Abstract
Lactobacilli are common inhabitants of the gastrointestinal tracts of mammals and have received considerable attention due to their putative health-promoting properties. Little is known about the traits that enhance the ability of these bacteria to inhabit the gastrointestinal tract. In this paper we describe the development and application of a strategy based on in vivo expression technology (IVET) that enables detection of Lactobacillus reuteri genes specifically induced in the murine gut. A plasmid-based system was constructed containing ′ermGT (which confers lincomycin resistance) as the primary reporter gene for selection of promoters active in the gastrointestinal tract of mice treated with lincomycin. A second reporter gene, ′bglM (β-glucanase), allowed differentiation between constitutive and in vivo inducible promoters. The system was successfully tested in vitro and in vivo by using a constitutive promoter. Application of the IVET system with chromosomal DNA of L. reuteri 100-23 and reconstituted lactobacillus-free mice revealed three genes induced specifically during colonization. Two of the sequences showed homology to genes encoding xylose isomerase (xylA) and peptide methionine sulfoxide reductase (msrB), which are involved in nutrient acquisition and stress responses, respectively. The third locus showed homology to the gene encoding a protein whose function is not known. Our IVET system has the potential to identify genes of lactobacilli that have not previously been functionally characterized but which may be essential for growth of these bacteria in the gastrointestinal ecosystem.
The gastrointestinal tracts of vertebrate animals are colonized by a complex microflora containing several hundred different species, some of which are present at high levels (29). Bacteria belonging to the genus Lactobacillus are common inhabitants of this ecosystem and have received considerable attention due to their putative health-promoting properties when they are ingested as probiotics (19, 36). Lactobacillus species comprise only a minor part of the bacterial community in human feces (19, 25), but in animals such as pigs, chickens, mice, and rats lactobacilli are the predominant bacteria in proximal regions of the gut. They can also be detected as minority members of the large bowel ecosystem in these animals (19). In order to persist in the gastrointestinal tract, which is a lotic and highly competitive ecosystem, the bacteria must have appropriate multiplication rates and metabolic activities. Ecological studies have suggested that only a minority of the lactobacilli found in intestinal samples meet these requirements and are truly autochthonous, but the bacterial factors that allow lactobacilli to become established and persist in the gastrointestinal tract are unknown (22, 34, 37).
In the last decade, promoter-trapping technologies have been developed to overcome the limitations of in vitro models for studying the traits that enhance ecological performance in complex ecosystems. In vivo expression technology (IVET) was developed by Mahan and coworkers (16) to study gene expression of the pathogen Salmonella enterica serovar Typhimurium during infection of mice. IVET has been used to identify in vivo induced (ivi) genes for a number of other pathogens (for a review see reference 21), and mutations within a subset of these ivi genes resulted in a decrease in virulence. IVET has been scarcely used to study bacterial ecology apart from the pioneering studies of the adaptation of Pseudomonas fluorescens to the plant rhizophere conducted by Rainey (20).
In this paper we describe the use of IVET to study Lactobacillus reuteri 100-23 in the gastrointestinal tract of reconstituted lactobacillus-free (RLF) mice (32). This Lactobacillus strain originated in the digestive tract of a rat (38), colonizes the murine digestive tract at a high level, and is amenable to genetic modification by electrotransformation (17). It harbors an indigenous plasmid, pGT232, whose replication region confers stable maintenance of recombinant plasmids in lactobacilli inhabiting the gastrointestinal tract of the murine host (11). Based on the replication region of pGT232, a plasmid-based IVET system was developed that allowed identification of three promoters that are specifically induced upon colonization of the mouse intestine in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli was grown at 37°C in Luria-Bertani medium (23). Lactobacilli were cultured anaerobically at 37°C in MRS medium (Difco Laboratories). Solidified media contained 1.5% (wt/vol) agar. When required, ampicillin was added to the culture media at a concentration of 50 μg/ml, erythromycin was added at a concentration of 500 μg/ml (E. coli) or 100 μg/ml (lactobacilli), and chloramphenicol was added at a concentration of 10 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5αF′ | Cloning host | 5 |
| L. reuteri 100-23 | Rodent gastrointestinal isolate | 38 |
| L. reuteri 100-23C | Plasmid-free derivative of L. reuteri 100-23 | 17 |
| L. sakei LTH 677 | Source of ldhL promoter, formerly designated strain Ls8 | 9 |
| L. reuteri JW100 | L. reuteri 100-23C harboring pJW100 | This study |
| L. reuteri JW200 | L. reuteri 100-23C harboring pJW200 | This study |
| Plasmids | ||
| pUC29 | Cloning vector, Apr, 2.7 kb | 1 |
| pGT232 | Indigenous cryptic plasmid from L. reuteri 100-23, 5.1 kb | 31 |
| pFX3 | Source of chloramphenicol resistance gene (cat-194), Cmr, 4.3 kb | 40 |
| pTZ19-Mac | Source of endo-1,3-1,4-β-glucanase gene (bglM), Apr, 3.7 kb | 7 |
| pTRK95 | Source of erythromycin resistance gene (ermGT), Apr Cmr Emr, 7.5 kb | 33 |
| p29T | pUC29 containing the rrnBT1T2 terminator cassette, Apr, 3.2 kb | 10 |
| pUC29cat | pUC29 in which the β-lactamase gene is replaced by cat-194, Cmr, 3.3 kb | This study |
| p29cat232 | pUC29cat containing the 2.7 kb SalI-NcoI fragment from pGT232, Cmr, 5.6 kb | This study |
| p29cat232small | p29cat232 with deletion of the nonessential 0.5-kb SalI fragment, Cmr, 5.1 kb | This study |
| p29Tbglm | p29T containing the promoterless bglM gene, Apr, 4.0 kb | This study |
| p29TIVET | p29Tbglm containing the promoterless ermGT gene, Apr, 4.8 kb | This study |
| pJW100 | Promoter trap vector, Cmr, 7.4 kb | This study |
| pJW200 | Derivative of pJW100 containing the ldhL promoter in multiple cloning site, Cmr, 7.5 kb | This study |
Genetic techniques.
DNA manipulations and agarose gel electrophoresis were performed by using standard protocols described by Sambrook et al. (23). Plasmid DNA from E. coli and lactobacilli were isolated with a QIAprep Spin Miniprep kit (Qiagen). For lactobacilli, cells in 6 ml of an overnight culture were harvested by centrifugation (9,000 × g for 3 min) and washed with 1 ml of 10 mM Tris-HCl (pH 8.0). The cell pellets were resuspended in 250 μl of buffer P1 containing lysozyme (10 mg/ml) and mutanolysin (250 U/ml) and were incubated at 37°C for 1 h. Further steps were performed according to the manufacturer's instructions. Restriction endonucleases, Klenow DNA polymerase (for end-filling reactions), and T4 DNA ligase were purchased from Roche and used according to the supplier's recommendations. Recombinant DNA molecules were introduced into E. coli and lactobacilli by electrotransformation (4, 17).
Construction of the promoter trap vector pJW100.
To construct the basic Lactobacillus-E. coli shuttle vector p29cat232small, the β-lactamase (Apr) gene of plasmid pUC29 (removed by digestion with DraI and SspI) was replaced with the 1.5-kb blunt-ended ClaI-Sau3AI fragment of pFX3 containing the chloramphenicol resistance (cat-194) gene to create pUC29cat. To make the plasmid functional in L. reuteri 100-23C, the lacZ promoter of pUC29cat was removed by digestion with PvuII and replaced with the blunt-ended 2.7-kb SalI-NcoI fragment from pGT232, which contained the double-strand origin of replication, the gene encoding the replication initiation protein (repA), and the putative single-strand origin necessary for stable maintenance of plasmid constructs in vivo (11). The resulting plasmid was designated p29cat232. As a unique SalI site was required for subsequent steps in construction of the IVET vector, p29cat232 was digested with SalI and religated to remove a 0.5-kb nonessential fragment, yielding p29cat232small. The functionality of p29cat232small was tested by electrotransformation of L. reuteri 100-23C.
For construction of the IVET cassette, the promoterless endo-1,3-1,4-β-glucanase-encoding bglM gene of Bacillus macerans (′bglM) was amplified by using primers bglm1forw (5′-TACTCGAGT CGACCCGGGTGATCATAGATAGCTAGTCATTTTGGAGGTGTATTATG-3′) and bglm2rev (5′-AGCTGGATCCATGCCCAGCTGCAATC-3′) with plasmid pTZ19-Mac as the template DNA. Primer bglm1forw contained recognition sites for the restriction endonucleases XhoI, SalI, SmaI, and BclI (italics), stop codons in all three reading frames (boldface type), and the ribosome-binding site (RBS) (underlined) of bglM. Primer bglm2rev contained a BamHI restriction site (italics). Amplification was carried out with a GeneAmp 9600 thermocycler (Perkin-Elmer). The reaction mixture (50 μl) contained 25 pmol of each primer, each deoxyribonucleoside triphosphate at a concentration of 0.2 mM, 1× reaction buffer, 2.5 U of Pwo polymerase (Roche), and 1 μl of template DNA. The amplification parameters were as follows: initial denaturation at 94°C for 2 min; 30 cycles of 94°C for 15 s, 52°C for 30 s, and 72°C for 1 min; and final extension at 72°C for 7 min. The resulting PCR product was purified with a QIAquick PCR purification kit (Qiagen), digested with XhoI and BamHI, and cloned into the corresponding sites of plasmid p29T, generating p29Tbglm. To complete the IVET cassette, the promoterless erythromycin resistance gene (′ermGT) was amplified by using primers ermGT1forw (5′-CATCGGATCCAAAAAGGAGAATTAATATATGGGG-3′) and ermGT2rev (5′-AATCAAGCTTCAAGATTGTTTAAGCAAAATAAG-3′) with plasmid pTRK95 as the template DNA. Primer ermGT1forw contained a BamHI restriction site (italics) and the RBS (underlined) of ermGT. Primer ermGT2rev contained a HindIII restriction site (italics). Primer ermGT2rev was based on the sequence downstream of the terminator of ermGT, which was determined by sequencing pTRK95 with primer ermGTterminator (5′-ATATGCAAGAATTGACG-3′). The amplification conditions used for ′ermGT were identical to those described above for ′bglM except that the annealing temperature was 50°C. The PCR product was purified, digested with BamHI and HindIII, and inserted into the corresponding sites in p29Tbglm to create p29TIVET. The nucleotide sequences of ′bglM and ′ermGT in p29Tbglm and p29TIVET, respectively, were confirmed by sequencing.
To construct the promoter trap vector pJW100, p29TIVET was digested with EcoRI and HindIII, and the 2.3-kb fragment containing the IVET cassette was end filled and inserted into the blunt-ended SalI restriction site of p29cat232small.
Test of functionality and stability of pJW100 in vitro.
The promoter of the ldhL lactate dehydrogenase gene of Lactobacillus sakei LTH667 was amplified by using primers LDHL1 (5′-AGTCGACGTTTGTTATTGC-3′) and LDHL2 (5′-CAATTCTTCATTTCGAAAAC-3′). Amplification was carried out with a GeneAmp 9600 thermocycler (Perkin-Elmer). The reaction mixture (50 μl) contained 12.5 pmol of each primer, each deoxyribonucleoside triphosphate at a concentration of 0.2 mM, 1× reaction buffer, 2.5 U of Pwo polymerase (Boehringer Mannheim), and 0.5 μl of template DNA. The amplification program was as follows: 94°C for 5 min (initial denaturation); 30 cycles of 95°C for 45 s, 49°C for 1 min, and 72°C for 1 min; and finally 72°C for 7 min. It should be noted that a hot-start amplification procedure was employed in which reaction mixtures were kept at 80°C after the initial denaturation until Pwo polymerase was added. The resulting PCR product was inserted into the dephosphorylated SmaI site of pJW100 to create pJW200, and the desired orientation was confirmed by digestion with SalI and BamHI.
The MIC of erythromycin for L. reuteri strains 100-23C, JW100, and JW200 was determined by using dilution series in MRS broth in microtiter plates. Experiments to determine the in vitro stability of plasmids maintained in L. reuteri were carried out as described by Heng et al. (11).
Construction of the genomic library.
Chromosomal DNA of L. reuteri 100-23C was extracted as described previously (24), partially digested with Sau3AI to obtain fragments that were ca. 0.1 to 1.5 kb long, and partially end filled with dATP and dGTP by using Klenow polymerase. Plasmid pJW100 was digested with SalI and partially end filled with dCTP and dTTP by using Klenow polymerase. DNA fragments were inserted into pJW100, and heat-inactivated (65°C, 10 min) ligation mixtures were used to transform E. coli DH5αF′. After aerobic incubation of plates for 48 h, the plasmid sizes of randomly picked clones were checked by using a fast method for plasmid isolation (Promega). To establish the genomic library in lactobacilli, ca. 40,000 colonies were pooled from the agar plates by using sterile saline, and plasmid DNA was extracted with a QIAprep Miniprep plasmid isolation kit (Qiagen). Transformation of L. reuteri 100-23C was performed with ca. 1 μg of pooled plasmid DNA per transformation mixture. Transformants were plated on MRS agar containing 10 μg of chloramphenicol per ml to obtain ca. 40,000 CFU. To determine the sizes of inserts in the recombinant plasmids, PCR was performed as described below by using 1-μl portions of cell suspensions for E. coli and 1-μl portions of plasmid DNA for L. reuteri.
Inoculation and treatment of RLF mice.
L. reuteri 100-23C, JW100, and JW200 and the pools of fusion strains containing the genomic library were cultivated on MRS medium supplemented with chloramphenicol (and erythromycin for JW200). After 48 h, cells were recovered from agar plates by using 3 ml of sterile water per plate or were concentrated by centrifugation (liquid media) and resuspended in sterile water. Each cell suspension was adjusted to an optical density at 600 nm of 20, and an aliquot (100 μl; ca. 5 × 108 cells) was administered by intragastric gavage to each mouse in a group of anesthetized mice that were obtained from our colony of RLF mice and were maintained in isolators (32). These animals harbor a complex microflora equivalent to that of conventional mice, but lactobacilli are not present. Lincomycin was added to the drinking water. The following antibiotic treatments (ABT) were used: for ABT 1, lincomycin treatment (19 mg/liter) was started 3 days before inoculation of the mice with lactobacilli and was continued throughout the experiment; for ABT 2, lincomycin treatment (19 mg/liter) was started 24 h after inoculation of the mice with lactobacilli and was continued throughout the experiment; and ABT 3 was the same as ABT2 except that 9.5 mg of lincomycin per liter was used. Mice were killed after an appropriate time, and lactobacilli were cultured quantitatively by previously described methods (17) from the forestomach, jejunum, and cecum of each mouse on Rogosa agar (Difco) containing 10 μg of chloramphenicol per ml.
Screening of the genomic library for putative ivi genes.
Pools of fusion strains obtained by electroporation of L. reuteri 100-23C with the genomic library (representing ca. 40,000 clones) were screened for promoters active in vivo by inoculating RLF mice treated with lincomycin (see above). In vitro promoter activity was determined by an agar plate assay in which the surface of PHB agar supplemented with lichenan (Sigma) was inoculated with bacterial cells (10). Briefly, colonies recovered from the gastrointestinal compartments on Rogosa agar containing chloramphenicol (10 μg/ml) were picked with sterile toothpicks and streaked on PHB-lichenan agar containing chloramphenicol (10 μg/ml) and erythromycin (1 μg/ml). The low concentration of erythromycin ensured plasmid stability during strain purification without preventing the growth of plasmid-containing lactobacilli. After overnight anaerobic incubation at 37°C, the surface of each plate was flooded with a Congo red solution. β-Glucanase activity was indicated by a yellow halo surrounding the bacterial growth on an otherwise red plate (35). Strains that did not produce a halo or produced a small halo were purified, and the plasmid DNA was extracted. The plasmids were used to electrotransform L. reuteri 100-23C, and the cells were plated on MRS agar containing chloramphenicol (10 μg/ml) and different concentrations of lincomycin (0, 1, 10, 50, and 250 μg/ml) to assess the approximate level of lincomycin resistance.
Sequence analysis of cloned fusions.
Inserts of the purified plasmids of putative ivi clones were amplified by using primers IVETrrnT1T2 (5′-CATTCAAATATGTATCCGCTC-3′) and IVETrev (5′-ACGCAAATGTGGTCACCAGTG-3′). The PCR was performed in 0.2-ml tubes with a PCR Express thermal cycling apparatus (Hybaid) by using a reaction mixture (50 μl) consisting of reaction buffer (final concentrations, 10 mM Tris-HCl, 2.5 mM MgCl2, and 50 mM KCl [pH 8.3]), each deoxynucleoside triphosphate at a concentration of 200 μM, 20 pmol of each primer, ca. 100 ng of plasmid DNA, and 3.75 U of Taq DNA polymerase (Boehringer Mannheim). The amplification program was as follows: 94°C for 2 min; 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min; and finally 72°C for 10 min. PCR products were purified with a QIAquick PCR purification kit (Qiagen) and were sequenced with IRD 800-labeled primers IVETrrnT1T2-800 (5′-TCAAATATGTATCCGCTC-3′) and IVETrev-800 (5′-GCAAATGTGGTCACCAGTG-3′) by using an AutoRead sequencing kit (Amersham Pharmacia Biotech) in combination with a LI-COR automated sequencing system (MWG Biotech). Homology searches were performed with the GenBank database by using the BLASTX algorithm (http://www.ncbi.nlm.nih.gov/BLAST). To obtain additional sequence information downstream of the promoter (clones Ivi127, Ivi146, and Ivi148), the Vectorette system (Sigma Genosys, Pampisford, United Kingdom) was used according to the manufacturer's instructions.
Induction of the xylA promoter.
MRS medium without glucose (pH 6.5) containing 0.05% lichenin and chloramphenicol (10 μg/ml) was prepared. After autoclaving, different filter-sterilized sugars were each added to a final concentration of 1%, and 2.5-μl portions of an overnight culture of L. reuteri Ivi139 and JW200 were used to inoculate the agar plates with a single drop. Plates with glucose and plates without glucose were incubated for 16 and 24 h, respectively, before β-glucanase activity was determined as described above.
Colonization of RLF mice with putative ivi clones.
Pure cultures of the putative ivi clones were used to investigate their abilities to colonize RLF mice treated with lincomycin. L. reuteri JW200 served as a positive control. A clone exhibiting weak β-glucanase activity was randomly picked after transformation of L. reuteri 100-23C with the genomic library and purified (strain Pre1). This strain, L. reuteri 100-23C, and JW100 served as negative controls. RLF mice (three animals per bacterial strain) were inoculated and treated by using ABT 3. Seven days after inoculation, lactobacilli were cultivated quantitatively from different compartments of the gastrointestinal tract as described above.
RESULTS
Implementation of an IVET selection strategy for L. reuteri.
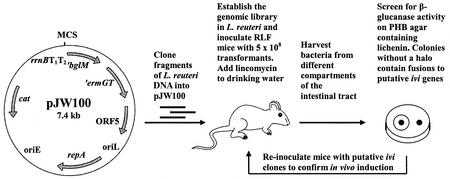
The IVET system developed for L. reuteri employs an antibiotic-based selection strategy (Fig. 1). Plasmid pJW100 was constructed to allow transcriptional fusions of random L. reuteri DNA fragments to a promoterless β-glucanase gene (′bglM) fused with a promoterless erythromycin-lincomycin resistance gene (′ermGT) (2, 33). Therefore, only lactobacilli containing pJW100 with an in vivo active promoter in the multiple cloning site should have been able to colonize lincomycin-treated RLF mice. To prevent transcriptional readthrough, pJW100 was constructed in such a way that strong rrnBT1T2 terminators preceded the IVET cassette. Additionally, pJW100 contained a chloramphenicol resistance gene and the replication region of pGT232 required for stable maintenance of plasmids in the Lactobacillus population in vivo (11). To test the suitability of pJW100 as a promoter trap vector, the constitutive ldhL promoter of L. sakei was cloned into the multiple cloning site of pJW100 to create pJW200 (Table 1). As expected, the host L. reuteri 100-23C and L. reuteri JW100 were sensitive to erythromycin (MICs, ca. 0.8 μg/ml and ca. 1.6 μg/ml, respectively) and did not exhibit β-glucanase activity. On the other hand, L. reuteri JW200 was highly resistant to erythromycin (MIC, >1,000 μg/ml) and lincomycin (MIC, >250 μg/ml) and showed marked β-glucanase activity (large halos surrounding the bacterial colonies). The stabilities of plasmids pJW100 and pJW200 in L. reuteri 100-23C were determined in vitro. Plasmid pJW100 showed high structural and segregational stability in the absence of antibiotics, and ca. 95% of the cells contained the plasmid after growth for ca. 50 generations. In contrast, plasmid pJW200 was less stable, and only ca. 30% of the cells contained the plasmid after ca. 32 generations. Instability of pJW200 was also observed in the presence of chloramphenicol and in the absence of erythromycin. In the presence of erythromycin (>1 μg/ml) pJW200 was structurally stable.
FIG. 1.
Selection of genes exhibiting elevated expression in the gastrointestinal tract of mice. Genes displaying elevated levels of expression in the intestinal tract were selected on the basis of their ability to drive the expression of a gene that is essential for colonization. Lactobacilli cannot become established in the guts of RLF mice treated with lincomycin administered in the drinking water. Colonization can occur only if an active promoter is inserted upstream of the promoterless ermGT gene. Recovery of strains from the gut after selection results in the detection of promoters that are either constitutive or ivi promoters. To distinguish between these two types of promoters, a promoterless marker gene (′bglM) is transcriptionally fused to ′ermGT, which enables screening for promoter activity in vitro. BglM− strains (no halo after agar plates containing lichenin are flooded with Congo red) or strains with low β-glucanase activity contain fusions to promoters activated in response to the gastrointestinal tract. ′ermGT, erythromycin resistance determinant; ′bglM, β-glucanase gene; cat, chloramphenicol resistance determinant; rrnBT1T2, E. coli rho-independent transcription terminators; repA, Lactobacillus replication gene; oriL, Lactobacillus origin of replication; oriE, E. coli origin of replication (pUC29); ORF5, ORF originating from pGT232.
To determine the in vivo stabilities of pJW100 and pJW200 and the suitability of the IVET system to select for promoters that were active in vivo, L. reuteri strains JW100 and JW200 were tested in RLF mice. In the case of strain JW100, lactobacilli were not detectable (<103 CFU/g) in the gut contents of RLF mice treated with lincomycin when ABT 1 was used 3 days after inoculation (four mice tested) or in the gut contents of RLF mice treated with lincomycin when ABT 3 was used 7 days after inoculation (three mice). L. reuteri JW200 was able to colonize the mouse gut at levels comparable to those observed in conventional rodents (30) under ABT 1 conditions after 4 days and under ABT 3 conditions after 7 days (five and three mice, respectively). The cell counts for JW200 in the forestomach, jejunum, and cecum were ca. 108, 106 to 107, and 107 to 108 bacteria per g of contents, respectively. For ABT 1, 120 colonies were randomly picked and tested. All isolates were resistant to chloramphenicol as well as erythromycin, and the β-glucanase activity was comparable to that of strain JW200. Purification of plasmids from two isolates showed that pJW200 was structurally stable. These colonization experiments indicated that pJW100 was suitable for use as a promoter trap vector and could identify promoters that were active in vivo.
Construction of the genomic library.
A library was constructed by cloning chromosomal fragments of L. reuteri 100-23C DNA into pJW100. The plasmid bank was first created in E. coli and then transferred by electroporation to L. reuteri 100-23C. Plasmid extraction from 25 clones randomly picked from the bank revealed plasmids of different sizes in both E. coli and L. reuteri. Ninety percent of the plasmids in L. reuteri were larger than pJW100 and therefore showed no structural instability. As determined by PCR, the average insert sizes were 540 and 380 bp for E. coli and L. reuteri, respectively, and the insert sizes ranged from 200 to 1,400 bp for both organisms. Phenotypic tests showed that ca. 4% of the clones were resistant to erythromycin (100 μg/ml) and ca. 10% showed at least low β-glucanase activity. Clones without β-glucanase activity could still grow on plates containing 1 μg of erythromycin per ml but not on plates containing lincomycin (1 μg/ml). One clone with low β-glucanase activity was selected for further investigation (strain Pre1).
Detection of putative L. reuteri genes induced in the gut of mice.
Experiments with different lincomycin treatments (ABT 1 with six mice and ABT 2 with 12 mice) were conducted to detect promoters which were specifically induced in vivo. L. reuteri 100-23C was electroporated with the genomic library, and the pool of transformants was used to inoculate RLF mice. In both experiments enrichment of β-glucanase-positive clones (yellow shift) was observed for each mouse, and 99.5 to 100% of the lactobacilli were highly β-glucanase positive after passage through the mice. One of the clones was selected for further investigation (strain Con1). Altogether, ca. 10,000 colonies were investigated, and five isolates that exhibited low levels of β-glucanase activity (putative ivi clones) were obtained; these five isolates were picked and purified. As pJW200 was structurally unstable without erythromycin in the medium and L. reuteri JW100 was able to grow on media containing low concentrations of erythromycin (MIC, ca. 1.6 μg/ml), a low concentration of erythromycin (1 μg/ml) was added to the medium to ensure plasmid stability during purification of the ivi clones. Plasmids of the ivi clones were again transferred to the host strain (L. reuteri 100-23C), and transformants were directly plated on agar plates containing different amounts of lincomycin. We observed that the purified ivi clones (Table 2) that exhibited low β-glucanase activity were also susceptible to lincomycin.
TABLE 2.
Characteristics of control strains and ivi clones recovered from the gastrointestinal tract of lincomycin-treated RLF-mice
| Clone or straina | Organ | Colonization time (days) | Sex of mouseb | ABT of mice | β-Glucanase activityc | Concn of lincomycin (μg/ml) at which growth occurredd | BLASTX resultse |
|---|---|---|---|---|---|---|---|
| ivi clones | |||||||
| Ivi127 | Forestomach | 3 | F | 1 | ± | 0 | Conserved hypothetical proteins of several bacteria (Chlamydophila pneumoniae, 49% identity) |
| Ivi130 | Cecum | 3 | F | 1 | + | 1 | Peptide Methionine sulfoxide reductases (MsrB) of several bacteria (Enterococcus faecalis, 60% identity) |
| Ivi139 | Cecum | 3 | F | 2 | + | 1 | Xylose isomerases (XylA) of several bacteria (L. pentosus, 68% identity) |
| Ivi146 | Forestomach | 4 | M | 2 | ± | 0 | Same as Ivi127 |
| Ivi148 | Jejunum | 4 | M | 2 | ± | 0 | Same as Ivi127 |
| Control strains | |||||||
| Con1 | Cecum | 4 | M | 1 | +++ | 250 | |
| Pre1 | ± | 0 | |||||
| JW100 | − | 0 | |||||
| JW200 | +++ | 250 |
Con1 contained pJW100 with a constitutive promoter and was randomly selected from a pool of clones recovered from mouse intestine; Pre1 was a preselected strain randomly picked from the fusion library.
F, female; M, male.
Determined on PHB agar plates supplemented with lichenan. Halo sizes: +++, 5 mm; + 1, 2 mm; ±, <1 mm; −, no halo.
Determined on MRS agar plates supplemented with different concentrations of lincomycin (0, 1, 10, 50, and 250 μg/ml).
The deduced amino acid sequence of the ivi gene was used as the query sequence; the hit with the highest homology is indicated in parentheses.
Sequence analysis of ivi promoters and genes.
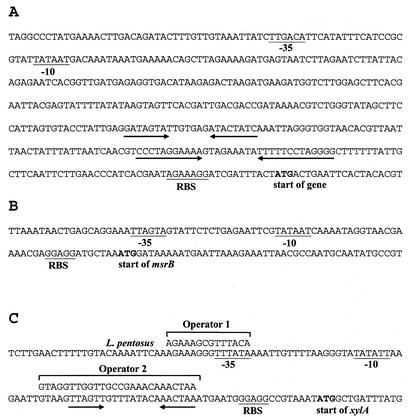
Sequence analysis of the plasmid inserts of the ivi clones Ivi127, Ivi130, Ivi139, Ivi146, and Ivi148 revealed putative promoters in the correct orientation to induce expression of the marker genes of the IVET cassette in pJW100. Additionally, for Ivi130 and Ivi139, an RBS and the corresponding open reading frame (ORF) were identified and showed homology to known genes. In the case of Ivi127, Ivi146, and Ivi148, we found that the sequences were identical and the start of an ORF was missing. Genomic walking with the genomic DNA of L. reuteri 100-23 provided additional sequence information for the region adjacent to the 3′ end of the plasmid insert. The similarities of the deduced amino acid sequences encoded by the putative ORFs to protein sequences contained in the databases are shown in Table 2. The sequences containing the putative promoters are shown in Fig. 2. A comparison of these sequences did not reveal similarities.
FIG. 2.
Promoter sequences of the ivi genes of L. reuteri 100-23. RBS and putative −10 and −35 sequences are underlined. The start of the genes is indicated. Inverted repeats are indicated by opposing arrows. (A) Promoter of the gene coding for a conserved hypothetical protein (Ivi127, Ivi146, and 148). (B) msrB promoter region. (C) Promoter of the xylA gene. Two putative cis-acting elements, operators 1 and 2, are shown together with the corresponding sequences of L. pentosus.
Further investigation of the promoter sequences of the xylA gene of Ivi139 revealed putative promoter sequences, including putative regulatory elements (Fig. 2C). Comparison of the sequences with those of the xylAB promoter of Lactobacillus pentosus (13) revealed high levels of homology to two putative operator-like elements involved in regulation of xylAB. To obtain insight into the regulation of the xylA gene of L. reuteri 100-23, strain Ivi139 was grown in the presence of different sugars. The promoter activity was indicated by the expression of the ′bglM gene of the IVET cassette. On MRS agar plates containing xylose or xylose plus galactose, Ivi139 exhibited high β-glucanase activity comparable to that of L. reuteri JW200 (Table 3). On agar plates containing galactose or xylose plus glucose, the activity was reduced, indicating that expression of the xylA gene in L. reuteri 100-23 was regulated in a manner similar to the manner of regulation in L. pentosus (i.e., induced by xylose and repressed by glucose). As expected, β-glucanase activity was negligible on agar containing glucose as the sole carbohydrate.
TABLE 3.
β-Glucanase activities of L. reuteri Ivi139 and JW200 in the presence of different sugars
| Strain | β-Glucanase activity on MRS agar plates containinga:
|
||||
|---|---|---|---|---|---|
| Glucose (1%) | Galactose (1%) | Glucose (1%) + xylose (1%) | Galactose (1%) + xylose (1%) | Xylose (1%) | |
| Ivi139 | + | ++ | ++ | +++ | +++ |
| JW200 | +++ | +++ | +++ | +++ | +++ |
Determined on MRS agar plates supplemented with lichenan. Halo sizes: +++, 5 mm; ++, 3.5; +, 2 mm.
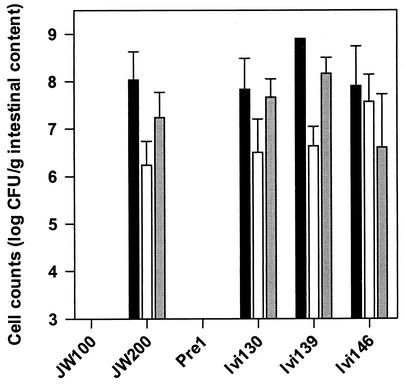
Colonization of RFL mice by ivi clones.
The ability of ivi clones containing DNA fragments of ivi genes to colonize lincomycin-treated RLF mice was investigated. As shown in Fig. 3, clones Ivi130, Ivi139, and Ivi146 were able to inhabit the gastrointestinal tracts of mice in the presence of lincomycin at levels similar to the level of L. reuteri JW200. This indicated that the promoters fused to the IVET cassette were induced in the gastrointestinal tract. On the other hand, the negative control strains L. reuteri 100-23C (data not shown), JW100, and Pre1 failed to colonize the mouse gut.
FIG. 3.
Populations of ivi clones and control strains in the different compartments of the gastrointestinal tract of RLF mice treated with lincomycin. The data are the means and standard errors for three animals. Solid bars, forestomach; open bars, jejunum; grey bars, cecum. The lower detection limit was 103 CFU/g. Pre1 is a BglM− strain obtained from the initial nonselected pools of transformants. For other strains of L. reuteri see Tables 1 and 2.
DISCUSSION
In this paper we describe successful application of an IVET strategy to identify ivi genes of L. reuteri in the murine gastrointestinal tract. The antibiotic-based strategy followed the classic IVET approach with a second reporter gene (′bglM) that had no role in vivo. Nevertheless, the secondary reporter was not inconsequential, since it provided a way to recognize constitutive promoters and to study the nature of the regulation, as described above for Ivi139 (xylA). Additionally, it provided a way to test whether the antibiotic treatment of the mice was adequate. This was indicated by a so-called yellow shift, which meant enrichment of bacteria containing constitutive promoters (BglM+). This provided the necessary indication that selection of strains that contained fusions to promoters that were transcriptionally active in vivo had taken place (16).
The vector pJW100 contains the complete origin of replication of pGT232 responsible for stable maintenance in the mouse intestine (11). In fact, instability of the plasmids was not observed during colonization of mice when lincomycin was added to the drinking water. Instability occurred in vitro in the absence of erythromycin. Addition of small amounts of erythromycin (1 μg/ml) to the culture medium was sufficient to stabilize the constructs without preventing growth of lactobacilli containing pJW100 with or without an insert and without promoter activity. The procedure used for recovery of lactobacilli from the gastrointestinal tract did not result in inherent selection of strains that were slightly erythromycin resistant because recovery was performed on Rogosa agar with no erythromycin added.
We identified three genes of L. reuteri 100-23 (Table 2) that were expressed at elevated levels in the gastrointestinal tract of mice. Peptide methionine sulfoxide reductase is a repair enzyme for proteins, which can reverse the loss of biological activity of proteins caused by oxidation of methionine to methionine sulfoxide (3). This enzyme protects bacteria against oxidative damage caused by reactive nitrogen intermediates (28) and reactive oxygen species (26). Other evidence suggests that the enzyme may be associated with bacterial adherence, as it contributes to the maintenance of adhesins in three major pathogens (39). It would be interesting to investigate the role of methionine sulfoxide reductase in the maintenance of adhesins of L. reuteri 100-23 because this strain forms a biofilm on the nonsecretory epithelium of the murine forestomach (30, 38). Two types of this enzyme (MsrA and MsrB) have been described, and all organisms that have been studied contain MsrA (3). Almost all free-living organisms except a few archaebacteria contain MsrB (6). Despite being functionally related, MsrA and MsrB failed to exhibit any overall sequence similarity (6). The deduced amino acid sequence of the cloned insert (Ivi130) showed the highest level of similarity to MsrB of Enterococcus faecalis, which is up-regulated in cells exposed to heavy metals (cadmium and mercury) (12). Additionally, msrB was required for cadmium resistance in E. coli (6). Elevated expression of msrB in L. reuteri indicates that lactobacilli colonizing the gastrointestinal tract of mice experience oxidative stress. This may arise from oxidative metabolism, from nutrient stress, or through exposure to the superoxide and hydroxyl radicals produced by other members of the microbial community.
Our study showed that the regulation of the xylA promoter of L. reuteri is similar to that of L. pentosus (13, 14). Transcription of the xylA promoter in Ivi139 is induced in the absence of glucose and in the presence of xylose, and the strength of transcription was comparable to that of the constitutive ldhL promoter in JW200. Since these two strains colonize the mouse gastrointestinal tract to similar degrees (Fig. 3), it can be assumed that in the gastrointestinal lumen, the level of glucose is rather limited and xylose is present. Xylose is a typical plant-derived sugar commonly found in straw and bran and is introduced into the gastrointestinal tract by feed. Additionally, xylose is a product of the breakdown of xylans and pectins by the gut microflora (15). Our results suggest that L. reuteri 100-23 meets its energy requirements in the gut at least in part by fermentation of xylose. Remarkably, xylose utilization has not been described previously for L. reuteri (8), indicating that this property is strain specific or may have escaped detection due to regulation or a long adaptation time. This might also be true for other autochthonous Lactobacillus species for which xylose utilization has not been described so far. It would be interesting to know which nutrients are available in intestinal ecosystems and which nutrients are utilized by autochthonous lactobacilli for maintenance in specific habitats.
The cloned inserts of three selected ivi clones (Ivi127, Ivi146, and Ivi148) contained putative promoter sequences but no putative start codons. Genomic walking revealed a gene whose deduced amino acid sequence showed the highest levels of homology (40 to 50% identity) to hypothetical proteins of several bacteria. To date, no function has been ascribed to these proteins. On the other hand, low levels of homology (<25% identity) to the cobalamin-independent methionine synthases of different bacteria were found, which suggests involvement in methylation processes. In fact, the amino acids His-641, Cys-643, and Cys-726 of the MetE methionine synthase of E. coli, which are involved in binding of zinc and in enzyme activity (41), are also present in the predicted L. reuteri homologue.
The sequences of the putative promoters of the ivi genes did not show any similarity, suggesting that each of the genes has a distinct mechanism of regulation. The promoter sequences of Ivi127, Ivi146, and Ivi148 contain pronounced inverted repeats able to form hairpin structures which are probably involved in regulation. The promoter of the xylA gene was similar to that described for L. pentosus (13, 14). Elements that might be involved in regulation were not detected in the putative promoter region of msrB.
In our IVET studies we used RLF mice which did not harbor lactobacilli but whose intestinal microflora was otherwise functionally similar to that of conventional animals (32). Thus, a high degree of colonization was ensured, and an added advantage was that there were considerable interactions between the lactobacilli and other members of the microflora. In fact, the presence of xylose in the gastrointestinal lumen could have been a result of the breakdown of polysaccharides by other members of the intestinal microflora and not by L. reuteri 100-23. Additionally, elevated expression of the methionine sulfoxide reductase might also have been induced by interactions with other members of the microflora.
The relatively small number of selected ivi clones in our study compared with the numbers in other IVET studies might have been due to instabilities of certain fusions. Furthermore, the lincomycin selection applied in vivo might have been too strong and favored selection of strong constitutive promoters. However, preliminary studies with a lower lincomycin concentration (6.5 mg/liter) revealed that the selection pressure was then too low (data not shown). Another reason for the low number of ivi clones might be the nature of the ecosystem. The gastrointestinal tract is an open system compared to the infection models used for IVET previously (21). It is likely that relatively high multiplication rates and hence high metabolic activity are necessary to maintain colonization in an open system. This would lead to selection of very strong promoters associated with metabolic genes that would be active most of the time. In fact, virtually all of the promoters (>99.5%) that we detected with our strategy showed a high degree of constitutive expression.
Lactobacilli are important members of the gastrointestinal microfloras of humans and other animals, and there is a growing body of evidence showing that these microbes provide benefits to the hosts in which they reside. The IVET system described here has the potential to identify genes of lactobacilli that have not previously been functionally characterized and might be essential for colonization in the gastrointestinal ecosystem. An understanding of the complex mechanisms by which bacteria colonize the gastrointestinal tract may contribute to more successful applications of probiotics. Additionally, specifically ivi promoters extend the spectra of known inducible gene expression systems in lactobacilli, and this should be useful for derivation of genetically modified lactobacilli for the delivery of immunogens (18) and other bioactive substances (27).
Acknowledgments
We thank M. Kranz for excellent technical assistance with DNA sequencing.
J. Walter's research period at the University of Otago was supported by a scholarship (Doktorandenstipendium im Rahmen des gemeinsamen Hochschulsonderprogramms III von Bund und Ländern) of the Deutscher Akademischer Austauschdienst. This work was financed in part by the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Benes, V., Z. Hostomsky, L. Arnold, and V. Paces. 1993. M13 and pUC vectors with new unique restriction sites for cloning. Gene 130:151-152. [DOI] [PubMed] [Google Scholar]
- 2.Borriss, R., K. Buettner, and P. Maentsaelae. 1990. Structure of the beta-1,3-1,4-glucanase gene of Bacillus macerans: homologies to other beta-glucanases. Mol. Gen. Genet. 222:278-283. [DOI] [PubMed] [Google Scholar]
- 3.Brot, N., and H. Weissbach. 2000. Peptide methionine sulfoxide reductase: biochemistry and physiological role. Biopolymers 55:288-296. [DOI] [PubMed] [Google Scholar]
- 4.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High-efficiency transformation of E. coli by high-voltage electroporation. Nucleic Acids Res. 16:6127-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant, S. G. N., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimaud, R., B. Ezraty, J. K. Mitchell, D. Lafitte, C. Briand, P. J. Derrick, and F. Barras. 2001. Repair of oxidized proteins. J. Biol. Chem. 276:48915-48920. [DOI] [PubMed] [Google Scholar]
- 7.Hahn, M., O. Olsen, O. Polity, R. Borriss, and U. Heinemann. 1995. Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-β-glucanase. J. Biol. Chem. 270:3081-3088. [DOI] [PubMed] [Google Scholar]
- 8.Hammes, W. P., and R. F. Vogel. 1995. The genus Lactobacillus, p. 19-54. In B. J. B. Wood and W. H. Holzapfel (ed.), The lactic acid bacteria, vol. 2. The genera of lactic acid bacteria. Blackie Academic and Professional, London, United Kingdom.
- 9.Hammes, W. P., A. Bantleon, and S. Min. 1990. Lactic acid bacteria in meat fermentation. FEMS Microbiol. Rev. 87:165-174. [Google Scholar]
- 10.Heng, N. C. K., H. F. Jenkinson, and G. W. Tannock. 1997. Cloning and expression of an endo-1,3-1,4-β-glucanase gene from Bacillus macerans in Lactobacillus reuteri. Appl. Environ. Microbiol. 63:3336-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heng, N. C. K., J. M. Bateup, D. M. Loach, X. Wu, H. F. Jenkinson, M. Morrison, and G. W. Tannock. 1999. Influence of different functional elements of plasmid pGT232 on maintenance of recombinant plasmids in Lactobacillus reuteri populations in vitro and in vivo. Appl. Environ. Microbiol. 65:5378-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laplace, J. M., A. Hartke, J. C. Giard, and Y. Auffray. 2000. Cloning, characterization and expression of an Enterococcus faecalis gene responsive to heavy metals. Appl. Microbiol. Biotechnol. 53:685-689. [DOI] [PubMed] [Google Scholar]
- 13.Lokmann, B. C., R. J. Leer, R. van Sorge, and P. H. Pouwels. 1994. Promoter analysis and transcriptional regulation of Lactobacillus pentosus genes involved in xylose catabolism. Mol. Gen. Genet. 245:117-125. [DOI] [PubMed] [Google Scholar]
- 14.Lokman, B. C., M. Heerikhuisen, R. J. Leer, A. van den Broek, Y. Borsboom, S. Chaillou, P. W. Postma, and P. H. Pouwels. 1997. Regulation of expression of the Lactobacillus pentosus xylAB operon. J. Bacteriol. 179:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macfarlane, S., M. J. Hopkins, and G. T. Macfarlane. 2000. Bacterial growth and metabolism on surfaces in the large intestine. Microb. Ecol. Health Dis. 2:S64-S72.
- 16.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 17.McConnell, M. A., A. A. Mercer, and G. W. Tannock. 1991. Transfer of plasmid pAMβ1 between members of the normal microflora inhabiting the murine digestive tract and modification of the plasmid in a Lactobacillus reuteri host. Microb. Ecol. Health Dis. 4:343-355. [Google Scholar]
- 18.Mercenier, A. 1999. Lactic acid bacteria as live vaccines, p. 113-127. In G. W. Tannock (ed.), Probiotics: a critical review. Horizon Scientific Press, Wymondham, United Kingdom.
- 19.Mitsuoka, T. 1992. The human gastrointestinal tract, p. 69-114. In B. J. B. Wood (ed.), The lactic acid bacteria, vol. 1. The lactic acid bacteria in health and disease. Elsevier Applied Science, London, United Kingdom.
- 20.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 21.Rainey, P. B., and G. M. Preston. 2000. In vivo expression technology strategies: valuable tools for biotechnology. Curr. Opin. Biotechnol. 11:440-444. [DOI] [PubMed] [Google Scholar]
- 22.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Havor, N.Y.
- 24.Schmidt, G., C. Hertel, and W. P. Hammes. 1999. Molecular characterisation of the dnaK operon of Lactobacillus sakei LTH681. Syst. Appl. Microbiol. 22:321-328. [DOI] [PubMed] [Google Scholar]
- 25.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skaar, E. P., D. M. Tobiason, J. Quick, R. C. Judd, H. Weiibach, F. Etienne, N. Brot, and H. S. Seifert. 2002. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc. Natl. Acad. Sci. USA 99:10108-10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steidler, L. 2001. Microbiological and immunological strategies for treatment of inflammatory bowel disease. Microbes Infect. 3:1157-1166. [DOI] [PubMed] [Google Scholar]
- 28.St. John, G., N. Brot, J. Ruan, H. Erdjument-Bromage, P. Tempst, H. Weissbach, and C. Nathan. 2001. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. USA 98:9901-9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tannock, G. W. 1995. Normal microflora. An introduction to microbes inhabiting the human body. Chapman and Hall, London, United Kingdom.
- 30.Tannock, G. W. 1997. Normal microbiota of the gastrointestinal tract of rodents, p. 187-215. In R. I. Mackie, B. A. White, and R. E. Isaacson (ed.), Gastrointestinal microbiology. Chapman and Hall, London, United Kingdom.
- 31.Tannock, G. W., and D. Savage. 1985. Detection of plasmids in gastrointestinal strains of lactobacilli. Proc. Univ. Otago Med. Sch. 63:29-30. [Google Scholar]
- 32.Tannock, G. W., C. Crichton, G. W. Welling, J. P. Koopman, and T. Midvedt. 1988. Reconstitution of the gastrointestinal microflora of lactobacillus-free mice. Appl. Environ. Microbiol. 54:2971-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tannock, G. W., J. B. Luchansky, L. Miller, H. Connell, S. Thode-Andersen, A. A. Mercer, and T. R. Klaenhammer. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid 31:60-71. [DOI] [PubMed] [Google Scholar]
- 34.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gobal. 2000. Analyses of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teather, R. M., and P. J. Wood. 1982. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43:777-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaughan, E. E., B. Mollet, and W. M. deVos. 1999. Functionality of probiotics and intestinal lactobacilli: light in the intestinal tract tunnel. Curr. Opin. Biotechnol. 58:505-510. [DOI] [PubMed] [Google Scholar]
- 37.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wesney, E., and G. W. Tannock. 1979. Association of rat, pig and fowl biotiypes of lactobacilli with the stomach of gnotobiotic mice. Microb. Ecol. 5:35-42. [DOI] [PubMed] [Google Scholar]
- 39.Wizemann, T. M., J. Moskovitz, B. J. Pearce, D. Cundell, C. G. Arvidson, M. So, H. Weissbach, N. Brot, and H. R. Masure. 1996. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc. Natl. Acad. Sci. USA 93:7985-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, F., L. Pearce, and P.-L. Yu. 1991. Construction of a family of lactococcal vectors for gene cloning and translational fusions. FEMS Microbiol. Lett. 77:55-60. [DOI] [PubMed]
- 41.Zhou, Z. S., K. Peariso, J. E. Penner-Hahn, and R. G. Matthews. 1999. Identification of the zinc ligands in cobalamin-independent methionine synthase (MetE) from Escherichia coli. Biochemistry 38:15915-15926. [DOI] [PubMed] [Google Scholar]