleotide similarity or 42 to 72% amino acid similarity with the B1711 strain in the VP4 region.
TABLE 2.
Nucleotide and amino acid sequence similarity of strain B1711 with VP4 sequences with different P genotype specificities
| P type | Strain | Origin | Similarity (%)
|
|
|---|---|---|---|---|
| Nucleotide | Amino acid | |||
| P[1] | A5 | Bovine | 65 | 62 |
| P[2] | SA11 | Simian | 64 | 59 |
| P[3] | HCR3 | Human | 65 | 61 |
| P[4] | RV5 | Human | 70 | 68 |
| P[5] | UK | Bovine | 63 | 58 |
| P[6] | mw131 | Human | 97 | 98 |
| P[6] | Se585 | Human | 96 | 98 |
| P[6] | Gottfried | Porcine | 80 | 81 |
| P[7] | OSU | Porcine | 65 | 58 |
| P[8] | Wa | Human | 72 | 70 |
| P[9] | AU1 | Human | 63 | 70 |
| P[10] | 69M | Human | 66 | 70 |
| P[11] | KK-3 | Bovine | 60 | 55 |
| P[12] | H2 | Equine | 66 | 61 |
| P[13] | MDR13 | Porcine | 63 | 55 |
| P[14] | Mc35 | Human | 62 | 54 |
| P[15] | LP14 | Ovine | 66 | 61 |
| P[16] | Eb | Murine | 60 | 52 |
| P[17] | 993/83 | Bovine | 58 | 42 |
| P[18] | L338 | Equine | 68 | 61 |
| P[19] | 4F | Porcine | 71 | 72 |
| P[20] | EHP | Murine | 64 | 59 |
| P[21] | Hg18 | Bovine | 65 | 61 |
FIG. 3.
Neighbor-joining phylogenetic tree based on the nucleotide sequences of the VP8* fragments of the VP4 genes for B1711 and other established rotavirus P types. The VP4 sequences (and their GenBank accession numbers) are as follows: A5 (D13395), SA11 (X14204), HCR3 (L19712), RV5 (M32559), UK (M22306), mw131 (AJ427322), Se585 (AJ311737), ST3 (L33895), Gottfried (M33516), OSU (X13190), Wa (L34161), AU1 (D10970), 69 M (M60600), KK-3 (D14367), H2 (L04638), MDR13 (L07886), Mc35 (D14032), Lp14 (L11599), Eb (L18992), 993/83 (D16352), L338 (D13399), 4F (L10359), EHP (U08424), Hg18 (AF237665). Abbreviations: BO, bovine; HU, human; PO, porcine; EQ, equine; CH, chicken; SI, simian; OV, ovine; MU, murine.
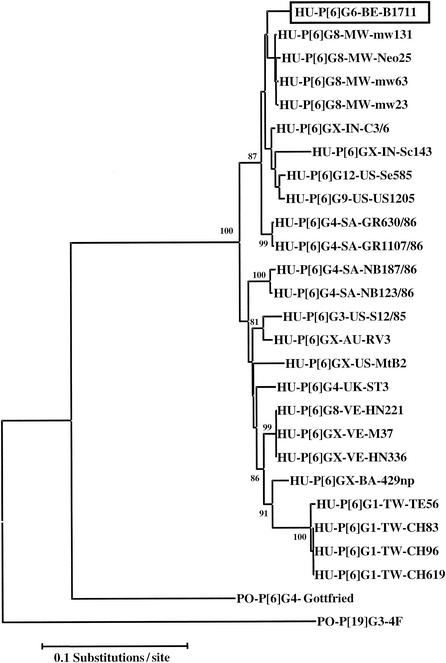
In a phylogenetic analysis of all known P[6] VP4 sequences that were available in the GenBank database (release 130.0) (Fig. 4), all human P[6] sequences clustered together and were only distantly related to the porcine Gottfried P[6]G4 strain.
FIG. 4.
Phylogenetic tree based on nucleotide sequences of VP8* fragments of VP4 genes for 1711 and other human P[6] rotaviruses. The VP4 sequences (and their GenBank accession numbers) are as follows: AC3/6 (U32619), Sc143 (AJ293721), US1205 (AF079356), mw23 (AJ278253), mw63 (AJ427320), Neo25 (AJ407319), Neo7 (AJ427318), GR630/86 (AF161825), GR1107/86 (AF161830), NB187/86 (AF161827), NB123/86 (AF161828), S12/85 (AF076925), RV3 (U16299), MtB2 (L25267), HN221 (L20878), M37 (L20877), HN336 (20879), 429np (U32166), TE56 (AF183869), CH619 (AF183867), CH83 (AF183864), CH96 (AF183865). The numbers adjacent to the nodes represent the percentage of bootstrap support (of 1,000 replicates) for the clusters to the right of the node. Bootstrap values lower than 75% were not shown. Abbreviations: BE, Belgium; IN, India; US, United States; MW, Malawi; SA, Republic of South Africa; UK, United Kingdom; AU, Australia; VE, Venezuela; BA, Bangladesh; TW, Taiwan.
Origin of the G6P[6] rotavirus strain.
The most-related VP7 sequences of the Belgian-ex-Mali G6P[6] B1711 strain are two G6P[9] strains from Italy and the United States (Fig. 2), and the most-related VP4 sequences are four Malawi G8P[6] strains (Fig. 4). G6P[6] rotaviruses might therefore be the result of a natural reassortment event between a G6P[9] strain with a G8P[6] strain, although reassortment between other G6- and P[6]-bearing rotaviruses cannot be ruled out.
Our patient had traveled to Mali in West Africa, and developed the rotavirus diarrhea immediately upon returning to Belgium. Rotavirus P[6] strains are currently a common P genotype in humans in African countries such as Guinea-Bissau, Nigeria, Ghana, Malawi, and South Africa but are uncommon in European countries such as France (1.3% during 1997 to 1998) (1, 6, 8, 16, 21, 52). These findings support the hypothesis that an African P[6] strain may be the donor of the VP4 gene of our strain.
Domestic animals and humans live closely together in many African rural regions and often share the same source of water, increasing the chance of animal-human transmission and mixed rotavirus infections. Bovine rotavirus G6 strains are the predominant genotype in African cattle and may have been the presumed donor of the VP7 gene in the G6P[6] strain that our patient carried to Belgium. It is most likely that, although isolated in Belgium, the B1711 isolate is a bona fide African rotavirus strain.
A study undertaken in Guinea-Bissau between 1996 and 1998, reported that 30% of the P[6] strains could not be G genotyped by the multiplex RT-PCR with primers that would have amplified G1, G2, G3, G4, G8, and G9 genotypes (21). It is possible that G6 rotaviruses constituted a major portion of these P[6] strains. Therefore, G6P[6] might already be prevalent in West Africa. It would be of interest to further characterize these nontypeable human West African strains.
The B1711 isolate is to our knowledge the first G6P[6] rotavirus described. Continued surveillance of rotavirus strains in both developed and developing countries, and in both humans and animals, will provide novel insights into the interspecies transmission processes of rotaviruses. Such molecular epidemiological studies will also be necessary in the planning, introduction, and postmarketing surveillance of rotavirus vaccines.
REFERENCES
- 1.Adah, M. I., A. Wade, and K. Taniguchi. 2001. Molecular epidemiology of rotaviruses in Nigeria: detection of unusual strains with G2P[6] and G8P[1] specificities. J. Clin. Microbiol. 39:3969-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert, Z. K., and M. C. Robert. 1996. Rotaviruses, p. 1657-1688. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 3.Altschul, S., F. W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Araujo, I. T., M. S. Ferreira, A. M. Fialho, R. M. Assis, C. M. Cruz, M. Rocha, and J. P. Leite. 2001. Rotavirus genotypes P[4]G9, P[6]G9, and P[8]G9 in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Clin. Microbiol. 39:1999-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armah, G. E., C. T. Pager, R. H. Asmah, F. R. Anto, A. R. Oduro, F. Binka, and D. Steele. 2001. Prevalence of unusual human rotavirus strains in Ghanaian children. J. Med. Virol. 63:67-71. [PubMed] [Google Scholar]
- 6.Asmah, R. H., J. Green, G. E. Armah, C. I. Gallimore, J. J. Gray, M. Iturriza-Gomara, F. Anto, A. Oduro, F. N. Binka, D. W. Brown, and F. Cutts. 2001. Rotavirus G and P genotypes in rural Ghana. J. Clin. Microbiol. 39:1981-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, R. F. 1996. Natural history of human rotavirus infection. Arch. Virol. 12:119-128. [DOI] [PubMed] [Google Scholar]
- 8.Bon, F., C. Fromantin, S. Aho, P. Pothier, E. Kohli, and the AZAY Group. 2000. G and P genotyping of rotavirus strains circulating in France over a three-year period: detection of G9 and P[6] strains at low frequencies. J. Clin. Microbiol. 38:1681-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browning, G. F., R. M. Chalmers, T. A. Fitzgerald, and D. R. Snodgrass. 1991. Serological and genomic characterization of L338, a novel equine group A rotavirus G serotype. J. Gen. Virol. 72:1059-1064. [DOI] [PubMed] [Google Scholar]
- 10.Browning, G. F., T. A. Fitzgerald, R. M. Chalmers, and D. R. Snodgrass. 1991. A novel group A rotavirus G serotype: serological and genomic characterization of equine isolate FI23. J. Clin. Microbiol. 29:2043-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buesa, J., C. O. de Souza, M. Asensi, C. Martinez, J. Prat, and M. T. Gil. 2000. VP7 and VP4 genotypes among rotavirus strains recovered from children with gastroenteritis over a 3-year period in Valencia, Spain. Eur. J. Epidemiol. 16:501-506. [DOI] [PubMed] [Google Scholar]
- 12.Chang, K. O., A. V. Parwani, and L. J. Saif. 1996. The characterization of VP7 (G type) and VP4 (P type) genes of bovine group A rotaviruses from field samples using RT-PCR and RFLP analysis. Arch. Virol. 141:1727-1739. [DOI] [PubMed] [Google Scholar]
- 13.Chang, K. O., A. V. Parwani, and L. J. Saif. 2000. Comparative sequence analysis of the VP7 genes of G6, G8 and G10 bovine group A rotaviruses and further characterization of G6 subtypes. Arch. Virol. 145:725-737. [DOI] [PubMed] [Google Scholar]
- 14.Cooney, M. A., R. J. Gorrell, and E. A. Palombo. 2001. Characterisation and phylogenetic analysis of the VP7 proteins of serotype G6 and G8 human rotaviruses. J. Med. Microbiol. 50:462-467. [DOI] [PubMed] [Google Scholar]
- 15.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999: predominance of novel P[6]G8 strains. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunliffe, N. A., S. Rogerson, W. Dove, B. D. Thindwa, J. Greensill, C. D. Kirkwood, R. L. Broadhead, and C. A. Hart. 2002. Detection and characterization of rotaviruses in hospitalized neonates in Blantyre, Malawi. J. Clin. Microbiol. 40:1534-1537. [DOI] [PMC free article] [PubMed]
- 17.Estes, M. K. 1996. Rotaviruses and their replication, p. 1625-1648. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 18.Estes, M. K., and J. Cohen. 1989. Rotavirus gene structure and function. Microbiol. Rev. 53:410-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falcone, E., M. Tarantino, L. Di. Trani, P. Cordioli, A. Lavazza, and M. Tollis. 1999. Determination of bovine rotavirus G and P serotypes in Italy by PCR. J. Clin. Microbiol. 37:3879-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang, Z. Y., H. Yang, J. Qi, J. Zhang, L. W. Sun, J. Y. Tang, L. Ma, Z. Q. Du, A. H. He, J. P. Xie, Y. Y. Lu, Z. Z. Ji, B. Q. Zhu, H. Y. Wu, S. E. Lin, H. P. Xie, D. D. Griffin, B. Ivanoff, R. I. Glass, and J. R. Gentsch. 2002. Diversity of rotavirus strains among children with acute diarrhea in China: 1998-2000 surveillance study. J. Clin. Microbiol. 40:1875-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, T. K., H. Steinsland, K. Molbak, R. Ca, J. R. Gentsch, P. Valentiner-Branth, P. Aaby, and H. Sommerfelt. 2000. Genotype profiles of rotavirus strains from children in a suburban community in Guinea-Bissau, Western Africa. J. Clin. Microbiol. 38:264-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerna, G., A. Sarasini, M. Parea, S. Arista, P. Miranda, H. Brussow, Y. Hoshino, and J. Flores. 1992. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J. Clin. Microbiol. 30:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerna, G., A. D. Steele, Y. Hoshino, M. Sereno, D. Garcia, A. Sarasini, and J. Flores. 1994. A comparison of the VP7 gene sequences of human and bovine rotaviruses. J. Gen. Virol. 75:1781-1784. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert, J. M., and H. B. Greenberg. 1997. Virus-like particle-induced fusion from without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J. Virol. 71:4555-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorziglia, M., K. Green, K. Nishikawa, K. Taniguchi, R. Jones, A. Z. Kapikian, and R. M. Chanock. 1988. Sequence of the fourth gene of human rotaviruses recovered from asymptomatic or symptomatic infections. J. Virol. 62:2978-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorziglia, M., G. Larralde, A. Z. Kapikian, and R. M. Chanock. 1990. Antigenic relationships among human rotaviruses as determined by outer capsid protein VP4. Proc. Natl. Acad. Sci. USA 87:7155-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green, K. Y., Y. Hoshino, and N. Ikegami. 1989. Sequence analysis of the gene encoding the serotype-specific glycoprotein (VP7) of two new human rotavirus serotypes. Virology 168:429-433. [DOI] [PubMed] [Google Scholar]
- 30.Griffin, D. D., T. Nakagomi, Y. Hoshino, O. Nakagomi, C. D. Kirkwood, U. D. Parashar, R. I. Glass, and J. R. Gentsch. 2002. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6],G12) and rare P3[9] strains related to bovine rotaviruses. Virology 294:256-269. [DOI] [PubMed] [Google Scholar]
- 31.Huang, J. A., H. S. Nagesha, and I. H. Holmes. 1993. Comparative sequence analysis of VP4s from five Australian porcine rotaviruses: implication of an apparent new P type. Virology 196:319-327. [DOI] [PubMed] [Google Scholar]
- 32.Husain, M., P. Seth, L. Dar, and S. Broor. 1996. Classification of rotavirus into G and P types with specimens from children with acute diarrhea in New Delhi, India. J. Clin. Microbiol. 34:1592-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iizuka, M., E. Kaga, M. Chiba, O. Masamune, G. Gerna, and O. Nakagomi. 1994. Serotype G6 human rotavirus sharing a conserved genetic constellation with natural reassortants between members of the bovine and AU-1 genogroups. Arch. Virol. 135:427-432. [DOI] [PubMed] [Google Scholar]
- 34.Iturriza-Gomara, M., J. Green, D. W. Brown, M. Ramsay, U. Desselberger, and J. J. Gray. 2000. Molecular epidemiology of human group A rotavirus infections in the United Kingdom between 1995 and 1998. J. Clin. Microbiol. 38:4394-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain, V., B. K. Das, M. K. Bhan, R. I. Glass, and J. R. Gentsch. 2001. Great diversity of group A rotavirus strains and high prevalence of mixed rotavirus infections in India. J. Clin. Microbiol. 39:3524-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang, G., J. Green, C. I. Gallimore, and D. W. Brown. 2002. Molecular epidemiology of rotaviral infection in South Indian children with acute diarrhea from 1995-1996 to 1998-1999. J. Med. Virol. 67:101-105. [DOI] [PubMed] [Google Scholar]
- 37.Kapikian, A. Z., Y. Hoshino, R. M. Chanock, and I. Perez-Schael. 1996. Efficacy of a quadrivalent rhesus rotavirus-based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young children. J. Infect. Dis. 174:65-72. [DOI] [PubMed] [Google Scholar]
- 38.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 39.Martella, V., A. Pratelli, G. Greco, M. Tempesta, M. Ferrari, M. N. Losio, and C. Buonavoglia. 2001. Genomic characterization of porcine rotaviruses in Italy. Clin. Diagn. Lab. Immunol. 8:129-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miren, I. G., J. Green, and J. Gray. 2000. Methods of rotavirus detection, sero- and genotyping, sequencing, and phylogenetic analysis. Methods Mol. Med. 34:189-216. [DOI] [PubMed] [Google Scholar]
- 41.Mummidi, S., P. S. Paul, and R. E. Holland. 1996. Sequence and phylogenetic analysis of the VP7 gene of a bovine rotavirus with G6 subtype. Virus Genes 12:203-204. [DOI] [PubMed] [Google Scholar]
- 42.Nicholas, K. B., H. B. Nicholas, and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. Embnet News 4:14. [Google Scholar]
- 43.O'Halloran, F., M. Lynch, B. Cryan, H. O'Shea, and S. Fanning. 2000. Molecular characterization of rotavirus in Ireland: detection of novel strains circulating in the population. J. Clin. Microbiol. 38:3370-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okada, J., T. Urasawa, N. Kobayashi, K. Taniguchi, A. Hasegawa, K. Mise, and S. Urasawa. 2000. New P serotype of group A human rotavirus closely related to that of a porcine rotavirus. J. Med. Virol. 60:63-69. [PubMed] [Google Scholar]
- 45.Okada, N., and Y. Matsumoto. 2002. Bovine rotavirus G and P types and sequence analysis of the VP7 gene of two G8 bovine rotaviruses from Japan. Vet. Microbiol. 84:297-305. [DOI] [PubMed] [Google Scholar]
- 46.Palombo, E. A., and R. F. Bishop. 1995. Genetic and antigenic characterization of a serotype G6 human rotavirus isolated in Melbourne, Australia. J. Med. Virol. 47:348-354. [DOI] [PubMed] [Google Scholar]
- 47.Parashar, U. D., J. S. Bresee, J. R. Gentsch, and R. I. Glass. 1998. Rotavirus. Emerg. Infect. Dis. 4:561-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao, C. D., K. Gowda, and B. S. Reddy. 2000. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 276:104-113. [DOI] [PubMed] [Google Scholar]
- 49.Santos, N., E. M. Volotao, C. C. Soares, M. C. Albuquerque, F. M. da Silva, T. R. de Carvalho, C. F. Pereira, V. Chizhikov, and Y. Hoshino. 2001. Rotavirus strains bearing genotype G9 or P[9] recovered from Brazilian children with diarrhea from 1997 to 1999. J. Clin. Microbiol. 39:1157-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sereno, M. M., and M. I. Gorziglia. 1994. The outer capsid protein VP4 of murine rotavirus strain Eb represents a tentative new P type. Virology 199:500-504. [DOI] [PubMed] [Google Scholar]
- 51.Snodgrass, D. R., T. Fitzgerald, I. Campbell, F. M. Scott, G. F. Browning, D. L. Miller, A. J. Herring, and H. B. Greenberg. 1990. Rotavirus serotypes 6 and 10 predominate in cattle. J. Clin. Microbiol. 28:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steele, A. D., M. C. van Niekerk, and M. J. Mphahlele. 1995. Geographic distribution of human rotavirus VP4 genotypes and VP7 serotypes in five South African regions. J. Clin. Microbiol. 33:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 37:1885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vende, P., R. Karoum, G. Manet, C. Rizet, F. Schelcher, J. Cohen, and H. Navetat. 1999. Molecular epidemiology of bovine rotaviruses from the Charolais area. Vet. Res. 30:451-456. [PubMed] [Google Scholar]