Abstract
To gain insight into the molecular mechanisms that direct position-dependent gene expression in the simple and evolutionarily old metazoan Hydra, we have examined DNA–protein interactions in the 1.5-kb cis regulatory region of the head-specific gene ks1. In vitro footprinting and gel-retardation techniques have been used to map the location of all protein-binding sites. To our surprise, we found substantially more proteins binding to ks1 promoter elements in nuclear extract from basal (gastric) than from apical (head- and tentacle-formation zone) cells. One of these proteins is the homeobox protein Cnox-2. In the head regeneration-deficient mutant reg-16, an increased level of nuclear protein binds to ks1 promoter elements. Treatment of polyps with the ks1-inducing phorbol ester 12-O-tetradecanoylphorbol 13-acetate (TPA) resulted in reduced binding of nuclear proteins to the ks1 cis regulatory region. As activation of ks1 transcription is correlated with the absence of nuclear proteins binding to the ks1 promoter, we propose that the majority of these proteins act as transcriptional repressors. In this view, the gradient of head activation along the Hydra body axis is caused by a decreasing amount of inhibitory factors, rather than an increasing amount of activators, toward the head. Thus, inhibitory mechanisms might have played a crucial role in regulating position-dependent gene activation during early metazoan evolution.
Hydra has a simple body plan consisting of a two-layered radially symmetrical tube with two differentiated structures, head and foot, at the ends. Hydra tissue grows constantly, and as cells divide they are displaced along the column toward the head or the foot (1, 2), where the cells differentiate and form the appropriate structures. The signals and molecular mechanisms regulating localized cell differentiation in Hydra are largely unknown. Numerous morphological and cellular studies suggest, however, that gradients of head-activating and -inhibiting substances are important in Hydra pattern formation processes (2). Transplantation experiments have shown, for example, that the apical end of the body axis is defined by a high level of head activation potential (3). Defects in these patterning systems result in various mutant phenotypes. One example is mutant Hydra magnipapillata reg-16 (4), which has a low capacity to regenerate head structures because of an altered level of head activation potential (5). Recently, some observations have provided insight into the molecular mechanisms involved in position-dependent cell differentiation and axis formation in Hydra: (i) the positional value can be increased by treatment of Hydra tissue with protein kinase C activators such as diacylglycerol or 12-O-tetradecanoylphorbol 13-acetate (TPA), resulting in the formation of ectopic heads along the gastric column (6); (ii) peptides have been isolated that function as positional signals (7); and (iii), HOM/HOX genes have been shown to be associated with apical-basal axis formation in Hydra (8, 9). One of those genes is the deformed-like gene Cnox-2, which is expressed predominantely in basal tissue, is suppressed whenever head structures are formed (9), and is sensitive to changes in the positional value (10).
ks1 is one of the target genes for head-specific signals (11). As shown in Fig. 1A, it is expressed in ectodermal tentacle epithelial cells and in about 10% of ectodermal epithelial cells in the upper gastric region (tentacle formation zone; TFZ) (12). Cells expressing ks1 in this region are undifferentiated but determined to tentacle-specific differentiation. Treatment of polyps with TPA results in both increased expression of ks1 in cells of apical tissue and ectopic expression in basal epithelial cells (11). The head-specific expression of ks1 can be explained in several ways: (i) ks1 transcription in head tissue is induced by high concentrations of activators; (ii) ks1 transcription is repressed in basal tissue by the presence of inhibitors; or (iii) a combination of both mechanisms regulates the precisely localized expression of ks1.
Figure 1.
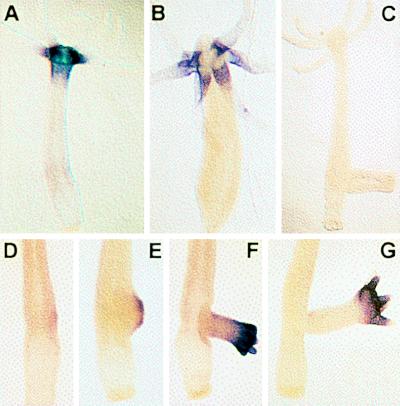
Expression pattern of ks1 in H. vulgaris. (A) ks1 is expressed as a gradient in apical tissue (head and TFZ). Substrate reaction for 40 min. (B) Strong ks1 expression is confined to base of tentacles (substrate reaction for 20 min). (C) antisense control. (D–G) ks1 expression in various stages of bud formation. A digoxigenin-labeled ks1 RNA probe was used as probe for in situ hybridization.
To gain insight into the molecular mechanisms regulating position-dependent gene expression in Hydra, we have examined the DNA–protein interactions at the ks1 cis regulatory region by DNaseI footprinting and electrophoretic mobility-shift assay (EMSA). We detected 47 target sites for nuclear proteins. Ten are binding sites for general DNA-binding proteins, and the remaining 37 ks1 promoter elements are binding sites for nuclear proteins from basal tissue. In TFZ and in head tissue, the amount and complexity of proteins binding to the ks1 promoter were greatly reduced. The same reduction in complexity was observed after TPA treatment. No promoter element was found to bind protein from apical tissue only. We therefore assume that transcription of the head-specific gene ks1 is induced by the absence of inhibitory factors. In this view, the postulated gradient of head activation along the body axis may be caused by a decreasing amount of transcriptional repressors.
MATERIALS AND METHODS
Animals and TPA Treatment.
Hydra vulgaris and H. magnipapillata mutant reg-16 (4) were cultured according to standard procedure at 18°C. TPA treatment was performed as described (11).
Isolation of the 1.5-kb Regulatory Region of the ks1 Gene using PCR.
To isolate the ks1 5′ flanking sequence, H. vulgaris genomic DNA was digested in parallel with different restriction enzymes and ligated to 2 μg of vector (pBluescript). A portion (10%) of the ligated products were used as template for PCR by using one primer specific for the ks1 promoter (5′-GTATAACGCAGCAGTCAACC-3′) pointing toward the unknown 5′ region and one vector-specific primer (Universal/T3). PCR reactions with T3 primer and Universal primer were performed in parallel (ks1-specific primer, 50 pmol; Universal/T3, 50 pmol; 1 min at 94°C, 1 min at 48°C, 5 min at 74°C; 30 cycles). Multiple DNA fragments were amplified from all genomic pools. Those appearing in both PCRs that used primer T3 or Universal were considered to be nonspecific. From the genomic pool generated by digestion with ClaI, one prominent DNA fragment of 1,350 bp could be amplified and was confirmed by Southern hybridization using the known 324-bp ks1 promoter sequence as a probe. Sequence analysis of the cloned fragment showed 100% identity to the known ks1 promoter sequence in the 150-bp overlap.
Preparation of Nuclear Protein Extract.
All steps for preparing nuclear protein extracts were done at 4°C. Animals were cut according to the ks1 expression pattern in tissue from the apical (head plus TFZ) and the basal (gastric) regions. Tissue was dissociated in 1 ml of dissociation medium (3.6 mM KCl/6 mM CaCl2/1.2 mM MgSO4/6 mM soium citrate/6 mM sodium pyruvate/6 mM glucose/12.5 mM TES/50 mg/ml rifampicin, pH 6.9) plus 5% Nonidet P-40 in a 2-ml homogenizer. Cell debris was separated from nuclei by centrifuging for 8 min at 85 × g. After pelleting the nuclei by centrifuging for 12 min at 8,000 × g, the nuclear pellet was washed in buffer A (20 mM Hepes⋅KOH, pH 7.9/75 mM KCl/1.5 mM MgCl2/0.1 mM EDTA/20% glycerol/4 mM Pefabloc). Nuclei were lysed by incubating in buffer C (20 mM Hepes/KOH ph 7, 9; 420 mM KCl; 1, 5 MgCl2 mM; 0.1 mM EDTA; 20% Glycerol; 4 mM Pefabloc) for 45 min on ice. Debris was removed by centrifuging at 14,000 × g for 30 min. Proteins were kept frozen at −80°C.
In Vitro Footprinting (DNaseI Protection Analysis).
From the 1,538-bp 5′ flanking sequence, 13 DNA fragments (A–M) of 150–220 bp were produced by PCR using one 32P end-labeled primer. For DNaseI footprinting reactions, 10–15 μg of nuclear extract was incubated in binding buffer (20 mM Hepes, pH 8.0/5 mM MgCl2/4 mM Pefabloc/1.5 μg poly(dI-dC)/1 mM DTT/1 μg of BSA/120 mM KCl) with 20,000 cpm-labeled probe in a volume of 20 μl. After incubation for 40 min on ice, 20 μl of buffer P2 (5 mM MgCl2/10 mM CaCl2) was added to the binding reaction. The DNA was digested by DNaseI (1–2.5 units) for 1 min and stopped with 100 μl of stop solution (100 mM Tris⋅HCl, pH 7.5/100 mM NaCl/1% SDS/10 μg of proteinase K). After incubation for 30 min at 37°C, the reaction products were extracted with phenol/chloroform and precipitated with ethanol and 150 μl of carrier DNA solution (66 ng/ml). The precipitate was washed with 70% ethanol, resuspended in 10 μl of distilled H2O, and 8,000–10,000 cpm per lane was loaded onto a denaturing 8% polyacrylamide gel.
EMSA.
Double-stranded oligonucleotides of 20–42 bp were used for EMSA. Nuclear protein (1.5 μg) was incubated with 5–10 fmol of 32P-labeled oligonucleotide (10,000 cpm) in binding buffer (20 mM Hepes, pH 8.0/5 mM MgCl2/4 mM Pefabloc/1.5 μg poly(dI-dC)/1 mM DTT/1 μg of BSA/120 mM KCl). Unlabeled competitor DNA was added to the reaction in 10- to 100-fold molar excess. For unspecific competition, oligonucleotide 5′-TTGATCTGACGTTGTTTTAACTAAAA-3′ was used. The total volume was 20 μl. In supershift experiments, antisera or preimmune sera were added last to the binding reaction. The reactions were separated on a 4% native polyacrylamide gel.
Molecular Techniques.
Nucleic acid isolation, Southern blotting, and sequence analysis were carried out following standard procedures. Whole-mount in situ hybridization was performed as described by Martinez et al. (13). For analysis of the ks1 5′ flanking sequence, the tfsearch computer program (version 1.3) was used (14).
RESULTS
ks1 as an Early Target Gene for Head-Specific Signals.
We previously reported that ks1 expression is sensitive to head-patterning signals (11). However, at that time we were unable to detect ks1-expressing cells in undifferentiated tissue of the TFZ and in the tissue of the budding zone, although in both of these regions head patterning signals are active (1). Here, by using RNA probes for in situ hybridization (13), we show that in mature polyps, ks1 is expressed as a gradient in the TFZ, with most of the ks1-expressing cells close to the tentacles (Fig. 1A). Strong expression of ks1 is confined to the base of the tentacles (Fig. 1B). Analysis of polyps in various stages of budding demonstrates that ks1 is expressed early during bud formation and even before any bud protrusion can be seen (Fig. 1 D–G).
Isolation of 1,538-bp Upstream Sequence of the ks1 Gene.
In our previous study (11) we isolated 324 bp of the ks1 5′ flanking sequence by using inverse PCR. To obtain sequence further upstream of the ks1 gene, a 1,538-bp fragment upstream of the transcription start was isolated by using a PCR approach. The complete sequence is shown in Fig. 2. Initial analysis of the ks1 5′ flanking sequence by using tfsearch revealed the presence of a large number of putative binding sites for known transcription factors, including dorsal-, Oct-1-, hunchback-, deformed-, and kruppel-related sequences. As shown below, the 5′ flanking region also contains several binding sequences for proteins not yet found in databases. They were termed Hyko-1 to Hyko-11 in Fig. 2.
Figure 2.
Diversity of protein-binding sites at the ks1 1,538-bp 5′ flanking sequence. Boxes indicate sequence elements that have been identified by in vitro footprinting and EMSA (see examples in Figs. 3–5). Light boxes, sequences related to known transcription-factor binding sites; dark boxes, Hyko sites, not related to known consensus sequences. ∗ indicates initiation site for transcription (11).
Localization of Specific Protein-Binding Sites in the ks1 5′ Flanking Region.
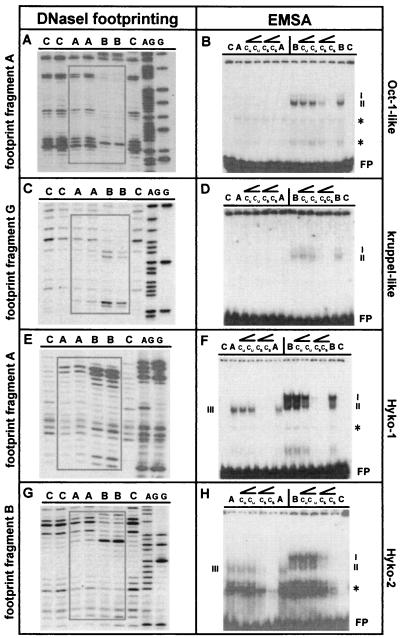
To quantitatively identify all binding sites for putative regulatory proteins at the 1.5-kb ks1 promoter, we used in vitro DNaseI footprinting. Overlapping subfragments of the complete 1.5 kb were incubated separately with nuclear extract from apical or basal tissue to identify differences in DNA–protein interactions along the body axis. Target sites differentially bound by nuclear extracts were analyzed by tfsearch to determine their similarity to known transcription factor-binding sequences. Examples of results are shown in Fig. 3. DNaseI-resistant and -hypersensitive regions indicated binding of nuclear proteins to each fragment. In all cases, nuclear extract from basal tissue caused greater changes in the DNaseI fragment pattern compared with nuclear extract from apical tissue, denoting that the ks1 promoter was preferentially bound by proteins from the basal region. In Fig. 3 A and C, two results are shown with fragments containing binding sequences that are related to known transcription-factor target sites. In vitro footprinting and sequence analysis revealed that the ks1 cis regulatory region contains at least 11 protein-binding sites that are not related to any known consensus sequence (Fig. 2). These Hyko sites could represent unique binding sites for Hydra-specific transcription factors. Similar to the results shown above, in vitro footprinting indicates preferential binding of proteins from basal region to those fragments. Examples are shown in Fig. 3 E and G.
Figure 3.
Identification of ks1 promoter elements as specific binding sites for nuclear protein from apical (head tissue and TFZ) or basal (gastric) tissue. (A–D) Examples of two sequence elements that are related to known transcription-factor binding sites are shown. (E–H) Two examples for elements not related to known transcription-factor binding sites are shown. C, control (without protein); A, nuclear extract from apical tissue; B, nuclear extract from basal tissue; AG/G, sequencing reaction; cu, addition of unspecific competitor; cs, addition of specific competitor; ∠, rising concentrations of unspecific or specific competitior (10- or 100-fold molar excess); FP, free probe; ∗, binding of general nuclear protein(s) to oligonucleotide; I–IV, specific oligonucleotide/protein complexes. Boxes indicate DNaseI-resistant and/or -hypersensitive regions in footprint experiments. Sequences of oligonucleotides used are: kruppel-like, 5′-GTAAAAATCGGGTTTATTCG-3′; Oct-1-like, 5′-GTGCCAAAGAATGCAAAGAAAAGTTCTA-3′; Hyko-1, 5′-TTCAAAAAAACCTCAAAGATAAAAGAAGCAAAC-3′; Hyko-2, 5′-GTTTTTTGTAGCTCTACTTTTTAAAGCCAAGGTAGA-3′.
To confirm that binding of nuclear factors to ks1 promoter elements is specific, EMSA was performed using oligonucleotides directed against the DNaseI-resistant and -hypersensitive regions. Examples of the data obtained are shown in Fig. 3. Corresponding to the results obtained by using DNaseI footprinting, all oligonucleotides tested showed strong shifts with nuclear protein(s) from basal tissue. With protein extract from apical tissue either no shifts or only weak shifts were observed. Moreover, most sequence elements migrated faster when incubated with protein extract from apical tissue compared with basal tissue (Fig. 3 F and H). Specificity of binding was determined by competition experiments using unlabeled sequence-specific or unrelated oligonucleotides in 10- to 100-fold molar excess.
Complete analysis of the 1.5-kb ks1 5′ flanking sequence using footprinting revealed a total of 47 binding sites for nuclear proteins. Ten of these sites were identified as general binding sites (data not shown), as they were bound equally by basal and apical nuclear extracts. Of the remaining 37 promoter elements, 11 are Hyko elements, whereas 26 elements show sequence similarity to known transcription factor binding sites. All 37 sequence elements were tested in EMSA and found to be specific binding sites, with the exception of the 4 cAMP response element (CRE)-like sites (see Fig. 2). Binding to these CRE-like elements could be prevented by addition of unrelated oligonucleotide (data not shown). Additionally, all specific target sites were preferentially bound by nuclear protein from basal region. In contrast, with nuclear protein from apical tissue—where a high level of ks1 transcripts can be found—either no or only weak shifts could be observed. Thus, the activation of ks1 transcription is directly correlated to the absence of proteins binding to the ks1 promoter.
DNA–Protein Interactions at the dfd-1 Site.
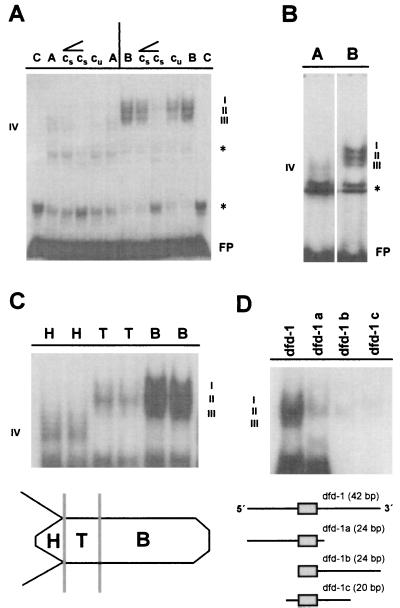
The 1.5-kb ks1 cis regulatory region contains five sequence elements (dfd-1 to dfd-5 in Fig. 2) related to deformed response elements present in a number of developmentally regulated genes in Drosophila (15). Because DNase I footprinting revealed protein binding to all five dfd-related elements, the dfd-1 site was chosen to characterize the DNA–protein interactions in more detail. Fig. 4 indicates that according to EMSA, the dfd-1 element is a specific binding site primarily for nuclear proteins from basal tissue. Three complexes (labeled I, II, and III in Fig. 4) were identified when the dfd-1 oligonucleotide was incubated with nuclear extract from basal tissue. With nuclear extract from apical tissue, only one weak shift (labeled IV in Fig. 4) was observed. To determine the localization of these dfd-1-binding proteins along the body axis, we isolated nuclear proteins from head, TFZ, and basal-region tissue. Fig. 4C shows a sharp decrease in the amount of proteins binding to the dfd-1 site in tissue of the TFZ compared with tissue of the basal region. Qualitatively, complexes I and III are not present in tissue of the TFZ. Complex IV, present in head tissue only, appears to be smaller than the complexes formed in basal tisssue and TFZ (see Discussion). To test whether complexes I to III represent three nuclear proteins binding to distinct recognition sequences at the dfd-1 site, we synthesized three subfragments of the 42-bp dfd-1 element. These subfragments (dfd-1a, dfd-1b, and dfd-1c in Fig. 4D) were used as probes in EMSA by using nuclear extract from basal tissue. Surprisingly, there was only weak or no binding of nuclear protein to each of the three subfragments compared with the full-length dfd-1 element, suggesting that protein–protein interactions are involved in the regulation of ks1 expression.
Figure 4.
DNA–protein interactions at the dfd-1 sequence element of the ks1 promoter. (A) Mobility shift experiment showing differential binding of nuclear protein from apical and basal tissue. (B) Higher resolution of DNA–protein interactions at the dfd-1 sequence element demonstrating formation of three complexes with nuclear proteins from basal tissue. (C) Gradient of nuclear proteins binding to the dfd-1 element along the body axis with sharp decrease of DNA–protein interactions in TFZ and head tissue. (D) Upper, DNA–protein interactions at subfragments dfd-1 a, dfd-1 b, and dfd-1 c of the dfd-1 site. Lower, Schematic representation of Dfd core sequence. C, control (without protein); A, nuclear extract from apical tissue; B, nuclear extract from basal tissue; H, nuclear extract from head tissue; T, nuclear extract from TFZ; cu, addition of unspecific competitor; cs, addition of specific competitor; ∠, rising concentrations of unspecific or specific competitior (10- or 100-fold molar excess); FP, free probe; ∗, binding of general nuclear protein(s) to oligonucleotide; I–IV, dfd-1–protein complexes. Sequence of the dfd-1 oligonucleotide is 5′-TTTGATTTGCGTATTAATTACAATTTAATAGATTTGAT-3′ (Dfd core sequence in italics).
Interaction Between Hydra Homeobox Protein Cnox-2 and the dfd-1 Site of the ks1 Promoter.
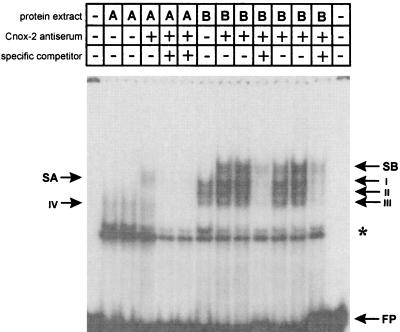
One of the Hydra homeobox-containing genes suggested to be involved in axis formation is the deformed-like protein Cnox-2 (9). To analyze whether Cnox-2 protein binds to ks1 dfd-1 element, we performed supershift experiments using a polyclonal antiserum directed against Cnox-2 (9). Fig. 5 shows that by coincubating the dfd-1 element and nuclear protein from basal tissue with Cnox-2-specific antiserum, a supershifted band can be observed (SB in Fig. 5). Coincubation of the dfd-1 element and nuclear protein from apical tissue with Cnox-2 antiserum also results in a supershifted band (SA in Fig. 5). Thus, Cnox-2, present in about 10% of the nuclei of the head (9), also binds to the dfd-1 target site in apical tissue. Interestingly, the shift observed in apical tissue is qualitatively different from the shift in basal tissue, indicating a modification or loss of cofactors of Cnox-2 in head tissue (see Discussion). Coincubation with Cnox-2 preimmune serum did not result in any supershift (data not shown).
Figure 5.
Supershift experiments with the 42-bp dfd-1 element and Cnox-2 antiserum (gift from H. Bode, University of California, Irvine, CA). A, nuclear extract from apical tissue; B, nuclear extract from basal tissue; FP, free probe; ∗, unspecific binding to oligonucleotide; I–IV, dfd-1–protein complexes using nuclear extract from apical and basal tissue; SA, supershift obtained by using nuclear extract from apical tissue; SB, supershift obtained by using nuclear extract from basal tissue.
Increase of Positional Value in Basal Tissue Is Accompanied by Decrease of Proteins Binding to dfd-1.
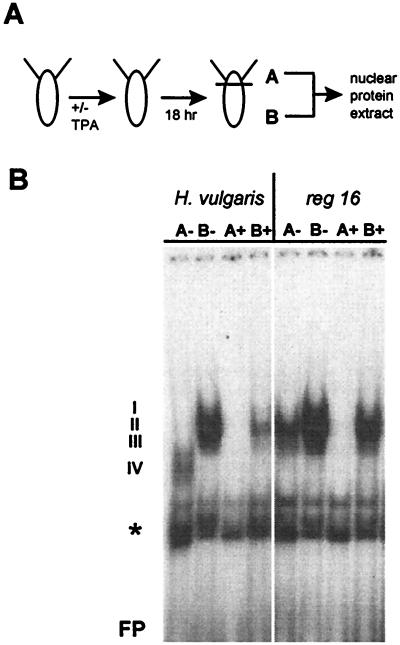
To examine the influence of changes in positional value on the protein interactions at the ks1 cis regulatory region, two sets of experiments were done. In the first approach, H. vulgaris polyps were treated with TPA to increase the positional value. Nuclear extract from apical and basal tissue was prepared and used for EMSA with the dfd-1 oligonucleotide (Fig. 6A). Fig. 6B shows that in contrast to untreated tissue, there was no binding of proteins to the dfd-1 site with nuclear extract of TPA-treated apical tissue. In nuclear extract from TPA-treated basal tissue, the binding was greatly reduced compared with nontreated tissue. Qualitatively, only complex II was present, similar to the observation made with nuclear extract from untreated tissue of the TFZ (Fig. 4C). Thus, increasing the positional value of tissue by TPA is correlated with reduced binding of nuclear proteins to ks1 cis regulatory elements.
Figure 6.
DNA–protein interactions at the ks1 dfd-1 element are sensitive to changes in positional value. (A) Procedure to analyze the influence of TPA on DNA–protein interactions in H. vulgaris and mutant H. magnipapillata reg-16. (B) EMSA experiments showing reduction in binding of nuclear protein from apical and basal tissue to the dfd-1 site after TPA treatment of H. vulgaris and mutant H. magnipapillata reg-16. “+” indicates addition of 30 nM TPA; I–IV, dfd-1–protein complexes formed by using nuclear extract from apical or basal tissue of wild-type or mutant animals; FP, free probe; ∗, nonspecific binding to oligonucleotide.
In the second approach, a mutant of H. magnipapillata that has a strongly reduced head-regeneration capacity (4) was used. The head-regeneration potential of mutant reg-16 can be partially rescued by treatment with TPA (J.U.L. and T.C.G.B., unpublished data). Presuming that the reduced capacity to regenerate head structures is correlated with a high level of nuclear proteins binding to the ks1 promoter, we examined the dfd-1 element by using extracts from reg-16 polyps. The results are shown in Fig. 6B: (i) in reg-16 polyps, more protein binds to the dfd-1 element compared with H. vulgaris polyps using extract from apical tissue. With extract from basal tissue, only minor differences were observed between wild type and mutant. In contrast to H. vulgaris, in mutant reg-16, complex II was formed with extract from apical tissue, similar to when nuclear extract of the TFZ from wild-type tissue was used (Fig. 4C). (ii) Treatment of reg-16 polyps with TPA leads to complete disappearance of binding proteins in apical tissue, similiar to the effect of TPA seen in H. vulgaris. Binding of nuclear protein from basal tissue is slightly reduced in mutant reg-16 after TPA treatment. In summary, our results clearly show that in both mutant reg-16 and in H. vulgaris the DNA–protein interactions at the ks1 promoter are sensitive to changes in positional value.
DISCUSSION
The ks1 5′ flanking region contains multiple binding sites for nuclear proteins (Fig. 2). Several observations indicate that the majority of these binding sites is functionally significant and that the proteins binding to them function as transcriptional repressors. (i) The proteins binding to the ks1 promoter are asymmetrically distributed along the apical–basal axis with a low level of nuclear proteins binding in apical tissue where ks1 is expressed (Figs. 3 and 4). Analysis of the dfd-1 site revealed a gradient in the distribution of nuclear proteins along the body axis with a significant decrease in the complexity of DNA–protein interactions at the gastric column–TFZ transition (Fig. 4C). This decrease is correlated with the onset of ks1 transcription in undifferentiated cells of the TFZ (see Fig. 1A). (ii) All of the sequence elements are highly specific binding sites with the exception of the four CRE-like sites. (iii) DNA–protein interactions at the ks1 regulatory region are sensitive to changes in positional value and can be reduced by protein kinase C activators, such as TPA (see Fig. 6B). Earlier observations that TPA ectopically induces the expression of ks1 in basal tissue (11) can now be explained by the capacitity of TPA to reduce binding of inhibitory factors to the ks1 promoter region. (iv) In mutant reg-16, a high level of nuclear proteins binds to ks1 cis regulatory elements (Fig. 6B). We therefore propose that the low capacity of mutant reg-16 to regenerate head structures is the result of a high level of inhibitory factors binding to the ks1 promoter, especially in apical tissue. Increasing the positional value of mutant tissue by treatment with TPA decreases the complexity of DNA–protein interactions at the dfd-1 site (Fig. 6B), providing a first molecular explanation for the partial rescue of this mutant by TPA. (v) One of the putative transcriptional repressors binding to the dfd-1 element is the homeobox protein Cnox-2 (Fig. 5). Taken together, we presume that the elements identified are specific binding sites for inhibitory factors playing a major role in head-specific regulation of ks1 expression. Because initiation of transcription by RNA polymerase II requires activators (16), the sites bound equally by apical and basal extracts could represent targets for such positive regulators. The observation that binding to all four CRE-like elements occurs unspecifically was surprising, because the CRE-binding protein CREB was thought to be involved in Hydra head formation (17).
We observed accelerated migration of DNA–protein complexes with nuclear proteins from apical tissue compared with complexes formed with protein from basal tissue (Figs. 3–5). This difference may be the result of modifications of nuclear protein present in basal tissue or to the loss of cofactor(s) in apical tissue. Evidence that protein–protein interactions are indeed important for binding to ks1 cis regulatory elements was obtained by analysis of the dfd-1 element (Fig. 4D). Very weak or no binding to subfragments of this target site suggests that cofactors alone cannot bind to the dfd-1 target site. Furthermore, the Dfd core sequence seems insufficient for effective Cnox-2 binding (Fig. 4C), indicating that Cnox-2 acquires its binding specificity through coselective binding with cofactors (18). In Drosophila, it was shown recently that autoactivation of the deformed gene requires not only the Dfd protein but also the coactivator extradenticle (19).
Nuclear proteins were found to bind specifically to both sequence elements related to known transcription factor-binding sites as well as to sequence elements not related to any known consensus sequence (Fig. 2 and 3). The latter elements could represent binding sites for Hydra-specific transcription factors. Such unique regulatory proteins are thought to code for species- or group-specific characters (20). Hydra-specific developmental features such as tentacle formation may be regulated by nuclear proteins binding to the Hyko sites shown in Fig. 2.
Localized ks1 Gene Expression Is Inversely Correlated to the Number of Nuclear Factors Binding to the 5′ Flanking Sequence.
Current views on head formation in Hydra are based on a gradient concept (1). Head tissue is thought to represent the gradient peak of head activation with hypostome and tentacles defined by separate reaction diffusion systems that are hierarchically linked (21). Based on the data presented above, we propose that localized ks1 expression is regulated by a gradient in the distribution of inhibitory factors along the apical–basal body axis, with a sharp decrease at the body column–TFZ transition (Fig. 7). This gradient of putative negative regulators is inversely oriented to the hypothetical head-activation gradient. The mechanism used to establish and maintain this repressor gradient is unknown. It seems possible that factors localized in apical tissue transform repressor proteins from an active into an inactive state as the tissue gets displaced toward the apical end. The results shown in Fig. 6 suggest that phosphorylation by protein kinase C may be involved in this process. Alternatively, a high positional value could locally inhibit expression of negative regulators. Thereby transcriptional repressors were present only in tissue of low positional value at a distance from the head. The fact that expression of Cnox-2 is actively suppressed in head tissue (9) agrees well with this latter possibility.
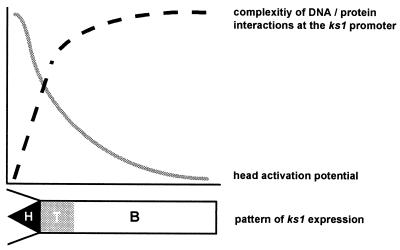
Figure 7.
Schematic summary of data and model of head-specific gene activation in Hydra. Data in Figs. 2–6 indicate that the amount and complexity of ks1 regulatory proteins (dashed line) is inversely correlated to the head activation potential (solid line). Lower, schematic diagram of the ks1 expression pattern along the body axis. H, head tissue; T, tissue of the TFZ; B, basal tissue. The model suggests that transcription of target genes for head-specific signals requires the absence of negative regulators.
Conclusion.
The results presented here demonstrate that regulation of position-dependent gene activation in one of simplest metazoan animals involves enormously complex DNA–protein interactions. As complexity is drastically reduced in ks1-expressing tissue, our results suggest that most proteins binding to the ks1 promoter act as transcriptional repressors and not, as previously assumed, activators. In this view, inhibitory mechanisms have played a crucial role in the regulation of position-dependent gene activation during early metazoan evolution.
Acknowledgments
We thank Hans Bode for providing Cnox-2 antiserum, G. Praetzel and G. Kumpfmueller for technical assistance, Charles N. David for discussion, Klaus Gellner, Uri Frank, Wolfgang Nellen, and Luis M. Salgado for critically reading the manuscript, and two anonymous referees for helpful comments. This work was supported by the Deutsche Forschungsgemeinschaft.
ABBREVIATIONS
- TFZ
tentacle formation zone
- TPA
12-tetradecanoylphorbol-13-acetate
- EMSA
electrophoretic mobility-shift assay
- CRE
cAMP response element
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF119101).
References
- 1.Bode P M, Bode H R. In: Pattern Formation. Malacinski G, Bryant S V, editors. New York: MacMillan; 1984. pp. 213–241. [Google Scholar]
- 2.Bosch T C G. In: Cellular and Molecular Basis of Regeneration: From Invertebrates to Humans. Ferretti P, Géraudie J, editors. New York: Wiley; 1998. pp. 111–134. [Google Scholar]
- 3.MacWilliams H K. Dev Biol. 1983;96:239–257. doi: 10.1016/0012-1606(83)90325-1. [DOI] [PubMed] [Google Scholar]
- 4.Sugiyama T, Fujisawa T. J Embryol Exp Morphol. 1977;42:65–77. [Google Scholar]
- 5.Acherman J, Sugiyama T. Dev Biol. 1985;107:13–27. doi: 10.1016/0012-1606(85)90371-9. [DOI] [PubMed] [Google Scholar]
- 6.Müller W A. Development (Cambridge, UK) 1989;105:306–316. [Google Scholar]
- 7.Hoffmeister S A. Development (Cambridge, UK) 1996;122:1941–1948. doi: 10.1242/dev.122.6.1941. [DOI] [PubMed] [Google Scholar]
- 8.Schummer M, Scheurlen I, Schaller C, Galliot B. EMBO J. 1992;11:1815–1823. doi: 10.1002/j.1460-2075.1992.tb05233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shenk M A, Bode H R, Steele R E. Development (Cambridge, UK) 1993;117:657–667. doi: 10.1242/dev.117.2.657. [DOI] [PubMed] [Google Scholar]
- 10.Shenk M A, Gee L, Steele R E, Bode H R. Dev Biol. 1993;160:108–118. doi: 10.1006/dbio.1993.1290. [DOI] [PubMed] [Google Scholar]
- 11.Weinziger R, Salgado L M, David C N, Bosch T C. Development (Cambridge, UK) 1994;120:2511–2517. doi: 10.1242/dev.120.9.2511. [DOI] [PubMed] [Google Scholar]
- 12.Hobmayer E, Holstein T W, David C N. Development (Cambridge, UK) 1990;109:897–904. [Google Scholar]
- 13.Martinez D E, Dirksen M L, Bode P M, Jamrich M, Steele R E, Bode R E. Dev Biol. 1997;192:523–536. doi: 10.1006/dbio.1997.8715. [DOI] [PubMed] [Google Scholar]
- 14.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel A E, Kel O V, Ignatieva E V, Ananko E A, Podkolodnaya O A, Kolpakov F A, et al. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regulski M, Dessain S, McGinnis N, McGinnis W. Genes Dev. 1991;5:278–286. doi: 10.1101/gad.5.2.278. [DOI] [PubMed] [Google Scholar]
- 16.Nikolov D B, Burley S K. Proc Natl Acad Sci USA. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galliot B, Welschof M, Schuckert O, Hoffmeister S, Schaller H C. Development (Cambridge, UK) 1995;121:1205–1216. doi: 10.1242/dev.121.4.1205. [DOI] [PubMed] [Google Scholar]
- 18.Biggin M D, McGinnis W. Development (Cambridge, UK) 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- 19.Pinsonneault J, Florence B, Vaessin H, McGinnis W. EMBO J. 1997;16:2032–2042. doi: 10.1093/emboj/16.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolittle R F. Nature (London) 1998;392:339–342. doi: 10.1038/32789. [DOI] [PubMed] [Google Scholar]
- 21.Meinhardt H. Dev Biol. 1993;157:321–333. doi: 10.1006/dbio.1993.1138. [DOI] [PubMed] [Google Scholar]