Abstract
Mutations within the Presenilin-2 (PS-2) gene are associated with early onset familial Alzheimer’s disease. The gene encodes a polytopic transmembrane protein that undergoes endoproteolytic processing resulting in the generation of N-terminal and C-terminal fragments (CTFs). PS-2 is also cleaved by proteases of the caspase family during apoptotic cell death. CTFs of PS-2 were shown to inhibit apoptosis, suggesting an important role in the regulation of programmed cell death. Recently, we found that the CTF of PS-2 is phosphorylated in vivo. We mapped the in vivo phosphorylation sites of PS-2 to serine residues 327 and 330, which are localized immediately adjacent to the cleavage sites of caspases after aspartate residues 326 and 329. Phosphorylation of PS-2 inhibits its cleavage by caspase-3. This effect can be mimicked by substitutions of serines 327 and 330 by aspartate or glutamate. In addition, the uncleavable form of PS-2 CTF was found to enhance its antiapoptotic properties, leading to a slower progression of apoptosis. These results demonstrate that PS-2 cleavage as well as its function in apoptosis can be regulated by protein phosphorylation. Alterations in the phosphorylation of PS-2 may therefore promote the pathogenesis of AD by affecting the susceptibility of neurons to apoptotic stimuli.
Alzheimer’s disease (AD) is defined by the invariable accumulation of senile plaques and neuronal loss in certain areas of the brain (1). Senile plaques are composed predominantly of the amyloid β-peptide (Aβ), which is derived from the β-amyloid precursor protein by proteolytic processing (2, 3). Whereas most Aβ molecules terminate at amino acid 40 (Aβ40), about 10% of the peptides are elongated by two additional amino acids at their C termini (Aβ42). The elongated form of Aβ aggregates much faster and induces neuronal cell death (4, 5). Mutations within the two homologous presenilin (PS) genes, PS-1 and PS-2, are a frequent cause of early onset familial AD (6–8). The mutations increase the production of Aβ42 (9–13), thus strongly supporting a central role of Aβ42 for AD pathology. PS proteins are membrane proteins with 6–8 transmembrane domains (14, 15), which are endoproteolytically processed within the large cytoplasmic loop (conventional proteolytic processing pathway) generating ≈20-kDa C-terminal (CTF) and ≈30-kDa N-terminal fragments (13, 16–18). The resulting fragments are the predominant forms in vivo, whereas very little if any full-length protein can be detected (13, 16–18). In addition, CTFs of PS-2, similar to those derived from proteolytic processing of the full-length protein, can also be generated in vivo by alternative transcription (19) and alternative splicing (20, 21). Increasing evidence suggests that the proteolytic fragments are biologically and pathologically active (22–26).
PS proteins are involved in apoptotic cell death (19, 27–29) and are substrates of proteases of the caspase superfamily (19, 30–32). PS-2 is cleaved by caspases during apoptosis between Asp-329 and Ser-330 and to a minor extent between Asp-326 and Ser-327 in vivo (Fig. 1a; refs. 19, 30, and 31). PS-1 is also cleaved by a member of the caspase superfamily after Asp-345 (31, 32). Caspases appear to be important for the progression of apoptotic cell death. These enzymes are activated by specific proteolysis, which occurs either autocatalytically or by other members of the caspase superfamily (33, 34). The activated enzymes then cleave a number of proteins. Studies analyzing the substrate specificity of caspases revealed that substrate recognition depends on the primary amino acid sequence N-terminal of the cleavage site as well as the critical aspartate residue at P1 (33–35).
Figure 1.
The PS-2 CTF is phosphorylated on serine residues 327 and 330 in vivo. (a) Amino acid sequence 300–348 of the PS-2 hydrophilic loop domain. The sites for conventional proteolytic processing (18) and the caspase-mediated cleavage (19, 30, 31) are indicated. Caspases cleave PS-2 predominantly between Asp-329 and Ser-330. A minor cleavage occurs between Asp-326 and Ser-327 (31). The in vivo phosphorylation sites identified in this study are shown in bold. (b) COS-7 cells were transiently transfected with cDNA constructs encoding either the PS-2 CTFwt, the indicated Ser → Ala substitutions, or a deleted derivative (36). Cells were labeled in vivo with [32P]orthophosphate, and phosphorylation was analyzed by autoradiography after transfer of proteins to a poly(vinylidene difluoride) membrane. (c) Expression of the respective cDNA constructs was monitored by immunoblotting with the monoclonal anti-Flag M2 antibody by using the same membrane as in b. Longer exposure times revealed the presence of PS-2 CTFcasp (the lower molecular weight CTF generated by caspase-mediated cleavage; data not shown), similar to that observed in Figs. 2 and 3.
In this study we mapped the in vivo phosphorylation sites of the PS-2 CTF to serine residues that are located immediately adjacent to the critical aspartate residues. Phosphorylation at these sites inhibits the caspase mediated cleavage of PS-2 in vitro and in vivo, demonstrating that caspase-mediated cleavage of PS-2 not only depends on its primary amino acid sequence N-terminal of the critical aspartate but also depends on its phosphorylation status. Furthermore, phosphorylation of the PS-2 CTF appears to enhance its antiapoptotic properties in cell culture.
MATERIALS AND METHODS
Cell Culture, Transfection, and Generation of Stable Cell Lines.
HeLa cells and COS-7 cells were maintained in DMEM containing Glutamax and 10% fetal calf serum (GIBCO). Cells were transfected by using Dotap (Boehringer Mannheim) according to the supplier’s instructions. HeLa cells stably expressing PS-2 cDNAs cloned in pcDNA3 vector (Invitrogen) were selected and maintained in the above mentioned medium containing 400 μg/ml G418 (GIBCO).
Construction of PS-2 cDNAs and Fusion Proteins.
Tagging of the PS-2 cDNA with a sequence encoding the Flag epitope at the C terminus and introduction of the respective point mutations by oligonucleotide-directed mutagenesis was carried out as described (36). The cDNAs encoding the respective PS-2 CTFs were generated by PCR using the full length PS-2 cDNA as template. Mutations were confirmed by DNA sequencing. The sequences of oligonucleotides used for mutagenesis in this study are available on request. All constructs were cloned into the pcDNA3 vector (Invitrogen). Fusionproteins of the respective PS-2 loop domains (amino acids 297–356) and maltose binding protein (MBP) were generated as described (36). The coding regions were amplified by PCR by using the primers 5′-CCGAATTCGGGATGGTGTGGACG-3′ and 5′-ACGCGTCGACCTCTTCTTCCAGCTC-3′, and the resulting fragments were cloned into the EcoRI/SalI restriction sites of pMAL-c2 (New England Biolabs). Fusion proteins were expressed in Escherichia coli DH5α and purified with amylose resin (New England Biolabs) according to the supplier’s instruction.
In Vivo Phosphorylation and Immunoprecipitation.
In vivo phosphorylation was carried out as described (36). Briefly, cells transiently expressing PS-2 cDNA constructs were incubated for 30 min in phosphate-free medium (GIBCO). The media were aspirated and fresh medium was added, containing 13 MBq/ml [32P]orthophosphate (Amersham), and cells were incubated for 2 h at 37°C. The cells were washed twice with ice-cold PBS and immediately lysed on ice with lysis buffer containing 1% Nonidet P-40 for 10 min. Cell lysates were centrifuged 10 min at 14,000 × g, and supernatants were immunoprecipitated with the specific anti-PS-2 antibody 3711 (36) or with the monoclonal M2 anti-FLAG antibody (Kodak). Immunoprecipitates were separated by using SDS/PAGE, transferred to poly(vinylidene difluoride) membranes (Imobilon, Millipore), and radiolabeled proteins were visualized by using autoradiography.
In Vitro Cleavage by Caspases.
PS-2 CTFs, immunoprecipitated from cell lysates, or 1 μg of the respective fusionprotein PS-2Loop-MBP were incubated for 4 h at 37°C in 25 μl of caspase assay buffer (20 mM Hepes/100 mM sodium chloride/10 mM DTT/10 mM magnesium chloride/1 mM EDTA/0.1% CHAPS/10% sucrose, pH 7.2) in the presence or absence of 20 ng of recombinant active caspase-3 (PharMingen). Reactions were terminated by the addition of SDS-containing electrophoresis sample buffer.
Induction of Apoptosis and Analysis of PS-2, poly(ADP-ribose) polymerase (PARP), and Caspase-3.
Apoptosis in HeLa cells was induced by treatment with staurosporine (STS) alone or in combination with okadoic acid (OA) as indicated. For analysis of caspase-mediated cleavage of PS-2, the protease inhibitor N-acetyl-leucinyl-leucinyl-norleucinal (LLnL) (50 μM) was added to the incubation media to protect the cleavage products from degradation (26). After various time periods, cells were lysed in sample buffer and subjected to SDS/PAGE. The PS-2 CTF and PARP were detected by immunoblotting using the monclonal M2 anti-Flag (Kodak) or a polyclonal anti-PARP antibody (Boehringer Mannheim). The p11 subunit of caspase-3 was detected by the polyclonal anti-CPP32 antibody p11 (Santa Cruz Biotechnology). Specifically bound antibodies were detected by using the enhanced chemiluminescence technique. For quantitation of PARP cleavage, Western blots were incubated with a 125I-labeled secondary anti-rabbit antibody (Amersham Pharmacia) and analyzed by using PhosphorImaging (Molecular Dynamics).
Measurement of DNA Fragmentation.
Quantitation of apoptotic cells was carried out with in situ detection of nuclear DNA fragmentation by using the TnT FragEL kit (Calbiochem). Apoptotic cells vs. nonapoptotic cells were counted in randomly selected fields by using a light microscope. Five to six fields (containing 35–120 cells) of the respective cell lines were analyzed in each experiment.
RESULTS
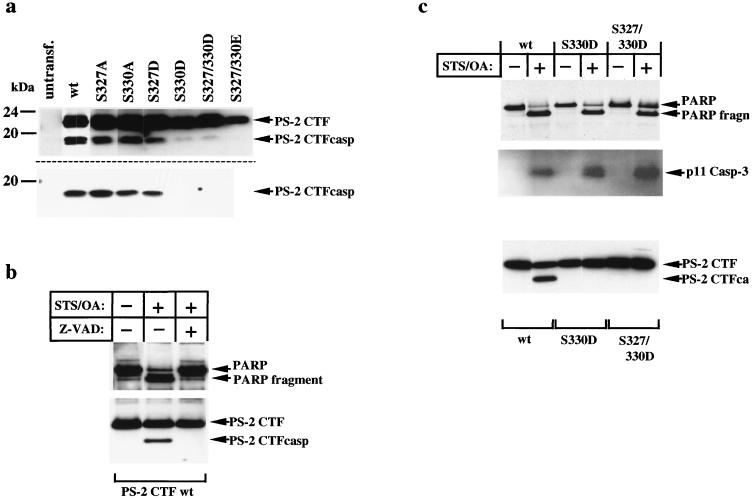
Recently, we demonstrated that the CTF of PS-2 is phosphorylated in vivo in the region between amino acids 327 and 335 in the large hydrophilic loop (ref. 36; see also Fig. 1a). This domain contains three potential phosphoryl acceptor sites. To identify the phosphorylation sites, we mutagenized serine to alanine at positions 327, 330, and 335 within recombinant cDNA constructs corresponding to the authentic PS-2 CTF. Expression of these constructs results in the generation of PS-2 CTFs similar to those produced in vivo by proteolytic processing, alternative splicing, and alternative transcription (13, 18–21). In vivo phosphorylation was analyzed in COS-7 cells that were transfected with the respective PS-2 cDNAs by labeling with [32P]orthophosphate. Cell lines such as COS, HeLa, and kidney 293 are frequently used to analyze proteolytic processing of presenilins and β-amyloid precursor protein. Results derived from biochemical experiments in such transfected cell lines were completely confirmed in neuronal cell lines as well as brain tissue (9–18, 22, 25, 30). The PS-2 CTFwt (wild type) and the PS-2 CTFS335A are phosphorylated to similar extents (Fig. 1b). In contrast, phosphorylation of PS-2 CTFS327A was reduced and that of CTFS330A was almost completely suppressed (Fig. 1b). As reported previously (36), the deletion mutant PS-2 CTFΔ327–335 completely abolished phosphorylation. Immunoblot analysis revealed that the constructs used were expressed at similar levels (Fig. 1c). Very similar data were obtained in experiments analyzing the phosphorylation of the respective PS-2 CTF derived from proteolytic processing of full length PS-2 (data not shown). These results indicate that serine residues 327 and 330 are in vivo phosphorylation sites of the PS-2 CTF. Both serine residues, Ser-327 and Ser-330, are located immediately adjacent to the essential Asp-326 and Asp-329, which are required for caspase recognition (Fig. 1a; refs. 30 and 31).
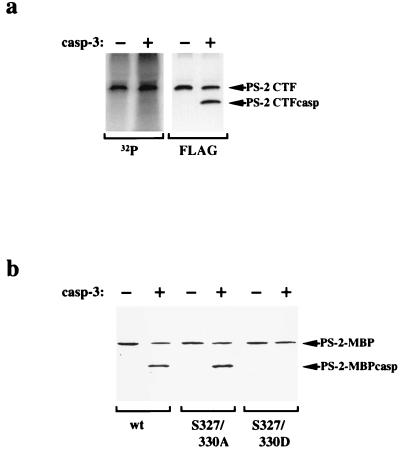
We therefore investigated whether phosphorylation of the PS-2 CTF at these sites could affect its cleavage by caspases. PS-2 CTFs were immunoprecipitated from HeLa cells labeled with [32P]orthophosphate in vivo and then incubated in the presence or absence of purified active caspase-3, which was shown previously to cleave PS-2 in vitro (19). Treatment with caspase-3 resulted in decreased amounts of PS-2 CTF and in the generation of a smaller cleavage product (PS-2 CTFcasp; Fig. 2a). In contrast, the phosphorylated form of PS-2 CTF did not decrease on treatment with caspase-3. Furthermore, PS-2 CTFcasp is not phosphorylated. These data indicate that caspase-3 cleaves selectively unphosphorylated forms of PS-2 CTF, whereas phosphorylated forms of PS-2 CTF are protected against caspase-mediated cleavage. To determine whether this effect of phosphorylation could be mimicked by substitution of the in vivo phosphorylation sites with a charged amino acid, we mutagenized serines 327 and 330 to aspartates. Fusion proteins of PS-2Loop and MBP (PS-2-MBP) were incubated in the presence or absence of caspase-3. As shown in Fig. 2b, fusion proteins carrying the PS-2 wt sequence or serine-to-alanine substitutions (S327/330A) are readily cleaved by caspase-3. In contrast, a fusion protein carrying the corresponding serine-to-aspartate substitutions (S327/330D) was not cleaved by caspase-3 in vitro. Therefore, the introduction of aspartate residues can be used to mimic phosphorylated serine residues as it has been shown for the functional analysis of other phosphoproteins (37, 38).
Figure 2.
Phosphorylation of PS-2 inhibits its cleavage by caspase-3. (a) HeLa cells stably expressing PS-2 CTFwt were labeled with [32P]orthophosphate, and the PS-2 CTFs were immunoprecipitated with the anti-Flag antibody. The immunoprecipitate was divided into two aliquots and incubated in the presence or absence of purified caspase-3. Proteins were separated by SDS/PAGE and transferred to a poly(vinylidene difluoride) membrane. Radiolabeled proteins were visualized by PhosphorImaging (Left). Subsequently, the same membrane was probed with the anti-Flag antibody (Right). Note that caspase-3 cleaves selectively the unphosphorylated form of PS-2 CTF. (b) Mimicking phosphorylation by substitution of serines 327 and 330 by aspartate residues also inhibits cleavage by caspase-3. Fusion proteins of MBP with the respective PS-2Loop domain (wt, S327/330A, or S327/330D) were incubated in the presence or absence of caspase-3. Full-length fusion proteins (PS-2-MBP) and cleavage products (PS-2-MBPCasp) were detected by immunoblotting with antibody 3711. Fusion protein carrying the PS-2Loop wt or the serine-to-alanine mutations were cleaved by caspase-3. In contrast, the introduction of negatively charged aspartate residues, which mimic phosphorylated serines, inhibits caspase-mediated cleavage. MBP itself was not cleaved by caspase-3 (data not shown). Similar results were obtained with purified active caspase-7 (although caspase-7 cleaves PS-2 with less efficiency than caspase-3; data not shown).
We next investigated whether mimicking phosphorylation by the introduction of negatively charged amino acids also protects PS-2 CTF from caspase-mediated cleavage in vivo. Serine residues 327 and 330 were substituted either by alanine residues to generate unphosphorylatable forms or by aspartate or glutamate residues. Transient overexpression of the PS-2 CTFwt resulted in the generation of the smaller CTF (CTFcasp), indicating proteolytic processing by caspases (Fig. 3a, Upper). This is consistent with previous results demonstrating the induction of apoptosis upon transient overexpression of PS-2 (28, 31). Substitution of Ser-327 or Ser-330 to alanine residues had no influence on this cleavage (Fig. 3a, Upper). In contrast, substitution of Ser-330 by aspartate (CTFS330D) efficiently inhibited the generation of CTFcasp, whereas substitution of Ser-327 (CTFS327D) reduced caspase-mediated cleavage to some extent (Fig. 3a, Upper). The substantially weaker effect of the S327D mutation is consistent with previous studies demonstrating that caspase-mediated cleavage occurs predominantly after Asp-329 (see Fig. 1a; refs. 30 and 31). The corresponding double mutations CTFS327/330D and CTFS327/330E almost completely inhibited caspase-mediated cleavage (Fig. 3a, Upper). Very similar data were obtained on transient overexpression of full-length PS-2wt or the corresponding serine mutations (Fig. 3a, Lower).
Figure 3.
Phosphorylation of PS-2 CTF inhibits caspase-mediated cleavage during apoptosis. (a) cDNA constructs encoding either the wt or the indicated phosphorylation site mutations of the PS-2 CTF (Upper) or the PS-2 full-length protein (Lower) were transiently transfected into COS-7 cells. PS-2 CTFs were immunoprecipitated with antibody 3711 and detected on immunoblots with the anti-Flag M2 antibody. (b) HeLa cells stably expressing the PS-2 CTFwt were incubated for 6 h either in the presence or absence of 1 μM STS/1 μM OA ± Z-VAD-fmk (50 μM). The combination of STS and OA induced apoptosis more potently than either drug alone (data not shown). As a control, cells were incubated with the carrier alone. PARP and the PS-2 CTF were detected by immunoblotting by using anti-PARP and anti-Flag antibodies, respectively. Note that on stable expression of PS derivatives, apoptosis is not induced, which is consistent with previous results (40, 41). (c) HeLa cells stably expressing PS-2 CTFwt, CTFS330D, or CTFS327/330D were incubated in the absence or presence of 1 μM STS/1 μM OA for 6 h. Analysis of PARP and PS-2 CTF was carried out as described in b. The p11 subunit of caspase-3, which is generated during its proteolytic activation (33, 35), is detected by immunoblotting using antibody anti-CPP32p11 (Santa Cruz Biotechnology). Note that capase-3 was activated in all three cell lines treated with STS/OA.
These results indicate that phosphorylation of the PS-2 CTF at serine residues 327 and 330, which are located immediately adjacent to the caspase recognition sites, inhibits caspase-mediated cleavage of PS-2.
To prove this phenomenon under defined apoptotic stimuli, HeLa cells stably expressing the PS-2 CTFwt were incubated in the presence or absence of STS and OA, two inducers of apoptosis (30–32, 39). In untreated cell cultures, no caspase-mediated cleavage of PS-2 CTFwt was observed (Fig. 3b). This is consistent with previous results (40, 41) demonstrating that stable expression of PS-2 does not induce apoptosis (in contrast to the high level expression on transient transfection; see also Fig. 3a). As a control, we analyzed caspase-mediated cleavage of PARP, a marker of apoptosis (30). No cleavage of PARP was observed in these cells, also demonstrating that apoptosis was not induced by stable overexpression of the PS-2 CTFwt under control conditions. On treatment with STS/OA for 6 h, the PS-2 CTFwt was cleaved to yield the smaller CTFcasp (Fig. 3b). Under these conditions, PARP was also cleaved, demonstrating that the cells underwent apoptosis. Cleavage of both PS-2 CTFwt and PARP was blocked by addition of the specific caspase inhibitor Z-VAD-fmk (Fig. 3b), indicating that PS-2 CTFwt is specifically cleaved by caspase activity during apoptosis. We then investigated the effect of substitutions that mimic phosphorylation on caspase-mediated cleavage of the PS-2 CTF in HeLa cells. Again, PS-2 CTFwt was readily cleaved on induction of apoptosis (Fig. 3c). In contrast, PS-2 CTF mutants mimicking either the single phosphorylated form (CTFS330D) or the double phosphorylated form (CTFS327/330D) were completely protected against caspase-mediated cleavage (Fig. 3c). PARP has been cleaved in each of these cell lines, demonstrating that the cells underwent apoptosis (Fig. 3c). Moreover, caspase-3, which has been demonstrated to cleave PS-2 in vitro (Fig. 2; ref. 19) also was activated under these conditions (as determined by the detection of the p11 cleavage product; Fig. 3c). These results therefore demonstrate that apoptotic cleavage of the PS-2 CTF is specifically inhibited during apoptosis by mimicking phosphorylation at its caspase recognition site.
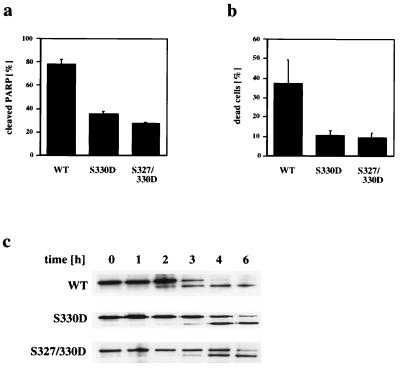
Because CTFs of PS-2 were shown to inhibit apoptosis (19, 27), we investigated the effect of phosphorylation of the PS-2 CTF on its apoptotic properties. Apoptosis was induced in HeLa cell lines stably expressing the PS-2 CTFwt or mutations mimicking phosphorylated PS-2 CTF (CTFS330D or CTFS327/330D) and was monitored either by analyzing the cleavage of PARP or the fragmentation of nuclear DNA. Quantitation of PARP cleavage 4 h after induction of apoptosis with STS revealed that ≈80% of cellular PARP was cleaved in cells stably transfected with PS-2 CTFwt (Fig. 4a). In contrast, in cells transfected with PS-2 CTFS330D or with CTFS327/330D, <40% of cellular PARP was cleaved (Fig. 4a). Quantitating apoptotic cell death by labeling nuclear DNA fragments in situ also demonstrated that expression of PS-2 CTFS330D or PS-2 CTFS327/330D inhibits apoptosis (Fig. 4b), whereas expression of the unphosphorylatable PS-2 CTFS330A had no significant effect (data not shown). As observed on prolonged treatment with STS, PARP also is cleaved in cells expressing PS-2 CTFs, which mimic phosphorylated molecules (see Fig. 3c). Therefore, to determine whether phosphorylation of the PS-2 CTF does not entirely block STS-induced apoptosis, but rather retards its progression, PARP cleavage was analyzed at various time points after induction of apoptosis. Cleavage of PARP occurred after about 2 h in HeLa cells expressing the PS-2 CTFwt. After 3 h, ≈50% of cellular PARP was cleaved, and after 4 h PARP was almost completely proteolyzed (Fig. 4c). In contrast, cells stably expressing PS-2 CTFS330D or CTFS327/330D, revealed a significant retardation of PARP cleavage (Fig. 4c).
Figure 4.
The phosphorylation site mutants inhibit the progression of apoptosis. (a) Four hours after induction of apoptosis, HeLa cells stably expressing PS-2 CTFwt, CTFS330D, or CTFS327/330D were lysed and cleavage of cellular PARP was analyzed by immunoblotting by using a 125I-labeled secondary antibody and quantitated by using PhosphorImaging. (b) Nuclear DNA fragmentation was analyzed 4 h after induction of apoptosis. Values represent means ± SD of triplicate experiments. The CTFwt itself showed a trend to inhibited apoptosis as compared with untransfected HeLa cells (data not shown), which is consistent with previous results (19, 27). (c) Cells were lysed after induction of apoptosis at the time points indicated. Cellular PARP was analyzed by immunoblotting. Note that caspase mediated cleavage of PARP is significantly delayed in cells expressing the PS-2 CTFS330D or CTFS327/330D as compared with cells expressing the CTFwt.
DISCUSSION
In this study, we demonstrate that phosphorylation of the PS-2 CTF on serine residues at the caspase recognition sites not only blocks its cleavage by caspases but also alters its functional properties during apoptosis.
Caspase recognition was previously shown to depend on the primary amino acid sequence preceding the essential aspartate residue (33–35). However, our findings indicate that phosphorylation of PS-2 on serine residues 327 and 330 can regulate apoptotic cleavage. Phosphorylation of the PS-2 loop domain clearly inhibits caspase-mediated cleavage in vitro and in vivo. Substitution of the in vivo phosphorylation sites of PS-2 by unphosphorylatable alanine residues did not inhibit its caspase-mediated cleavage. In contrast, mimicking phosphorylated residues by aspartate or glutamate substitutions inhibits cleavage of PS-2 by caspases during apoptosis. It is important to note that a number of other caspase substrates, such as retinoblastoma protein, fodrin, and focal adhesion kinase (33, 34), contain potential phosphorylation sites within their caspase-recognition motif. As demonstrated previously, the PS-1 CTF is also phosphorylated in vivo (42, 43). Interestingly, serine residue 346 adjacent to aspartate 345, which is required for caspase recognition of PS-1, is located within a consensus sequence for protein kinases (PK) A and C. Because the PS-1 CTF is known to be phosphorylated by PKA and PKC (42, 43), it is tempting to speculate that phosphorylation of PS-1 also regulates its cleavage by caspases. Phosphorylation of caspase substrates may therefore represent a novel in vivo regulatory mechanism to protect these proteins from proteolytic degradation. This conclusion is supported by a recent in vitro study, demonstrating the inhibition of caspase-mediated cleavage of IkBα by phosphorylation (44). Although the functional role of caspase cleavage during apoptosis remains to be clarified in detail, it is believed that it either activates proapoptotic or inactivates antiapoptotic proteins (33, 34). Antiapoptotic effects of the PS-2 CTF have been described previously (19, 27, 28). Because phosphorylation of the PS-2 CTF was found to protect against caspase-mediated cleavage, this will result in stabilization of the antiapoptotic protein. Indeed, we could demonstrate that cellular expression of PS-2 CTF mutant proteins, which mimic constitutively phosphorylated forms, results in a marked inhibition of apoptosis.
Increasing evidence suggests that protein phosphorylation/dephosphorylation plays a role in the regulation of apoptosis. Agents affecting the activities of protein kinases (e.g., STS) or phosphatases (e.g., OA), can modulate apoptotic cell death (45, 46). In addition, PKCδ (47), MEKK-1 (48), focal adhesion kinase (49), and PKC-related kinase-2 (50) are cleaved by caspases during apoptosis. Recently, it was found that protein phosphatase 2A is also activated by caspase-mediated cleavage during apoptosis (51), suggesting that changes in the phosphorylation status of certain proteins modulate the progression of apoptosis. Indeed, PS-2 could be such a candidate, because we demonstrated that its antiapoptotic properties depend on its phosphorylation status. To our knowledge this is the first example demonstrating that phosphorylation of a “death substrate” at the caspase recognition site modulates its (anti)apoptotic properties.
AD is characterized by massive neuronal cell loss in susceptible regions of the brain (52), and increasing evidence suggests that this neuronal loss may be initiated by apoptosis (53). Furthermore, it was shown that the Volga German mutation (N141I) of PS-2 enhances the proapoptotic activity of PS-2 (27). In addition, an increased level of the PS-2 caspase fragment was found in brain of a familial AD patient carrying the same mutation (30). Both results suggest that caspase-mediated cleavage of PS-2 plays an important role in AD. As found in this study, phosphorylation regulates the cleavage of PS-2 during apoptosis and enhances its antiapoptotic properties. Alterations in the phosphorylation of PS-2 could therefore influence the pathogenesis of AD by affecting the susceptibility of neurons for apoptotic stimuli. Thus, PS-2 may play a role not only in early-onset familial AD but in the development of sporadic AD as well.
Acknowledgments
We thank Drs. M. Peter, D. Teplow, and members of our laboratory for critically reading this manuscript. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB317) and funds from Boehringer Ingelheim.
ABBREVIATIONS
- Aβ
amyloid β-peptide
- AD
Alzheimer’s disease
- CTF
C-terminal fragment
- CTFcasp
caspase generated CTF
- MBP
maltose-binding protein
- PK
protein kinase
- PARP
poly(ADP-ribose) polymerase
- PS
presenilin
- STS
staurosporine
- OA
okadaic acid
- wt
wild type
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Selkoe D J. Neuron. 1991;6:487–498. doi: 10.1016/0896-6273(91)90052-2. [DOI] [PubMed] [Google Scholar]
- 2.Haass C, Selkoe D J. Cell. 1993;75:487–498. doi: 10.1016/0092-8674(93)90312-e. [DOI] [PubMed] [Google Scholar]
- 3.Checler F. J Neurochem. 1995;65:1431–1444. doi: 10.1046/j.1471-4159.1995.65041431.x. [DOI] [PubMed] [Google Scholar]
- 4.Jarret J T, Lansbury P T., Jr Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 5.Yankner B A. Neuron. 1996;16:921–932. doi: 10.1016/s0896-6273(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 6.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Nature (London) 1995;375:754–760. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 7.Levy-Lahad E, Wasco W, Poorkaj P, Romano D M, Oshima J, Pettingell W H, Yu C-E, Jondro P D, Schmidt S D, Wang K, et al. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 8.Rogaev E I, Sherrington R, Rogaeva E A, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, et al. Nature (London) 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 9.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird T D, Hardy J, Hutton M, Kukull W, et al. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 10.Borchelt D, Thinakaran G, Eckman C, Lee M, Davenport F, Ratovitsky T, Prada C-M, Kim G, Seekins S, et al. Neuron. 1996;17:1005–10013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 11.Duff K, Eckman C, Zehr C, Yu X, Prada C M, Perez-Tur J, Hutton M, Buee L, Harigaya Y, Yager D, et al. Nature (London) 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 12.Citron M, Westaway D, Xia W, Carlson G, Diehl T S, Levesque G, Johnson-Wood K, Lee M, Seubert P, Davies A, et al. Nat Med. 1997;3:67–72. doi: 10.1038/nm0197-67. [DOI] [PubMed] [Google Scholar]
- 13.Tomita T, Maruyama K, Saido T C, Kume H, Shinozaki K, Tokuhiro S, Capell A, Walter J, Grünberg J, Haass C, et al. Proc Natl Acad Sci USA. 1997;94:2025–2030. doi: 10.1073/pnas.94.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doan A, Thinakaran G, Borchelt D R, Slunt H H, Ratovitsky T, Podlisny M, Selkoe D J, Seeger M, Gandy S E, Price D L, Sisodia S S. Neuron. 1996;17:1023–1030. doi: 10.1016/s0896-6273(00)80232-9. [DOI] [PubMed] [Google Scholar]
- 15.DeStrooper B, Beullens M, Contreras B, Levesque L, Craessaerts K, Cordell B, Moechars D, Bollen M, Fraser P, St George-Hyslop P H, van Leuven F. J Biol Chem. 1997;272:3590–3598. doi: 10.1074/jbc.272.6.3590. [DOI] [PubMed] [Google Scholar]
- 16.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, et al. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 17.Kim T-W, Pettingell W H, Hallmark O G, Moir R D, Wasco W, Tanzi R E. J Biol Chem. 1997;272:11006–11010. doi: 10.1074/jbc.272.17.11006. [DOI] [PubMed] [Google Scholar]
- 18.Shirotani K, Takahashi K, Ozawa K, Kunishita T, Tabira T. Biochem Biophys Res Comm. 1997;240:728–731. doi: 10.1006/bbrc.1997.7730. [DOI] [PubMed] [Google Scholar]
- 19.Vito P, Ghayur T, D’Adamio L. J Biol Chem. 1997;271:31025–31028. doi: 10.1074/jbc.271.49.31025. [DOI] [PubMed] [Google Scholar]
- 20.Prihar G, Fuldner R A, Perez-Tur J, Lincoln S, Duff K, Crook R, Hardy J, Philips C A, Venter C, Talbot C, et al. NeuroReport. 1996;7:1680–1684. doi: 10.1097/00001756-199607080-00031. [DOI] [PubMed] [Google Scholar]
- 21.Grünberg J, Walter J, Eckman C, Capell A, Schindzielorz A, Younkin S, Mehta N, Hardy J, Haass C. NeuroReport. 1998;9:3293–3299. doi: 10.1097/00001756-199810050-00027. [DOI] [PubMed] [Google Scholar]
- 22.Lee M K, Borchelt D R, Kim G, Thinakaran G, Slunt H H, Ratovitski T, Martin L J, Kittur A, Gandy S, Levey A I, et al. Nat Med. 1997;3:756–760. doi: 10.1038/nm0797-756. [DOI] [PubMed] [Google Scholar]
- 23.Capell A, Grünberg J, Pesold B, Diehlmann A, Citron M, Nixon R, Beyreuther K, Selkoe D J, Haass C. J Biol Chem. 1998;273:3203–3211. doi: 10.1074/jbc.273.6.3205. [DOI] [PubMed] [Google Scholar]
- 24.Thinakaran G, Regard J B, Bouton C M L, Harris C L, Sabo S, Price D L, Borchelt D R, Sisodia S S. Neurobiol Dis. 1998;4:438–453. doi: 10.1006/nbdi.1998.0171. [DOI] [PubMed] [Google Scholar]
- 25.Yu G, Chen F, Levesque G, Nishimura M, Zhang D M, Levesque L, Rogaeva E, Xu D, Liang Y, Duthie M, et al. J Biol Chem. 1998;273:16470–16475. doi: 10.1074/jbc.273.26.16470. [DOI] [PubMed] [Google Scholar]
- 26.Steiner H, Capell A, Pesold B, Citron M, Kloetzel P-M, Selkoe D, Romig H, Mendla K, Haass C. J Biol Chem. 1998;273:32322–32331. doi: 10.1074/jbc.273.48.32322. [DOI] [PubMed] [Google Scholar]
- 27.Vito P, Lacaná E, D’Adamio L. Science. 1996;271:521–525. doi: 10.1126/science.271.5248.521. [DOI] [PubMed] [Google Scholar]
- 28.Wolozin B, Iwasaki K, Vito P, Ganjei J K, Lacana E, Sunderland T, Zhao B, Kusiak J W, Wasco W, D’Adamio L. Science. 1996;274:1710–1713. doi: 10.1126/science.274.5293.1710. [DOI] [PubMed] [Google Scholar]
- 29.Roperch J-P, Alvaro V, Prieur S, Tuynder M, Nemani M, Lethrosne F, Piouffre L, Gendron M-C, Israeli D, Dausset J, et al. Nat Med. 1998;4:835–838. doi: 10.1038/nm0798-835. [DOI] [PubMed] [Google Scholar]
- 30.Kim T-W, Pettingel W H, Jung Y-K, Kovacs D M, Tanzi R E. Science. 1997;277:373–376. doi: 10.1126/science.277.5324.373. [DOI] [PubMed] [Google Scholar]
- 31.Loetscher H, Deuschle U, Brockhaus M, Reinhardt D, Nelboeck, Mous J, Grünberg J, Haass C, Jacobsen H. J Biol Chem. 1997;272:20655–20659. doi: 10.1074/jbc.272.33.20655. [DOI] [PubMed] [Google Scholar]
- 32.Grünberg J, Walter J, Loetscher H, Deuschle U, Jacobsen H, Haass C. Biochemistry. 1998;273:3205–3211. doi: 10.1021/bi972106l. [DOI] [PubMed] [Google Scholar]
- 33.Nicholson D W, Thornberry N A. Trends Biochem Sci. 1997;22:299–306. doi: 10.1016/s0968-0004(97)01085-2. [DOI] [PubMed] [Google Scholar]
- 34.Cohen G M. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thornberry N A, Rano T A, Peterson E P, Rasper D M, Timkey T, Garcia-Calvo M, Houtzager V M, Nordstorm P A, Roy S, Vaillancourt J P, et al. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 36.Walter J, Grünberg J, Schindzielorz A, Haass C. Biochemistry. 1998;37:5961–5967. doi: 10.1021/bi971763a. [DOI] [PubMed] [Google Scholar]
- 37.Egelhoff T T, Lee R J, Spudich J A. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- 38.Huang W, Erikson R L. Proc Natl Acad Sci USA. 1994;91:8960–8963. doi: 10.1073/pnas.91.19.8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haldar S, Jena N, Crocce C M. Proc Natl Acad Sci USA. 1995;92:4507–4511. doi: 10.1073/pnas.92.10.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oyama F, Sawamura N, Kobayashi K, Morishima-Kawashima M, Kuramochi T, Ito M, Tomita T, Maruyama, Saido T-C, Iwatsubo T, et al. J Neurochem. 1998;71:313–322. doi: 10.1046/j.1471-4159.1998.71010313.x. [DOI] [PubMed] [Google Scholar]
- 41.Tomita T, Tokuhiro S, Hashimoto T, Aiba K, Saido T C, Maruyama K, Iwatsubo T. J Biol Chem. 1998;273:21153–21160. doi: 10.1074/jbc.273.33.21153. [DOI] [PubMed] [Google Scholar]
- 42.Seeger M, Nordstedt C, Petanceska S, Kovacs D M, Gouras G K, Hahne S, Fraser P, Levesque L, Czernik A J, St. George-Hyslop P, et al. Proc Natl Acad Sci USA. 1997;94:5090–5094. doi: 10.1073/pnas.94.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walter J, Grünberg J, Capell A, Pesold B, Schindzielorz A, Citron M, Mendla K, St. George-Hyslop P, Multhaup G, Selkoe D J, Haass C. Proc Natl Acad Sci USA. 1997;94:5349–5354. doi: 10.1073/pnas.94.10.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barkett M, Xue D, Horvitz H R, Gilmore T D. J Biol Chem. 1997;272:29419–29422. doi: 10.1074/jbc.272.47.29419. [DOI] [PubMed] [Google Scholar]
- 45.Morana S J, Wolf C M, Li J, Reynolds J E, Brown M K, Eastman A. J Biol Chem. 1996;271:118263–182271. doi: 10.1074/jbc.271.30.18263. [DOI] [PubMed] [Google Scholar]
- 46.Yan Y, Shay J W, Wright W E, Mumby M C. J Biol Chem. 1997;272:15220–15226. doi: 10.1074/jbc.272.24.15220. [DOI] [PubMed] [Google Scholar]
- 47.Emoto Y, Manome Y, Meinhard G, Kisaki H, Kharbanda S, Robertson M, Ghayur T, Wong W W, Kamen R, Weichselbaum R, Kufe D. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cardone M H, Salvesen G S, Widman C, Johnson G, Frisch S M. Cell. 1997;90:315–323. doi: 10.1016/s0092-8674(00)80339-6. [DOI] [PubMed] [Google Scholar]
- 49.Wen L-P, Fahrni J A, Troie S, Guan J-L, Orth K, Rosen G D. J Biol Chem. 1997;272:26056–26061. doi: 10.1074/jbc.272.41.26056. [DOI] [PubMed] [Google Scholar]
- 50.Cryns V L, Byun Y, Rana A, Mellor H, Lustig K D, Ghanem L, Parker P J, Kirschner M W, Yuan J. J Biol Chem. 1997;272:29449–29453. doi: 10.1074/jbc.272.47.29449. [DOI] [PubMed] [Google Scholar]
- 51.Santoro M F, Annand R R, Robertson M M, Peng Y-W, Brady M J, Mankovich J A, Hackett M C, Ghayur T, Walter G, Wong W W, Giegel D A. J Biol Chem. 1998;273:13119–13128. doi: 10.1074/jbc.273.21.13119. [DOI] [PubMed] [Google Scholar]
- 52.Gomez-Isla T, Price J L, McKeel D W, Morris J C, Growdon J H, Hyman B T. J Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson A, Su J H, Cotman C W. J Neurosci. 1996;16:1710–1719. doi: 10.1523/JNEUROSCI.16-05-01710.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]