Abstract
The DNA-dependent protein kinase (DNA-PK) consists of a heterodimer DNA-binding complex, Ku70 and Ku80, and a large catalytic subunit, DNA-PKcs. To examine the role of DNA-PKcs in lymphocyte development, radiation sensitivity, and tumorigenesis, we disrupted the mouse DNA-PKcs by homologous recombination. DNA-PKcs-null mice exhibit neither growth retardation nor a high frequency of T cell lymphoma development, but show severe immunodeficiency and radiation hypersensitivity. In contrast to the Ku70−/− and Ku80−/− phenotype, DNA-PKcs-null mice are blocked for V(D)J coding but not for signal-end joint formation. Furthermore, inactivation of DNA-PKcs leads to hyperplasia and dysplasia of the intestinal mucosa and production of aberrant crypt foci, suggesting a novel role of DNA-PKcs in tumor suppression.
Severe combined immunodeficiency (SCID) mice are hypersensitive to radiation, deficient in DNA double-strand break repair, and impaired in variable (diversity) joining [V(D)J] recombination. Recent studies strongly suggest that the SCID defect lies in the gene encoding the catalytic subunit of DNA-dependent protein kinase (DNA-PK) (1–3). DNA-PK is a serine/threonine kinase consisting of a 465-kDa catalytic subunit (DNA-PKcs) and a heterodimeric regulatory complex termed Ku, which is composed of a 70-kDa (Ku70) and an 86-kDa (Ku80) polypeptide. Although it is generally believed that Ku helps to recruit DNA-PKcs to DNA in vitro and is likely to be required for the physiological activation of DNA-PK at the site of DNA damage (4, 5), there is evidence at least in vitro that DNA-PKcs can itself bind to linear DNA fragments and become activated for kinase activity in the absence of Ku (6, 7). It has been shown that the SCID phenotype correlates with a nonsense mutation at Tyr-4046 in the extreme C-terminal region of the DNA-PKcs gene (8–10). This T to A transversion results in the substitution of an ocher termination codon and a loss of 83 amino acids from the extreme C-terminal end (9, 10). Therefore, one plausible reason for the “leaky” phenotype of SCID is that the truncated DNA-PKcs protein has activity that is weak but functionally sufficient for some T cell development. Recently, Jhappan et al. (11) generated homozygous mice from the transgenic mice harboring the yeast cAMP phosphodiesterase gene (designated the Sra5–1 or slip mouse). The Sra-1 homozygotes were found to be immunodeficient, lacking mature lymphocytes, suggesting that the transgene had integrated into a gene required for the normal development of T and B cells. The integration of the transgene was subsequently shown to occur directly into the DNA-PKcs locus, as suggested by chromosomal localization of the transgene, the complementation experiments with SCID mice, and the depleted levels of DNA-PK activity. The most striking difference from the SCID phenotype, however, is the strong predisposition to thymic lymphoblastic lymphomas that arise in slip mice with complete penetrance. In contrast, lymphomas develop in only about 15% of CB-17 SCID mice (12) and have not been reported for Ku80 null mice. Integration of these data to generate a global model for the role of the DNA-PK complex in tumorigenesis or tumor suppression is difficult. First, assuming that DNA-PK activity requires the assembly of Ku and DNA-PKcs on DNA breaks, then comparison between the Ku80−/− (no tumor development) and slip phenotype (100% penetrance of tumor development) suggests that DNA-PK kinase activity is not required for tumor suppression. Perhaps other distinct functions for this kinase molecule, independent of Ku, are involved in tumor suppression, inactivation of which leads to the predisposition of thymic lymphoma as seen in the slip mice. It is also plausible that in the generation of slip mice, the multiple copies of transgenes incorporated in the DNA-PKcs locus may affect the adjacent gene(s) expression, for example, by means of a methylation or positioning effect. One of these cis-activated/inactivated genes may function as an oncogene/tumor-suppressor gene.
To elucidate the function of the individual components of DNA-PK in vivo, we have previously generated Ku70−/− and Ku80−/− mice (1, 13, 16). In the present study, we disrupted the DNA-PKcs gene by means of homologous recombination. In the resultant DNA-PKcs−/− mice, T and B lymphocyte development was arrested and V(D)J coding-end rearrangement was deficient, but V(D)J signal-end joining ability was intact. DNA-PKcs-null mice exhibit neither growth retardation nor a high frequency of T cell lymphoma development. Furthermore, inactivation of DNA-PKcs leads to hyperplasia and dysplasia of the intestinal mucosa and production of aberrant crypt foci, suggesting a role of DNA-PKcs in tumor suppression.
MATERIALS AND METHODS
Targeted Disruption of DNA-PKcs and Generation of DNA-PKcs−/− Mice.
The mouse genomic DNA-PKcs gene was isolated from a sCos-I cosmid library constructed from a mouse strain 129 embryonic stem (ES) cell line. The targeting vector was constructed by substituting half of exon 3 and part of intron 3 with the PGK-neo gene. The targeting construct was linearized with NotI and transfected into CJ7 ES cells by electroporation. Four hundred clones were screened and eight positive pools were initially identified by PCR. One positive ES clone carrying the targeted mutation of DNA-PKcs was identified by second-round PCR and further confirmed by Southern blot analysis. This positive ES clone was injected into C57BL/6 blastocysts and surgically implanted into pseudopregnant females to generate chimeric mice. The chimeras were crossed with C57BL/6 females, resulting in five mice with germline transmission of seven males screened. The DNA-PKcs−/− mice were obtained by intercrossing DNA-PKcs+/− mice. CB-17 SCID mice were obtained from Taconic Farms.
The genotype of the mice was determined by PCR, which distinguishes the endogenous from the targeted DNA-PKcs allele. PCR reaction contains 1 μg genomic DNA; 0.6 μM (each) of primers MD-20: TATCCGGAAGTCGCTTAGCATTG; MD-21: AAGACGGTTGAAGTCAGAAGTCC; and POL-8: TTCACATACACCTTGTCTCCGACG; 0.2 mM (each) dNTP; 1.5 mM MgCl2 and 2.5 units of Taq polymerase. Primers MD-20 and MD-21 give a product of the wild-type allele that is 264 bp; primers MD-20 and Pol-8 yield a product of the targeted allele that is 360 bp.
Establishment of Primary and SV40-Transformed Cell Lines.
Primary lung fibroblast cells were isolated from 4-week-old DNA-PKcs wild-type (+/+), heterozygous (+/−), homozygous (−/−) mice and a CB-17 SCID mouse. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2 in air by using alpha-MEM medium supplemented with 10% fetal calf serum, 100 unit/ml penicillin and 100 μg/ml streptomycin. SV40-transformed lung fibroblasts were obtained by transfecting the SV40 T antigen expression plasmid by using a calcium phosphate transfection system (Catalogue no. 18306–019, GIBCO/BRL).
Reverse Transcription–PCR (RT-PCR), Western Blot Analysis, and in Vitro Kinase Assay.
For RT-PCR assay, total RNA was prepared from SV40-transformed lung fibroblast cells by using the Qiagen RNeasy kit (Qiagen, Chatsworth, CA). After digestion of contaminated genomic DNA by DNase I (Ambion, Austin, TX), cDNA synthesis was carried out with the Superscript preamplification system (GIBCO/BRL) according to the included protocol. PCR primers used for RT-PCR were MD-3: ATCAGAAGGTCTAAGGCTGGAAT, MD-5: CGTACGGTGTTGGCTACTGC for amplification between exons 1 and 4 of DNA-PKcs, MD-28: CACTGAGGGCTTTCCGCTCTTGT, MD-29: GCTCTTGTGCACGAATGTTGTAG for PI-3 kinase domain, and GA-5: AGAAGACTGTGGATGGCCCC, GA-3: AGGTCCACCACCCTGTTGC for control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplification.
Whole-cell extracts were prepared as described previously (15). Protein concentration of the extracts was determined by Bradford analysis by using BSA as a standard. Western blotting analysis of DNA-PKcs and Ku70 was performed as described previously (1, 16) by using the DNA-PKcs monoclonal antibody [42–26] (17) and anti-mouse Ku70 goat-polyclonal antibody M-19 (Santa Cruz Biotechnology).
Histology, Cell Preparation, and Flow Cytometric Analysis.
To determine the pathological changes, histological sections of various organs of DNA-PKcs−/− and wild-type littermate mice were prepared and examined as previously described (16, 18). For flow cytometry, single-cell suspensions from lymphoid organs of 4- to 9-week-old mutants, their littermate controls, and CB-17 SCID mice were prepared for staining as described previously (16) and analyzed on FACScan with cell quest software (Becton Dickinson). Cells were stained with combinations of phycoerythrin (PE)-labeled anti-CD4 and fluorescein isothiocyanate (FITC)-labeled anti-CD8, or PE-labeled anti-B220 and FITC-labeled anti-CD43, or FITC-anti-IgM and PE-anti-B220 (PharMingen), as needed. Bone marrow (BM) cells were harvested from femurs by syringe lavage, and cells from thymus and spleen were prepared by homogenization. Cells were collected and washed in PBS plus 5% fetal calf serum and counted by using a hemacytometer. Samples from individual mice were analyzed separately. Dead cells were gated out by forward and side-scatter properties. Experiments were performed at least three times and yielded consistent results.
DNA Preparation and Analysis of V(D)J Recombination Products.
T cell antigen receptor (TCR) and Ig recombination in T and B lymphocytes were measured by amplifying rearranged DNA fragments by using PCR. Genomic DNAs were isolated from thymus, spleen, and BM from 4- to 9-week-old DNA-PKcs heterozygous (+/−), homozygous (−/−), and SCID mice. Oligonucleotides for PCR primers and probes are as follow. For TCRβ Vβ8-Jβ2 rearrangement (16), Vβ8.1: GAGGAAAGGTGACATTGAGC, Jβ2.6: GCCTGGTGCCGGGACCGAAGTA, and Vβ8 probe: GGGCTGAGGCTGATCCATTA. For TCRδ Dδ2-Jδ1 rearrangement (16), DR6: TGGCTTGACATGCAGAAAACACCTG, DR53: TGAATTCCACAGTCACTTGGGTTC, and DR2 probe: GACACGTGATACAAAGCCCAGGGAA. For TCRδ Dδ2-Jδ1 signal joint (19), DR21: GTCATATCTTGTCCAGTCAACTTCC, DR162:GATGAGCCAGCTGGATGAGTAACAC, and DR161 probe: GCCCTCTAGCCATGACATCAGAGC. For Ig VH7183-JH4 rearrangement (19), DR214: CGCGAAGCTTCGTGGAGTCTGGGGGA, DR217: GGGGAATTCCTGAGGAGACGGTGACT, and DR218 probe: ACCCCAGTAGTCCATAGCATAGTAAT. For control GAPDH amplification, the same primers were used as for the RT-PCR experiment. Probe DNA for mouse GAPDH was purchased from Ambion (Catalogue no. 7330). Amplified PCR products were resolved on 2% of agarose gel in 0.5× TBE (45 mM Tris/45 mM boric acid/1 mM EDTA, pH 8.3) and transferred to Hybond N+ nylon membrane. By using radiolabeled oligonucleotide or DNA probes, PCR products were hybridized and visualized by autoradiography.
Radiation Survival Assays.
Survival curves for each cell line were obtained by measuring the colony-forming ability of irradiated cell populations. Cells were plated on 60-mm plastic Petri dishes and irradiated with 137Cs γ-rays at the rate of 2.2 Gy/min to achieve a cumulative dose of 1, 2, 3, or 5 Gy 2 hr after plating. After 7 days, cells were fixed and stained with 1% crystal violet in a 70% ethanol solution, colonies that contained more than 20 cells were scored, and the mean value for triplicate culture dishes was determined. Cell survival was normalized to plating efficiency of untreated controls for each cell type.
RESULTS
Targeted Disruption of the DNA-PKcs Gene.
To determine the roles of DNA-PKcs in vivo, we targeted DNA-PKcs in mice by means of homologous recombination. The DNA-PKcs gene was inactivated by substituting the 3′-half of the exon 3 and part of the intron 3 with the PGK-neo gene (Figs. 1 A and B). Mice heterozygous for the targeted DNA-PKcs allele did not show any detectable defects compared with wild-type littermates. These PKcs+/− heterozygotes were subsequently bred with each other, generating PKcs−/− homozygotes in 25% of the offspring. Therefore, disruption of the DNA-PKcs gene did not result in embryonic lethality. Adult PKcs−/− mice are fertile and give comparable litter size (about six pups) relative to PKcs+/− or PKcs+/+ mice (about eight pups). In contrast to the 50% smaller body size of Ku70−/− and Ku80−/− mice (13, 16), PKcs−/− mice were about the same size as their PKcs+/− and PKcs+/+ littermates.
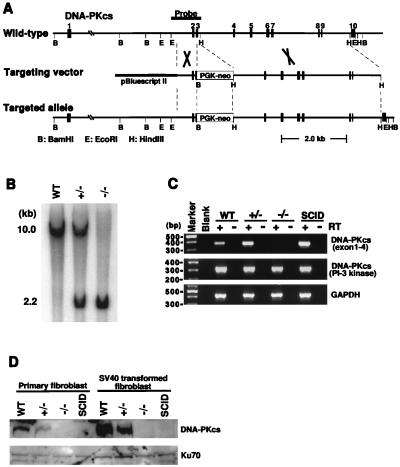
Figure 1.
Inactivation of DNA-PKcs by homologous recombination. (A) Schematic diagram of the murine DNA-PKcs locus from exons 1 to 10 and hybridization probe (Top), the targeting construct (Middle), and the targeted allele. BamHI (B), EcoRI (E), and HindIII (H) restriction sites are indicated. (B) Southern blot of the BamHI-digested tail DNA from control wild-type (WT), heterozygous (+/−), and homozygous (−/−) DNA-PKcs-targeted mice. The wild-type and mutant fragments are 10 and 2.2 kb, respectively. (C) RT-PCR of 5′-(exon 1–4) and 3′-(PI-3 kinase domain) regions of DNA-PKcs RNA from wild-type, DNA-PKcs-targeted, and SCID mouse cells. Total RNA was isolated from SV40-transformed lung fibroblast cells. PCR reactions were performed with (+) or without (−) reverse transcriptase (RT). RT-PCR for GAPDH was performed to ensure RNA integrity. (D) Western blot analysis of the various cells. Whole-cell extracts were prepared from primary and SV40-transformed lung fibroblast cells. Anti-DNA-PKcs monoclonal antibody and anti-Ku70 polyclonal antibody were used for detection. Note that there is another gene, MCM4, which is located about 700 bp upstream of DNA-PKcs (32). The transcription of DNA-PKcs and MCM4 is independently controlled by two distinct promoters located in this 700-bp region. We have carefully designed the DNA-PKcs knockout vector in exon 3, which is about 10 kb away from the promoter region, thus avoiding any possibility of interfering with the expression of the MCM4 gene. Furthermore, we have shown that aberrant DNA-PKcs mRNA is expressed in DNA-PKcs−/− mice, confirming that the promoter region of the DNA-PKcs gene is not affected by our knockout construct.
To confirm that the disruption produced a null mutation, DNA-PKcs mRNA and protein expression were analyzed by RT-PCR, Western blotting, and in vitro DNA-PK kinase assay. It is clearly shown in Fig. 1C that the RT-PCR products between exon 1 and exon 4 were absent in DNA-PKcs−/− cells. DNA-PKcs immunoreactivity was undetectable (Fig. 1D), and there was no kinase activity in DNA-PKcs−/− fibroblasts (data not shown). The levels of the DNA-binding component, the Ku70 and Ku80 proteins, were similar to that of the wild-type controls (Fig. 1D and data not shown).
Development of T and B Lymphocytes Is Blocked at the Early Stage in DNA-PKcs−/− Mice.
To determine whether there were specific pathological changes in the targeted mice, we examined the histology of various organs (Fig. 2A). With the exception of their lymphoid organs and gastrointestinal tract, DNA-PKcs−/− mice appeared normal. Spleen and lymph nodes were disproportionately smaller by 5- to 10-fold relative to controls and were devoid of lymphocytes. The DNA-PKcs−/− thymus was also disproportionately smaller, had no cortical-medullary boundary, and contained 50- to 100-fold fewer thymocytes than wild-type littermates (2–6 × 106 and 2 × 108, respectively). In addition, the gut-associated lymphoid tissue, specialized structures called Peyer’s patches in the small intestine, was drastically reduced or absent.
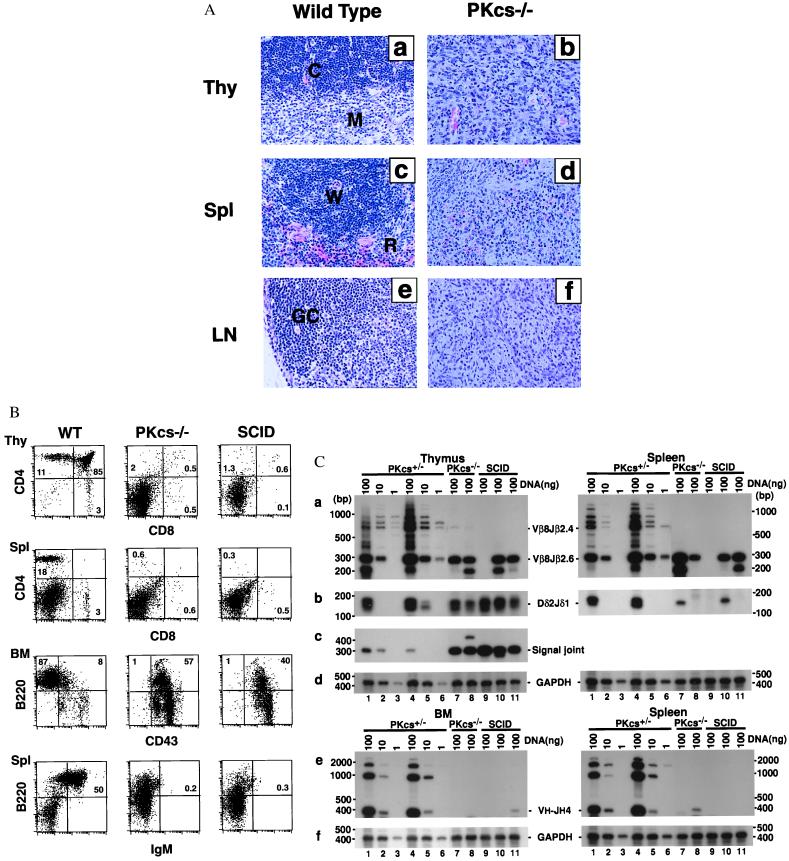
Figure 2.
Development of lymphocytes is blocked at early stages in DNA-PKcs−/− mice. (A) Histological analysis of thymus (Thy), spleen (Spl) and lymph node (LN) from wild-type and DNA-PKcs−/− mice (×200). Tissue sections were stained with hematoxylin and eosin. In tissue samples from DNA-PKcs-deficient mice, we observed effacement of normal histology and replacement by immature cells. The abbreviations are as follows: C, cortex; M, medulla; W, white pulp; R, red pulp; GC, germinal center. (B) Flow cytometric analysis of cells from the thymus (Thy), bone marrow (BM), and spleen (Spl) for the presence of precursor and mature T and B cells. Thymocytes and splenocytes were stained with fluorochrome-conjugated antibodies to CD4 and CD8; splenocytes and BM cells were stained with fluorochrome-conjugated antibodies to B220 and IgM or CD43. Profiles shown are representative results from a 4- to 5-week-old DNA-PKcs−/− mouse, its heterozygous littermate, and an age-matched CB-17 SCID mouse. (C) TCR and Ig gene rearrangement in DNA-PKcs−/− mice. (a) TCRβ rearrangement by PCR analysis. Thymus and spleen DNA were assayed for recombination of Vβ8-Jβ2.6. Both the quantity and the diversity of TCRβ rearrangement were reduced in DNA-PKcs−/− and SCID mice. (b) Coding joint of TCRδ rearrangement. Thymus and spleen DNA were assayed for recombination of Dδ2-Jδ1. (c) Signal joint of TCRδ rearrangement. Thymus DNA was assayed for Dδ2-Jδ1 circular signal joint products. There is more amplified signal for both DNA-PKcs−/− and SCID mice than for heterozygous control mice. (e) Ig heavy-chain rearrangement by PCR analysis. BM and spleen DNA were used for recombination of VH7183-JH4. Rearrangement in DNA-PKcs−/− and SCID is severely reduced in both BM and spleen. (d) and (f) Control GAPDH amplification from thymus, spleen, and BM DNA. DNA (100, 10, or 1 ng) from thymus, spleen, and BM or from a 5-week-old DNA-PKcs+/− mouse (lanes 1–3), of a 9-week-old DNA-PKcs+/− mouse (lane 4–6), and 100 ng DNA of three individual DNA-PKcs−/− mice (lanes 7–9) and three individual SCID mice (lanes 10–12). DNA-PKcs−/− and SCID mice analyzed were also between 4 and 9 weeks of age.
To examine the immunological defect in DNA-PKcs−/− mice, cells from thymus, BM, and spleen were labeled with monoclonal antibodies specific for lymphocyte surface markers and analyzed by using multiparameter flow cytometry. Consistent with the histological data, there was a complete absence of mature B cells in the spleen (Fig. 2B). Examination of the BM showed that B-cell development was blocked at early progenitor B220+ CD43+ stage.
DNA-PKcs−/− thymus displayed variable contents of cells expressing CD4+CD8+ thymocytes (1–7%), although CD4−CD8− cells usually made up the majority population (≈95%). The spleen cells from DNA-PKcs−/− mice contained detectable CD4+ single positive T cells (1–5%), which was slightly more than that reported for SCID mice. Taken together, the immunological phenotype in DNA-PKcs−/− mice closely resembles that of SCID, but differs from those of Ku80−/− and Ku70−/− mice (13, 16). In terms of successful T cell development, the rank order is wild type, Ku70−/−, DNA-PKcs−/−, SCID, Ku80−/−, with Ku80−/− being the most deficient.
T Cell Receptor and Ig Gene Rearrangement.
To determine whether a null mutation in DNA-PKcs affects rearrangements of antigen-receptor gene segments in T and B lymphocytes in vivo, DNA from the BM was amplified with primers specific to Ig V-DJH rearrangements, and DNA from the thymus was amplified with primers that detect V-DJβ and Dδ-Jδ rearrangements (Fig. 2C). Similar to that found in SCID mice, V-DJH rearrangements were not detected in DNA-PKcs−/− B cells, possibly accounting for the absence of mature B cells in these mutant mice.
DNA-PKcs −/− T cells in the thymus and spleen do undergo Dδ2-Jδ1 recombination at a level that is similar to that found in SCID mice and in the heterozygous littermates. However, the V-DJβ rearrangements were significantly reduced in both quantity and diversity (Fig. 2C). Signal joint formation of Dδ-Jδ rearrangements in both DNA-PKcs−/− and SCID mice shows, however, much higher signals than control heterozygous littermates. In conclusion, our results demonstrate that DNA-PKcs is required for coding, but not for signal joint formation in mice, a phenotype that closely resembles that found in SCID mice but is distinctly different from the Ku70−/− or Ku80−/− mice.
Absence of DNA-PKcs Confers Radiation Hypersensitivity.
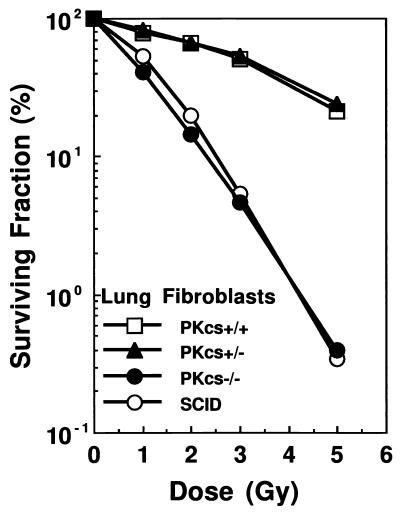
To demonstrate that inactivation of DNA-PKcs leads to hypersensitivity to ionizing radiation, monolayers of DNA-PKcs−/−, DNA-PKcs+/−, and DNA-PKcs+/+ lung fibroblasts were exposed to graded doses of γ-irradiation (0–5 Gy), and survival was determined by the colony formation assay. Fig. 3 clearly shows that DNA-PKcs−/− cells were much more radiosensitive than were the heterozygous and wild-type controls, with a >100-fold difference in survival after 5 Gy of γ-irradiation. The radiation dose–response curve of DNA-PKcs−/− cells was, however, nearly identical to that of the SCID lung fibroblast cells.
Figure 3.
Radiation-dose response of DNA-PKcs−/− cells. Clonogenic survival was measured on SV40-transformed mouse lung fibroblasts irradiated with graded doses of ionizing radiation. DNA-PKcs−/− cells show similar sensitivity to ionizing radiation as SCID and are much more sensitive than wild-type (+/+) and heterozygous (+/−) cells.
Preneoplastic Lesions in DNA-PKcs−/− Mice.
Recently, Jhappan et al. (11) reported that the integration of a transgene at the DNA-PKcs locus resulted in strong predisposition to thymic lymphoblastic lymphomas, which arise in slip mice with complete penetrance. To examine whether our DNA-PKcs−/− mice are also susceptible to tumor development, we randomly assigned litters arising from heterozygous intercrosses (e.g., PKcs+/+, PKcs+/−, and PKcs−/−) as well as homozygous crosses and monitored the mice daily for tumor development and survival. None of the DNA-PKcs+/+ (n = 59) and DNA-PKcs+/− (n = 102) littermates developed tumors through an observation period of 12 months. Among 120 DNA-PKcs−/− mice, only three developed thymic lymphomas between 3 to 12 months of age, in sharp contrast to the observation in slip mice.
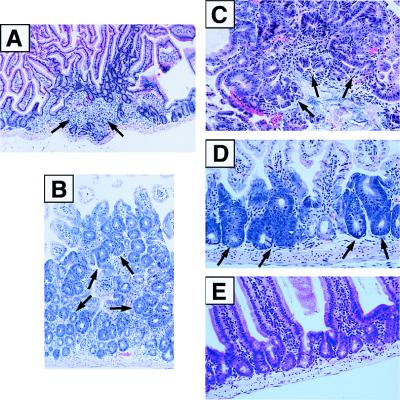
Autopsy examination of the lower gastrointestinal tract revealed the lack of mature Peyer’s patches in DNA-PKcs−/− mice. In addition, we observed an increase in cellularity in the colonic glands, which was confirmed by the Ki67 proliferative index (data not shown). In each of 21 randomly selected healthy DNA-PKcs−/− mice (ages between 1 and 6 months), we found intestinal segments with inflammatory infiltrates composed of polymorphonuclear cells, resulting in histopathological changes compatible with inflammatory polyps (Fig. 4A). In addition, in 15 of these 21 null mice, we detected the presence of hyperplastic polypoid lesions, composed of well differentiated colonic epithelial cells with foci of mild to moderate dysplasia (Fig. 4B). In eight cases we found areas of moderate to severe dysplasia. In the cases with severe dysplasia, we further identified areas of loose connective tissue stroma and dysplastic cells intruding the core of the stalk, suggesting invasion into the lamina propria (Fig. 4C). Furthermore, three of these cases revealed segments of colonic mucosa replaced by flat lesions composed of dysplastic cells, reminiscent of the so-called aberrant crypt foci (Fig. 4D). In two of these cases, these changes were observed along all intestines, including the small bowel.
Figure 4.
Preneoplastic lesions in DNA-PKcs−/− mice. Intestinal tissue samples from 6-week- to 6-month-old DNA-PKcs−/− mice were sectioned, stained with hematoxylin and eosin, and photographed. (A) Section of intestinal tissue showing inflammation and mild epithelial hyperplasia (×100). (B) Photomicrograph of colonic mucosa showing crypt hyperplasia with mild to moderate dysplasia (×200). (C) Adenomatous polyp of the colon showing areas of severe dysplasia (×400). (D) Aberrant crypt foci along the intestinal mucosa showing severe dysplasia (×400). (E) Section of intestinal tissue from a wild-type mouse showing normal morphology (×250).
DISCUSSION
In summary, we carried out targeted disruption of the DNA-PKcs−/− gene in mice by means of homologous recombination. In the resultant DNA-PKcs−/− mice, both T and B lymphocyte development was arrested at early progenitor stages, V(D)J coding-end rearrangement deficient, but V(D)J signal-end joining ability intact. DNA-PKcs−/− fibroblasts are hypersensitive to radiation and deficient in the repair of DNA double-strand breaks (data not shown). Taken together, our data conclusively demonstrate that DNA-PKcs−/− is essential for the development of T and B lymphocytes. We have also provided direct and definitive genetic evidence that the SCID phenotype is caused by the alteration of DNA-PKcs protein. The striking similarity between DNA-PKcs−/− and SCID mice in terms of their lymphocyte development and V(D)J recombination suggests that the “leaky” phenotype frequently observed in the lymphocyte development of SCID mice may not be due to the “leakiness” of DNA-PKcs expression. Thus, there may exist alternate, perhaps less efficient, pathways in V(D)J recombination and lymphocyte development.
Of significant interest are three other findings. First, during a 12-month observation period, only three of 120 DNA-PKcs−/− mice developed thymic lymphoma. This low frequency of thymic lymphoma is similar to that observed in Ku80−/− and SCID mice, but distinctly different from Ku70−/− mice and slip mice in which the DNA-PKcs locus was disrupted by the integration of a transgene (11). The marked difference between DNA-PKcs−/− (with a less than 3% incidence of spontaneous tumor development) and slip mice (which show strong predisposition to thymic lymphoblastic lymphomas) raises a question for the role of DNA-PKcs in lymphomagenesis. Although DNA-PKcs plays a crucial role in DNA double-strand break repair and V(D)J recombination, our data suggest that the DNA-PK catalytic subunit is not essential for T cell tumor suppression. Differences in genetic background are unlikely to contribute to the different phenotypes of Ku70−/−, Ku80−/−, and DNA-PKcs−/− in the development of tumors. All of our Ku70−/−, Ku80−/−, and DNA-PKcs−/− strains were in a mixed 129/SV × C57BL/6 background and were generated in the transgenic mouse core facility at Memorial Sloan–Kettering Cancer Center by using identical protocols. Furthermore, an independently derived line of DNA-PKcs−/− mice had a phenotype essentially identical to that we described (20). And to date the propensity for lymphoma development has not been reported in DNA-PKcs-deficient mice generated by means of targeted disruption (20, 21).
Second, that DNA-PKcs−/− mice are able to carry out signal-end rejoining and exhibit no growth retardation, in contrast to Ku70−/− and Ku80−/− animals (13, 16, 22), strongly suggests that Ku proteins may have functions in V(D)J recombination and DNA damage repair that are independent of DNA-PKcs.
Third, and perhaps most interesting, is the propensity of DNA-PKcs−/− mice for development of hyperplastic polyps and aberrant crypt foci in the intestine. These changes are considered preneoplastic lesions and carcinoma in situ-like lesions in carcinogen-treated rodents and in humans with a high risk for developing colorectal malignancy (23–27). Our results clearly show that inactivation of DNA-PKcs leads to hyperplasia, dysplasia of intestinal mucosa, and production of aberrant crypt foci, suggesting a role of DNA-PKcs in tumor suppression.
Carcinogenesis is a complex multistep process, involving multiple events occurring at molecular, cellular, and morphological levels. Because colon tumors evolve through well-defined morphological stages, an elegant model for colorectal tumorigenesis has been established (26). The development of colorectal tumors appears to be initiated by mutations at the APC tumor-suppressor gene, which leads to the formation of benign adenomas. Sequential mutations in RAS, DCC, and p53 tumor-suppressor genes appear to complete the process, which finally results in progression from the benign to malignant state. Recent studies of two distinct hereditary syndromes, familial adenomatous polyposis (FAP) and hereditary nonpolyposis colorectal cancer (HNPCC) (27), suggest that the genetic defect in familial adenomatous polyposis affects the rate of tumor initiation by disrupting the “gatekeeper” function of the APC gene. In contrast, the defect in HNPCC largely affects tumor progression by targeting the genome guardian function of DNA mismatch repair genes (MMR). It is plausible that mutation in DNA-PKcs, in addition to alterations in the APC gene, may affect the initiation of a colorectal tumor or result in a predisposition to such tumors. Alternatively, defects in DNA-PKcs may affect tumor progression, a “caretaker” role similar to that proposed for the MMR genes. It has been shown that DNA-PKcs phosphorylates many transcription factors in vitro (28–31), suggesting the involvement of DNA-PKcs in transcription regulation. Although it remains to be proven, it is likely that the potential tumor-suppressor function of DNA-PKcs may be related to the transcription control activity of this kinase molecule. Further investigations should reveal how DNA-PKcs exerts its effect and why mutations in different components of the DNA-PK complex result in discrete phenotypes.
Acknowledgments
We thank T. Deloherey for the FACS analysis, P. Krechmer for word processing, and C. C. Ling and Z. Fuks for valuable suggestions and support. This work was supported in part by National Institutes of Health Grants CA-31397, CA-56909, and CA-78497 (G.C.L.) and CA-50519 (D.J.C.) and Department of Energy Office of Health and Environmental Research (D.J.C.). H. Ouyang is a postdoctoral fellow supported in part by National Institutes of Health Training Grant CA-61801.
ABBREVIATIONS
- DNA-PK
DNA-dependent protein kinase
- SCID
severe combined immunodeficiency
- V(D)J
variable (diversity) joining
- DNA-PKcs
catalytic subunit of DNA-dependent protein kinase
- ES
embryonic stem
- RT-PCR
reverse transcription–PCR
- TCR
T cell antigen receptor
- BM
bone marrow
References
- 1.Peterson S R, Kurimasa A, Oshimura M, Dynan W S, Bradbury E M, Chen D J. Proc Natl Acad Sci USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, et al. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 3.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 4.Chan D W, Lees-Miller S P. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 5.Suwa A, Hirakata M, Takeda Y, Jesch S A, Mimori T, Hardin J A. Proc Natl Acad Sci USA. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaneva M, Kowalewski T, Lieber M R. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammarstein O, Chu G. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danska J S, Holland D P, Mariathasan S, Williams K M, Guidos C J. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Araki R, Fujimori A, Hamatani K, Mita K, Saito T, Mori M, Fukumura R, Morimyo M, Muto M, Itoh M, et al. Proc Natl Acad Sci USA. 1997;94:2438–2443. doi: 10.1073/pnas.94.6.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blunt T, Gell D, Fox M, Taccioli G E, Lehman A R, Jackson S P, Jeggo P A. Proc Natl Acad Sci USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jhappan C, Morse III H C, Fleischmann R D, Gottesman M M, Merlino G. Nat Genet. 1997;17:483–486. doi: 10.1038/ng1297-483. [DOI] [PubMed] [Google Scholar]
- 12.Custer R P, Bosma G C, Bosma M J. Am J Pathol. 1985;120:464–477. [PMC free article] [PubMed] [Google Scholar]
- 13.Nussenzweig A, Chen C, da Costa Soares V, Sanchez M, Sokol K, Nussenzweig M C, Li G C. Nature (London) 1996;382:551–555. doi: 10.1038/382551a0. [DOI] [PubMed] [Google Scholar]
- 14.Li G C, Ouyang H, Li X, Nagasawa H, Little J B, Chen D J, Ling C C, Fuks Z, Cordon-Cardo C. Mol Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- 15.Peterson S R, Stackhouse M, Waltman M J, Chen F, Sato K, Chen D J. J Biol Chem. 1997;272:10227–10231. doi: 10.1074/jbc.272.15.10227. [DOI] [PubMed] [Google Scholar]
- 16.Ouyang H, Nussenzweig A, Kurimasa A, da Costa Soares V, Li X, Cordon-Cardo C, Li W, Cheong N, Nussenzweig M, Iliakis G, et al. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhu C, Bogue M A, Lim D-S, Hasty P, Roth D B. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver D T, Alt F W. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 21.Taccioli G E, Amatucci A G, Beamish H J, Gell D, Xiang X H, Torres Arzayus M I, Priestly A, Jackson S P, Rothstein A M, Jeggo P A, et al. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 22.Gu Y, Seidl K J, Rathbun G A, Zhu C, Manis J P, van der Stoep N, Davidson L, Cheng H-L, Sekiguchi J M, Frank K, et al. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 23.Moen C J A, van der Valk M A, Bird R P, Augustinus A M, Demant P. Cancer Res. 1996;56:2382–2386. [PubMed] [Google Scholar]
- 24.Roncussi L, Pedroni M, Fante R, Di Gregorio C, Ponz de Leon M. Cancer Res. 1993;53:3726–3729. [PubMed] [Google Scholar]
- 25.Bird R P. Cancer Lett. 1995;93:55–71. doi: 10.1016/0304-3835(95)03788-X. [DOI] [PubMed] [Google Scholar]
- 26.Vogelstein B, Kinzler K W. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 27.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 28.Yumoto Y, Shirakawa H, Yoshida M, Suwa A, Watanabe F, Teraoka H. J Biochem (Tokyo) 1998;124:519–527. doi: 10.1093/oxfordjournals.jbchem.a022143. [DOI] [PubMed] [Google Scholar]
- 29.Peterson S R, Jesch S A, Chamberlin T N, Dvir A, Rabindran S K, Wu C, Dynan W S. J Biol Chem. 1995;20:1449–1454. doi: 10.1074/jbc.270.3.1449. [DOI] [PubMed] [Google Scholar]
- 30.Pan Z Q, Amin A A, Gibbs E, Niu H, Hurwitz J. Proc Natl Acad Sci USA. 1994;91:8343–8347. doi: 10.1073/pnas.91.18.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson C W. Trends Biochem Sci. 1993;18:433–437. doi: 10.1016/0968-0004(93)90144-c. [DOI] [PubMed] [Google Scholar]
- 32.Saito T, Matsuda Y, Ishii H, Watanabe F, Mori M, Hayashi A, Araki R, Fujimori A, Fukumura R, Morimyo M, et al. Mamm Genome. 1998;9:769–772. doi: 10.1007/s003359900861. [DOI] [PubMed] [Google Scholar]