Abstract
The response to damage is crucial for cellular survival, and eukaryotic cells require a broad array of proteins for an intact damage response. We have found that the YPL170W (DAP1 [for damage response protein related to membrane-associated progesterone receptors]) gene is required for growth in the presence of the methylating agent methyl methanesulfonate (MMS). The DAP1 open reading frame shares homology with a broadly conserved family of membrane-associated progesterone receptors (MAPRs). Deletion of DAP1 leads to sensitivity to MMS, elongated telomeres, loss of mitochondrial function, and partial arrest in sterol synthesis. Sensitivity of dap1 strains to MMS is not due to loss of damage checkpoints. Instead, dap1 cells are arrested as unbudded cells after MMS treatment, suggesting that Dap1p is required for cell cycle progression following damage. Dap1p also directs resistance to itraconazole and fluconazole, inhibitors of sterol synthesis. We have found that dap1 cells have slightly decreased levels of ergosterol but increased levels of the ergosterol intermediates squalene and lanosterol, indicating that dap1 cells have a partial defect in sterol synthesis. This is the first evidence linking a MAPR family member to sterol regulation or the response to damage, and these functions are probably conserved in a variety of eukaryotes.
Eukaryotic cells are constantly exposed to exogenous and endogenous damage. Cells respond to DNA damage by delaying cell cycle progression (27), repairing the damage, and activating or repressing transcription of a number of genes (19, 28). Mutants that are unable to activate the checkpoint pathway continue to replicate damaged DNA and die (56). Checkpoint proteins include the protein kinase Mec1p (1, 34, 40, 57) and Rad9p (56). Some yeast damage repair mutants have a functioning checkpoint response and are capable of delaying the cell cycle but cannot repair the damaged lesion. In general, yeast DNA repair mutants fall into three categories: mutants affected in nucleotide excision repair, postreplication repair, and recombinational repair (18).
Yeast requires a large number of processes to respond to damage. Diploid yeast contains 130 nonessential genes that are required for the response to ionizing radiation, and 100 of these genes are required for the response to methyl methanesulfonate (MMS) (3). The total number of genes required for the response to MMS is not known, and it is not clear which of the radiation-sensitive diploids are sensitive as haploids. In addition to proteins regulating the damage checkpoint and recombinational repair, diploid yeasts require proteins directing replication, chromatin silencing, and mitotic chromosome transmission for the response to MMS (3). Genes with less-direct roles in DNA metabolism are also required for the response to MMS, including genes encoding the nuclear pore complex; vacuolar, Golgi, and endocytosis components; and members of the ubiquitin degradation pathway and genes regulating transcription and RNA metabolism (3). Finally, deletion of the ERG3 gene, encoding C-5 sterol desaturase, leads to MMS sensitivity, and erg28 (17) and arv1 (for ARE2 required for viability) (53) strains are sensitive to a lesser extent (3). Thus, the response to ionizing radiation and MMS is complex and requires the function of a number of genes, including some genes that function in sterol synthesis.
In yeast, several proteins that regulate the DNA damage response are also required for telomere length maintenance (reviewed in reference 4). Damage repair proteins probably serve protective functions for telomeres, distinguishing them from a double-stranded DNA break. Ku70p localizes to telomeres in the absence of damage and then translocates to sites of damage following double-stranded DNA breaks (35, 36, 38). The telomere binding proteins Sir3p, Sir4p, and Rap1p also diffuse from the telomere after damage. Following translocation from the telomere, these proteins probably serve to cap broken DNA ends and block replication and transcription in the DNA adjacent to the break, facilitating the processing and repair of these regions. A large number of proteins that contribute to checkpoint or damage repair also regulate telomere length, including members of the Mre11p-Rad50p-Xrs2p complex (5), Mec1p (43), Mec3p (6), Rad27p (41), and Tel1p (21, 33).
We have searched for novel yeast genes that regulate the damage response. The murine 25-Dx protein is induced following exposure of the carcinogen 2,3,7,8-tetrachloro-p-dioxin, suggesting a role in the response to cellular stress and in carcinogenesis (46). 25-Dx is part of a highly conserved family of proteins that is collectively called the membrane-associated progesterone receptor (MAPR) family. The porcine MAPR binds to progesterone and was isolated from liver membrane fractions (14, 37). However, the biological function of MAPRs in progesterone signaling is unknown.
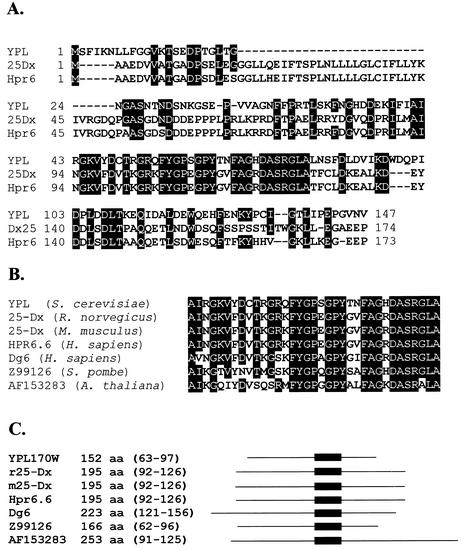
The yeast open reading frame YPL170W encodes a protein that is part of the MAPR family (Fig. 1). The predicted protein product from the YPL170W open reading frame shares homology throughout its coding sequence with its human and rodent counterparts (Fig. 1A) and has a region identical to proteins from a broad variety of organisms, including Arabidopsis thaliana and Schizosaccharomyces pombe (Fig. 1B). Many of these proteins are similar in molecular weight and contain the homologous region near the center of the coding sequence (Fig. 1C).
FIG. 1.
DAP1 is homologous to the MAPR family of proteins. (A) Identity between YPL170W/Dap1p (YPL) and the full-length rat and human MAPR family members, 25-Dx and Hpr6.6, respectively. Identical residues are indicated by a black background. (B) Identity in the most highly conserved region between YPL170W/Dap1p and related proteins from rat (rat 25-Dx), mouse (mouse 25-Dx), humans (Hpr6.6 and Dg6), fission yeast (Z99126), and Arabidopsis (AF153283). (C) List of YPL170W/Dap1p and related proteins along with the total amino acids (aa) of the predicted proteins and the corresponding amino acids of each protein that share the greatest identity with Dap1p (indicated by shading in panel B). A schematic diagram of each protein is to the right of the list, with the region of homology indicated by a dark box.
Because of the role of 25-Dx in carcinogenesis, we deleted the YPL170W open reading frame and have characterized the phenotypes of the mutant strain. We have found that the phenotypes of the YPL170WΔ strain are pleiotropic, affecting growth in response to damage, telomere length, and sterol regulation. Our results are the first genetic analysis of a MAPR family member and indicate new roles for these proteins in the damage response and cellular metabolism.
MATERIALS AND METHODS
Yeast strains and growth conditions.
All strains were isogenic with W303 (leu2-3,112 his3-11,15 ura3-1 ade2-1 trp1-1 can1-100 rad5-535) (52) except for the alterations described in Table 1. In most of the strains, the rad5-535 allele was replaced with the wild-type RAD5 gene by crossing and tested by PCR as described previously (9). In some strains, the type X subtelomeric repeat element on the left telomere of chromosome XV was replaced with URA3 (designated XVL-TEL-URA3) as described previously (8).
TABLE 1.
Strains
| Strain | Relevant genotype | Reference |
|---|---|---|
| W303 | aade2-1 can1-100 his3-11,15 leu2-3,112 rad5-535 trpI-1 ura3-1 | 52 |
| HLK1042-1C | α CAN1 hom3-10 RAD5 | |
| JMY327-1d | α RAD53-13myc (::KanMX) | |
| RCY23 | tel1::ura3 VL-TEL-URA3 | 8 |
| RCY61-1b | α rif1 Δ::KanMX rif2Δ::HIS3 tel1::ura3 | |
| RCY90 | dap1Δ::LEU2 VL-TEL-URA3 | |
| RCY93-2a | dap1Δ::LEU2 tel1::ura3 | |
| RCY93-4c | α dap1Δ::LEU2 | |
| RCY93-7c | Wild type a | |
| RCY269-3d | mec1-21 RAD5 XVL-TEL-URA3 | 9 |
| RCY278-1a | α mec1-21 RAD5 tel1Δ::KanMX XVL-TEL-URA3 | 9 |
| RCY300-6a | α dun1-100::HIS3 RAD5 XVL-TEL-URA3 (listed incorrectly as scs2::HIS3 in reference 9) | 9 |
| RCY307-4a | CAN1 RAD5 rad9Δ::KanMX | 10 |
| RCY327-3c | hom3-10 RAD5 rad9Δ::KanMX | 10 |
| RCY344-2b | CAN1 dap1Δ::LEU2 RAD5 VL-TEL-URA3 | |
| RCY344-4d | α dap1Δ::LEU2 RAD5 | |
| RCY406-1d | RAD5 rad9Δ::KanMX | |
| RCY406-5a | dap1Δ::LEU2 RAD5 rad9Δ::KanMX | |
| RCY407-1d | CAN1 dap1Δ::LEU2 RAD5 | |
| RCY407-3a | α CAN1 dap1Δ::LEU2 dun1Δ::HIS3 RAD5 XVL-TEL-URA3 | |
| RCY407-12d | RAD5 | |
| RCY409-2a | Wild type a | |
| RCY409-4b | α CAN1 dap1Δ::LEU2 RAD5 | |
| RCY409-5c | α CAN1 dun1Δ::HIS3 RAD5 XVL-TEL-URA3 | |
| RCY429-1a | α RAD5 | |
| RCY429-1d | dap1Δ::LEU2 RAD5 RAD53-13myc (::KanMX) | |
| RCY429-2b | α RAD5 RAD53-13myc (::KanMX) | |
| SPY40 | tel1::URA3 | 42 |
The DAP1 gene was replaced with the LEU2 gene by a one-step transplacement. The LEU2 gene was amplified from plasmid pRS305 (49) by using the primers DAP1-KOF and DAP1-KOR, so that the PCR product contained the entire LEU2 gene with flanking homology to DAP1. Strains were grown at 30°C in yeast extract-peptone-dextrose (YPD) or synthetic media (22). All primer sequences are available on request.
All genetic manipulations were performed essentially as described previously (22). The haploid strains used in these studies were primarily derivatives of diploid strains. The diploids were constructed by crossing the indicated strains (genotypes are listed in Table 1), as follows: RCY93 (RCY61-1b × RCY90), RCY344 (RCY90 × HLK1042-1C), RCY406 (RCY327-3c × RCY344-4d), RCY407 (RCY344-2b × RCY300-6a), RCY409 (RCY337-26a × RCY407-3a), RCY412 (RCY278-1a × RCY407-1d), and RCY429 (JMY327-1d × RCY407-1d). For transformations with plasmids or PCR products, we used the standard lithium acetate-polyethylene glycol method.
Drug sensitivity assays and cell cycle analysis.
Overnight cultures were serially diluted 1:10 in water. Five microliters of the diluted cells was then spotted onto plates containing 20 mM hydroxyurea (Sigma) or 0.01% MMS (Sigma). For the response to antifungal agents, 100 μl of itraconazole (0.1 mg/ml) (Ortho Biotech) or fluconazole (1 mg/ml) (Pfizer) was spread on YPD plates or plates made of synthetic media lacking histidine, as indicated. Itraconazole and fluconazole were purchased from the University of North Carolina Hospitals Pharmacy Storeroom. Once the plates were dry, serially diluted cells were spotted on the plates. Cells were grown for 2 to 3 days and photographed.
For cell cycle analysis, log-phase cells were arrested with 0.5 mM α-factor (Sigma) for 2 h. The cells were then centrifuged, washed once in YPD, and then diluted in fresh YPD with or without 0.05% MMS. At the indicated time points, 1 ml of each culture was removed and fixed in 3.7% formaldehyde at room temperature for 1 h. The cells were then centrifuged, washed once in phosphate-buffered saline, and resuspended in 0.25 ml of phosphate-buffered saline. Budding was monitored by light microscopy. For each series shown, cells were synchronized and released at least two separate times with the same result.
Protein analysis.
Log-phase cells were centrifuged, washed once in distilled water (dH2O) containing 1 mM phenylmethylsulfonyl fluoride, and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer containing 1 mM phenylmethylsulfonyl fluoride. The samples were then boiled for 10 min and further disrupted by mixing on a vortex mixer with an equal volume of glass beads (Sigma). Cells were vortexed three times for 1 min each with 1 min intervening at 0°C. Samples were then centrifuged at the maximum setting in a microcentrifuge at 4°C. For each sample, 25 μl was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 15% acrylamide gel, transferred to HyBond C (Amersham), and probed with an antibody to the Myc (9B11; Cell Signaling) epitope tag sequence.
Southern blot analysis.
Genomic DNA was isolated from 5-ml cultures grown in YPD or selective media overnight at 30°C. DNA purification was performed as previously described (22). DNA was digested with PstI, separated by 1% agarose gel electrophoresis, and transferred to HyBond N+ membranes (Amersham). Blots were probed with a DNA fragment of the Y′ subtelomeric repeat, amplified from plasmid pYT14 by PCR with the primers TELO-5′ and TELO-3′, as described previously (7).
Assay for mitochondrial maintenance, canavanine resistance, membrane permeability, and cation pulse resistance.
For assays of mitochondrial maintenance, cells were grown on YPD plates for 2 to 3 days. Colonies were picked, suspended in 0.5 ml of water, and diluted 104, and 100 μl was plated on YPD plates. The plates were incubated at 30°C for 2 days and then left at 4°C for 3 to 7 days, and the numbers of red and white colonies were counted. Colonies were verified as petite by patching at least 100 colonies on plates containing 2% glycerol as a carbon source. For each assay, at least 15 colonies were plated, and the percentage of petite colonies was calculated for each plate and averaged.
Canavanine resistance assays were performed as previously described (10). Briefly, overnight cultures were diluted in water and plated onto plates lacking arginine, with or without 60 mg of l-canavanine (Sigma) per liter. Twenty independent cultures were analyzed in two separate assays. The rate of canavanine resistance was calculated by using the method of the median (32).
Membrane permeability assays were performed essentially as described previously (2). Yeast cultures were grown to a density of 5 × 107 cells/ml and incubated at various temperatures for 5 min, and crystal violet (Sigma) (preincubated to the same temperature as the culture) was added to a final concentration of 5 μg/ml. Cells were then incubated at various temperatures for 10 min and centrifuged, and the dye concentration in the supernatant was measured by optical density (OD) at 595 nm. The percent crystal violet uptake was calculated by subtracting the absorbance of cell supernatants from the absorbance of the starting solution and then dividing by the absorbance of the starting solution.
To assay resistance to a cation pulse, yeast cultures were grown to an OD of 1, and 1 ml of cells was removed and centrifuged. Cell pellets were mixed with 50 μl of a 2 M solution of CaCl2, CrCl3, KCl, MgCl2, or NiCl2 (all were purchased from Sigma) and incubated at 30°C for 10 min. Cells were then centrifuged, resuspended in phosphate-buffered saline, and plated for viability on YPD plates. All assays were performed in triplicate and duplicated in a separate analysis.
Sterol analysis.
To determine the percentage of total cellular mass represented by sterol, quantitative sterol analysis was performed as described by Molzahn and Woods (39). Briefly, cells were grown in YPD to an OD at 660 nm of 0.8 to 0.9. Cells were pelleted, washed once with dH2O, and then resuspended in 10 ml of alcoholic KOH (4.5 M KOH in 60% ethanol) and transferred to a clean round-bottom flask. The suspension was allowed to reflux at 88 to 90°C for 60 min. One milliliter of 95% ethanol was added down the condensor, refluxing was continued for another 60 min, and then the apparatus was cooled to room temperature. The condensor was removed, and 4 ml of dH2O and 10 ml of n-heptane (Sigma) were added to the flask before shaking vigorously for 2.5 min.
Two milliliters of the n-heptane layer (containing sterol) was transferred to a clean vial, and 3 μl of sample was analyzed by gas chromatography as described below. To calculate the percentage of sterols per milligram of cell mass, 20 ml of the original culture was harvested by vacuum filtration onto a preweighed 0.45-μm-pore-size Millipore filter. The filters were predried in a 105°C oven overnight and subsequently placed in a dessiccator for 4 h. After vacuum filtration of harvested cells, the heating-desiccation step was repeated, and the weight of the cells was determined. The amount of individual sterols was calculated based on the area under each peak of the chromatograph relative to a known amount of ergosterol loaded onto the gas chromatograph, using the Hewlett-Packard sterol quantitation program. Each sample was injected twice, and the value reported for each sterol quantity was the average of those for two injections.
Sterols were analyzed by gas chromatography with a Hewlett-Packard HP5890 series II chromatograph equipped with the Hewlett-Packard CHEMSTATION software package. The capillary column (DB-1; J&W Scientific) was programmed from 195 to 280°C (1 min at 195°C and then an increase at 20C°/min to 240°C, followed by an increase at 2°C/min until the final temperature of 280°C was reached). The linear velocity was 30 cm/s, nitrogen was the carrier gas, and all injections were run in the splitless mode.
RESULTS
Identification of DAP1.
YPL170W is a 456-bp open reading frame encoding a 152-amino-acid protein with a predicted molecular mass of 16.7 kDa. There is little homology between the YPL170W open reading frame and other yeast open reading frames, except for a 52-amino-acid region with 39% identity with the anaphase checkpoint protein Pds1p (not shown). However, YPL170W contains strong homology with a group of proteins that is collectively called the MAPR family (Fig. 1A). The rat and human homologues of YPL170W resemble the YPL170W open reading frame in size and contain homology to YPL170W throughout their coding sequences. The MAPR family includes 25-Dx (37, 46), the human 25-Dx homologues Hpr6.6 and Dg6 (20) (Fig. 1B), and uncharacterized family members in S. pombe, A. thaliana (Fig. 1B), and Caenorhabditis elegans.
We deleted the entire YPL170W open reading frame by one-step transplacement with the LEU2 gene. The following sections describe the phenotypes associated with loss of YPL170W function: damage sensitivity, telomere elongation, partial loss of ergosterol synthesis, and defects in mitochondrial biogenesis. Because YPL170WΔ mutants are damage sensitive and YPL170W contains homology to the MAPR family, we propose the name DAP1 (for damage response protein related to the membrane-associated progesterone receptor) for YPL170W.
Cells lacking DAP1 are sensitive to MMS.
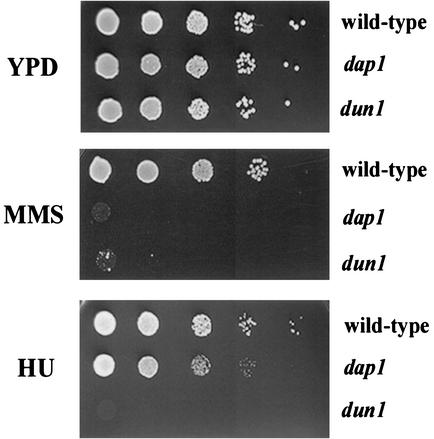
As part of our characterization of dap1Δ mutants, we tested the ability of dap1Δ strains to grow in the presence of damaging agents. We found that dap1Δ strains were unable to grow on plates containing 0.01% MMS (Fig. 2, middle panel, second row). The sensitivity of dap1Δ strains to MMS was completely reversed by a single-copy plasmid containing the entire DAP1 open reading frame.
FIG. 2.
Dap1p regulates the response to MMS-induced damage. Log-phase cells were diluted 1:10 and spotted onto YPD plates (upper panel), YPD plates containing 0.01% MMS (middle panel), or YPD plates containing 20 mM hydroxyurea (HU) (lower panel). The strains tested were wild-type (RCY409-2a, top rows), dap1 (RCY409-4b, second rows), and dun1 (RCY409-5c, third rows). Strains harboring the dap1 mutation grew poorly on hydroxyurea and did not grow on plates containing MMS. The dun1 strain RCY409-5c is included as a damage-sensitive control strain.
The dap1Δ strains were only mildly sensitive to the ribonucleotide reductase inhibitor hydroxyurea, with reduced growth at 20 mM (Fig. 2, lower panel, second row). This dose of hydroxyurea was toxic to cells lacking the Dun1p damage repair protein (Fig. 2, lower panel, bottom row). UV light and gamma irradiation had no effect on growth of haploid dap1Δ strains, even at doses that killed checkpoint mutant strains. However, diploid dap1Δ strains were approximately 10-fold more sensitive to ionizing and UV radiation than a comparable diploid wild-type strain, suggesting that Dap1p contributes to the growth response to radiation in diploid cells.
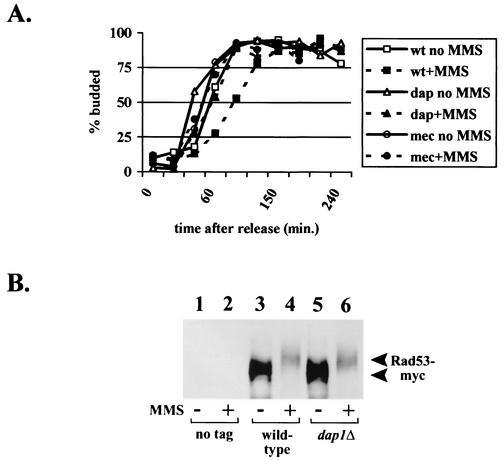
We surmised that MMS sensitivity in dap1Δ strains might be due to a checkpoint defect. Yeast cells arrested in G1 delay budding when damaged (47), and this budding checkpoint is MEC1 dependent (48). We synchronized log-phase wild-type and dap1Δ strains in G1 with α-factor and then released the cells from G1 arrest and followed progression of the cell cycle by analyzing budding morphology. Both wild-type and dap1Δ strains delayed budding when released into media containing MMS (Fig. 3A), suggesting that the cell cycle checkpoint is intact in dap1Δ cells. In contrast, mec1-21 cells did not delay budding under similar conditions (Fig. 3A).
FIG. 3.
Dap1p does not regulate the damage checkpoint in MMS-treated cells. (A) Wild-type (wt) (RCY409-2a), dap1Δ (dap) (RCY407-1d), or mec1-21 (mec) (RCY269-3d) cells were synchronized in the G1 phase of the cell cycle with α-factor and then released in the presence or absence of 0.05% MMS. Samples of cells were taken at various time points and fixed, and the number of budded cells were counted at each time point. (B) Rad53p phosphorylation after treatment with MMS is not Dap1p dependent. Rad53p was fused in frame with 13 copies of the Myc epitope tag and crossed into the dap1Δ mutant background. Wild-type and mutant cells were then analyzed by Western blotting with an anti-Myc antibody before (lanes 1, 3, and 5) and after (lanes 2, 4, and 6) treatment with MMS. The strains analyzed were RAD53 DAP1 (RCY429-1a) (lanes 1 and 2), RAD53-Myc DAP1 (RCY429-2b) (lanes 3 and 4), and RAD53-Myc dap1Δ (RCY429-1d) (lanes 5 and 6).
We examined a molecular end point for the cell cycle checkpoint, monitoring the phosphorylation of the Rad53p checkpoint protein kinase. Following treatment with MMS, Rad53p becomes hyperphosphorylated (44, 51), and this phosphorylation is dependent on an intact Mec1p/Tel1p pathway (44). We used a strain with 13 copies of the Myc epitope tag integrated at the 3′ end of the genomic copy of RAD53; the epitope-tagged Rad53p was readily detectable in strains harboring the integrated tag (Fig. 3B, lanes 3 to 6) but was undetectable in cells harboring the native RAD53 allele (Fig. 3B, lanes 1 and 2). Treatment of wild-type cells with 0.05% MMS resulted in a marked increase in Rad53p phosphorylation, as expected (Fig. 3B, compare lanes 3 and 4), and the same shift in mobility was detected in dap1Δ cells (Fig. 3B, compare lanes 5 and 6). Thus, Dap1p is not required for Rad53p hyperphosphorylation following treatment with MMS. Using two separate criteria, we conclude that Dap1p is not required for the MMS-initiated cell cycle checkpoint.
Next, unsynchronized wild-type or dap1Δ cells were treated with MMS and cell cycle progression was determined by microscopic analysis of budding and by flow cytometry. We found that dap1Δ cells frequently develop an aberrant shmoo morphology when arrested with α-factor (R. J. Craven, unpublished observations). However, these elongated shmoo-shaped cells resembled budded cells, making the early stages of the cell cycle difficult to distinguish, so we resorted to analysis of unsynchronized cells.
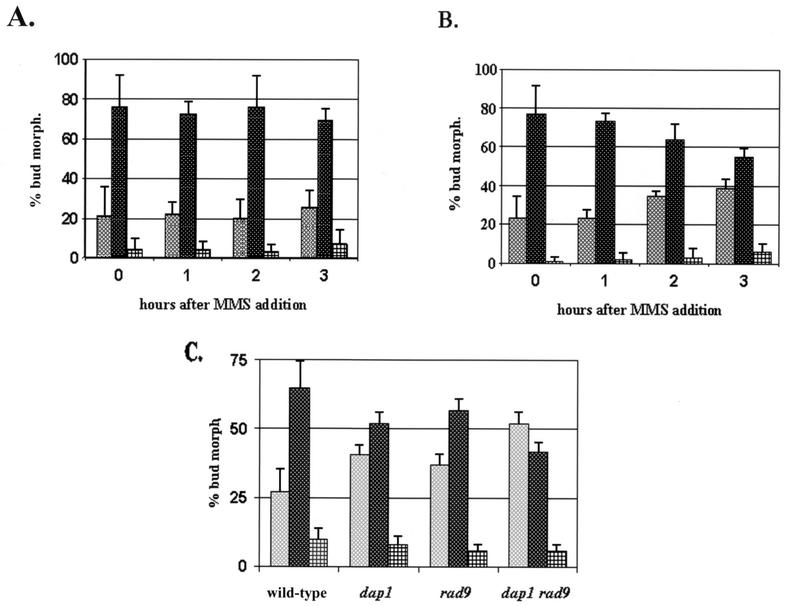
In unsynchronized log-phase cultures, the cell cycle profiles of wild-type and dap1Δ cells were similar. Following 2 h of MMS treatment, the vast majority of wild-type cells had small to medium-sized buds (Fig. 4A), while dap1Δ cultures began to accumulate unbudded cells (Fig. 4B). The difference in unbudded cells between wild-type and dap1Δ cells following 3 h of MMS treatment was significant (P = 0.002) when analyzed by the Student t test. Failure to bud indicates an arrest in the G1 phase of the cell cycle. Although the G1-arrested cells failed to grow, they were not dead, because they excluded the dead-cell marker phloxine B.
FIG. 4.
Dap1p regulates cell cycle progression following DNA damage. (A and B) Unsynchronized wild-type (A) or dap1Δ (B) cells were treated with 0.05% MMS and then fixed and counted at the time points indicated. Cells were scored microscopically as unbudded (left bars), small budded (center bars), or doublets (right bars). In dap1Δ cells, there was a twofold increase in the number of unbudded cells and a sixfold increase in the percentage of large-budded cells within 2 h of MMS addition. The strains analyzed were RCY409-2a (wild type) and RCY409-4b (dap1Δ). (C) The budding profiles of four different strains were compared at 2 h following the addition of 0.05% MMS. The strains were RCY409-2a (wild type), RCY409-4b (dap1), RCY406-1d (rad9), and RCY406-5a (dap1 rad9). Deletion of the RAD9 gene did not reverse the G1 arrest observed in dap1 mutants following MMS-induced damage. Error bars indicate standard deviations.
One explanation for the accumulation of dap1Δ cells in G1 following MMS exposure is that Dap1p is required for adaptation to MMS exposure. Following prolonged damage, cells can override cell cycle arrest and continue to grow, a process called adaptation (54). If Dap1p is required for adaptation to a checkpoint, we expected that loss of Dap1p in combination with loss of checkpoint function would suppress the G1 arrest. To test this idea, we constructed a strain lacking both DAP1 and the RAD9 checkpoint gene, and we found that dap1Δ rad9Δ cells had an even stronger G1 arrest than either single mutant (Fig. 4C). We conclude that loss of Dap1p function inhibits cell cycle progression in G1 following MMS-induced damage and that this arrest is not due to an inability to adapt to the Rad9p-mediated checkpoint.
Dap1p regulates telomere length.
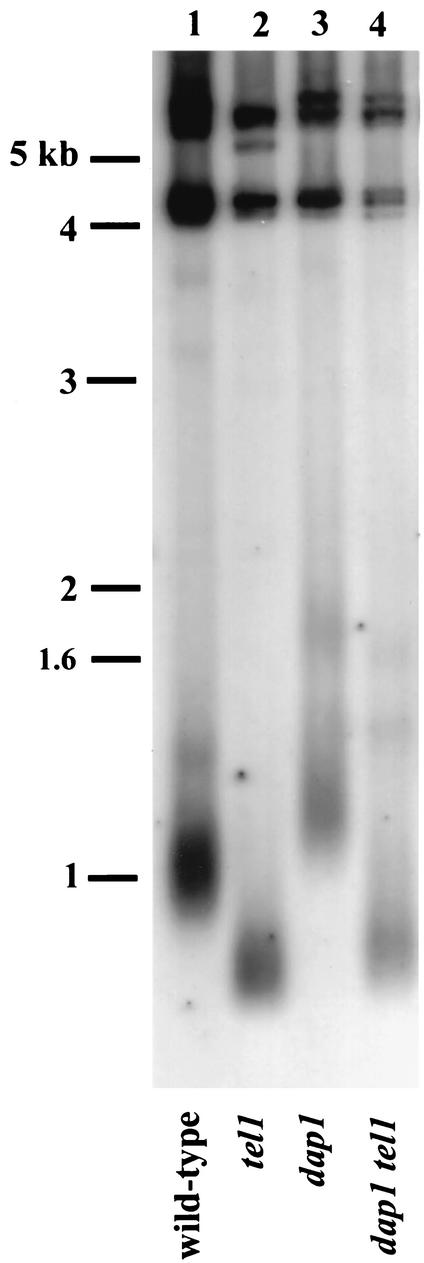
A number of genes required for damage repair also regulate telomere length. We analyzed the telomeres of dap1Δ strains by Southern blotting. Telomeres in the dap1Δ mutant were elongated by approximately 100 bp compared to those in wild-type strains (Fig. 5, lane 3). While there was a small effect on telomere length in dap1Δ strains, telomeric silencing in dap1Δ strains was unaffected.
FIG. 5.
DAP1 regulates telomere length. Southern blot analysis of telomere lengths in the strains RCY93-7c (wild type) (lane 1), RCY23 (tel1) (lane 2), RCY93-4c (dap1) (lane 3), and RCY93-2a (dap1 tel1) (lane 4) is shown. In all cases, DNA was purified according to standard protocols, digested with PstI, blotted, and probed with a labeled fragment of the Y′ subtelomeric repeat. Migration of molecular size standards is indicated to the left.
Tel1p is a high-molecular-weight protein kinase related to the human Atm protein, and Tel1p is a central regulator of telomere length. To test whether DAP1 and TEL1 are in the same genetic pathway, we compared telomere lengths of dap1Δ tel1 double mutants with those of the single dap1Δ and tel1 mutants. A combined dap1Δ tel1 mutant had telomeres that were slightly longer than those of the comparable tel1 strain (Fig. 5, compare lanes 2 and 4). In general, combined mutations in two genes in the same genetic pathway result in a phenotype of one or the other single mutant. Because the dap1Δ tel1 phenotype was not intermediate between those of the dap1Δ and tel1 single mutants, we conclude that TEL1 is epistatic to DAP1 and that they function in the same genetic pathway.
Cells lacking DAP1 are sensitive to drugs inhibiting sterol synthesis.
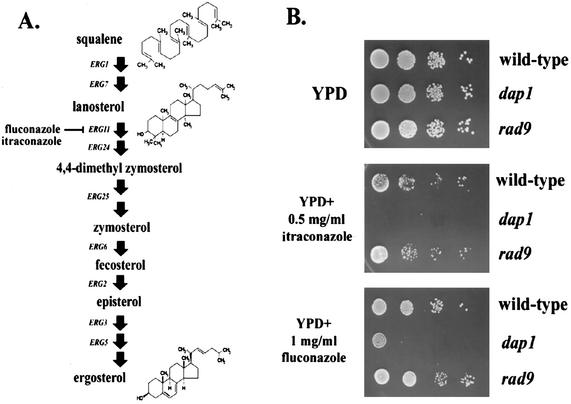
Microarray analyses demonstrated altered expression of the DAP1 transcript either in strains with repressed ERG11 expression or in strains treated with the ergosterol inhibitor itraconazole (26). ERG11 encodes lanosterol C-14 demethylase (29), a key enzyme in ergosterol synthesis. Erg11p is inhibited by azole compounds, which inhibit Erg11p by forming a complex with the heme iron component in the cytochrome group (Fig. 6) (55, 58). In addition, the homology between Dap1p and known steroid binding proteins (Fig. 1) suggested that Dap1p might regulate sterol synthesis in yeast.
FIG. 6.
Dap1p is required for growth in response to inhibitors of ergosterol synthesis. (A) The ergosterol biosynthetic pathway, with the step inhibited by itraconazole and fluconazole indicated. (B) Itraconazole and fluconazole inhibit the growth of dap1Δ strains. Wild-type (RCY407-12d, top rows), dap1Δ (RCY407-1d, middle rows), and rad9Δ (RCY307-4a, bottom rows) strains were diluted 1:10 serially and spotted on YPD plates without drug (upper panel) or layered with itraconazole (middle panel) or fluconazole (lower panel).
We tested the ability of dap1Δ mutants to grow on itraconazole by directly applying the drug to the surfaces of YPD plates, allowing them to dry, then spotting series of 10-fold dilutions to the plates. Wild-type cells grew readily on plates coated with 0.1 mg of itraconazole per ml, while dap1Δ mutants did not (Fig. 6B, middle panel, top two rows). Itraconazole sensitivity is not a general property of damage checkpoint mutants, because rad9Δ mutants grow readily on itraconazole. Interestingly, a mec1-21 mutant strain was moderately sensitive to itraconazole, although the reason for this is unclear, as no sterol synthetic phenotypes have been attributed to mec1 mutants. A second Erg11p inhibitor, fluconazole, was also toxic to dap1Δ strains (Fig. 6B, lower panel, middle row).
Partial loss of sterol synthesis in dap1Δ mutants.
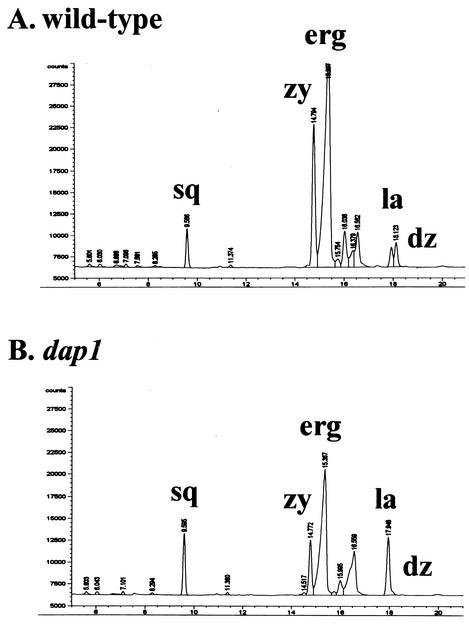
Because dap1Δ cells were sensitive to itraconazole, we analyzed the profile of sterols in dap1Δ cells by gas chromatography. In dap1Δ cells, we detected a 9% decrease in overall sterol concentrations, with a 5% decrease in ergosterol and 43 and 34% decreases in the ergosterol intermediates zymosterol and fecosterol, respectively (Fig. 7). In addition, the ergosterol intermediate 4,4-dimethylzymosterol was undetectable in dap1Δ cells but was present at 3.3% in wild-type cells. In contrast, there was a 17.5% increase in squalene and a 4.7-fold elevation in the ergosterol intermediate lanosterol (eluting at 17.948 min in Fig. 7B). For these analyses, each sample was injected twice and the value reported for each sterol quantity was the average of two injections. Lanosterol metabolism is the target of the azole inhibitors itraconazole and fluconazole. We conclude that dap1Δ strains have a partial lesion in sterol synthesis that leads to decreases in products downstream of Erg11p (lanosterol-14-demethylase).
FIG. 7.
Dap1p regulates sterol synthesis in yeast. Gas chromatography sterol accumulation profiles in the wild-type (RCY409-2a) (A) and dap1 (RCY409-4b) (B) strains from the same genetic cross are shown. The peaks for squalene (sq), zymosterol (zy), ergosterol (erg), lanosterol (la), and 4,4-dimethylzymosterol (dz) are indicated. Loss of Dap1p led to an increase in squalene and lanosterol and a decrease in zymosterol and ergosterol.
Membrane permeability in dap1Δ strains.
Loss of ergosterol synthesis can lead to elevated membrane permeability. Mutants lacking the ERG6 gene have a 35-fold increase in uptake of crystal violet compared to wild-type strains at low temperatures, although the elevation is modest at 30°C (2). We examined whether the sensitivity of dap1Δ strains to MMS could be due to altered membrane permeability. In principal, elevated membrane permeability could lead to increased local concentrations of MMS, causing a greater loss of viability than in wild-type strains.
Wild-type and dap1Δ strains were grown to log phase and incubated with 5 μg of crystal violet per ml at four different temperatures. The cells were then centrifuged, and the concentration of crystal violet in the supernatant was measured and compared with the beginning concentration to calculate uptake. Surprisingly, dap1Δ cells had a decreased crystal violet uptake relative to wild-type strains at each of the temperatures analyzed (Table 2), ranging from 1.3- to 13-fold lower than in the wild type. Our levels of uptake for the wild-type strain W303 were similar to those reported previously for the wild-type strain A184D at each of the temperatures analyzed (2). At all temperatures tested, the uptake of crystal violet in dap1Δ strains was significantly lower than that in wild-type cells, suggesting alterations in the endocytic pathway or transmembrane diffusion in dap1Δ mutants. To test whether dap1Δ strains are defective in uptake of compounds other than crystal violet from the media, we plated wild-type and dap1Δ strains on plates containing the selective drug Geneticin. Both wild-type and dap1Δ strains were sensitive to Geneticin, indicating that under physiological conditions, dap1Δ strains are capable of transporting compounds to the interior of the cell.
TABLE 2.
Membrane permeability in dap1Δ cells determined by crystal violet uptake
| Strain | Uptakea at the following temp (°C):
|
|||
|---|---|---|---|---|
| 0 | 22 | 30 | 37 | |
| Wild type (RCY409-2a) | 6.5 ± 0.8 | 24.9 ± 2.4 | 23.6 ± 1.3 | 31.8 ± 0.8 |
| dap1Δ (RCY409-2b) | 0.5 ± 0.5b | 16.2 ± 2.1b | 18.1 ± 1.0b | 25.3 ± 1.4b |
Crystal violet uptake was measured as described by Bard et al. (2) and in Materials and Methods. Cells were incubated with crystal violet for 10 min at the various temperatures and centrifuged. The A595 of the supernatant (Af) was measured and compared with the absorbance of the starting solution (Ai); percent uptake equals (Ai − Af)/Ai. The results from four separate reactions were averaged, and the standard deviation is shown.
Significantly different from the result for the wild-type strain by the Student t test (P < 0.05).
We then measured the sensitivity of dap1Δ strains to a cationic pulse. Cells were exposed to 2 M solutions of the chloride salts of calcium, chromium, potassium, magnesium, and nickel for 10 min and then plated for viability. Mutants with hyperpermeable membranes are sensitive to a pulse of cations at high concentrations (2). We found that the response of dap1Δ strains to these five cations was indistinguishable from that of wild-type strains (Table 3). We conclude that the altered sterol composition of dap1Δ strains does not disrupt the function of the cell membrane and does not allow a major influx of dyes or salts. Thus, it is unlikely that dap1Δ strains are sensitive to MMS purely because of overly permeable membranes, in contrast to phenotypes associated with the erg6 mutation.
TABLE 3.
Membrane permeability in dap1Δ cells determined by viability after a cation pulse
| Strain | % Viabilitya after exposure to:
|
||||
|---|---|---|---|---|---|
| Ca2+ | Cr+ | K+ | Mg2+ | Ni+ | |
| Wild type (RCY409-2a) | 96 ± 8 | 18 ± 3 | 86 ± 6 | 91 ± 8 | 80 ± 17 |
| dap1Δ (RCY409-2b) | 80 ± 11 | 22 ± 5 | 94 ± 4 | 93 ± 3 | 88 ± 18 |
Viability compared to that in a saline control after 10 min of exposure to 2 M solutions of the various chloride salts. Values reported are the means of three separate determinations ± standard deviations, and all determinations were repeated one to three times.
Dap1p regulates mitochondrial maintenance.
Mutant dap1Δ cells have elevated levels of petite colony formation (Table 4). The W303 strain background contains the ade2 mutation, which causes a red colony color due to the accumulation of adenine precursors. Cells that have lost the ability to respire grow as small white colonies and are inviable on plates containing a nonfermentable carbon source such as glycerol (12). Plate cultures of dap1Δ isolates contained increased numbers of small, white colonies. We measured the frequency with which these white colonies emerge in the population and found that dap1Δ strains have a 4.4-fold elevation in petite formation (19.6%, compared to 4.5% for the wild-type strain [Table 4]). Thus, Dap1p is required for wild-type levels of mitochondrial maintenance. The elevated level of petite formation may be due to the partial arrest of ergosterol synthesis in dap1 strains, because other ergosterol mutants, such as erg3 and erg6 mutants, have elevated petite formation (45; M. Bard, unpublished observations).
TABLE 4.
Mitochondrial stability in dap1Δ cellsa
| Strain (genotype) | % Petite colonies (mean ± SD)b | Avg % petite colonies (fold vs wild type) |
|---|---|---|
| RCY407-12d (wild type) | 6.1 ± 1.3, 2.8 ± 3.6 | 4.5 (1) |
| RCY407-1d (dap1Δ) | 16.9 ± 7.4, 22.3 ± 8.8 | 19.6 (4.4)c |
| RCY409-5c (dun1Δ) | 16.1 ± 5.6, 11.8 ± 4.6 | 14.0 (3.1)c |
Twenty separate colonies were picked, diluted, and then plated on YPD plates. After 3 days of growth at 30°C and an additional 3 days of incubation at 4°C, the numbers of red and white colonies were counted, and the percentage of small, white colonies was calculated. One hundred white colonies from each genotype were then patched onto medium containing glycerol as a carbon source to confirm that the colonies were petite.
Results from two 20-plate assays are shown.
Statistically significant (t test; P < 0.05) difference compared to wild-type cells.
Petite colony formation is also characteristic of strains with mutations within RNR genes (13). Dun1p is required for transcriptional activation of RNR genes in response to damage (59), and dun1 mutants are damage sensitive, have elevated rates of petite formation (16) and mitotic recombination (15), and have diminished telomeric silencing (8). We used a dun1 strain as a positive control for elevated petite colony formation, and as expected, dun1 strains had an elevated level of petite colonies (Table 4).
These results suggest that Dap1p regulates mitochondrial stability. Petite colonies can arise through loss of mitochondrial biogenesis or through mutations in nuclear genes required for mitochondrial maintenance. Thus, it was possible that dap1Δ strains lose mitochondria because of an elevated mutation frequency in the nuclear genome. We measured the rate of mutation at the CAN1 locus in dap1Δ strains in duplicate assays of 20 independent cultures. We did not detect a significant elevation in mutation frequency in dap1Δ cells compared to wild-type strains (2.8 × 10−7 in dap1Δ cells versus 2.4 × 10−7 in wild-type cells) (10).
DISCUSSION
We have found that the Dap1p protein is required for the damage response, telomere length maintenance, mitochondrial biogenesis, and sterol regulation. Mutants lacking Dap1p are particularly sensitive to the methylating agent MMS. MMS methylates DNA, causing the replication fork to stall during S phase, leading to double-stranded DNA breaks. Following cell cycle arrest and repair of the broken DNA, the cell cycle resumes. Our data indicate that yeast requires Dap1p in order to progress through the G1-S phase transition following MMS-induced damage.
Another interpretation of these data is that MMS induces a second type of damage aside from induction of double-stranded DNA breaks and that Dap1p is required for the G1-S transition following this type of damage. One potential secondary type of damage could arise from MMS-induced oxidative stress, which is associated with MMS (19). We pursued the possibility that MMS-induced oxidative stress might cause cell cycle arrest by damaging the cellular pool of sterols. Thus, the requirement for Dap1p to exit G1 might be related to the “sparking” requirement for ergosterol, in which diminished ergosterol levels prevent the G1-S transition (11). To test this idea, we treated wild-type cells for 3 h with various doses of itraconazole and then plated the cells on MMS. Surprisingly, itraconazole pretreatment had no effect on damage responsiveness in wild-type cells. This suggests that decreased sterol synthesis coupled with MMS-induced damage does not lead to an irreversible G1 arrest.
Cells exposed to chronic damage arrest and repair the damage. If the damage persists, cells attempt to override the arrest checkpoint in a process called adaptation (54). Our results indicate that it is unlikely that Dap1p functions by overriding a Rad9p-mediated G1 checkpoint. We prefer a model in which Dap1p performs an essential function in the G1-S transition following MMS-induced damage. The molecular function of Dap1p in exiting G1 after damage is not clear. However, Dap1p binds indirectly to a network of proteins that regulate cell cycle progression and the G1-S phase transition (25). We propose that protein interactions between Dap1p and its binding partners are required for the G1-S phase transition following MMS-induced damage.
We have shown that Dap1p is required for wild-type ergosterol levels. Ergosterol is a key component of the yeast cell membrane and resembles cholesterol structurally. Ergosterol levels regulate membrane fluidity and permeability, endocytosis, secretion, vacuole fusion, mitochondrial respiration, oxygen sensing, gene expression (50), and a cell cycle sparking function (reviewed in reference 11). We have demonstrated that dap1 strains are sensitive to azole inhibitors of ergosterol synthesis and have increased levels of sterol intermediates. However, dap1 mutants are distinct from erg mutants in that dap1 is not a sterol auxotroph, and dap1 mutants are capable of synthesizing ergosterol, albeit at reduced levels compared to the wild type.
Proteins that direct the synthesis of ergosterol have been previously implicated in damage repair. Deletion of the ERG3 gene, encoding C-5 sterol desaturase, leads to MMS sensitivity, and erg28 and arv1 strains are MMS sensitive, but to a lesser extent (3). In addition, erg6 mutants are sensitive to the chemotherapeutic agents dactinomycin and doxorubicin (24). It is likely that some of these strains are sensitive to MMS because of increased membrane permeability (2), increasing the effective concentration of damaging agent in the cell. However, we found that dap1 strains do not have elevated membrane permeability by two separate assays, indicating that the deficiency of dap1 strains in damage repair goes beyond simple changes in membrane structure. Interestingly, loss of ergosterol synthesis has been linked to decreased endocytosis in some mutants (23), consistent with the decreased uptake of crystal violet in dap1Δ cells.
Mutants lacking Dap1p are defective in mitochondrial mitogenesis. We propose that this phenotype is due to abnormal ergosterol synthesis in dap1 cells. The relationship between ergosterol synthesis and mitochondrial regulation is poorly understood, because ergosterol is not a major constituent of mitochondrial membranes relative to other membranes. However, several ergosterol synthetic mutants have decreased mitochondrial function, including erg3 (45) and erg6 (unpublished observation) strains. In addition, overexpression of the mitochondrial COX3 gene confers resistance to azole compounds (31), further suggesting a link between altered respiration and ergosterol regulation.
Dap1p is homologous to the MAPR family. The term MAPR is based on the properties of individual family members, because only one member is a known membrane protein and only one has probable progesterone binding activity. The porcine MAPR was isolated biochemically from liver membrane extracts based on its binding to tritiated progesterone (37, 14). A rat homologue of MAPR, called 25-Dx, was cloned at the same time in a phage display screen for elevated transcripts in rat liver following treatment with 2,3,7,8-tetrachlorodibenzo-p-dioxin (46). Following treatment, expression of 25-Dx was elevated three- to sevenfold (46), suggesting a role in the response to dioxin, a potent carcinogen. A murine MAPR homologue (also called 25-Dx) was subsequently cloned based on its reduced expression during lordosis, a female rodent mating behavior, and its negative regulation by progesterone (30). Green fluorescent protein-tagged murine 25-Dx localized to the cell periphery (30), and a number of MAPR family members have putative membrane-spanning sequences near their amino termini. The homology between Dap1p and membrane-associated progesterone binding proteins is consistent with a similar localization for Dap1p. Because MAPR proteins are putative progesterone binding proteins, we examined the toxicity of progesterone to strains lacking Dap1p but detected no difference between dap1Δ and wild-type cells with respect to progesterone-mediated toxicity.
Dap1p is the first member of the MAPR family to be directly linked to the response to damage and sterol synthesis. Some of these functions are probably conserved in other organisms. DAP1 homologues are present in many model organisms, including budding and fission yeasts, Drosophila, C. elegans, Arabidopsis, and mice. Our results also indicate that an improved understanding of this gene family may lead to novel approaches for antifungal compounds or strategies to increase the efficacy of existing antifungal agents.
Acknowledgments
We thank Julia Mallory and Bettina Meier for helpful comments on the manuscript and Vytas Bankaitis and members of the T. D. Petes and W. G. Cance labs for helpful discussions. We also thank XiHui Yang for technical assistance, Amos McKenzie and John Pringle for plasmids, and an anonymous reviewer for suggestions for the Discussion section.
This work was funded by start-up funds from the University of North Carolina Medical School. R.J.C. is a Scholar of the Building Interdisciplinary Research Careers in Women's Health Program from the NIH, grant K12HD001441. M.B. was funded by NIH grant R01 GM62104.
REFERENCES
- 1.Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2416-2428. [DOI] [PubMed] [Google Scholar]
- 2.Bard, M., N. D. Lees, L. S. Burrows, and F. W. Kleinhaus. 1978. Differences in crystal violet uptake and cation-induced death among yeast sterol mutants. J. Bacteriol. 135:1146-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 5.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corda, Y., V. Schramke, M. P. Longhese, T. Smokvina, V. Paciotti, V. Brevet, E. Gilson, and V. Geli. 1999. Interactions between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat. Genet. 21:204-208. [DOI] [PubMed] [Google Scholar]
- 7.Craven, R. J., and T. D. Petes. 1999. Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics 152:1531-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craven, R. J., and T. D. Petes. 2000. Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:2378-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craven, R. J., and T. D. Petes. 2001. The Saccharomyces cerevisiae suppressor of choline sensitivity (SCS2) gene is a multicopy suppressor of mec1 telomeric silencing defects. Genetics 158:145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven, R. J., P. W. Greenwell, M. Dominska, and T. D. Petes. 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologues of the mammalian ATM and ATR genes. Genetics 161:493-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daum, G., N. D. Lees, M. Bard, and R. Dickson. 1998. Biochemistry, cell biology, and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14:1471-1510. [DOI] [PubMed] [Google Scholar]
- 12.Dujon, B. 1981. Mitochondrial genetics and functions, p. 505-653. In J. N. Strathern, F. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Elledge, S. J., and R. W. Davis. 1987. Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol. Cell. Biol. 7:2783-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkenstein, E., C. Meyer, C. Eisen, P. C. Scriba, and M. Wehling. 1996. Full-length cDNA sequence of a progesterone membrane-binding protein from vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 229:86-89. [DOI] [PubMed] [Google Scholar]
- 15.Fasullo, M., J. Koudelik, P. AhChing, P. Giallanza, and C. Cera. 1999. Radiosensitive and mitotic recombination phenotypes of the Saccharomyces cerevisiae dun1 mutant defective in DNA damage-inducible gene expression. Genetics 152:909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fikus, M. U., P. A. Mieczkowski, P. Koprowski, J. Rytka, E. Sledziewska-Gojska, and Z. Ciesla. 2000. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics 154:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gachotte, D., J. Eckstein, R. Barbuch, T. Hughes, C. Roberts, and M. Bard. 2001. A novel gene conserved from yeast to humans is involved in sterol biosynthesis. J. Lipid Res. 42:150-154. [PubMed] [Google Scholar]
- 18.Game, J. C. 2000. The Saccharomyces repair genes at the end of the century. Mutat. Res. 451:277-293. [DOI] [PubMed] [Google Scholar]
- 19.Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge, and P. O. Brown. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12:2987-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerdes, D., M. Wehling, B. Leube, and E. Falkenstein. 1998. Cloning and tissue expression of two putative steroid membrane receptors. Biol. Chem. 379:907-911. [DOI] [PubMed] [Google Scholar]
- 21.Greenwell, P. W., S. L. Kronmal, S. E. Porter, J. Gassenhuber, B. Obermaier, and T. D. Petes. 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82:823-829. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 23.Heese-Peck, A., H. Pichler, B. Zanolari, R. Watanabe, G. Daum, and H. Riezman. 2002. Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell 13:2664-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemenway, C. S., and J. Heitman. 1996. Immunosuppressant target protein FKBP12 is required for P-glycoprotein function in yeast. J. Biol. Chem. 271:18527-18534. [DOI] [PubMed] [Google Scholar]
- 25.Ho, Y., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, T. R., et al. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 27.Jackson, S. P. 1996. The recognition of DNA damage. Curr. Opin. Genet. Dev. 6:19-25. [DOI] [PubMed] [Google Scholar]
- 28.Jelinsky, J. A., and L. D. Samson. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 96:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalb, V. F., C. W. Woods, T. G. Turi, C. R. Dey, T. R. Sutter, and J. C. Loper. 1987. Primary structure of the P450 lanosterol demethylase gene from Saccharomyces cerevisiae. DNA 6:529-537. [DOI] [PubMed] [Google Scholar]
- 30.Krebs, C. J., E. D. Jarvis, J. Chan, J. P. Lydon, S. Ogawa, and D. W. Pfaff. 2000. A membrane-associated progesterone-binding protein, 25-Dx, is regulated by progesterone in brain regions involved in female reproductive behaviors. Proc. Natl. Acad. Sci. USA 97:12816-12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Launhardt, H., A. Hinnen, and T. Munder. 1998. Drug-induced phenotypes provide a tool for the functional analysis of yeast genes. Yeast 14:935-942. [DOI] [PubMed] [Google Scholar]
- 32.Lea, D. E., and C. A. Coulson. 1949. The distribution of the number of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 33.Lustig, A. J., and T. D. Petes. 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83:1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallory, J. M., and T. D. Petes. 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97:13749-13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, S. G., T. Laroche, N. Suka, M. Grunstein, and S. M. Gasser. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97:621-633. [DOI] [PubMed] [Google Scholar]
- 36.McAinsh, A. D., S. Scott-Drew, J. A. H. Murray, and S. P. Jackson. 1999. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr. Biol. 9:963-966. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, C., R. Schmid, P. C. Scriba, and M. Wehling. 1996. Purification and partial sequencing of high-affinity progesterone binding sites from porcine liver membranes. Eur. J. Biochem. 239:726-731. [DOI] [PubMed] [Google Scholar]
- 38.Mills, K. D., D. A. Sinclair, and L. Guarente. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97:609-620. [DOI] [PubMed] [Google Scholar]
- 39.Molzahn, S. W., and R. A. Woods. 1972. Polyene resistance and the isolation of sterol mutants in Saccharomyces cerevisiae. J. Gen. Microbiol. 72:339-348. [DOI] [PubMed] [Google Scholar]
- 40.Paciotti, V., M. Clerici, M. Scotti, G. Lucchini, and M. P. Longhese. 2001. Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol. Cell. Biol. 21:3913-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parenteau, J., and R. J. Wellinger. 2000. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porter, S. E., P. W. Greenwell, K. B. Ritchie, and T. D. Petes. 1996. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 24:582-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritchie, K. B., J. C. Mallory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, C. L., M. Grey, M. Schmidt, M. Brendel, and J. A. P. Henriques. 1999. Allelism of Saccharomyces cerevisiae genes PSO6, involved in survival after 3-CPs + UVA induced damage, and ERG3, encoding the enzyme sterol C-5 desaturase. Yeast 15:1503-1510. [DOI] [PubMed] [Google Scholar]
- 46.Selmin, O., G. W. Lucier, G. C. Clark, A. M. Tritscher, J. P. Vanden Heuvel, J. A. Gastel, N. J. Walker, T. R. Sutter, and D. A. Bell. 1996. Isolation and characterization of a novel gene induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin in rat liver. Carcinogenesis 17:2609-2615. [DOI] [PubMed] [Google Scholar]
- 47.Siede, W., A. S. Friedberg, and E. C. Friedberg. 1993. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:7985-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siede, W., J. B. Allen, S. J. Elledge, and E. C. Friedberg. 1996. The Saccharomyces cerevisiae MEC1 gene, which encodes a homolog of the human ATM gene product, is required for G1 arrest following radiation treatment. J. Bacteriol. 178:5841-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, S. J., J. H. Crowley, and L. W. Parks. 1996. Transcriptional regulation by ergosterol in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5427-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 53.Tinkelenberg, A. H., Y. Liu, F. Alcantara, S. Khan, Z. Guo, M. Bard, and S. L. Sturley. 2000. Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1. J. Biol. Chem. 275:40667-40670. [DOI] [PubMed] [Google Scholar]
- 54.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 55.Vanden Bosshe, H. G., G. Willemsens, W. Cools, P. Marichal, and W. Lauwers. 1983. Hypothesis on the molecular basis of the antifungal activation of the N-substituted imidazoles and triazoles. Biochem. Soc. Trans. 11:665-667. [DOI] [PubMed] [Google Scholar]
- 56.Weinert, T. A., and L. H. Hartwell. 1990. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol. Cell. Biol. 10:6554-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida, Y., and Y. Aoyama. 1987. Interaction of azole antifungal agents with cytochrome P-45014DM purified from Saccharomyces cerevisiae microsomes. Biochem. Pharmacol. 36:229-235. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, Z., and S. J. Elledge. 1993. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75:1119-1127. [DOI] [PubMed] [Google Scholar]