Abstract
We have prepared a molecular map of the Chlamydomonas reinhardtii genome anchored to the genetic map. The map consists of 264 markers, including sequence-tagged sites (STS), scored by use of PCR and agarose gel electrophoresis, and restriction fragment length polymorphism markers, scored by use of Southern blot hybridization. All molecular markers tested map to one of the 17 known linkage groups of C. reinhardtii. The map covers approximately 1,000 centimorgans (cM). Any position on the C. reinhardtii genetic map is, on average, within 2 cM of a mapped molecular marker. This molecular map, in combination with the ongoing mapping of bacterial artificial chromosome (BAC) clones and the forthcoming sequence of the C. reinhardtii nuclear genome, should greatly facilitate isolation of genes of interest by using positional cloning methods. In addition, the presence of easily assayed STS markers on each arm of each linkage group should be very useful in mapping new mutations in preparation for positional cloning.
Studies using the unicellular eukaryotic alga Chlamydomonas reinhardtii have yielded important insights into many cellular processes including photosynthesis (45, 123), flagellar assembly and motility (24, 28, 86, 112, 114, 124, 137, 140), basal body assembly and positioning (115), gametogenesis and fertilization (35, 42, 167), DNA repair (109), phototaxis (49, 139), cell wall assembly (1), circadian rhythms (91, 162), and the regulation of metabolic pathways (3, 14, 31, 50).
A major strength of C. reinhardtii as an experimental system is its usefulness for genetic experiments (45, 47, 74). Vegetative cells are haploid, facilitating the analysis of mutant phenotypes, but stable diploid strains can be easily produced for dominance and complementation tests. Gametes can be crossed to yield diploid zygotes that sporulate to produce four products of meiosis, allowing routine tetrad analysis. Over the past 50 years, hundreds of mutations have been isolated; more than 200 genetic loci have been mapped to 17 linkage groups (28, 29, 45, 46, 52). Mutations induced by chemical or UV mutagenesis have been supplemented recently by mutations induced by transposition of one of several transposable element families in the genome (17, 34, 130, 133) or by insertional mutagenesis (13, 101, 149).
Insertional mutagenesis has become the favored method for generating mutations since the development of procedures for efficient transformation of the nuclear genome (59, 132). Upon transformation, plasmid DNA inserts in random positions into the nuclear genome, facilitating cloning of affected genes by using the transforming plasmid as a hybridization probe. This method of gene tagging has led to the isolation of numerous genes identified by mutation over the past several years. Despite its usefulness, the insertional-mutagenesis approach has drawbacks, including the inability to clone essential genes, difficulty in analyzing the large deletions that occur in some cases, and a limitation in the types of phenotypes that can be found by using a method that generates mostly null mutations.
To increase the power of molecular genetic approaches using C. reinhardtii, we have developed a molecular map aligned with the genetic map. In this paper, we present a detailed map of the C. reinhardtii nuclear genome based on the analysis of restriction fragment length polymorphism (RFLP) and sequence-tagged site (STS) markers. The availability of such a physical map will facilitate the cloning of genes identified by any type of mutation in C. reinhardtii.
(A preliminary version of the molecular map of C. reinhardtii was published previously [133].)
MATERIALS AND METHODS
C. reinhardtii strains, growth conditions, and genetic crosses.
The C. reinhardtii standard laboratory wild-type strain 21gr mt+ (CC-1690) and the interfertile field isolate strain S1-C5 mt− (CC-1952) were used as parental strains. The 21gr strain and the other commonly used laboratory strain, 137c, are very closely related (64); almost all PCR amplifications of genomic DNA using primers predicted from the sequence of one of the strains amplify DNA from both strains. The S1-C5 strain is identical to the S1-D2 mt− strain (CC-2290) (44); the two strains were isolated from the same soil sample. The 21gr and S1-C5 strains were crossed as described previously (75). Tetrad progeny from the resulting zygotes were separated; a total of 136 random progeny from 136 complete tetrads were used in the mapping experiments. Cells were grown in TAP medium (43) or M medium (125) by using the modification described by Schnell and Lefebvre (130).
Molecular markers.
Several types of molecular markers were mapped in this study. Markers designated GP were obtained by digesting C. reinhardtii genomic DNA (strain 137c) with the restriction enzyme PstI, size fractionating the DNA on an agarose gel, and preparing minilibraries of cloned fragments (0.5 to 6.0 kb) in plasmid vector pUC119 (119). Random cDNA clones constituting the CNA, CNB, and CNC series of markers were obtained from a C. reinhardtii cDNA library (143). Additional markers consisted of genomic DNA clones or cDNA clones provided by other laboratories and genomic DNA, cDNA, or expressed sequence tag (EST) sequences obtained from the GenBank database.
Scoring markers by RFLP detection.
Genomic DNA was isolated by the method of Schnell and Lefebvre (130). DNA (1 μg per lane) was digested with restriction enzymes (PstI, PvuII, EcoRI plus XhoI, or HindIII). DNA fragments were separated by electrophoresis on a 1% agarose gel (12.7 by 20 by 0.5 cm) at 35 V for 18 to 20 h in TBE buffer (0.45 M Tris, 0.44 M boric acid, 0.01 M EDTA [pH 8.0]). The DNA was denatured and transferred to a MagnaGraph nylon membrane (Micron Separations Inc., Westborough, Mass.) by using the protocol of Sambrook et al. (126). DNA was cross-linked to the membrane by using a model 1800 UV Stratalinker (Stratagene, La Jolla, Calif.) at 1,200 μJ for 30 s. The membrane was baked at 80°C for 2 h in a vacuum oven. Membranes containing digested DNA from each of the 136 random progeny mapping strains were incubated with hybridization solution (50% formamide, 5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 10× Denhardt's solution, 4% sodium dodecyl sulfate [SDS], and 300 μg of single-stranded salmon sperm DNA/ml) for 1 h at 42°C. Denatured, labeled probe (see below) was added, and the hybridization reaction mixture was incubated overnight at 42°C. Filters were washed with 2× SSPE-1% SDS for 20 min, followed by three washes in 0.2× SSPE-0.2% SDS for 20 min each at 68°C. Digoxigenin-labeled probes were detected according to the protocol from Roche Molecular Biochemicals (Indianapolis, Ind.) except that Tween 20 was increased to 0.3% in buffer A and 5% powdered milk (Carnation) was used in buffer B instead of Blocking Powder.
Preparation of hybridization probes.
Hybridization probes were labeled using the digoxigenin nonradioactive system from Roche Molecular Biochemicals. Plasmids or purified plasmid inserts were labeled by random priming according to the manufacturer's instructions with the following modifications. Probe DNA in 13 μl of H2O (200 to 500 ng of plasmid DNA or 50 to 75 ng of purified insert DNA) was denatured by boiling, and 4 μl of 5× OLB (0.225 M Tris-HCl [pH 8.0], 0.025 M MgCl2, 0.02 M dithiothreitol, 1.36 A260 units of hexanucleotides [Pharmacia Biotech, Piscataway, N.J.]), 2 μl of DIG DNA labeling mixture (Roche), and 1 μl (2 U) of Klenow enzyme (Roche) were added. The reaction mixture was incubated overnight at 37°C, and the reaction was stopped with 2 μl of 0.2 M EDTA. The probe was mixed with 20 μl of 5% Blue Dextran and column purified by using Bio-Gel P-60 agarose (Bio-Rad Laboratories, Hercules, Calif.). PCR labeling of cloned DNA fragments utilized sets of plasmid-specific primers and the PCR DIG Probe Synthesis kit (Roche). The PCR mixture (50 μl) contained 5 ng of plasmid DNA, 200 mM digoxigenin deoxynucleoside triphosphates, 1× PCR Mg2+ buffer, 25 pmol of primers, 9% dimethyl sulfoxide, and 2.5 U of Taq polymerase. PCR program steps were as follows: (i) 94°C for 5 min, (ii) 29 cycles of 94°C for 1 min, 56°C for 45 s, and 72°C for 3 min, and (iii) 94°C for 1 min, 56°C for 45 s, and 72°C for 10 min. For amplification of cDNA clones in the λExlox vector, phage DNA was prepared by resuspending a plaque in 200 μl of distilled water, freezing in liquid nitrogen, and thawing at room temperature. The freeze-thaw cycle was repeated, and the sample was boiled for 5 min. The crude DNA (25 μl) was used as template in a 100-μl PCR mixture containing PCR buffer with Mg2+ (Roche), 2 μl of DIG DNA labeling mixture, 20 pmol of primers, 2.5% dimethyl sulfoxide, and 2.5 U of Taq polymerase (Roche). PCR program steps were as follows: (i) 94°C for 4 min, 55°C for 2 min, and 72°C for 3 min, (ii) 35 cycles of 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min, and (iii) 94°C for 1 min, 55°C for 2 min, and 72°C for 10 min.
Scoring markers by use of PCR.
The PRIMER program (79) was used to design primers for amplification of fragments from 3′ untranslated regions (3′ UTR) of gene, cDNA, or EST sequences. The primer criteria included an optimal length of 22 nucleotides (range, 20 to 24), an optimal melting temperature (Tm) of 73°C (range, 71 to 75°C), and a GC content of 60 to 65%. The default settings of the program were used for other criteria. PCR mixtures (25 μl) contained buffer A (Fisher Biotech), 10% glycerol, 5% formamide, 200 μM each deoxynucleoside triphosphate, 12.5 pmol of each primer, 25 ng of genomic DNA, and 0.55 U of Taq DNA polymerase (Fisher Biotech). PCR program steps were as follows: (i) 94°C for 1 min, (ii) 30 cycles of 94°C for 1 min, 55°C (annealing temperature) for 1 min, and 72°C for 1 min, and (iii) 72°C for 10 min, followed by a 10°C hold. Amplified DNA fragments obtained from template DNA from the 21gr and the S1-C5 strain were sequenced by the Advanced Genetics Analysis Center, University of Minnesota. Nucleotide sequence polymorphisms between 21gr DNA and S1-C5 DNA provided the basis for manual design of allele-specific primers by using principles described by Dieffenbach et al. (25) and Kwok et al. (68). For some markers, ESTs obtained from strain S1-D2 (CC-2290) were available in the database; these sequences provided a source of nucleotide polymorphisms used in allele-specific primer design. The annealing temperature for PCRs using the allele-specific primers was optimized by using a gradient thermal cycler (DNA Engine Dyad; MJ Research, Waltham, Mass.) to amplify DNA from the 21gr and S1-C5 parent strains. DNA from progeny strains was amplified in 96-well format by using the optimal annealing temperature. Reaction products (7 μl) were fractionated on 1.5% agarose gels by using the Sunrise 96 apparatus (Life Technologies, Rockville, Md.) and visualized by ethidium bromide staining.
Linkage analysis.
For all loci scored in this study, the data consisted of a scorable hybridization fragment or PCR product derived from each of the two parental strains. No plus-minus data sets were used in the analysis. Data were entered and annotated by using the Map Manager QTX program (version b12) (82) and were then exported to the mapping Mapmaker/QTL program (version 3.0) (69, 78) for linkage analysis and map construction. The F2 backcross function of the program was used for map construction in this haploid organism by classifying one genotype as “homozygous” and the other as “heterozygous.” The “group” command, at a Lod score threshold of 4.0, was used to place the markers in linkage groups. All loci mapped to one of the 17 known linkage groups in the C. reinhardtii genome. Map order within linkage groups was determined by using the multipoint mapping functions of the “order” and “build” commands. Markers placed with a confidence of at least Lod 2.0 are presented on the map (Fig. 1). For markers that could not be placed on the map at a Lod score greater than or equal to 2.0, the “try” command was used to establish the most likely position of the marker on the map. The Kosambi function was used to assign map distances in centimorgans.
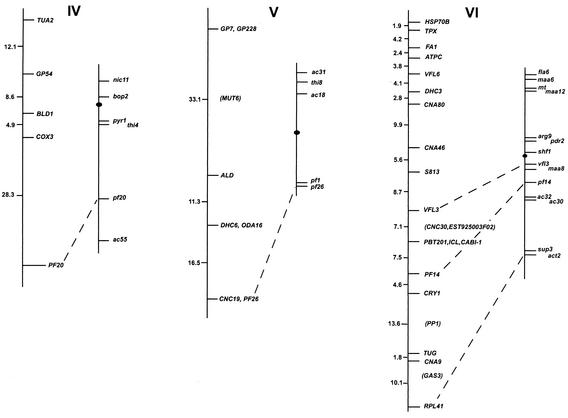
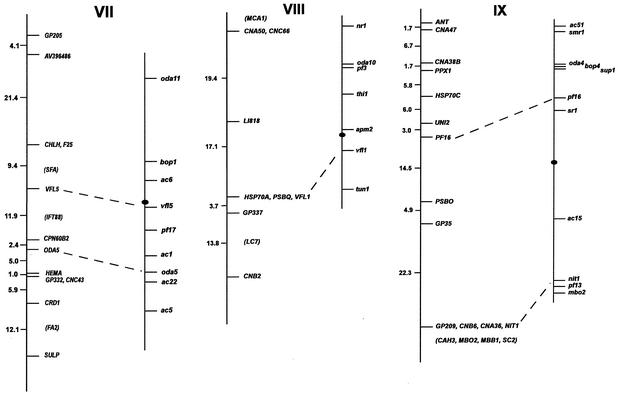
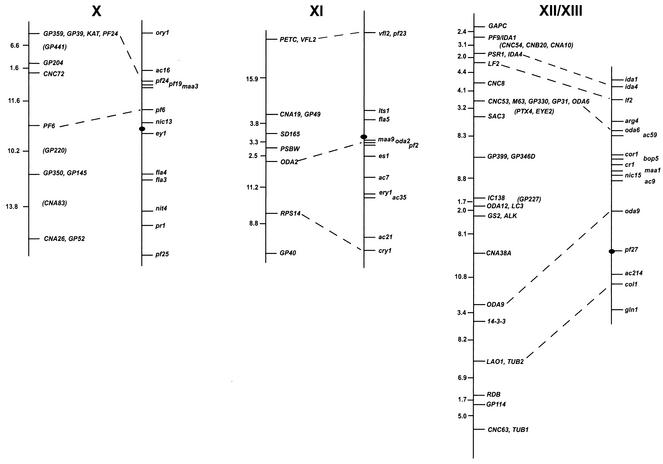
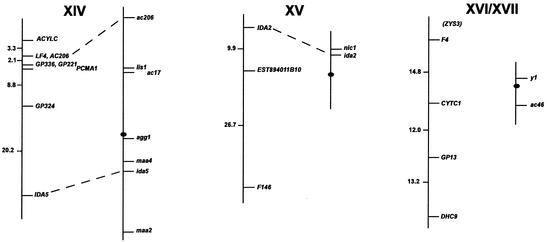
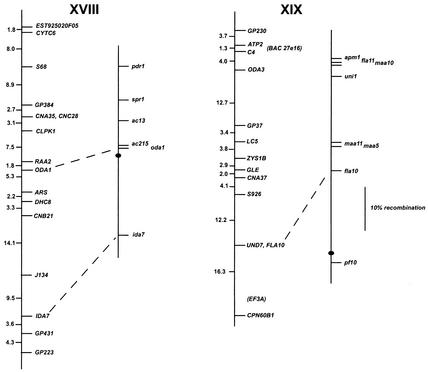
FIG. 1.
Chlamydomonas molecular and genetic maps. For each of the 17 linkage groups, the genetic map (adapted from the work of Harris [46]) is shown on the right, with the centromere represented by the black oval, and the molecular map is shown on the left. For genetic maps, spaces between markers represent recombination units (percent recombination); the scale bar is to the right of the maps for linkage group XIX. For molecular maps, numbers to the left of the vertical line indicate centimorgans (Kosambi units). The order of molecular markers placed on the map is predicted to be accurate with a Lod score of at least 2.0 by use of MAPMAKER/QTL 3.0 (78). For markers that could not be ordered with a Lod score of at least 2.0, the “try” command was used to determine the most likely map position. Such markers are enclosed in parentheses. For markers separated by commas on the same line, the order was indistinguishable by recombination. Dashed lines connecting the genetic and molecular maps indicate a molecular marker corresponding directly to a previously mapped phenotypic marker. The orientation of the molecular map with respect to the genetic map has not been confirmed for linkage groups VIII, XV, and XVI/XVII. Further information on the markers is available at http://www.biology.duke.edu/chlamydb/.
RESULTS
Using a combination of RFLP and PCR-based markers, we have placed 264 molecular markers on the 17 linkage groups of the C. reinhardtii genome (Fig. 1; Table 1). These markers were mapped on a panel of 136 random progeny from a cross of strain 21gr (mt+) with the field isolate S1-C5 (mt−) (44). All of the markers map to the 17 known linkage groups, indicating that if other linkage groups exist in the C. reinhardtii genome, they must be very small. The total length of the molecular map (in Kosambi units) is 1,025 centimorgans (cM). Any point on the C. reinhardtii genome is, on average, 2 cM from one of the 264 mapped molecular markers. Given that the size of the genome is approximately 108 bp (46), 1 cM in C. reinhardtii should correspond to about 100,000 bp. This number is consistent with the centimorgan-to-base pair ratio found during the positional cloning of the LF1 gene (R. Nguyen and P. A. Lefebvre, unpublished data).
TABLE 1.
Description of markers
| Linkage group | Locus | Gene or marker description | Source or reference(s) | Accession no. |
|---|---|---|---|---|
| I | CYP1 | Cyclophilin 1 | 145 | AF052206 |
| I | OPSIN1 | Chlamyopsin 1 | 21 | Z48968 |
| I | TCTEX1 | 14-kDa dynein light chain | 48 | AF039437 |
| I | LF3 | LF3/ULF1 gene | L.-W. Tam and P. Lefebvre, unpublished data | |
| I | GP387 | Random genomic fragment | This study | AY219891 |
| I | CNA13 | cDNA fragment | This study | |
| I | GP123 | Random genomic fragment | This study | AF525919 |
| I | GP396B | Random genomic fragment | This study | |
| I | PC1 | NADPH: protochlorophyllide oxidoreductase | 76 | U36752 |
| I | ARG7 | Argininosuccinate lyase | 19 | X16619 |
| I | PBT302 | Genomic fragment | B. Tailon and J. Jarvik, unpublished data | |
| I | CNC41 | cDNA corresponding to EST 1031062D08 | This study | BI722495 |
| I | CNA79 | cDNA corresponding to EST 1031013A09 | This study | BI994521 |
| I | GBP1 | G-strand binding protein | 110 | U10442 |
| I | RB47 | Poly(A) binding protein | 173 | AF043297 |
| I | CNA73 | cDNA corresponding to EST 894090B12 | This study | BE726259 |
| I | GAS96 | Gene expressed during cell differentiation | C. F. Beck, unpublished data | |
| I | COX2B | Cytochrome c oxidase subunit II | 104 | AF305540 |
| I | LC4 | LC4, 17-kDa dynein light chain | 60 | U34345 |
| II | PKA | Protein kinase A | N. Wilson and P. Lefebvre, unpublished data | AV390434 |
| II | KLP1 | Kinesin-like protein | 6 | X78589 |
| II | GP15 | Random genomic fragment | This study | |
| II | GP36 | Random genomic fragment | This study | |
| II | GP130 | Random genomic fragment | This study | |
| II | CNC17 | cDNA corresponding to EST 1031071E11 | This study | B1724441 |
| II | S641 | pcf 6-41 cDNA, upregulated after deflagellation | 129 | |
| II | CPX1 | Coproporphyrinogen III oxidase precursor | 117 | AF133671 |
| II | ALAD | Porphobilinogen synthase | 84 | U19876 |
| II | DIC8 | Genomic fragment | B. Williams and J. Rosenbaum, unpublished data | |
| II | RB60 | Protein disulfide isomerase | 153 | AF036939 |
| II | GP226 | Random genomic fragment | This study | |
| II | S6175 | pcf 6-175 cDNA, upregulated after deflagellation | 129 | |
| II | LC1 | LC1, 22-kDa dynein light chain | 4 | AF112476 |
| II | CIA5 | Regulator of carbon concentrating mechanism | 170 | AF317732 |
| II | S6135 | pcf 6-135 cDNA, upregulated after deflagellation | 129 | |
| II | CNB8 | cDNA corresponding to EST 1024021G10 | This study | BG848462 |
| II | GP366 | Random genomic fragment | This study | AY220530 |
| II | GP225 | Random genomic fragment | This study | AY220531 |
| II | DHC4 | Dynein heavy chain 4 | 113 | U81367 |
| II | GSP1 | Gamete specific protein 1 | 66 | AF108140 |
| II | CNA72 | cDNA fragment | This study | |
| II | CNA45 | cDNA corresponding to EST BI724982 | This study | BI724982 |
| II | ZYMC16 | Gene expressed in zygotes | 161 | |
| II | RBCS2 | Ribulose biphosphate carboxylase small subunit 2 | 41 | X04472 |
| II | DHC2 | Dynein heavy chain 2 | 113 | U61365 |
| II | GAS18 | Gene expressed during sexual differentiation | 157 | |
| II | GP383 | Random genomic fragment | This study | |
| II | S321 | pcf 3-21 cDNA, upregulated after deflagellation | 129 | |
| II | YPTC4 | Small G protein | 26 | U13167 |
| II | CNA43 | cDNA fragment | This study | |
| III | CNC24 | cDNA fragment | This study | AF525918 |
| III | DHC α | Dynein heavy chain alpha | 87, 88 | L26049 |
| III | EST 925021G06 | EST | 132a | BE442506 |
| III | LRG5 | Gene involved in blue light signaling | 40 | U73818 |
| III | COX2A | Cytochrome c oxidase subunit II | 104 | AF305080 |
| III | GSAT | Glutamate-1-semialdehyde aminotransferase | 83 | UC3632 |
| III | SAC1 | Sulfur limitation gene | 15 | U47541 |
| III | PF15 | Component of the central pair microtubule apparatus | E. F. Smith and P. Lefebvre, unpublished data | |
| III | MAA7 | Tryptophan synthase beta-subunit | 97 | AF047024 |
| III | CNB4 | cDNA corresponding to EST 833013E07 | This study | AW721386 |
| III | GP108 | Random genomic fragment | This study | |
| III | GP219 | Random genomic fragment | This study | |
| III | EF12E | Genomic fragment | E. Fernandez, unpublished data | |
| III | GAR1 | Gamete activation-regulated cobalamin-independent methionine synthase | 67 | U36197 |
| III | NAP | Novel actin-like protein | 57, 72 | U68060 |
| III | NIT2 | NIT2 gene | 130 | |
| III | RIB43A | Microtubule ribbon protein | 96 | AF196576 |
| III | FLA14 | LC8, 10-kDa dynein light chain | 61, 102 | U19490 |
| III | AC208 | Apoplastocyanin | 116 | L07282 |
| III | CPC1 | Central pair-associated complex 1 protein | 90 | |
| III | CNC21 | Random cDNA fragment | This study | |
| III | PTX2 | Phototaxis-deficient gene | 101 | |
| III | YPTC6 | YPTC6, small G protein | 26 | U13169 |
| III | LC6 | LC6, 13-kDa dynein light chain | 61 | U19484 |
| III | UNI3 | δ-Tubulin | 30 | AF013108 |
| III | TUA1 | α-Tubulin | 134 | M11447 |
| III | GP201 | Random genomic fragment | This study | |
| III | GP317 | Random genomic fragment | This study | |
| III | GP32 | Random genomic fragment | This study | |
| III | CNA34 | cDNA corresponding to EST 1031065B08 | This study | BI722955 |
| III | GP437 | Random genomic fragment | This study | |
| III | RSB1 | Radial spokehead polypeptide 1; corresponds to EST 1031064E02 | 165; this study | B1722838 |
| III | PHOT1 | Phototropin-like protein | 52a | AJ416557 |
| IV | TUA2 | α2-Tubulin | 134 | M11448 |
| IV | GP54 | Random genomic fragment | This study | |
| IV | BLD1 | Intraflagellar transport protein 52 | 9, 18 | AF397450 |
| IV | COX3 | Cytochrome c oxidase subunit III | 105 | AF233515 |
| IV | PF20 | Protein required for flagellar central pair microtubule assembly | 144 | U78547 |
| V | GP7 | Random genomic fragment | This study | AF467707 |
| V | GP228 | Genomic fragment; corresponds to EST 1024061H08 | This study | BG859190 |
| V | MUT6 | DEAH Box RNA helicase involved in gene silencing | 168 | AF305070 |
| V | ALD | Plastid fructose-1,6-bisphosphate aldolase | 103 | X85495 |
| V | DHC6 | Dynein heavy chain 6 | 113 | U61369 |
| V | ODA16 | Genomic fragment | D. Mitchell, unpublished data | |
| V | CNC19 | cDNA corresponding to EST AV621283 | This study | AV621283 |
| V | PF26 (S6187) | pcf 6-187 cDNA, upregulated after deflagellation; radial spoke protein 6 | 12, 120, 129 | M87526 |
| VI | HSP70B | Chloroplast-localized heat shock protein | 27 | X96502 |
| VI | TPX | Thioredoxin peroxidase | Y. Lee, S. H. Miller, and L. Keller, unpublished data | AF312025 |
| VI | FA1 | Flagellar autotomy protein | 36, 37 | AF246990 |
| VI | ATPC | Chloroplast ATP synthase gamma subunit | 142 | M73493 |
| VI | VFL6 | VFL6 gene | K. Iyadurai and C. Silflow unpublished data | |
| VI | DHC3 | Dynein heavy chain 3 | 113 | U61366 |
| VI | CNA80 | cDNA corresponding to EST 1031002F08 | This study | B1816487 |
| VI | CNA46 | cDNA fragment | This study | |
| VI | S813 | G protein beta subunit-like protein | 127 | X53574 |
| VI | VFL3 | VFL3 gene | Iyadurai and Silflow, unpublished | |
| VI | CNC30 | cDNA corresponding to cab II-1 | 54; this study | M24072 |
| VI | EST 925003F02 | EST | 132a | BE441254 |
| VI | ICL | Isocitrate lyase | 111 | U18765 |
| VI | CABI-1 | Light harvesting complex protein I-20 | 53 | X65119 |
| VI | PBT201 | Genomic fragment | B. Tailon and J. Jarvik, unpublished data | |
| VI | PF14 | Radial spoke polypeptide 3 | 166 | X14549 |
| VI | CRY1 | DNA photolyase/blue light photoreceptor | 141 | L07561 |
| VI | PP1 | Axonemal type-1 phosphatase | 172 | AF156101 |
| VI | TUG | γ-Tubulin | 138 | U31545 |
| VI | CNA9 | cDNA corresponding to EST 1024003B10 | This study | BG843541 |
| VI | GAS3 | Gene expressed during sexual differentiation | 157 | |
| VI | RPL41 | Ribosomal protein L41 (ACT2 locus) | 146 | AF130727 |
| VII | GP205 | Random genomic fragment | This study | AF467706 |
| VII | AV396486 | EST | 2 | AV396486 |
| VII | CHI H | Magnesium chelatase H subunit | 10 | AJ307055 |
| VII | F25 | Class IV zygote-specific cDNA | 33 | |
| VII | SFA | SF-assemblin | 70 | U56982 |
| VII | VFL5 | VFL5 gene | Iyadurai and Silflow, unpublished data | |
| VII | IFT88 | Intraflagellar transport protein 88 | 100 | AF298884 |
| VII | CPN60B2 | Chloroplast chaperonin beta-like subunit | 152 | L27473 |
| VII | ODA5 | Outer dynein arm protein | M. Blomberg-Wirschell and G. Witman unpub- lished data | |
| VII | HEMA | Glutamyl-tRNA reductase | R. D. Willows et al.; unpublished data | AF305613 |
| VII | GP332 | Random genomic fragment | This study | |
| VII | CNC43 | cDNA corresponding to EST 1024039D03 | This study | BG854228 |
| VII | CRD1 | Copper response target 1 protein | 92 | AF226628 |
| VII | FA2 | Flagellar autotomy protein | 81 | AF479588 |
| VII | SULP | Chloroplast sulfate transport system permease | H.-C. Chen, K. Yokthongwattana, and A. Melis, unpublished data | AF481828 |
| VIII | CNB2 | cDNA corresponding to EST AV626610 | This study | AV626610 |
| VIII | LC7 | LC7, 11-kDa dynein light chain | 8 | AF140239 |
| VIII | GP337 | Random genomic fragment | This study | |
| VIII | HSP70A | 70-kDa heat shock protein | 93 | M76725 |
| VIII | PSBQ | OEE3 protein of photosystem II | 85 | X13832 |
| VIII | VFL1 | VFL1 gene | 136 | AF154916 |
| VIII | LI818 | Polypeptide related to CAB proteins | 121 | X95326 |
| VIII | CNA50 | cDNA corresponding to EST 833007A09 | This study | AW676510 |
| VIII | CNC66 | cDNA fragment | This study | |
| VIII | MCA1 | RNA stability factor | A. Watson et al., unpublished data | AF330231 |
| IX | ANT | Mitochondrial ADP/ATP translocator | 131 | X65194 |
| IX | CNA47 | cDNA corresponding to EST 1031051H10 | This study | BI997794 |
| IX | CNA38B | cDNA fragment | This study | |
| IX | PPX1 | Protoporphyrinogen oxidase precursor | 118 | AF068635 |
| IX | HSP70C | 68-kDa heat shock protein | 158 | |
| IX | UNI2 | UNI2 gene | W.-C. Wu and C. Silflow, unpublished data | |
| IX | PF16 | Protein in C1 microtubule of flagellar central apparatus | 143 | U40057 |
| IX | PSBO | OEE1 protein of photosystem II | 85 | X13826 |
| IX | GP35 | Random genomic fragment | This study | AF467704 |
| IX | GP209 | Random genomic fragment | This study | |
| IX | CNB6 | cDNA corresponding to EST 1024027D07 | This study | BG849900 |
| IX | CNA36 | cDNA fragment | This study | |
| IX | NIT1 | Nitrate reductase | 32 | AH001336 |
| IX | CAH3 | Carbonic anhydrase, alpha type | 39 | U73856 |
| IX | MBO2 | MBO2 gene | 150 | AF394181 |
| IX | MBB1 | Required for expression of psbB/psbT/psbH | 155 | AJ296291 |
| IX | SC2 | Genomic fragment | A. Nguyen and W. Dentler, unpublished data | |
| X | GP359 | Random genomic fragment | This study | AY219892 |
| X | GP39 | Random genomic fragment | This study | |
| X | KAT | p60 katanin subunit | 80 | AF205377 |
| X | PF24 | PF24 gene | P. Yang and W. Sale, unpublished data | |
| X | GP441 | Random genomic fragment | This study | |
| X | GP204 | Random genomic fragment | This study | |
| X | CNC72 | cDNA fragment | This study | |
| X | PF6 | PF6 gene | 124a | AF327876 |
| X | GP220 | Random genomic fragment | This study | AF525922 |
| X | GP350 | Random genomic fragment | This study | AF525921 |
| X | GP145 | Random genomic fragment | This study | AF525920 |
| X | CNA83 | cDNA corresponding to EST 1031072D11 | This study | BI724522 |
| X | CNA26 | cDNA corresponding to EST AV634482 | 2; this study | AV634482 |
| X | GP52 | Random genomic fragment | This study | |
| XI | PETC | Chloroplast Rieske Fe-S precursor protein | 22, 23 | X76299 |
| XI | VFL2 | Centrin/caltractin | 73 | X57973 |
| XI | CNA19 | cDNA fragment | This study | AF503637 |
| XI | GP49 | Random genomic fragment | This study | |
| XI | SD165 | Genomic fragment | S. Dutcher, unpublished data | |
| XI | PSBW | Core subunit of photosystem II | 7 | AF170026 |
| XI | ODA2 | Dynein heavy chain gamma | 163 | U15303 |
| XI | RPS14 | Ribosomal protein S14 (CRY1 locus) | 95 | U06937 |
| XI | GP40 | Random genomic fragment | This study | AF525923 |
| XII/XIII | GAPC | Glyceraldehyde-3-phosphate dehydrogenase, (NAD) cytosolic, subunit C | 58 | L27669 |
| XII/XIII | PF9/IDA1 | Dynein heavy chain 1 | 94 | U61364 |
| XII/XIII | CNC54 | cDNA fragment | This study | AF174532 |
| XII/XIII | CNB20 | cDNA fragment | This study | AF486824 |
| XII/XIII | CNA10 | cDNA fragment | This study | |
| XII/XIII | PSR1 | Phosphorus metabolism regulatory protein | 169 | AF174532 |
| XII/XIII | IDA4 | p28, dynein inner arm light chain | 71 | Z48059 |
| XII/XIII | LF2 | LF2/ULF2 gene | C. Amundsen and P. Lefebvre, unpublished data | |
| XII/XIII | CNC8 | cDNA corresponding to EST 963109F12 | This study | B1873562 |
| XII/XIII | CNC53 | cDNA corresponding to EST 894081D12 | This study | BE725171 |
| XII/XIII | M63 | Gene involved in cytokinesis | J. Larsen, L.-W. Tam and C. Silflow, unpublished data | |
| XII/XIII | GP330 | Random genomic fragment | This study | |
| XII/XIII | GP31 | Random genomic fragment | This study | |
| XII/XIII | ODA6 | IC70 dynein intermediate chain | 89 | X55382 |
| XII/XIII | PTX4 | Gene involved in phototaxis | G. Pazour, unpublished data | |
| XII/XIII | EYE2 | Gene required for eyespot assembly | 122 | AF233430 |
| XII/XIII | SAC3 | Kinase regulating response to sulfur limitation | 16 | AF100162 |
| XII/XIII | GP399 | Random genomic fragment | This study | |
| XII/XIII | GP346D | Random genomic fragment | This study | |
| XII/XIII | IC138 | Inner arm dynein I1 subunit | P. Yang and W. Sale, unpublished data | |
| XII/XIII | GP227 | Random genomic fragment | This study | |
| XII/XIII | ODA12 | LC2, 19-kDa outer arm dynein light chain | 102 | U89649 |
| XII/XIII | LC3 | LC3, 17-kDa dynein light chain | 99 | U43610 |
| XII/XIII | GS2 | Glutamine synthetase 2 (GS2) | 11 | U46208 |
| XII/XIII | ALK | Chlamydomonas aurora-like kinase | 98 | AF199021 |
| XII/XIII | CNA38A | cDNA fragment | This study | |
| XII/XIII | ODA9 | IC78 dynein intermediate chain | 164 | U19120 |
| XII/XIII | 14-3-3 | 14-3-3 protein | 77 | X79445 |
| XII/XIII | LAO1 | L-Amino acid oxidase catalytic subunit | 156 | U78797 |
| XII/XIII | TUB2 | β2-Tubulin | 174 | K03281 |
| XII/XIII | RDB | Roadblock protein, EST 1024060G02 | S. King, unpublished data | BG858985 |
| XII/XIII | GP114 | Random genomic fragment | This study | |
| XII/XIII | CNC63 | cDNA fragment | This study | |
| XII/XIII | TUB1 | β1-Tubulin | 174 | M10064 |
| XIV | ACYLC | EST 832001D08 | 132a | AW676021 |
| XIV | LF4 | Genomic fragment | S. Berman and P. Lefebvre, unpublished data | |
| XIV | AC206 | Ccs1, required for chloroplast C-type holocytochrome formation | 55 | U70999 |
| XIV | GP336 | Random genomic fragment | This study | AF467702 |
| XIV | GP221 | Genomic fragment; corresponds to EST 1031037H01 | 132a | BI996414 |
| XIV | PCMA1 | Genomic fragment | C. Asleson and P. Lefebvre, unpublished data | |
| XIV | GP324 | Random genomic fragment | This study | AF467703 |
| XIV | IDA5 | Actin | 147 | D50838 |
| XV | IDA2 | Dynein heavy chain 1-β of I1 complex | 108 | AJ242525 |
| XV | EST 894011B10 | EST | 132a | BE056715 |
| XV | F146 | Class VI zygote-specific cDNA | 33 | |
| XVI/XVII | ZYS3 | Zygote-specific cDNA | 65 | AB004043 |
| XVI/XVII | F4 | Class I zygote-specific cDNA | 33 | |
| XVI/XVII | CYTC1 | Mitochondrial cytochrome c1 | A. Atteia et al., unpublished data | AF245393 |
| XVI/XVII | GP13 | Random genomic fragment | This study | AF467701 |
| XVI/XVII | DHC9 | Dynein heavy chain 9 | 113 | U61372 |
| XVIII | EST 925020F05 | EST | 132a | BE442454 |
| XVIII | CYTC6 | Cytochrome c6 | 51 | M67448 |
| XVIII | S68 | pcf6-8 cDNA, upregulated after deflagellation | 129 | |
| XVIII | GP384 | Random genomic fragment | This study | |
| XVIII | CNA35 | cDNA corresponding to EST AV622556 | This study | AV622556 |
| XVIII | CNC28 | cDNA fragment | This study | |
| XVIII | CLPK1 | Cyclic nucleotide-dependent protein kinase | U. Kawabata et al., unpublished data | AB042714 |
| XVIII | RAA2 | Trans-splicing factor | 106 | AJ243394 |
| XVIII | ODA1 | cDNA corresponding to EST BG854636 | 148 | AY039618 |
| XVIII | ARS | Arylsulfatase | 20 | X52304 |
| XVIII | DHC8 | Dynein heavy chain 8 | 113 | U61371 |
| XVIII | CNB21 | cDNA corresponding to EST 833007B10 | This study | AW676519 |
| XVIII | J134 | Genomic fragment | J. Jarvik, unpublished data | |
| XVIII | IDA7 | 140-kDa inner arm dynein | 107, 171 | AF159260 |
| XVIII | GP431 | Random genomic fragment | This study | |
| XVIII | GP223 | Random genomic fragment | This study | AF467708 |
| XIX | GP230 | Random genomic fragment | This study | |
| XIX | ATP2 | ATP synthase mitochondrial F1 beta subunit | 38 | X61624 |
| XIX | BAC 27e16 | BAC end sequence | This study | |
| XIX | C4 | Genomic fragment | W. Dentler, unpublished data | |
| XIX | ODA3 | Genomic fragment | 63 | AF001309 |
| XIX | GP37 | Random genomic fragment | This study | |
| XIX | LC5 | LC5, 14-kDa dynein light chain | 99 | U43609 |
| XIX | ZYS 1B | Protein expressed during zygote formation | 154 | X76117 |
| XIX | GLE | Gamete lytic enzyme | 62 | D90503 |
| XIX | CNA37 | cDNA corresponding to Lhcb4 | 151; this study | AB051211 |
| XIX | S926 | cDNA, upregulated after deflagellation | 128 | X62135 |
| XIX | UND7 | Genomic fragment | P. J. Ferris and U. W. Goodenough, unpublished data | |
| XIX | FLA10 | Kinesin-homologous protein | 160 | L33697 |
| XIX | EF3A | Genomic fragment | E. Fernandez, unpublished data | |
| XIX | CPN60B1 | Chloroplast chaperonin beta-like subunit | 152 | L27471 |
Frequency of polymorphism.
Previous efforts to develop an extensive molecular map for C. reinhardtii were hampered by the low frequency of DNA polymorphism observed for molecular markers by using C. reinhardtii and the interfertile strain Chlamydomonas smithii (120, 135). In this study, the laboratory strain 21gr and an interfertile field isolate strain (S1-C5) showed a high degree of polymorphism for many molecular markers. When DNA from the two strains was digested with PstI or PvuII, 94% of hybridization probes tested (n = 204) showed an RFLP with one or both of the enzymes. With three additional restriction enzymes, EcoRI plus XhoI and HindIII, the RFLP rate increased to 98%. Thus, it was possible to map almost all markers by using a set of standard filters prepared with genomic DNA digested with only a few different restriction enzymes.
The underlying variation in DNA sequence responsible for the high level of RFLP was confirmed by direct sequencing of a large number of 3′ UTR sequences from S1-C5 DNA. We chose to examine 3′ UTR sequences because they exhibit greater sequence variation than do coding sequences. In addition, 3′ UTR sequences are likely to be unique, even among genes in a multigene family. To obtain 3′ UTR sequences from S1-C5 genes, primers were designed to amplify 3′ UTR fragments from C. reinhardtii genes available in the GenBank database. Of 100 reactions that produced products by using 21gr DNA as a template, 82 also produced products by using S1-C5 DNA. Among these products, 16% showed a length polymorphism with the product obtained from the 21gr template DNA. We sequenced the amplified 3′ UTR regions from 62 S1-C5 genes, for a total of 29,053 bp. When these sequences were compared with the equivalent regions from strain 21gr or 137c (available in GenBank), single nucleotide substitutions were found at 793 positions (447 transitions and 346 transversions), for an average of 2.7 base substitutions per 100 bp of sequence. In addition, at 159 sites we found insertions or deletions of bases at a frequency of 0.54 per 100 bp.
The high level of sequence polymorphism in the S1-C5 gene sequences made it possible to design allele-specific primers based on single nucleotide polymorphisms (SNPs). We designed primers that yield PCR products of different lengths when template DNAs from the 21gr and S1-C5 parental strains are used (Table 2). These primer sets reproducibly generated reaction products when uniform reaction components and optimized annealing temperatures were used. The primer sets, corresponding to loci distributed over each of the linkage groups, were used to amplify PCR products ranging from 100 to 600 bp by using template DNA from the random progeny strains. The lengths of the resulting products were analyzed and scored by using agarose gel electrophoresis.
TABLE 2.
Mapping primersa
| Linkage group | Gene (3′ UTR) | Accession no.b | Primersc | C. reinhardtii allele (bp) | S1-C5 allele (bp) | Annealing temp (°C) |
|---|---|---|---|---|---|---|
| I | CYP1 | AF052206 | cyp1-R, GCATGCATTCCACAACGCACGC | 232 | 378 | 56.0 |
| cyp1-F2, GGCGCGTACGCTCCGCGCd | ||||||
| cyp1-F3, GGCTAATGGCTGTGCGGCTGGe | ||||||
| I | ARG7 | X16619 | ARG7-R, CGTCCCACACCTCCAAACGCCA | 367 | 236 | 56.0 |
| ARG7-F2, GCCTTGACGTGAGGCTGCGCTGe | ||||||
| ARG7-F3, TGGGGTACAAGGCGCTGTGAGAGAd | ||||||
| I | CNA73 | BE726259 | CNA73-R, CTTCTGCAGCCGTAGAAACCCGGC | 436 | 379 | 63.9 |
| CNA73-F2, GCATAGGGGCTGTGCGGCGe | ||||||
| CNA73-F3, TCTGTATGTGCCCCATGCGCACAd | ||||||
| I | COX2B | AF305540 | cox2B-R, ATGGCTACGCCACCGCCGGTTT | 386 | 547 | 63.9 |
| cox2BDNA-F3, TTGCTGGGCTGTGGCCGCGd | ||||||
| cox2BDNA-F4, CAGCGGTCCCTCAGGGACGTTACAe | ||||||
| I | LC4 | U34345 | LC4-F, GCCGCGCGAGCTGGAAGAGTTT | 565 | ∼590 | 62.7 |
| LC4-R, GTGCCGCCCACGAAACGTTCTG | ||||||
| II | CPX1 | AF133671 | Cpx1-F, TTGCGTGCTAGCAGGCGTGGTG | 329 | 393 | 61.0 |
| Cpx1-R2, GCTCCAAACCTGCTGCGGTCAGTCe | ||||||
| Cpx1-R3, GCACGCACACACGCACACGAd | ||||||
| II | RB60 | AF036939 | RB60-R, GCGTTCGGAACCACGCACATCC | 229 | 433 | 55.0 |
| RB60-F2, ACCAGCAGCAGCGCGTGATCCGGd | ||||||
| RB60-F3, GCCAAAGAGGGACGCTGTCCACAGe | ||||||
| II | CIA5 | AF317732 | CIA5-F, TGGCTGCGTGCCACGACCGT | 354 | 314 | 55.0 |
| CIA5-R2, GCTGAAGGTGAGTGCGGCAGCGd | ||||||
| CIA5-R3, CCAGCTGCTGTTGGCGCCAGCe | ||||||
| II | CNA45 | B1724982 | CNA45-R, CGTGGTTCTTACATCACCCCAGCG | 244 | 328 | 58.9 |
| CNA45-F2, TGTGGTGGGTGTTGATGGAGGAATGd | ||||||
| CNA45-F3, TTGCGCGCATTGACAGATGTACAGe | ||||||
| II | YPTC4 | U13167 | YptC4-R, CGCCGTGATCAGCAGCAACAAGC | 360 | 269 | 56.4 |
| YptC4-F2, TCCACATGATGGCTAGTGCGGACGe | ||||||
| YptC4-F3, CCGTCAGCTACTGGGAAGGCCCGd | ||||||
| III | DHCa | L26049 | DHC-alpha-F, AGGACATGCCCGCCAAGTGGGT | 309 | ∼290 | 58.9 |
| DHC-alpha-R2, GCGGCACCTGGCTACTGCTGTACA | ||||||
| III | COX2A | AF305080 | cox2A-F, TGCGGAGAAGGCGCTGGTCAAG | 522 | ∼490 | 55.0 |
| cox2A-R, GGCGTCTTGCGCCATTGCTGAA | ||||||
| III | GSAT | U03632 | GSAT-R, GAGGGTGCAATCAGAGCCCCCTTG | 561 | 389 | 55.0 |
| GSAT-F2, CGCGTGCACAGCTTGCAGCAAAe | ||||||
| GSAT-F3, CGGGCGGTGCCTGGTTCTTCGd | ||||||
| III | MAA7 | AF047024 | MAA7-F, CGGCGACAAGGACGTCAACAACG | 114f | 349 | 55.0 |
| MAA7-F2, TGTGGGAGCGGGAGTGACTGCA | 349 | |||||
| MAA7-R, TGCAACCATCTCCCTTCGGCCC | 473 | |||||
| MAA7-R2, TAATCCGCCTCAGCCCCAACCG | ||||||
| III | GAR1 | U36197 | MethSyn-R, GCAATGCGTTGGGTTACAAGCAGC | 339 | 179 | 61.0 |
| MethSyn-F2, GCGAGCGGTACCGACTAGGCAGAe | ||||||
| MethSyn-F3, GCTGAATTGTGTACGGTGCACACGGd | ||||||
| III | FLA14 | U19490 | LC8-F, TTCAAGTCGGGCTAAGCGGCCG | 189 | 97 | 55.0 |
| LC8-R2, CATCCCTCCCCGCTATGTCCCGd | ||||||
| LC8-R3, CCAGAGACCGCGCTCCGCCe | ||||||
| III | CNA34 | B1722955 | CNA34-F, GCAGCTGCCTGTCAATGCGCCT | 376 | ∼350 | 55.0 |
| CNA34-R, GTCTGCGTAGCCGTACACGCGTCA | ||||||
| III | RSB1 | B1722838 | RSB1-R3, CCGCCACCCATGTCACGGCd | 109 | 146 | 55.0 |
| RSB1-F2, TGTTCCCGCCGGAGGAGG | ||||||
| RSB1-R4, CACACCACACGCTGCCTACAGGe | ||||||
| III | PHOT1 | AJ416557 | PHOT1-F, CGCGAGGAAGGGTTTGAGGTGCTGd | 338 | 407 | 55.0 |
| PHOT1-R, CGGATAACAGCTGCGTCCTTCCCC | ||||||
| PHOT1-F2, CCGCCCCGGCTGCAGCTAAe | ||||||
| IV | TUA2 | M11448 | a2-tub-R, GCCAATAGAGGCACGGTCGTGGA | 130 | 150 | 55.0 |
| a2-tub-F2, GGCGTGATCTGAGGCTTCGTTGG | ||||||
| IV | COX3 | AF233515 | cox3-F, ACGGCATCATCTACGTCGGCCAG | 470 | ∼500 | 55.0 |
| cox3-R, ACATAAACCGTCCACGCGGCTGC | ||||||
| IV | PF20 | U78547 | PF20-F, TGTCTCTCCGTTCCCTTGCGCGe | 328 | 223 | 59.3 |
| PF20-F4, GGACCCCGGTCCTCTGCTACCGd | ||||||
| PF20-R2, ACACACCAAACCGCCCATGACCC | ||||||
| V | GP228 | BG859190 | GP228-F, CAACATGGTGGAGGAGCAGGAGGG | 244 | 369 | 63.9 |
| GP228-R2, GGCAGCCATCACCTCACACCAe | ||||||
| GP228-R3, GCTTCTCATCACCCCCTGCTCTTAAd | ||||||
| V | ALD | X85495 | ald-F, CTGCCCAGGGCATGTACGAGAAGG | 588 | 122 | 60.0 |
| ald-R, TCCCGACGCTCGATGGATAGGAGGe | ||||||
| ald-R4, GTCGCAGGCTGCTGCGGCTGd | ||||||
| V | PF26 | M87526 | RSP6-R, AGATGAACCTCGTGCCTCAGCGGC | 206 | 529 | 61.0 |
| RSP6-F2, GAAGAACGGATCAGAGCGGTGTGGGd | ||||||
| RSP6-F3, GGAACGTGGGGGACATACCCGGe | ||||||
| VI | FA1 | AF246990 | FA1-F, ACGAGGAGGACATTCGGGAGCTGC | 182 | 331 | 64.3 |
| FA1-R2, GCTCAGCCGTTCCAAGGAAGCAATG | ||||||
| FA1-R3, GTCCAAGCAGGTCCAAGCCGTCAA | ||||||
| VI | ATPC | M73493 | ATPC-R, TCGCCTCATGTCGGCACACAGG | 578 | 411 | 55.0 |
| ATPC-F2, TAAATGCCTGGGCTCTTGGGCTCGe | ||||||
| ATPC-F3, CGCCGTGAATTTGCGTGGCGd | ||||||
| VI | S8-13 | X53574 | S8-13-F, GCGCCCCGAGTTCAACATCACC | 331 | 285 | 55.0 |
| S8-13-R2, CCTCAACACACCGCGACGCAGAd | ||||||
| S8-13-R3, GCATCAACGCGTTACAGATCGCCAe | ||||||
| VI | CRY1 | L07561 | DNAph-R, GAGACCGAAAGGCAAGGCACAGGC | 443 | 443 | 63.5 |
| DNAph-F3, AGCTAACCATGTCGGCCGGTCGd | 147f | |||||
| DNAph-F4, GCTGCATTGGGCGCACATGGe | ||||||
| VI | TUG | U31545 | g-tub-R, GTCGCCAGGAATTTTGCCCCTGG | 281 | 443 | 61.0 |
| g-tub-F2, GCGCGCCTGGCGGTAGCACATAd | ||||||
| g-tub-F3, AGCAGCGCTATGTTCGCTTCCCCe | ||||||
| VI | RPL41 | AF130727 | RPL41-R, TGCAACTTGCAATCCATCCGTTGC | 262 | 107 | 55.0 |
| RPL41-F2, GCAACTAAACGTGGCGGCCTACCGe | ||||||
| RPL41-F3, GGTAACCGATTCGAGCGTTCTGGAd | ||||||
| VII | CHLH | AJ307055 | chlh-F, TTGGCGGGTTGTGGTTGGACTAGG | 127 | 375 | 62.4 |
| chlh-R2, TCCTCGCGGAGCGCTCTCGe | ||||||
| chlh-R3, CACAGCTCACACACACACGCACAAd | ||||||
| VII | SFA | U56982 | SF-assem-R, ACAGCATGCCCTGCAAGCTCGC | 330 | 211 | 65.0 |
| SF-assem-F2, TTGCATGGGCAGCACTGGTCGAe | ||||||
| SF-assem-F3, GCCGTATAAATTCAGGGCAGGCGCd | ||||||
| VII | IFT88 | AF298884 | IFT88-F, TGCTGAGCTTTGGCTCGGCTGG | 191 | 91 | 55.0 |
| IFT88-R2, ACATACACAAATGGCGGGCTGCAGd | ||||||
| IFT88-R3, CTGGGGACCCCTGCAGCCAAAAe | ||||||
| VII | CPN60B2 | L27473 | cr2-R2, AGCTGCTTGGCAGCGGCTGTTG | 260 | 621 | 61.0 |
| cr2-F4, TGGAATTGGCGGTGCGAGCGd | ||||||
| cr2-F5, TGCAGCACAACTCCCGGCTGCe | ||||||
| VII | FA2 | AF479588 | fa2-F, GCACGTCGTACTACACCAGCGCCA | 140 | 396 | 55.0 |
| fa2-R2, CCCCGTCAACCTGGGCCAATCAe | ||||||
| fa2-R3, CCGTCAACACCTCGAGTGGACACGAd | ||||||
| VII | SULP | AF481828 | SulP-R, TGCGTCCTTCGCTCAATCCCTGC | 339 | 193 | 55.0 |
| SulP-F2, GTGGGAGGGGTGGGACTTTGGGe | ||||||
| SulP-F3, GGTATGGGGATGTCCGCACGCTTCd | ||||||
| VIII | MCA1 | AF330231 | MCA1-F, CGCGGGCGAGTTTGCTGTTGCT | 113 | 226 | 55.0 |
| MCA1-R2, CGGATCCCGAACAGCGGCAGe | ||||||
| MCA1-R3, CCCCGTGACTCAATCAAGTCCCTGd | ||||||
| VIII | LI818 | X95326 | LI818-R, TCCGATGCACTCACGCTCACAGC | 334 | 141 | 55.0 |
| LI818-F2, TGGGCATGCGGAAATGCGTGTCe | ||||||
| LI818-F3, CTTGCTTGGCCGGCACGGGd | ||||||
| VIII | PSBQ | X13832 | OEE3-R, CGTGCTGTTGCGAGCCACTCCA | 347 | 535 | 65.0 |
| OEE3-F2, GCCGAGTTCTCAACCCTCTCGGCd | ||||||
| OEE3-F3, GGGTGCAACCTCCGGTGGCCTAe | ||||||
| IX | PPX1 | AF068635 | Ppx1-R, CATGGCACTTATGGGCGAAGCCG | 526 | 181 | 55.0 |
| Ppx1-F2, GGGCAAGCGGAGTGGAGGCGAe | ||||||
| Ppx1-F3, TCGAAGTGCCTTCGAAAGTGGGCAd | ||||||
| IX | PSBO | X13826 | OEE1-R, CGCATGCACGACGAGAAGCGAG | 510 | 161 | 55.0 |
| OEE1-F2, GTCGACCGCTGCGAGGAGGAGAe | ||||||
| OEE1-F3, CGAGCGCCGTATCATCCGGCTTAd | ||||||
| IX | MBO2 | AF394181 | MBO2-F, CGTTAACAGCCCTGAACTCGGCCG | 516 | 406 | 65.0 |
| MBO2-R2, TCACGCCACACCTGTACGTGCAAd | ||||||
| MBO2-R3, ATGCGCCAAACCCGGAGCTACCe | ||||||
| IX | CAH3 | U73856 | CAH3-F, CATGCAGCCCATCAAGGTGCCC | 307 | 359 | 55.0 |
| CAH3-R2, TCCCCACCGTGGGCCAAACCe | ||||||
| CAH3-R3, CGTGCAGGCGATGCCTCCAd | ||||||
| IX | MBB1 | AJ296291 | mbb1 F, GGATGAGGCGGTGGAGGCACTACA | 382 | 192 | 55.0 |
| mbb1-R2, GCGTGCCGCGTGTCACCAAGTAd | ||||||
| mbb1-R3, GGCCATACGCCATCATAACCGAGGe | ||||||
| X | KATANIN | AF205377 | Katan-F, ACGAGGAGTGGCTCAGCGTGTTCG | 394 | ∼410 | 55.0 |
| Katan-R, GGACGCCCAAGCTTCAAATCCACG | ||||||
| X | PF6 | AF327876 | PF6-R, GACAAACCCGTGTACCATCCGGCC | 362 | 229 | 55.0 |
| PF6-F2, GCACAAGCAATGCATCGGTGTGCe | ||||||
| PF6-F3, CGTTCCAGCGGCACTCACGGGd | ||||||
| X | CNA83 | B1724522 | CNA83-R, TGCACATACCTCTGCCGCTCCACC | 278 | 324 | 55.0 |
| CNA83-F3, CGGATCGGGTATGCGGATGCCAd | ||||||
| CNA83-F4, CGGCCGCTGAAGCTGCTGTGAe | ||||||
| XI | VFL2 | X57973 | vfl2-R, CCGCAGGCTGGCGATGGGAATA | 463 | 283 | 58.9 |
| vfl2-F2, GACGCCGGGGCTTGCTTTCACCe | ||||||
| vfl2-F4, TGCTGTGAAGGGTGGACACCCTGGd | ||||||
| XI | ODA2 | U15303 | ODA2-R, CACGCAGTGGCATCCTGCGC | 298 | 197 | 55.0 |
| ODA2-F2, TTAGGGAGGCGGCACTGACGCAe | ||||||
| ODA2-F3, AGCGTGCGATTGGCGTACGAGATTAd | ||||||
| XI | RPS14 | U06937 | CRY1-F, CATCCCCACCGACTCCACCCG | 268 | 248 | 54.2 (C. reinhardtii), 58.9 (S1-C5) |
| CRY1-R2, CCCGCCGCCGCCACCTAd | ||||||
| CRY1-R3, CCAGCCGCCAGGCGGGCe | ||||||
| XII/XIII | GAPC | L27669 | gapC-F, CAGATTGCTTCAGGGCTTCGGCG | 572 | ∼500 | 56.0 |
| gapC-R, TTCACGCACCGTGTGGCAGTCC | ||||||
| XII/XIII | PSR1 | AF174532 | Psr1-R, AGCACCCGTCCACACACCGCAA | 344 | 189 | 62.0 |
| Psr1-F2, GCACCTGCGCATGCATCTGTTGe | ||||||
| Psr1-F3, AGACAGCGGTTGGCCCTTGCTTGd | ||||||
| XII/XIII | EYE2 | AF233430 | EYE2-F, CGCGCGAGCTGACAGCTGAAGA | 525 | 211 | 64.8 |
| EYE2-R2, TCACATACTGCGCAGCGCTCTCCd | ||||||
| EYE2-R3, CGGGGTTGCCACAAGTTTCCTTCCe | ||||||
| XII/XIII | SAC3 | AF100162 | Sac3-R, ACTGCACAGCTCTGGGATGTCGCC | 324 | 508 | 55.0 |
| Sac3-F2, ACGGAGCGCACTGGGTTCTTGCAAd | ||||||
| Sac3-F3, TCGCGGTCCGGTCCCAGTATGe | ||||||
| XII/XIII | IC138 | IC138-F, CGGGGCAGGCGTAGGACTGGAA | 287 | 171 | 55.0 | |
| IC138-R2, GCAAGCCTGGCCCCATCTGTTCd | ||||||
| IC138-R3, CCTGGGCATCAGCACAGCACTTGe | ||||||
| XII/XIII | ODA12 | U89649 | LC2-F, GAGTAATGGTGCGGCCAAGCTGCC | 426 | ∼450 | 55.0 |
| LC2-R, TTGCAACGGCAAGCCGCCAT | ||||||
| XII/XIII | 14-3-3 | X79445 | 14-3-3R, AGTGCGCTTCAACACGCCTCACG | 317 | 205 | 55.0 |
| 14-3-3F2, CGGCGTGAAGTGGCGTTACAGCTAe | ||||||
| 14-3-3F3, TGACATTGTGTTGGCCATCACGGAd | ||||||
| XII/XIII | TUB2 | K03281 | b2-tub-R, CACGTGCACGAGTGTGTGGCCA | 113 | 283 | 55.0 |
| b2-tub-F2, GGAGGGGGGCCCATTGCCCd | ||||||
| b2-tub-F3, CGGCAGGGGCAGGTAACTGCCe | ||||||
| XII/XIII | RDB | BG858985 | CRB-R, CGTCAATTTGGCCGACCTGACCG | 558 | 257 | 55.0 |
| CRB-F2, CCCGAAGCCATGGGCATCGAAe | ||||||
| CRB-F3, GCGTCGACAACCATCTGCGACCAd | ||||||
| XIV | AC206 | U70999 | CCS1-R, ACGAATGCTGGGTGGGCCAAGC | 483 | 156 | 64.7 |
| CCS1-F2, GGGGTCACGACAGGGGTAGGGTGe | ||||||
| CCS1-F3, CGTGCGGCAAACAAGCACCCTTGd | ||||||
| XIV | IDA5 | D50838 | ACTIN-R, AAACCCCAGCGCTTTGGCGC | 249 | 529 | 55.0 |
| ACTIN-F2, AAGCGCTTGTGAGTGCGCCAGAd | ||||||
| ACTIN-F3, ACGCAGGTGGCAGGCCGAGGe | ||||||
| XV | EST 894011B10 | BE056715 | BE056715-R, CCCCCAAAATCAGCATGGGGTCC | 316 | 237 | 55.0 |
| BE056715-F2, TGGATGAGGTGGGGTCGTTTGTCGe | ||||||
| BE056715-F3, ACTGGCGTCGCGTCTGCAGGd | ||||||
| XV | IDA2 | AJ242525 | dhc10-F, TGCTGCTGTCGCTGGCCACGTA | 514 | ∼550 | 55.0 |
| dhc10-R, TCACGGCAACCTGAAAGGACGCC | ||||||
| XVI/XVII | CYTC1 | AF245393 | Cytc1-F, GCCCATCAAGTCGCAGCGCATC | 131 | 436 | 66.0 |
| Cytc1-R3, CAGCTGAACAGCCTGTGCGGCAd | ||||||
| Cytc1-R4, GCAAAGACACTCAGGCCGCGCTCe | ||||||
| XVI/XVII | ZYS3 | AB004043 | Zys3-F, AGCCGCCACGTGTTTGTGGAGG | 344 | ∼530 | 55.0 |
| Zys3-R, ACTGCCTTCTGGCTCGTATGCGGG | ||||||
| XVIII | CYTC6 | M67448 | CytC6-R, AAGCGCGTTCATGGTTCGGCC | 221 | 319 | 55.0 |
| CytC6-F2, GTGTAGCTAGCTTTTGCCCCGGCAd | ||||||
| CytC6-F3, CATCACGCAAATGGACACGTTCCGe | ||||||
| XVIII | CLPK1 | AB042714 | cpk1-F, GGATGGCAGCGTACCAGCTGTCACd | 314 | 252 | 62.0 |
| cpk1-R, CACCGCATGTGTATTGGAGGCGC | ||||||
| cpk1-F2, ATGCAGCAACGGTAGGCGCTAGCGe | ||||||
| XVIII | RAA2 | AJ243394 | Maa2-F, ATTGACCACTGCGGCGCTAGCG | 595 | ∼530 | 59.0 |
| Maa2-R, TAGTAGGGGCATCCGTGGCTCTCG | ||||||
| XVIII | ODA1 | AY039618 | CDS-022-F, GACGCGGCGGTGATGGGCd | 160 | 277 | 55.0 |
| CDS-022-R, CCCCGAGCGGATTGAGGTAATGG | ||||||
| CDS-022-F2, CAAGGGTTCAGGGGCAGAATACCGe | ||||||
| XVIII | IDA7 | AF159260 | IC140-F, TGCTTATGGAAGGGCTGGGCGG | 174 | 395 | 65.0 |
| IC140-R2, CTTGCGCCCGCCTCAGACACGe | ||||||
| IC140-R3, GCGCAAGGCTTCGTCAGGCTGTCd | ||||||
| XIX | ATP2 | X61624 | atpB-R, TCGCAGTCCGTACCCTTGACACCG | 102 | 255 | 55.0 |
| atpB-F2, GGCAGGGCGGTGCAGGCTTAAd | ||||||
| atpB-F3, CGGGGCCATGTCAGCATGGGAe | ||||||
| XIX | BAC 27e16 | PTQ10139.x1 | BAC27e16-F, GTCTGTGCAGCGCTGCGCCTTT | 296 | ∼400 | 64.3 |
| BAC27e16-R, AATGGCCAGGATGTGCGGGTAGC | ||||||
| XIX | ZYS1B | X76117 | zys1B-F, GCTTTGAGTGGAGCGAGGCGCA | 466 | 303 | 61.0 |
| zys1B-R2, TAAATGCATCTCCGCAGTTTTCTCCGb | ||||||
| zys1B-R3, GCATTGGGCATAACCAGTATGTGCCAe | ||||||
| XIX | FLA10 | L33697 | FLA10-F, CTGCGCGCCAGCAAGCTCAAGT | 491 | 467 | 55.0 |
| FLA10-R, GGTAACAGCCCGTCTTCCAGGGCC | ||||||
| XIX | CNB60B1 | L27471 | cr40-F2, GATTGGGGGCAGTGGGCAGGe | 388 | 350 | 56.0 |
| cr40-R2, CACCGCCATGCGAAAGTGCC | ||||||
| cr40-F3, GCCTGTGCGGGATGGCGTGAGe |
All amplification reactions should be performed as described in Materials and Methods, at the annealing temperature given for each locus, and all of the primers listed should be mixed in a single tube for each amplification.
Accession numbers refers to entries in the GenBank database (http://www.ncbi.nlm.nih.gov/), except for marker BAC 27e16 on linkage group XIX, which is from the JGI Chlamydomonas BAC-end sequence database (http://bahama.jgi-psf.org/prod/bin/chlamy/home.chlamy.cqi).
When only two primers are listed they generate allele-specific product size differences.
S1-C5-specific primers.
21gr-specific primers.
The predominant product when more than two amplification products were observed.
Anchoring the molecular map to the genetic map.
For most linkage groups, the molecular map was anchored to the genetic map by using as mapping probes genes corresponding to mapped phenotypic markers. On linkage group I, for example, molecular probes for the LF3 gene (149) and for the ARG7 gene (19) were mapped, allowing the molecular map to be oriented relative to the genetically mapped lf3 and arg7 loci. Cloned genes corresponding to mapped mutations were not available for some linkage groups. For these six linkage groups, the molecular map was oriented relative to the preexisting genetic map by reference to earlier molecular mapping results that provided information about centromere linkage. The orientation of linkage group II, for example, is based on the observation that the centromere distances for markers S6175 and S6135 are 3 and 2 cM, respectively. For linkage group IV, the orientation of the map was supported by data from tetrads indicating that TUA2 (α2 tubulin) and PYR1 (the pyrithiamine resistance gene) are on opposite sides of the centromere and that TUA2 maps within 7 cM of its centromere (120). For linkage group V, the orientation of the map was determined by examining the data from tetrad progeny showing that DHC6 lies between the PF26 marker and the centromere (113). The correspondence of the molecular and genetic maps for linkage group XVI/XVII is based on the demonstration that DHC9 is linked to the phenotypic marker y1 (113). The orientation of the molecular map for linkage group XIX is supported by data showing that the EF3A marker maps within 3 cM of the centromere (120). For linkage groups VIII, XV, and XVI/XVII, the orientation of the anchored map relative to the genetic map is not known because there is only a single point of anchorage.
For most loci, cloned genes corresponding to previously mapped phenotypic loci were placed on the expected locations on the molecular map. The single exception was the ac21 locus on linkage group XI. The PETC gene (encoding the chloroplast Rieske iron-sulfur center protein) has been shown to be the gene affected by the ac21 mutation (5, 22, 23). When we mapped PETC, however, it was indistinguishable from VFL2 by recombination. The map location of ac21 was previously determined to be on the opposite arm of linkage group XI.
DISCUSSION
The availability of the C. reinhardtii molecular map should enable researchers to take advantage of rapid advances in C. reinhardtii genomics to identify genes corresponding to mapped mutations. Sequences from more than 100,000 cDNA clones are publicly available (Chlamydomonas Genetics Center, Duke University [http://www.biology.duke.edu/chlamy_genome/]; Kazusa DNA Research Institute, Kazusa, Japan [http://www.kazusa.or.jp/en/plant/chlamy/EST/]). The ends of 15,000 bacterial artificial chromosome (BAC) clones have been sequenced by the Joint Genome Institute (JGI; Walnut Creek, Calif.) and are available for searching by use of BLAST algorithms (http://bahama.jgi-psf.org/prod/bin/chlamy/home.chlamy.cgi). The complete sequence of the nuclear genome is being completed by the JGI and should be available early in 2003.
All of these resources taken together should allow positional cloning and candidate gene approaches to be used to clone the genes identified by mapped mutations. BAC clone contigs anchored on each of the molecular markers have already been prepared. The total length of genomic DNA contained in these contigs represents more than 20% of the nuclear genome. By comparing the sequence of the nuclear genome to the sequences of the BAC clone ends, it will be possible to rapidly complete the coverage of the nuclear genome with mapped BAC clones.
For positional cloning, C. reinhardtii offers many technical advantages for efficient testing of whether a particular BAC clone corresponds to a mutation of interest. Transformation of C. reinhardtii involves simple and fast procedures such as vortexing with glass beads (59) or electroporation (132). The efficiency of cotransformation with BAC clones and a selectable marker gene on a separate plasmid is usually in the range of 1 to 2%, and it is easy to generate hundreds of transformants for screening. A BAC clone can be tested for rescue of a mutant phenotype in less than 2 weeks. Because screening of BAC clones for phenotypic rescue is so rapid and efficient, it should not be necessary to do extensive genetic mapping of new mutations in preparation for cloning. Numerous BAC clones, covering a large genetic interval, can be readily tested for phenotypic rescue of a mutation of interest upon transformation. As the sequence of the nuclear genome is completed and annotated, it should be possible to accelerate positional cloning and transformation using a candidate gene approach.
To use positional cloning or candidate gene approaches to clone genes corresponding to mutations, it is necessary to place these mutations on the genetic map. Hundreds of mutations have been mapped using phenotypic markers and multiply marked strains, but genetic mapping using phenotypic markers is tedious and is limited by the availability of useful mutations. A serious limitation is that many potentially useful genetic markers produce a similar phenotype, such as acetate auxotrophy or paralyzed flagella, so that only one mutant of each type can be used in an individual mapping cross. The molecular markers described in this report should greatly facilitate genetic mapping of new mutations in preparation for positional cloning.
To map the mutation in a new mutant strain, it should first be crossed to the S1-C5 strain, and 20 to 50 random progeny from different tetrads should be scored for the phenotype of interest. Small quantities of DNA from each progeny strain can then be used as template DNA for PCR-based mapping strategies. The primers for SNP scoring reported in this study (Table 2), because they produce PCR products of distinguishable sizes from the two parent strains, facilitate scoring of molecular markers in the progeny strains by using readily available thermal cycler and gel electrophoresis equipment. The marker set covers most of the genome, with multiple loci representing both arms of most linkage groups. Previously, primers for the scoring of SNPs using high-throughput methods were defined for 186 loci, including many of those mapped in this study (159).
A hierarchical approach to marker selection for mapping can be used to limit the number of PCRs to be performed. For each linkage group, the first marker to be tested for linkage analysis should be one that maps near the center of the group of markers. More than 50% of the molecular markers we developed map to only five linkage groups (I, II, III, VI, and XII/XIII). That the density of molecular markers corresponds roughly to the underlying gene density is suggested by the fact that slightly more than 50% of previously mapped mutations map to these five linkage groups as well. The first markers to be tested, therefore, should be from these linkage groups. If no linkage is detected between the mutant phenotype and the central marker on each linkage group, additional tests with markers spaced 20 to 30 cM from the first marker may be carried out until linkage is detected.
As soon as a new mutation is mapped to an arm of one of the linkage groups, other molecular markers mapping to that arm can be used to attempt to place the mutation between pairs of flanking markers. Consulting the overlapping BAC contig map will then allow the choice of BAC clones to be tested for phenotypic rescue using cotransformation with a selectablemarker gene (http://www.biology.duke.edu/chlamy_genome/BAC/index.html). A number of selectable markers are available for transformation of C. reinhardtii (47). If a project requires fine structure mapping of a genetic region, and multiple STS markers are not available, it is possible to generate additional STS loci. The sequences of all the BAC ends are available, and reference to the BAC contig map places these sequences at intervals of tens of thousands of bases along the genome. A BAC end sequence can be turned into an STS locus by sequencing the same region of genomic DNA from the S1-C5 strain and using sequence polymorphisms to design informative primers for PCR. The high degree of sequence polymorphism between the laboratory strains of C. reinhardtii and S1-C5 makes the task of finding useful sequence polymorphisms routine. One source of sequence polymorphisms is the set of ESTs derived from CC-2290 (strain S1-D2), available in the National Center for Biotechnology Information (NCBI) database. Many of these EST sequences were used to generate the markers reported in this study. Another possible source for additional molecular markers for mapping is microsatellite repeat sequences, which have been shown to be abundant in the C. reinhardtii genome (56).
ADDENDUM IN PROOF
The sequence of the Chlamydomonas genome obtained by the DOE Joint Genome Institute is posted on the Internet at http://genome.jgi-psf.org/chlre1/chlre1.home.html.
Acknowledgments
This work was supported by grants from the NSF (MCB 9975765 and MCB 8819133), the NIH (GM 34437), and the University of Minnesota Experiment Station. W. J. Brazelton was supported by a Mabel and Arnold Beckman Foundation Award. C. reinhardtii strains were obtained from the Chlamydomonas Genetics Center at Duke University (supported by NSF grant DBI 9319941).
We thank Laura Ranum and Marilyn Kobayashi for contributions to the early stages of this work. We also thank our many colleagues in the Chlamydomonas community for providing molecular markers for mapping.
REFERENCES
- 1.Adair, W. S., and W. J. Snell. 1990. The Chlamydomonas cell wall: structure, biochemistry, and molecular biology, p. 15-84. In R. P. Mecham and W. S. Adair (ed.), Matrix organization and assembly of plant and animal extracellular matrix. Academic Press, Orlando, Fla.
- 2.Asamizu, E., K. Miura, K. Kucho, Y. Inoue, H. Fukuzawa, K. Ohyama, Y. Nakamura, and S. Tabata. 2000. Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 7:305-307. [DOI] [PubMed] [Google Scholar]
- 3.Ball, S. G. 1998. Regulation of starch biosynthesis, p. 549-567. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 4.Benashski, S. E., R. S. Patel-King, and S. M. King. 1999. Light chain 1 from Chlamydomonas outer dynein arm is a leucine-rich repeat protein associated with the motor domain of the gamma heavy chain. Biochemistry 38:7253-7264. [DOI] [PubMed] [Google Scholar]
- 5.Bendall, D. S., M. Sanguansermsri, J. Girard-Bascou, and P. Bennoun. 1986. Mutations of Chlamydomonas reinhardtii affecting the cytochrome bf complex. FEBS Lett. 203:31-35. [Google Scholar]
- 6.Bernstein, M., P. L. Beech, S. G. Katz, and J. L. Rosenbaum. 1994. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 125:1313-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, C. L., A. J. Cain, S. Purton, and J. H. A. Nugent. 1999. Molecular cloning and sequence analysis of the Chlamydomonas nuclear gene encoding the photosystem II subunit PsbW. Plant Physiol. 121:313.. [DOI] [PubMed] [Google Scholar]
- 8.Bowman, A. B., R. S. Patel-King, S. E. Benashski, M. McCaffery, L. S. B. Goldstein, and S. M. King. 1999. Drosophila roadblock and Chlamydomonas LC7: a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility and mitosis. J. Cell Biol. 146:165-179. [PMC free article] [PubMed] [Google Scholar]
- 9.Brazelton, W. J., C. D. Amundsen, C. D. Silflow, and P. A. Lefebvre. 2001. The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 11:1591-1594. [DOI] [PubMed] [Google Scholar]
- 10.Chekounova, E., V. Voronetskaya, J. Papenbrock, B. Grimm, and C. F. Beck. 2001. Characterization of Chlamydomonas mutants defective in the H subunit of Mg-chelatase. Mol. Genet. Genomics 266:363-373. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Q., and C. D. Silflow. 1996. Isolation and charaterization of glutamine synthetase genes in Chlamydomonas reinhardtii. Plant Physiol. 112:987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curry, A. M., B. D. Williams, and J. L. Rosenbaum. 1992. Sequence analysis reveals homology between two proteins of the flagellar radial spoke. Mol. Cell. Biol. 12:3967-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies, J. P., F. Yildiz, and A. R. Grossman. 1994. Mutants of Chlamydomonas reinhardtii with aberrant responses to sulfur deprivation. Plant Cell 6:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, J. P., and A. R. Grossman. 1998. Responses to deficiencies in macronutrients, p. 613-635. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 15.Davies, J. P., F. Yildiz, and A. R. Grossman. 1996. SacI, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 15:2150-2159. [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, J. P., F. H. Yildiz, and A. R. Grossman. 1999. Sac3, an Snf1-like serine/threonine kinase that positively and negatively regulates the responses of Chlamydomonas to sulfur limitation. Plant Cell 11:1179-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Day, A., M. Schirmer-Rahire, M. R. Kuchka, S. P. Mayfield, and J.-D. Rochaix. 1988. A transposon with an unusual arrangement of long terminal repeats in the green alga Chlamydomonas reinhardtii. EMBO J. 7:1917-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deane, J. A., D. G. Cole, E. S. Seeley, D. R. Diener, and J. L. Rosenbaum. 2001. Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr. Biol. 11:1586-1590. [DOI] [PubMed] [Google Scholar]
- 19.Debuchy, R., S. Purton, and J.-D. Rochaix. 1989. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8:2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Hostos, E. L., J. Schilling, and A. R. Grossman. 1989. Structure and expression of the gene encoding the periplasmic arylsulfatase of Chlamydomonas reinhardtii. Mol. Gen. Genet. 218:229-239. [DOI] [PubMed] [Google Scholar]
- 21.Deininger, W., P. Kroger, U. Hegemann, F. Lottspeich, and P. Hegemann. 1995. Chlamyrhodopsin represents a new type of sensory photoreceptor. EMBO J. 14:5849-5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Vitry, C. 1994. Characterization of the gene of the chloroplast Rieske iron-sulfur protein in Chlamydomonas reinhardtii: indications for an uncleaved lumen targeting sequence. J. Biol. Chem. 269:7603-7609. [PubMed] [Google Scholar]
- 23.de Vitry, C., G. Finazzi, F. Baymann, and T. Kallas. 1999. Analysis of the nucleus-encoded and chloroplast-targeted Rieske protein by classic and site-directed mutagenesis of Chlamydomonas. Plant Cell 11:2031-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiBella, L. M., and S. M. King. 2001. Dynein motors of the Chlamydomonas flagellum. Int. Rev. Cytol. 210:227-268. [DOI] [PubMed] [Google Scholar]
- 25.Dieffenbach, C. W., T. M. J. Lowe, and G. S. Dveksler. 1995. General concepts for PCR primer design, p. 133-142. In D. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Dietmaier, W., S. Fabry, H. Huber, and R. Schmitt. 1995. Analysis of a family of ypt genes and their products from Chlamydomonas reinhardtii. Gene 158:41-50. [DOI] [PubMed] [Google Scholar]
- 27.Dryzmalla, C., M. Schroda, and C. F. Beck. 1996. Light-inducible gene HSP70B encodes a chloroplast-localized heat shock protein in Chlamydomonas reinhardtii. Plant Mol. Biol. 31:1185-1194. [DOI] [PubMed] [Google Scholar]
- 28.Dutcher, S. K. 2000. Chlamydomonas reinhardtii: biological rationale for genomics. J. Eukaryot. Microbiol. 47:340-349. [DOI] [PubMed] [Google Scholar]
- 29.Dutcher, S. K., J. Power, R. E. Galloway, and M. E. Porter. 1991. Reappraisal of the genetic map of Chlamydomonas reinhardtii. J. Hered. 82:295-301. [DOI] [PubMed] [Google Scholar]
- 30.Dutcher, S. K., and E. C. Trabuco. 1998. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes δ-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell 9:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez, E., A. Galvan, and A. Quesada. 1998. Nitrogen assimilation and its regulation, pp. 15-84. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Fernandez, E., R. Schnell, L. P. W. Ranum, S. C. Hussey, C. D. Silflow, and P. A. Lefebvre. 1989. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 86:6449-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferris, P. J., and U. W. Goodenough. 1987. Transcription of novel genes, including a gene linked to the mating-type locus, induced by Chlamydomonas fertilization. Mol. Cell. Biol. 7:2360-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferris, P. J., J. P. Woessner, and U. W. Goodenough. 1996. A sex recognition glycoprotein is encoded by the plus mating-type gene fus1 of Chlamydomonas reinhardtii. Mol. Biol. Cell 7:1235-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferris, P. J., and U. W. Goodenough. 1997. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146:859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferris, P. J., E. V. Armbrust, and U. W. Goodenough. 2002. Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160:181-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finst, R. J., P. J. Kim, E. R. Griffis, and L. M. Quarmby. 2000. Fa1p is a 171 kDa protein essential for axonemal microtubule severing in Chlamydomonas. J. Cell Sci. 113:1963-1971. [DOI] [PubMed] [Google Scholar]
- 38.Franzen, L. G., and G. Falk. 1992. Nucleotide sequence of cDNA clones encoding the beta subunit of mitochondrial ATP synthase from the green alga Chlamydomonas reinhardtii: the precursor protein encoded by the cDNA contains both an N-terminal presequence and a C-terminal extension. Plant Mol. Biol. 19:771-780. [DOI] [PubMed] [Google Scholar]
- 39.Funke, R. P., J. L. Kovar, and D. P. Weeks. 1997. Intracellular carbonic anhydrase is essential to photosynthesis in Chlamydomonas reinhardtii at atmospheric levels of CO2. Demonstration via genomic complementation of the high-CO2-requiring mutant ca-1. Plant Physiol. 114:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glockner, G., and C. F. Beck. 1997. Cloning and characterization of LRG5, a gene involved in blue light signaling in Chlamydomonas gametogenesis. Plant J. 12:677-683. [DOI] [PubMed] [Google Scholar]
- 41.Goldschmidt-Clermont, M., and M. Rahire. 1986. Sequence, evolution and differential expression of the two genes encoding variant small subunits of ribulose biphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. J. Mol. Biol. 191:421-432. [DOI] [PubMed] [Google Scholar]
- 42.Goodenough, U., E. V. Armbrust, A. M. Campbell, and P. J. Ferris. 1995. Molecular genetics of sexuality in Chlamydomonas. Annu. Rev. Plant Physiol. 46:21-44. [Google Scholar]
- 43.Gorman, D. S., and R. P. Levine. 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 54:1665-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gross, C. H., L. P. W. Ranum, and P. A. Lefebvre. 1988. Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13:503-508. [DOI] [PubMed] [Google Scholar]
- 45.Harris, E. H. 1989. The Chlamydomonas sourcebook. Academic Press, New York, N.Y.
- 46.Harris, E. H. 1993. Chlamydomonas reinhardtii, p. 2.156-2.169. In S. J. O'Brien (ed.), Genetic maps: a compilation of linkage and restriction maps of genetically studied organisms. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Harris, E. H. 2001. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:363-406. [DOI] [PubMed] [Google Scholar]
- 48.Harrison, A., P. Olds-Clarke, and S. M. King. 1998. Identification of the t complex-encoded cytoplasmic dynein light chain tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J. Cell Biol. 140:1137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hegemann, P. 1997. Vision in microalgae. Planta 203:265-274. [DOI] [PubMed] [Google Scholar]
- 50.Hicks, G. R., C. M. Hironaka, D. Dauvillee, R. P. Funke, C. D'Hulst, S. Waffenschmidt, and S. G. Ball. 2001. When simpler is better. Unicellular green algae for discovering new genes and functions in carbohydrate metabolism. Plant Physiol. 127:1334-1338. [PMC free article] [PubMed] [Google Scholar]
- 51.Hill, K. L., H. H. Li, J. Singer, and S. Merchant. 1991. Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J. Biol. Chem. 266:15060-15067. [PubMed] [Google Scholar]
- 52.Holmes, J. A., D. E. Johnson, and S. K. Dutcher. 1993. Linkage group XIX of Chlamydomonas reinhardtii has a linear map. Genetics 133:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52a.Huang, K., T. Merkle, and C. F. Beck. 2002. Islation and characterization of a Chlamydomonas gene that encodes a putative blue-light photoreceptor of the phototropin family. Physiol. Plant. 115:613-622. [DOI] [PubMed] [Google Scholar]
- 53.Hwang, S., and D. L. Herrin. 1993. Characterization of a cDNA encoding the 20-kDa photosystem I light-harvesting polypeptide of Chlamydomonas reinhardtii. Curr. Genet. 23:512-517. [DOI] [PubMed] [Google Scholar]
- 54.Imbault, P., C. Wittemer, U. Johanningmeier, J. D. Jacobs, and S. H. Howell. 1988. Structure of the Chlamydomonas reinhardtii cabII-1 gene encoding a chlorophyll-a/b-binding protein. Gene 73:397-407. [DOI] [PubMed] [Google Scholar]
- 55.Inoue, K., B. W. Dreyfuss, K. L. Kindle, D. B. Stern, S. Merchant, and O. A. Sodeinde. 1997. Ccs1, a nuclear gene required for the post-translational assembly of chloroplast c-type cytochromes. J. Biol. Chem. 272:31747-31754. [DOI] [PubMed] [Google Scholar]
- 56.Kang, T. J., and M. W. Fawley. 1997. Variable (CA/GT)n simple sequence repeat DNA in the alga Chlamydomonas. Plant Mol. Biol. 35:943-948. [DOI] [PubMed] [Google Scholar]
- 57.Kato-Minoura, T., S. Uryu, M. Hirono, and R. Kamiya. 1998. Highly divergent actin expressed in a Chlamydomonas mutant lacking the conventional actin gene. Biochem. Biophys. Res. Commun. 251:71-76. [DOI] [PubMed] [Google Scholar]
- 58.Kersanach, R., H. Brinkmann, M. F. Liaud, D. X. Zhang, W. Martin, and R. Cerff. 1994. Five identical intron positions in ancient duplicated genes of eubacterial origin. Nature 367:387-389. [DOI] [PubMed] [Google Scholar]
- 59.Kindle, K. L. 1990. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87:1228-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.King, S. M., and R. S. Patel-King. 1995. Identification of a Ca2+-binding light chain within Chlamydomonas outer arm dynein. J. Cell Sci. 108:3757-3764. [DOI] [PubMed] [Google Scholar]
- 61.King, S. M., and R. S. Patel-King. 1995. The Mr = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J. Biol. Chem. 270:11445-11452. [DOI] [PubMed] [Google Scholar]
- 62.Kinoshita, T., H. Fukuzawa, T. Shimada, T. Saita, and Y. Matsuda. 1992. Primary structure and expression of a gamete lytic enzyme in Chlamydomonas reinhardtii: similarity of functional domains to matrix metalloproteases. Proc. Natl. Acad. Sci. USA 89:4693-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koutoulis, A., G. J. Pazour, C. G. Wilkerson, K. Inaba, H. Sheng, S. Takada, and G. B. Witman. 1997. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J. Cell Biol. 137:1069-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kubo, T., J. Abe, T. Saito, and Y. Matsuda. 2002. Genealogical relationships among laboratory strains of Chlamydomonas reinhardtii as inferred from matrix metalloprotease genes. Curr. Genet. 41:115-122. [DOI] [PubMed] [Google Scholar]
- 65.Kuriyama, H., H. Takano, L. Suzuki, H. Uchida, S. Kawano, H. Kuroiwa, and T. Kuroiwa. 1999. Characterization of Chlamydomonas reinhardtii zygote-specific cDNAs that encode novel proteins containing ankyrin repeats and WW domains. Plant Physiol. 119:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurvari, V., N. V. Grishin, and W. J. Snell. 1998. A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. J. Cell Biol. 28:1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kurvari, V., F. Qian, and W. J. Snell. 1995. Increased transcript levels of a methionine synthase during adhesion-induced activation of Chlamydomonas reinhardtii gametes. Plant Mol. Biol. 29:1235-1252. [DOI] [PubMed] [Google Scholar]
- 68.Kwok, S., S.-Y. Chang, J. J. Sninsky, and A. Wang. 1995. Design and use of mismatched and degenerate primers, p. 143-155. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 69.Lander, E., P. Green, J. Abrahamson, A. Barlow, M. Daly, S. E. Lincoln, and L. Newburg. 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174-181. [DOI] [PubMed] [Google Scholar]
- 70.Lechtreck, K. F., and C. D. Silflow. 1997. SF-assemblin in Chlamydomonas: sequence conservation and localization during the cell cycle. Cell Motil. Cytoskeleton 36:190-201. [DOI] [PubMed] [Google Scholar]
- 71.Ledizet, M., and G. Piperno. 1995. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol. Biol. Cell 6:697-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee, V. D., S. L. Finstad, and B. Huang. 1997. Cloning and characterization of a gene encoding an actin-related protein in Chlamydomonas. Gene 197:153-159. [DOI] [PubMed] [Google Scholar]
- 73.Lee, V. D., M. Stapleton, and B. Huang. 1991. Genomic structure of Chlamydomonas caltractin. Evidence for intron insertion suggests a probable genealogy for the EF-hand superfamily of proteins. J. Mol. Biol. 221:175-191. [PubMed] [Google Scholar]
- 74.Lefebvre, P. A., and C. D. Silflow. 1999. Chlamydomonas: the cell and its genomes. Genetics 151:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levine, R. P., and W. T. Ebersold. 1960. The genetics and cytology of Chlamydomonas. Annu. Rev. Microbiol. 14:197-216. [DOI] [PubMed] [Google Scholar]
- 76.Li, J., and M. P. Timko. 1996. The pc-1 phenotype of Chlamydomonas reinhardtii results from a deletion mutation in the nuclear gene for NADPH:protochlorophyllide oxidoreductase. Plant Mol. Biol. 30:15-37. [DOI] [PubMed] [Google Scholar]
- 77.Liebich, I., and J. Voigt. 1995. A Chlamydomonas homologue to the 14-3-3 proteins: cDNA and deduced amino acid sequence. Biochim. Biophys. Acta 1263:79-85. [DOI] [PubMed] [Google Scholar]
- 78.Lincoln, S., M. Daly, and E. Lander. 1992. Constructing genetic maps with MAPMAKER/EXP 3.0, 3rd ed. Whitehead Institute Technical Report. Whitehead Institute, Cambridge, Mass.
- 79.Lincoln, S. E., M. J. Daly, and E. S. Lander. 1991. PRIMER: a computer program for automatically selecting PCR primers. Whitehead Institute for Biomedical Research, Cambridge, Mass.
- 80.Lohret, T. A., L. Zhao, and L. M. Quarmby. 1999. Cloning of Chlamydomonas p60 katanin and localization to the site of outer doublet severing during deflagellation. Cell Motil. Cytoskeleton 43:221-231. [DOI] [PubMed] [Google Scholar]
- 81.Mahjoub, M., B. Montpetit, L. Zhao, R. J. Finst, B. Goh., A. C. Kim, and L. M. Quarmby. 2002. The FA2 gene of Chlamydomonas encodes a MINA family kinase with roles in cell cycle progression and microtubule severing during deflagellation. J. Cell Sci. 115:1759-1768. [DOI] [PubMed] [Google Scholar]
- 82.Manly, K. F., R. H. Cudmore, and J. M. Meer. 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12:930-932. [DOI] [PubMed] [Google Scholar]
- 83.Matters, G. L., and S. I. Beale. 1994. Structure and light-regulated expression of the gsa gene encoding the chlorophyll biosynthetic enzyme, glutamate 1-semialdehyde aminotransferase, in Chlamydomonas reinhardtii. Plant Mol. Biol. 24:617-629. [DOI] [PubMed] [Google Scholar]
- 84.Matters, G. L., and S. I. Beale. 1995. Structure and expression of the Chlamydomonas reinhardtii alad gene encoding the chlorophyll biosynthetic enzyme, δ-aminolevulinic acid dehydratase (porphobilinogen synthase). Plant Mol. Biol. 27:607-617. [DOI] [PubMed] [Google Scholar]
- 85.Mayfield, S. P., G. Schirmer-Rahire, H. Frank, H. Zuber, and J.-D. Rochaix. 1989. Analysis of the genes of the OEE1 and OEE3 proteins of the photosystem II complex of Chlamydomonas reinhardtii. Plant Mol. Biol. 12:683-693. [DOI] [PubMed] [Google Scholar]
- 86.Mitchell, D. R. 2000. Chlamydomonas flagella. J. Phycol. 36:261-273. [Google Scholar]
- 87.Mitchell, D. R., and K. S. Brown. 1994. Sequence analysis of the Chlamydomonas alpha and beta dynein heavy chain genes. J. Cell Sci. 107:635-644. [DOI] [PubMed] [Google Scholar]
- 88.Mitchell, D. R., and K. S. Brown. 1997. Sequence analysis of the Chlamydomonas reinhardtii flagellar alpha dynein gene. Cell Motil. Cytoskeleton 37:120-126. [DOI] [PubMed] [Google Scholar]
- 89.Mitchell, D. R., and Y. Kang. 1991. Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J. Cell Biol. 113:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mitchell, D. R., and W. S. Sale. 1999. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 144:293-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mittag, M. 2001. Circadian rhythms in microalgae. Int. Rev. Cytol. 206:213-247. [DOI] [PubMed] [Google Scholar]
- 92.Moseley, J., J. Quinn, M. Eriksson, and S. Merchant. 2000. The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19:2139-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Muller, F. W., G. L. Igloi, and C. F. Beck. 1992. Structure of a gene encoding heat-shock protein HSP70 from the unicellular alga Chlamydomonas reinhardtii. Gene 111:165-173. [DOI] [PubMed] [Google Scholar]
- 94.Myster, S. H., J. A. Knott, E. O'Toole, and M. E. Porter. 1997. The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol. Biol. Cell 8:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nelson, J. A., P. B. Savereide, and P. A. Lefebvre. 1994. The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14:4011-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Norrander, J. M., A. M. Decathelineau, J. A. Brown, M. E. Porter, and R. W. Linck. 2000. The Rib43a protein is associated with forming the specialized protofilament ribbons of flagellar microtubules in Chlamydomonas. Mol. Biol. Cell 1:201-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Palombella, A. L., and S. K. Dutcher. 1998. Identification of the gene encoding the tryptophan synthase beta-subunit from Chlamydomonas reinhardtii. Plant Physiol. 117:455-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pan, J., and W. J. Snell. 2000. Regulated targeting of a protein kinase into an intact flagellum. An aurora/Ipl1p-like protein kinase translocates from the cell body into the flagella during gamete activation in Chlamydomonas. J. Biol. Chem. 275:24106-24114. [DOI] [PubMed] [Google Scholar]
- 99.Patel-King, R. S., S. E. Benashski, A. Harrison, and S. M. King. 1996. Two functional thioredoxins containing redox-sensitive vicinal dithiols from the Chlamydomonas outer dynein arm. J. Biol. Chem. 271:6283-6291. [DOI] [PubMed] [Google Scholar]
- 100.Pazour, G. J., B. L. Dickert, Y. Vucica, E. S. Seeley, J. L. Rosenbaum, G. B. Witman, and D. G. Cole. 2000. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 151:709-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pazour, G. J., O. A. Sineschekov, and G. B. Witman. 1995. Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J. Cell Biol. 131:427-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pazour, G. J., C. G. Wilkerson, and G. B. Witman. 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141:979-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Peltzer-Reith, B., S. Freund, C. Schnarrenberger, H. Yatsuki, and K. Hori. 1995. The plastid aldolase gene from Chlamydomonas reinhardtii: intron/exon organization, evolution, and promoter structure. Mol. Gen. Genet. 248:481-486. [DOI] [PubMed] [Google Scholar]
- 104.Perez-Martinez, X., A. Antaramian, M. Vazquez-Acevedo, S. Funes, E. Tolkunova, J. d'Alayer, M. G. Claros, E. Davidson, M. P. King, and D. Gonzalez-Halphen. 2001. Subunit II of cytochrome c oxidase in chlamydomonad algae is a heterodimer encoded by two independent nuclear genes. J. Biol. Chem. 276:11302-11309. [DOI] [PubMed] [Google Scholar]
- 105.Perez-Martinez, X., M. Vazquez-Acevedo, E. Tolkunova, S. Funes, M. G. Claros, E. Davidson, M. P. King, and D. Gonzalez-Halphen. 2000. Unusual location of a mitochondrial gene. Subunit III of cytochrome c oxidase is encoded in the nucleus of chlamydomonad algae. J. Biol. Chem. 275:30144-30152. [DOI] [PubMed] [Google Scholar]
- 106.Perron, K., M. Goldschmidt-Clermont, and J.-D. Rochaix. 1999. A factor related to pseudouridine synthases is required for chloroplast group II intron trans-splicing in Chlamydomonas reinhardtii. EMBO J. 18:6481-6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Perrone, C. A., P. Yang, E. O'Toole, W. S. Sale, and M. E. Porter. 1998. The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol. Biol. Cell 9:3351-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perrone, C. A., S. H. Myster, R. Bower, E. T. O'Toole, and M. E. Porter. 2000. Insights into the structural organization of the I1 inner arm dynein from a domain analysis of the 1β dynein heavy chain. Mol. Biol. Cell 11:2297-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Petersen, J. L., D. W. Lang, and G. D. Small. 1999. Cloning and characterization of a class II DNA photolyase from Chlamydomonas. Plant Mol. Biol. 40:1063-1071. [DOI] [PubMed] [Google Scholar]
- 110.Petracek, M. E., L. M. Konkel, M. L. Kable, and J. Berman. 1994. A Chlamydomonas protein that binds single-stranded G-stranded telomere DNA. EMBO J. 13:3648-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Petridou, S., K. Foster, and K. Kindle. 1997. Light induces accumulation of isocitrate lyase mRNA in a carotenoid-deficient mutant of Chlamydomonas reinhardtii. Plant Mol. Biol. 33:381-392. [DOI] [PubMed] [Google Scholar]
- 112.Porter, M. E., R. Bower, J. A. Knott, P. Byrd, and W. Dentler. 1999. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10:693-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Porter, M. E., J. A. Knott, S. H. Myster, and S. J. Farlow. 1996. The dynein gene family in Chlamydomonas reinhardtii. Genetics 144:569-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Porter, M. E., and W. S. Sale. 2000. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 151:F37-F42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Preble, A. M., T. M. Giddings, Jr., and S. K. Dutcher. 2000. Basal bodies and centrioles: their function and structure. Curr. Top. Dev. Biol. 49:207-233. [DOI] [PubMed] [Google Scholar]
- 116.Quinn, J., H. H. Singer, J. Morimoto, L. Mets, K. Kindle, and S. Merchant. 1993. The plastocyanin-deficient phenotype of Chlamydomonas reinhardtii Ac-208 results from a frame shift mutation in the nuclear gene encoding preapoplastocyanin. J. Biol. Chem. 268:7832-7841. [PubMed] [Google Scholar]
- 117.Quinn, J. M., S. S. Nakamoto, and S. Merchant. 1999. Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J. Biol. Chem. 274:14444-14454. [DOI] [PubMed] [Google Scholar]
- 118.Randolph-Anderson, B. L., R. Sato, A. M. Johnson, E. H. Harris, C. R. Hauser, K. Oeda, F. Ishige, S. Nishio, N. W. Gillham, and J. E. Boynton. 1998. Isolation and characterization of a mutant protoporphyrinogen oxidase gene from Chlamydomonas reinhardtii conferring resistance to porphyric herbicides. Plant Mol. Biol. 38:839-859. [DOI] [PubMed] [Google Scholar]
- 119.Ranum, L. P. W. 1989. Mapping nuclear sequences of Chlamydomonas reinhardtii using restriction fragment length polymorphisms. Ph.D. thesis. University of Minnesota, St. Paul.
- 120.Ranum, L. P. W., M. D. Thompson, J. A. Schloss, P. A. Lefebvre, and C. D. Silflow. 1988. Mapping flagellar genes in Chlamydomonas using restriction fragment length polymorphisms. Genetics 120:109-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Richard, C., H. Ouellet, and M. Guertin. 2000. Characterization of the LI818 polypeptide from the green unicellular alga Chlamydomonas reinhardtii. Plant Mol. Biol. 42:303-316. [DOI] [PubMed] [Google Scholar]
- 122.Roberts, D. G. W., M. R. Lamb, and C. L. Dieckmann. 2001. Characterization of the EYE2 gene required for eyespot assembly in Chlamydomonas reinhardtii. Genetics 158:1037-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rochaix, J.-D. 2001. Assembly, function, and dynamics of the photosynthetic machinery in Chlamydomonas reinhardtii. Plant Physiol. 127:1394-1398. [PMC free article] [PubMed] [Google Scholar]
- 124.Rosenbaum, J. L., D. G. Cole, and D. R. Diener. 1999. Intraflagellar transport: the eyes have it. J. Cell Biol. 144:385-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124a.Rupp, G., E. O’Toole, and M. E. Porter. 2001. The Chlamydomonas PF6 locus encodes a large alanine/proline-rich polypeptide that is required for assembly of a central pair projection and regulates flagellar motility. Mol. Biol. Cell 12:739-751. [DOI] [PMC free article] [PubMed]
- 125.Sager, R., and S. Granick. 1953. Nutritional studies in Chlamydomonas reinhardtii. Ann. N. Y. Acad. Sci. 56:831-838. [DOI] [PubMed] [Google Scholar]
- 126.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 127.Schloss, J. A. 1990. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol. Gen. Genet. 221:443-452. [DOI] [PubMed] [Google Scholar]
- 128.Schloss, J. A., and H. B. Croom. 1991. Normal Chlamydomonas nuclear gene structure on linkage group XIX. J. Cell Sci. 100:877-881. [DOI] [PubMed] [Google Scholar]
- 129.Schloss, J. A., C. D. Silflow, and J. L. Rosenbaum. 1984. mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol. Cell. Biol. 4:424-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schnell, R. A., and P. A. Lefebvre. 1993. Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics 134:737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sharpe, J. A., and A. Day. 1993. Structure, evolution and expression of the mitochondrial ADP/ATP translocator gene from Chlamydomonas reinhardtii. Mol. Gen. Genet. 237:134-144. [DOI] [PubMed] [Google Scholar]
- 132.Shimogawara, K., S. Fujiwara, A. R. Grossman, and H. Usuda. 1998. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148:1821-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132a.Shrager, J., C. Hauser, C. W. Chang, E. H. Harris, J. Davies, J. McDermott, R. Tamse, Z. Zhang, and A. R. Grossman. 2003. Chlamydomonas reinhardtii genome project. A guide to the generation and use of the cDNA information. Plant Physiol. 131:401-408. [DOI] [PMC free article] [PubMed]
- 133.Silflow, C. D. 1998. Organization of the nuclear genome, p. 25-40. In J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant (ed.), The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 134.Silflow, C. D., R. L. Chisholm, T. W. Conner, and L. P. Ranum. 1985. The two alpha-tubulin genes of Chlamydomonas reinhardtii code for slightly different proteins. Mol. Cell. Biol. 5:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Silflow, C. D., P. Kathir, and P. A. Lefebvre. 1995. Molecular mapping of genes for flagella proteins in Chlamydomonas. Methods Cell Biol. 47:525-530. [DOI] [PubMed] [Google Scholar]
- 136.Silflow, C. D., M. LaVoie, L.-W. Tam, S. Tousey, M. Sanders, W. Wu, M. Borodovsky, and P. Lefebvre. 2001. The Vfl1 protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153:63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Silflow, C. D., and P. A. Lefebvre. 2001. Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiol. 127:1500-1507. [PMC free article] [PubMed] [Google Scholar]
- 138.Silflow, C. D., B. Liu, M. Lavoie, E. A. Richardson, and B. A. Palevitz. 1999. Gamma-tubulin in Chlamydomonas: characterization of the gene and localization of the gene product in cells. Cell Motil. Cytoskeleton 42:285-297. [DOI] [PubMed] [Google Scholar]
- 139.Sineshchekov, O. A., and E. G. Govorunova. 1999. Rhodopsin-mediated photosensing in green flagellated algae. Trends Plant Sci. 4:58-63. [DOI] [PubMed] [Google Scholar]
- 140.Sloboda, R. D. 2002. A healthy understanding of intraflagellar transport. Cell Motil. Cytoskeleton 52:1-8. [DOI] [PubMed] [Google Scholar]
- 141.Small, G. D., B. Min, and P. A. Lefebvre. 1995. Characterization of a gene encoding a protein of the DNA photolyase/blue light photoreceptor family. Plant Mol. Biol. 28:443-454. [DOI] [PubMed] [Google Scholar]
- 142.Smart, E. J., and B. R. Selman. 1993. Complementation of a Chlamydomonas reinhardtii mutant defective in the nuclear gene encoding the chloroplast coupling factor 1 (CF1) gamma-subunit (atpC). J. Bioenerg. Biomembr. 25:275-284. [DOI] [PubMed] [Google Scholar]
- 143.Smith, E. F., and P. A. Lefebvre. 1996. PF16 encodes a protein with armadillo repeats and localizes to a single microtubule of the central apparatus in Chlamydomonas flagella. J. Cell Biol. 132:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Smith, E. F., and P. A. Lefebvre. 1997. PF20 gene product contains WD repeats and localizes to the intermicrotubule bridges in Chlamydomonas flagella. Mol. Biol. Cell 8:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Somanchi, A., and J. V. Moroney. 1999. As Chlamydomonas reinhardtii acclimates to low-CO2 conditions there is an increase in cyclophilin expression. Plant Mol. Biol. 40:1055-1062. [DOI] [PubMed] [Google Scholar]
- 146.Stevens, D. R., A. Atteia, L.-G. Franzen, and S. Purton. 2001. Cycloheximide resistance conferred by novel mutations in ribosomal protein L41 of Chlamydomonas reinhardtii. Mol. Gen. Genet. 264:790-795. [DOI] [PubMed] [Google Scholar]
- 147.Sugase, Y., M. Hirono, K. L. Kindle, and R. Kamiya. 1996. Cloning and characterization of the actin-encoding gene of Chlamydomonas reinhardtii. Gene 168:117-121. [DOI] [PubMed] [Google Scholar]
- 148.Takada, S., C. G. Wilkerson, K. K. Wakabayashi, R. Kamiya, and G. B. Witman. 2002. The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol. Biol. Cell 13:1015-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tam, L.-W., and P. A. Lefebvre. 1993. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135:375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tam, L.-W., and P. A. Lefebvre. 2002. The Chlamydomonas MBO2 locus encodes a conserved coiled-coil protein important for flagellar waveform conversion. Cell Motil. Cytoskeleton 51:197-212. [DOI] [PubMed] [Google Scholar]
- 151.Teramoto, H., T. Ono, and J. Minagawa. 2001. Identification of Lhcb gene family encoding the light-harvesting chlorophyll-a/b proteins of photosystem II in Chlamydomonas reinhardtii. Plant Cell Physiol. 42:849-856. [DOI] [PubMed] [Google Scholar]
- 152.Thompson, M. D., C. D. Paavola, T. R. Lenvik, and J. S. Gantt. 1995. Chlamydomonas transcripts encoding three divergent plastid chaperonins are heat-inducible. Plant Mol. Biol. 27:1031-1035. [DOI] [PubMed] [Google Scholar]
- 153.Trebitsh, T., E. Meiri, O. Ostersetzer, A. Adam, and A. Danon. 2001. The protein disulfide isomerase-like RB60 is partitioned between stroma and thylakoids in Chlamydomonas reinhardtii chloroplasts. J. Biol. Chem. 276:4564-4569. [DOI] [PubMed] [Google Scholar]
- 154.Uchida, H., S. Kawano, N. Sato, and T. Kuroiwa. 1993. Isolation and characterization of novel genes which are expressed during the very early stage of zygote formation in Chlamydomonas reinhardtii. Curr. Genet. 24:296-300. [DOI] [PubMed] [Google Scholar]
- 155.Vaistij, F. E., E. Boudreau, S. D. Lemaire, M. Goldschmidt-Clermont, and J.-D. Rochaix. 2000. Characterization of mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 97:14813-14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vallon, O., and F. A. Wollman. 1997. cDNA sequence of Mα, the catalytic subunit of the Chlamydomonas reinhardtii L-amino acid oxidase (accession no. U78797). A new sequence motif shared by a wide variety of flavoproteins (PGR 97-171). Plant Physiol. 115:1729.9414571 [Google Scholar]
- 157.von Gromoff, E. D., and C. F. Beck. 1993. Genes expressed during sexual differentiation of Chlamydomonas reinhardtii. Mol. Gen. Genet. 241:415-421. [DOI] [PubMed] [Google Scholar]
- 158.von Gromoff, E. D., U. Treier, and C. F. Beck. 1989. Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol. Cell. Biol. 9:3911-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vysotskaia, V. S., D. E. Curtis, A. V. Voinov, P. Kathir, C. D. Silflow, and P. A. Lefebvre. 2001. Development and characterization of genome-wide single nucleotide polymorphism markers in the green alga Chlamydomonas reinhardtii. Plant Physiol. 127:386-389. [PMC free article] [PubMed] [Google Scholar]
- 160.Walther, Z., M. Vashishtha, and J. L. Hall. 1994. The Chlamydomonas Fla10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 126:175-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wegener, D., and C. F. Beck. 1991. Identification of novel genes specifically expressed in Chlamydomonas reinhardtii zygotes. Plant Mol. Biol. 16:937-946. [DOI] [PubMed] [Google Scholar]
- 162.Werner, R. 2002. Chlamydomonas reinhardtii as a unicellular model for circadian rhythm analysis. Chronobiol. Int. 19:325-343. [DOI] [PubMed] [Google Scholar]
- 163.Wilkerson, C. G., S. M. King, and G. B. Witman. 1994. Molecular analysis of the gamma heavy chain of Chlamydomonas flagellar outer-arm dynein. J. Cell Sci. 107:497-506. [DOI] [PubMed] [Google Scholar]
- 164.Wilkerson, C. G., S. M. King, A. Koutoulis, G. J. Pazour, and G. B. Witman. 1995. The 78,000 Mr intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 129:169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Williams, B. D., D. R. Mitchell, and J. L. Rosenbaum. 1986. Molecular cloning and expression of flagellar radial spoke and dynein genes of Chlamydomonas. J. Cell Biol. 103:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Williams, B. D., M. A. Velleca, A. M. Curry, and J. L. Rosenbaum. 1989. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J. Cell Biol. 109:235-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wilson, N. F., and W. J. Snell. 1998. Microvilli and cell-cell fusion during fertilization. Trends Cell Biol. 8:93-96. [DOI] [PubMed] [Google Scholar]
- 168.Wu-Scharf, D., B. Jeong, C. Zhang, and H. Cerutti. 2000. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science 290:1159-1162. [DOI] [PubMed] [Google Scholar]
- 169.Wykoff, D. D., A. R. Grossman, D. P. Weeks, H. Usuda, and K. Shimogawara. 1999. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. USA 96:15336-15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xiang, Y., J. Zhang, and D. P. Weeks. 2001. The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 98:5341-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yang, P., and W. S. Sale. 1998. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell 9:3335-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang, P., L. Fox, R. J. Colbran, and W. S. Sale. 2000. Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J. Cell Sci. 113:91-102. [DOI] [PubMed] [Google Scholar]
- 173.Yohn, C. B., A. Cohen, A. Danon, and S. P. Mayfield. 1998. A poly(A) binding protein functions in the chloroplast as a message-specific translation factor. Proc. Natl. Acad. Sci. USA 95:2238-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Youngblom, J., J. A. Schloss, and C. D. Silflow. 1984. The two beta-tubulin genes of Chlamydomonas reinhardtii code for identical proteins. Mol. Cell. Biol. 4:2686-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]