Abstract
Candida albicans mutants with mutations in mitogen-activated protein (MAP) kinase HOG1 displayed an increased sensitivity to agents producing reactive oxygen species, such as oxidants (menadione, hydrogen peroxide, or potassium superoxide), and UV light. Consistent with this finding, C. albicans Hog1 was activated not only in response to an increase in external osmolarity, as happens with its Saccharomyces cerevisiae homologue, but also in response to hydrogen peroxide. The Hog1-mediated response to oxidative stress was different from that of transcription factor Cap1, the homologue of S. cerevisiae Yap1, as shown by the different sensitivities to oxidants and the kinetics of cell death of cap1Δ, hog1, and hog1 cap1Δ mutants. Deletion of CAP1 did not influence the level of Hog1 phosphorylation, and deletion of HOG1 did not affect Cap1 nuclear localization. Moreover, we show that the HOG1 gene plays a role in chlamydospore formation, another oxygen-related morphogenetic event, as demonstrated by the fact that hog1 cells were unable to generate these thick-walled structures in several media through a mechanism different from that of the EFG1 regulator. This is the first demonstration of the role of the Hog1-mediated MAP kinase pathway in resistance to oxidative stress in pathogenic fungi, and it allows us to propose a molecular model for the oxidative stress response in C. albicans.
One of the situations that often challenges growing yeast cells is oxidative stress, which is characterized by an abnormally high oxidative potential. This characteristic may be determined by the normal respiratory metabolism (the mitochondrial respiratory chain) but may also be generated by environmentally elevated oxygen pressure or by exposure to ionizing radiation. Under these conditions, the concentration of reactive oxygen species (ROS), such as superoxide anion (O2˙−), the hydroxyl radical (OḢ), or hydrogen peroxide (H2O2), is augmented, damaging—either directly or indirectly—several components of the cell, such as DNA, proteins, and lipids (28, 68). These effects are toxic and eventually lead to cell death. It is therefore not surprising that yeast cells have developed antioxidant mechanisms to cope with these conditions (see references 42 and 43 for recent reviews). Among the protective mechanisms are the activities of detoxifying enzymes (e.g., superoxide dismutases and catalases) or other, nonenzymatic scavenging substances that prevent the action of ROS. While this response has been characterized in some detail for bacteria (9, 16, 40), knowledge of these mechanisms in lower eukaryotes is lacking. It is clear, however, that this cellular response is accomplished, at least partially, at the transcriptional level. In the model yeast Saccharomyces cerevisiae, Yap1 (27), a transcription factor with a well-defined role in resistance to several drugs (43, 59), has been shown to mediate the oxidative stress response, a conclusion inferred not only from the susceptibility of yap1 mutants to several oxidants but also from the Yap1-dependent transcription of different genes with a protective role (24, 31, 59, 63). Other elements, such as those encoded by the SKO1 (55), YAP2 (4, 63), MSN2/MSN4 (18, 33), HAL1 (72), or SKN7 (36, 39, 44) genes, play a role in this adaptive response. In Schizosaccharomyces pombe, two mechanisms seem to control the response to oxidative stress—the activation of the StyI/Spc1 mitogen-activated protein (MAP) kinase pathway and the function of the Yap1 homologue—although these phenomena seem to be interdependent. The former pathway in this microorganism, the homologue of the HOG pathway, is responsible for sensing both osmotic stress and oxidative stress; the MAP kinase StyI is activated by phosphorylation (13, 57) to phosphorylate the transcription factor Atf1 (54, 60, 69), which is responsible for the transcription of different oxidative response genes. Pap1, the homologue of Yap1, is necessary to respond to oxidative stress through the transcription of another set of oxidative response genes, but its activity depends on its nuclear localization, which is hampered in the absence of StyI (65).
Candida albicans is a pathogenic yeast (49) of great clinical interest due to the incidence (21) and severity of the infections that it causes, especially in immunocompromised individuals; it is therefore a preferred model of a fungal pathogen. It is evident that the mechanisms of defense against oxidative stress are especially relevant for many pathogenic fungi, such as C. albicans, since the neutrophil-macrophage system is crucial in the control and outcome of the infections that they cause (66) through oxidative killing mechanisms. A major limitation in defining the molecular determinants of resistance to oxidative stress in C. albicans is the lack of a powerful genetic system, only partially developed in the last few years (11, 51). There are some indications of the physiological response of C. albicans to oxidants in vitro (32), and recently, the roles of two C. albicans genes were analyzed in some detail. A YAP1 homologue named CAP1 was isolated and shown to play a role in multidrug and oxidative stress resistance (1, 2). Mutational analysis of this gene revealed a terminal cysteine residue essential for the regulated nuclear localization of the protein (71), similar to the carboxy-terminal cysteine-rich domain of Yap1 (15, 37). These authors also reported differences between Yap1 and Cap1, since CAP1 did not completely complement the H2O2 sensitivity of yap1 strains (71). The CAT1 gene, which encodes a protein with catalase activity, was also shown to be involved in oxidant susceptibility, and its deletion generated cells that were less virulent in the mouse acute systemic infection model (70).
In this work, we demonstrate that the Hog1 MAP kinase is essential for C. albicans resistance to oxidative stress through a mechanism different from those found in other yeast models, defining an additional role for this MAP kinase besides the previously described functions in osmoadaptation (58) and morphogenesis (3).
MATERIALS AND METHODS
Strains and growth conditions.
Yeast strains are listed in Table 1. For clarity, and unless otherwise stated, hog1 will always indicate the homozygous hog1/hog1 Ura+ strain (strain CNC13). Construction of his1Δ, ura3Δ his1Δ, cap1Δ, and hog1 cap1Δ strains is indicated below. Unless otherwise stated, RM100 was used as the wild-type strain. his1Δ strains behaved in the same manner as strain CAF2 or SC5314.
TABLE 1.
Strains used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| Escherichia coli DH5αF′ | K-12 Δ(lacZYA-argF)u169 supE44 thi-1 recA1 endA1 hsdR17 gyrA relA1 (φ80lacZΔM15) F′ | 26 |
| Candida albicans | ||
| SC5314 | Wild type (HOG1/HOG1) | 23 |
| CAF2 | URA3/ura3Δ::λimm434 | 19 |
| CAI4 | ura3Δ::λimm434/ura3Δ::λimm434 | 19 |
| RM1 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG-URA3-hisG/HISI | This work |
| RM10 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/HISI | This work |
| RM100 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG-URA3-hisG | This work |
| RM1000 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG | This work |
| CNC11 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG HOG1/hog1::hisG-URA3-hisG | 58 |
| CNC13 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG hog1::hisG-URA3-hisG/hog1::hisG | 58 |
| CNC15 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG hog1::hisG/hog1::hisG | 3 |
| CNC16 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG HOG1:URA3/hog1::hisG | 3 |
| CCC1 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG cap1Δ::URA3/cap1Δ::HIS1 | This work |
| CHC1 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG hog1::hisG/hog1::hisG cap1Δ::URA3/cap1Δ::HIS1 | This work |
| RCG1 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG CAP1/CAP1-GFP-URA3 | This work |
| HCG1 | ura3Δ::λimm434/ura3Δ::λimm434his1Δ::hisG/his1Δ::hisG hog1::hisG/hog1::hisG CAP1/CAP1-GFP-URA3 | This work |
Yeast strains were grown in YPD medium (1% yeast extract, 2% peptone, 2% glucose) or SD minimal medium (2% glucose, 0.67% yeast nitrogen base without amino acids) with the appropriate auxotrophic requirements. Histidine and uridine were routinely added to liquid and solid media for phenotypic assays. The growth temperature was 37°C unless otherwise indicated. Usually, overnight cultures were refreshed to an optical density at 600 nm (OD600) of 0.05, and experiments were performed when cultures reached an OD600 of 1 when exponential-phase cells were required. When stationary-phase cells were required, cells from a 3-day-old culture were routinely used. The viability of stationary-phase cultures was checked by staining with propidium iodide (14).
Deletion of the HIS1 gene.
A KpnI fragment from YEpHISX carrying the HIS1 gene (52) was subcloned in the KpnI site of pUC19 to generate pUCHISA. A 0.7-kbp EcoRV fragment of pUCHISA (carrying the HIS1 gene) was substituted with a blunt-ended BamHI hisG-URA3-hisG cassette (obtained from pCUB6-K [58]) to generate plasmid pHP4. pHP4 was linearized by using SacI and BamHI to obtain a 7-kbp fragment for HIS1 deletion in strain CAI4. Homologous recombination was achieved by using 2,170- and 730-bp flanking regions (5′ and 3′ fragments, respectively) to obtain strain RM1. Strains RM10, RM100, and RM1000 (Table 1) were obtained by using a previously described procedure (19). Correct disruptions were confirmed by PCR with primers oYBR7 (5′-AACAGTGTCGCCAGAATGTGCCG-3′) and oURA3 (5′-AGATCCAGATATTGAAGGTAAAAG-3′) for Ura+ strains or oYBR7 and oHG1 (5′-GTTTTCCGCCATCGCAATCAGGC-3′) for Ura− strains.
Deletion of the CAP1 gene.
The CAP1 gene was isolated in a functional screening for the identification of azole-resistant clones in S. cerevisiae (to be described elsewhere). For deletion of this gene, we used a strategy in which the 5′ and 3′ regions of the gene to be deleted were amplified by PCR with oligonucleotides oCAP1 (5′-AGAGCGTCGACGGGCCCATGCAGATATTAAAAGAAAT-3′) and oCAP2 (5′-AGGCAGTCGACAAGCTTCAACTCATTTTTCAACACGTCTAC-3′) for the 5′ region and oligonucleotides oCAP3 (5′-GCGGACTAGTGATTTTGTCAAGAATTCATTACCT-3′) and oCAP4 (5′-ATACCTGCGGCCGCTTAATGTTTTATACTTCGCTC-3′) for the 3′ region. These pieces of DNA were first independently subcloned in the pGEMT vector, generating plasmids pGEMT-5CAP1 and pGEMT-3CAP1. Then, the 5′ region was excised from pGEMT-5CAP1 by using the restriction sites SalI and ApaI, and the 3′ region was excised from pGEMT-3CAP1 by using the restriction sites SpeI and NotI. The fragments were then included in a four-fragment ligation in plasmid pCAPUCf2 flanking the URA3 marker. Similarly, the fragments were placed in plasmid pCAPHKn1 flanking the HIS1 marker (to be described elsewhere). To delete the CAP1 gene, strain RM1000 (ura3 his1) was cotransformed with plasmids pCAPUCf2 and pCAPHKn1 (digested with XbaI and NotI) by electroporation (34). Histidine and uracil prototrophs were isolated after 4 to 5 days of growth at 30°C and then replica plated on plates containing 4 mM hydrogen peroxide to detect knockouts. The correct integration of the construction in the sensitive clones was confirmed by PCR analysis with appropriate oligonucleotides from regions outside of the integration cassette. hog1 cap1Δ strains were obtained by deleting CAP1 in strain CNC15 (3). Deletion of the CAP1 gene was assessed by PCR with primers oCAP1 and oURA3 (described above) or oHIS1 (5′-GCTGTAACTTATTGAGTGGTGCCG-3′).
CAP1-GFP integration.
CAP1-green fluorescent protein (GFP)-expressing strains RCG1 and HCG1 were obtained through the integration of plasmid pDS569 (W. S. Moye-Rowley and D. Sanglard, unpublished data) in the CAP1 locus by using SphI to cut the CAP1-GFP construct. Strains showing correct integration were detected by PCR with oligonucleotides oCAP1 (see above) and oGFPLO (5′-CCAGTAGTACAAATAAATTTTAAGGTC-3′). Expression of the CAP1-GFP fusion was ascertained by Western blotting (see conditions below) with an anti-GFP monoclonal antibody (JL-8; Clontech, Palo Alto, Calif.).
Oxidative stress assays.
Hydrogen peroxide, menadione sodium bisulfite (MD), and potassium superoxide (KO2) were obtained from Sigma. Dilutions were performed by using sterile double-distilled H2O. Susceptibility to hydrogen peroxide was measured by using exponential- or stationary-phase growing cells in YPD medium at 37°C. A total of 107 cells were transferred to an Eppendorf tube, and hydrogen peroxide was added to a final concentration of 100 mM. The tube was incubated at 37°C, and 10-μl samples were collected at different times and spotted onto YPD plates. The plates were incubated for 24 h at 37°C and photographed. Susceptibility to MD and other oxidants at various concentrations was quantified in a similar way. Cell death was estimated by CFU counting or by propidium iodide staining and flow cytometry (14).
Protein extracts and immunoblot analysis.
Yeast strains were grown to an optical density of 1 at 37°C in YPD medium. NaCl or hydrogen peroxide was added to the medium at a final concentration of 1.2 M or 10 mM, respectively. Samples were taken 10 min after the challenge or at different times. Cell extracts were obtained by using glass beads in a Fast Prep cell breaker as indicated previously (41). Equal amounts of proteins were loaded onto gels, as assessed by 280-nm measurements of the samples and Ponceau red staining of the membranes prior to blocking and detection. Blots were probed with phospho-p38 MAP kinase (Thr180-Tyr182) monoclonal antibody 28B10 (Cell Signaling Technology, Inc.) and a polyclonal antibody to S. cerevisiae Hog1 (Santa Cruz Biotechnology) and developed with a Hybond ECL kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions. The anti-S. cerevisiae Hog1 signal was used both as an internal control in the experiments concerning the activation of C. albicans Hog1 and as a loading control. Image densitometry was performed by Bio-Rad Multi-Analyst 1.1 software analysis of the bands scanned with a Bio-Rad GS-690 imaging densitometer.
Fluorescence microscopy.
Yeast strains were grown at 30°C in SD minimal medium to an optical density of 0.8. Samples were centrifuged and washed twice with phosphate-buffered saline. Cells were resuspended in phosphate-buffered saline and maintained at 30°C for 15 min. For H2O2-treated cells, H2O2 was added to a final concentration of 0.4 mM. Cell fixation and nuclear staining (with propidium iodide) were performed as described previously (3). Fluorescence microscopy was carried out with a Nikon Eclipse TE2000-U microscope at a magnification of ×100. Images were captured by using a Hamamatsu ORCA-ER CCD camera with AquaCosmos 1.3 software. All images were processed in the same manner and mounted by using Adobe Photoshop 4.0.
Catalase assays.
Strains were grown to early exponential phase (OD600, 0.4), and H2O2 was added to a final concentration of 0.4 mM. Samples were taken at 0, 15, 30, and 60 min after H2O2 addition. Total protein extracts of different strains were obtained by glass bead breakage in the presence of protein inhibitors and used for measurement of catalase activity as indicated previously (8). The amounts of proteins in the extracts were quantified by measuring the absorbance at 280 nm.
Chlamydospore formation.
Chlamydospore formation was assayed essentially as indicated previously (62). The borders of more than 50 colonies were examined for each strain tested.
RESULTS
HOG1 mediates resistance to oxidative stress.
In order to understand the lack of virulence of C. albicans hog1 mutants in a systemic model of infection (3), we examined other phenotypic traits of this mutant. Invasion of epithelial cells was not impaired in hog1 strains (F. Navarro-García, F. García del Portillo, C. Nombela, and J. Pla, unpublished observations), suggesting that other defects should be responsible for the loss of virulence. Thus, we tested the sensitivity to oxidative stress of hog1 mutants, since ROS production is one of the mechanisms used by cells of the immune system for killing microbial pathogens (66). We subjected exponential- or stationary-phase growing cells to externally added hydrogen peroxide (100 mM H2O2) to test the viability of the cultures at different times. In both cases, the hog1 mutant strain displayed enhanced susceptibility to this compound compared to the wild-type strain. As shown in Fig. 1A, exponentially growing cells were found to be more susceptible to H2O2, the hog1 cells being unable to grow after a treatment of 0.5 to 1 min; in contrast, longer times (more than 2 min; data not shown) were required to reduce the viability of the wild-type cells to the same level. In stationary phase, the times required for the same reductions in viability were 8 min for the mutant and 30 min for the wild type (Fig. 1A). It has been shown that oxidant sensitivity diminishes when yeast cells enter stationary phase as a consequence of metabolic changes that enhance the endogenous production of ROS, thus generating an adaptive response to oxidants (42). Our results indicate that the entry of C. albicans cells into stationary phase, similar to what is seen for S. cerevisiae, increases cell resistance to oxidants. Although this is also true for hog1 mutant cells, they are still more sensitive than wild-type cells to H2O2 (Fig. 1A). The higher sensitivity of hog1 mutants was also observed when cells were spotted onto YPD plates containing 1 mM H2O2 (data not shown). We also found that pretreatment of the cells at 42°C for 30 min did not affect sensitivity to hydrogen peroxide (no to hypertonic medium) in assays on solid medium (data not shown), indicating that the types of responses generated to heat and oxidants are, to a certain extent, separate phenomena in this organism.
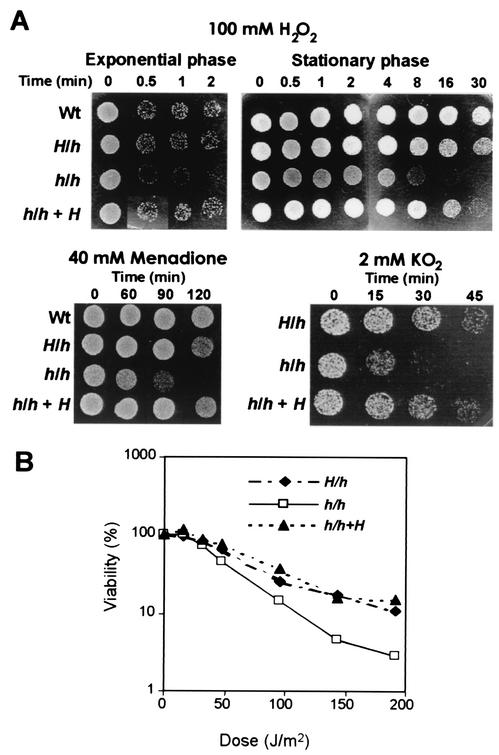
FIG. 1.
C. albicans hog1 mutants are sensitive to oxidative stress. (A) C. albicans cells were exposed to 100 mM hydrogen peroxide, 40 mM MD, and 2 mM KO2 in liquid YPD medium. Samples were taken at different times, diluted, and spotted onto YPD plates (105 cells in 10 μl). (B) A total of 100 to 200 C. albicans cells were plated on YPD plates and irradiated at 254 nm at different times by using a UV source. Plates were incubated at 37°C, and the percent survival was calculated by CFU counting after 24 h. The results are the means of three experiments. The standard error was always less than 10%. Strains used were as follows: Wt, HOG1/HOG1; H/h, HOG1/hog1; h/h, hog1/hog1; and h/h + H, HOG1 reintegrated in a hog1/hog1 strain (Table 1).
Oxidants can be generated inside the cell by different mechanisms based either on the cellular metabolism or the effect of external agents, such as UV light. These can generate free radicals (O2−) that are detoxified by the sequential action of superoxide dismutases (O2= generators) and catalases (O2 generators) (42). We therefore determined the effects of other agents different from peroxides on hog1 strains. To address the capacity of hog1 cells to detoxify superoxide anion (O2˙−), we tested sensitivity to MD (40 mM), a superoxide anion generator that affects S. cerevisiae but in a manner different from that of hydrogen peroxide (30). hog1 cells were found to be more sensitive than wild-type cells to this agent at a 40 mM concentration; control experiments showed that sodium bisulfite, either in liquid or on solid media, had no effect (data not shown). As shown in Fig. 1B, mutant cells were unable to grow after 120 min of treatment (growth defects were already evident at 90 min), while wild-type cells and the single-knockout strains grew normally under these conditions. Consistently, the effect on viability of another substance producing superoxide anion was qualitatively similar, as evidenced when cells were exposed to 2 mM KO2. While heterozygous strains could grow after 30 min of treatment, the hog1 mutant died (Fig. 1). As already described for other genes in C. albicans (35, 47, 58), the presence of one copy of the wild-type HOG1 gene, either in the heterozygous strain or in the strain with reintegrated HOG1, conferred an intermediate phenotype. The hog1 mutants also showed increased susceptibility to UV irradiation, as indicated by the faster loss of viability for these mutants than for the heterozygous mutants (which showed sensitivity similar to that of wild-type strains; data not shown) (Fig. 1). All of these findings demonstrate that Hog1 function is important for survival in the presenc of oxidative stress.
The HOG pathway in C. albicans is activated in response to oxidative stress.
The experiments described above indicated that Hog1 plays a role in the protective response against oxidants in C. albicans, suggesting that such a function for the HOG pathway would also exist in C. albicans. A likely—but not exclusive—explanation for the protective role of Hog1 is that oxidative stress itself could activate the pathway mediated by this MAP kinase. This is in fact the situation for the nonpathogenic fungus S. pombe (12, 57). We therefore analyzed the activation of this pathway by using a monoclonal antibody raised against phospho-p38, a mammalian homologue of the Hog1 MAP kinase (22, 25). This antibody recognizes the TGY motif characteristic of stress-activated MAP kinases activated by phosphorylation in threonine and tyrosine (6), thus providing a demonstration of the activation of the pathway. Protein extracts were obtained from C. albicans wild-type and hog1 cells exposed or not exposed to osmotic stress and oxidative stress and analyzed by Western blotting with the aforementioned antibody. We detected a band of about 43 kDa in response to osmotic stress (1.2 M NaCl, 10 min). No bands of the same size could be detected in extracts of hog1 mutants or in extracts of nontreated strains. Reintroduction of a functional copy of HOG1 into the genome of a hog1 mutant restored the detection of the same band in extracts of osmotically treated cells (Fig. 2A). After stripping of the anti-phospho-p38 antibody, we immunoblotted the same membrane with a commercial S. cerevisiae Hog1 polyclonal antibody, which recognized a band of the same size (≈43 kDa) in extracts of HOG1 strains but not hog1 strains (Fig. 2A). This was the only band that did not appear in hog1 strain protein extracts under basal or stress conditions when tested with the S. cerevisiae Hog1 polyclonal antibody. The detection of the band of phosphorylated Hog1 with the phospho-p38 monoclonal antibody under conditions of osmotic stress is consistent with the originally described role of the HOG pathway in S. cerevisiae and C. albicans (5, 58). The similarity of C. albicans Hog1 to the S. cerevisiae homologue (≈80%) is the reason for the cross-reactivity with the Hog1 polyclonal antibody.
FIG. 2.
Hog1 is phosphorylated in the presence of NaCl and H2O2 with similar kinetics. Western blots show the activation of Hog1 after H2O2 (10 mM) or NaCl (1.2 M) treatment for 10 min (A) or in a kinetic study at different times (B). Intermediate times were analyzed in the kinetic study, but only representative time points are shown. The same blots were assayed with a monoclonal antibody raised against phospho-p38 (Anti-TGYP) and a polyclonal antibody raised against S. cerevisiae Hog1 (Anti-ScHog1) (see Materials and Methods). See the legend to Fig. 1 for strain designations.
For oxidatively stressed cells (10 mM H2O2, 10 min), the anti-phospho-p38 antibody detected a 43-kDa band that cross-reacted with the S. cerevisiae Hog1 polyclonal antibody as well (Fig. 2A). Moreover, the phosphorylation signal increased significantly upon exposure of the cells to oxidative stress compared to osmotic stress (Fig. 2A). Image densitometry revealed that the intensities of the signals in sodium chloride stress and in oxidative stress were two times and three times higher, respectively, than that under basal conditions.
To explore the characteristics of the responses to both stimuli, we performed kinetic studies of the activation of the pathway by monitoring Hog1 phosphorylation. The results showed that the onset of activation was similar and fast in both cases; phosphorylation was detected after 1 min of treatment (Fig. 2B). In the presence of both stimuli, the signal extinction was also similar, although H2O2-induced phosphorylation was still high after 1 h of H2O2 treatment (Fig. 2B). We also found that Hog1 is phosphorylated over a broad range of H2O2 concentrations (from 0.4 to 100 mM; unpublished data). All of these results indicate the specificity of Hog1 phosphorylation induced by either osmotic or oxidative stress.
Deletion of CAP1 in a hog1 background leads to additive phenotypes in response to oxidative stress.
Yap1, a bZIP transcription factor, has been shown to be involved in resistance to oxidative stress in S. cerevisiae (59, 63) through the transcription of different genes in an oxidant-specific fashion (10), a phenomenon that is correlated with its nuclear location (37). The homologue of Yap1 in C. albicans, Cap1, has been cloned (1) and characterized. This protein also localizes in the nucleus in response to oxidative stress (71). Deletion of CAP1 renders the cells hypersensitive to different compounds—in particular, hydrogen peroxide—(2). In view of the facts that CAP1 is the best-characterized C. albicans gene involved in oxidative stress and that transcription factors are common targets of MAP kinase pathways, we addressed the possibility of the existence of an epistatic relationship of CAP1 with the Hog1 pathway. To this end, we deleted the CAP1 gene from both a wild-type background (RM1000) and a hog1 background. Strain RM1000, used for CAP1 deletion, was derived from CAI4 and carried a deletion of both alleles of the HIS1 gene constructed by URA3 gene Blaster methodology (19) (see Materials and Methods). Disruption of HIS1 did not affect growth rate (data not shown) or virulence (3). Deletion of CAP1 was accomplished by using the two-marker strategy (48), and the correct (homologous) integration of the genetic construction was checked by PCR with appropriate internal primers (see Materials and Methods). Strains with a deletion of the CAP1 gene did not show a significant reduction in growth either in normal minimal medium or in rich liquid medium at 37°C; the doubling times of these strains were similar to those of their parental strains (data not shown). Restoration of CAP1 in either cap1Δ or hog1 cap1Δ strains led to a recovery of the CAP1-deficient phenotype, that is, sensitivity to both cadmium stress and oxidative stress (data not shown).
Confirming previous observations (2), cap1Δ mutants were found to be hypersensitive to hydrogen peroxide and a variety of other oxidants in solid medium. In a standard diffusion method, cap1Δ and hog1 mutants behaved similarly; double mutants were more sensitive (data not shown). However, the differences between both types of mutants were evident in experiments in which a rapid adaptive response was required. Incubation of exponentially growing cells (optical density, 0.5) with 50 mM H2O2 resulted in a rapid death of hog1 cells, which were unable to grow after 4 min of incubation, while cells of heterozygous strains could grow longer (Fig. 3A). The cap1Δ mutant behaved in a manner similar to that of hog1 mutants; however, the hog1 cap1Δ double mutant died soon after treatment, indicating an increased sensitivity to H2O2, higher than that of hog1 or cap1Δ single mutants (Fig. 3A). A higher temperature aggravated this phenotype; the growth defect was more evident at 42 and 37°C than at 30°C, and the effect was additive for hog1 cap1Δ with respect to hog1 (data not shown).
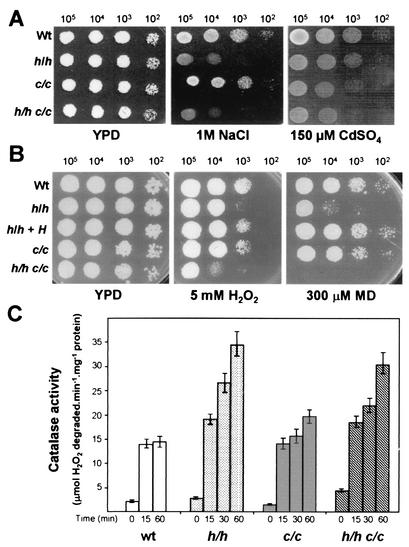
FIG. 3.
hog1 cap1Δ knockout strains are more sensitive to oxidative stress than hog1 or cap1Δ knockout strains. (A) C. albicans exponentially growing cells were exposed to 50 mM hydrogen peroxide in liquid YPD medium as indicated in Materials and Methods and spotted onto YPD plates. The plates were incubated at 37°C for 24 h. (B) The viability of different strains in YPD medium containing 100 mM hydrogen peroxide was determined by fluorescence-activated cell sorting analysis with propidium iodide as a cell death marker. Error bars indicate standard errors. c/c, cap1Δ/cap1Δ strain; h/h c/c, hog1/hog1 cap1Δ/cap1Δ strain; see the legend to Fig. 1 for other strain designations.
In order to define precisely the response to oxidative stress, we performed a detailed kinetic analysis of cell death when cells were exposed to hydrogen peroxide in liquid cultures. Viability was monitored by propidium iodide staining. As shown in Fig. 3B, the addition of 100 mM H2O2 resulted in a clear reduction after 4 h in the viability of hog1 mutants (34% viability) compared to cap1Δ mutants (50%) and wild-type and heterozygous strains (70 and 62%, respectively). The viability of the hog1 cap1Δ double mutant was lower (20%) than that of hog1 or cap1Δ mutants (Fig. 3B). These results indicate that there are clear differences in the adaptation to oxidative stress between both types of mutants; cap1Δ mutants are able to survive the challenge, probably through the production of an alternative response that hog1 or hog1 cap1Δ mutants cannot elicit. Since hog1 cap1Δ mutants showed exacerbated sensitivity to H2O2, the Cap1- and Hog1-mediated mechanisms seem to be at least partially independent, and each contributes to the susceptibility observed.
Hog1 and Cap1 mediate different oxidative stress response mechanisms.
The differences in the type and intensity of stress responses between cap1Δ and hog1 mutants also suggest differences in phenotypes for other types of stress.
We addressed the sensitivities of single and double mutants to specific types of stress previously described as harmful for each of the traits under consideration, namely, high osmolarity for hog1 and heavy metals for cap1Δ. As shown in Fig. 4A, only hog1 and hog1 cap1Δ mutants were hypersensitive to osmotic stress (1 M NaCl), whereas cap1Δ mutants behaved like the wild-type strain. In contrast, only cap1Δ and hog1 cap1Δ mutants did not grow in the presence of 150 μM cadmium (Fig. 4A). It follows that the responses to osmotic stress and this heavy metal do not overlap in C. albicans.
FIG. 4.
hog1 and cap1Δ knockout strains behave differently, the hog1 cap1Δ double mutant having some mixed phenotypes. (A) Responses to high osmolarity and heavy metal stress of wild-type and mutant strains. Serial 10-fold dilutions of exponentially growing cells of the strains indicated were spotted onto YPD plates containing 1 M NaCl or 150 μM CdSO4. (B) Responses to different types of oxidative stress of wild-type and mutant strains. Serial 10-fold dilutions of exponentially growing cells of the strains indicated were spotted onto YPD plates containing 5 mM H2O2 or 300 μM MD. (C) Catalase activities in hog1, cap1Δ, and hog1 cap1Δ mutants. The results are the means and standard deviations of three experiments.
We also studied the responses of these strains to two types of oxidative agents: hydrogen peroxide and MD. The responses to peroxide were similar in the wild type and cap1Δ mutants, which were less sensitive than hog1 and hog1 cap1Δ mutants to 5 mM H2O2 (Fig. 4B). On the other hand, the responses to MD were different; cap1Δ mutants were slightly sensitive but more resistant than hog1 mutants (Fig. 4B). Interestingly, the hog1 cap1Δ double mutant could not grow at all in the presence of the same concentration of MD, showing that its ability to overcome superoxide stress is markedly impaired (Fig. 4B). These results represent a clear indication that Cap1 and Hog1 operate through different mechanisms to overcome oxidative stress, since the double mutants were much more sensitive than the single-knockout mutants but maintained the defects of the individual mutants.
To search for potential targets of the Hog1 or Cap1 activity (as has been done for S. cerevisiae or S. pombe), we measured catalase activity, which is one of the enzymatic mechanisms that cells use to eliminate peroxides and which has also been indicated to be an important element in resistance to neutrophils in C. albicans (70). Catalase activity was induced similarly in cap1Δ and wild-type cells exposed to 0.4 mM H2O2 (Fig. 4C). Interestingly, the hog1 and hog1 cap1Δ strains displayed higher levels of catalase activity, in some cases twice the levels found in wild-type and cap1Δ strains (Fig. 4C). Basal catalase activity was also higher in the hog1 and hog1 cap1Δ strains (2.8 and 4.4 μmol of H2O2 degraded per min per mg of protein, respectively, versus 2.1 and 1.4 for wild-type and cap1Δ strains). It follows that catalase activity depends on neither Cap1 nor Hog1 function but is increased in the absence of Hog1.
In the preceding experiments, we showed that the defective phenotype of cap1Δ strains is not as dramatic as the hog1 mutant phenotype under conditions of oxidative stress. Regarding the possible influence of Cap1 in Hog1 activation, we must take into account the increase in oxidative radicals in the medium, which cannot be counteracted by the contribution of Cap1, so that it is feasible that it could enhance the response of Hog1 by a compensatory mechanism. To address this question, we measured the phosphorylation of Hog1 in different strains (Fig. 5A). This activation is very fast (1 min) (Fig. 2B), so that one could expect an increased level of phosphorylation in cap1Δ mutants only under basal conditions or if the stimulus were present. Neither of those possibilities was confirmed; in both cases, the levels of Hog1 phosphorylation seemed to be equal (Fig. 5A). Thus, the absence of Cap1 function does not seem to influence the Hog1 pathway in terms of activation.
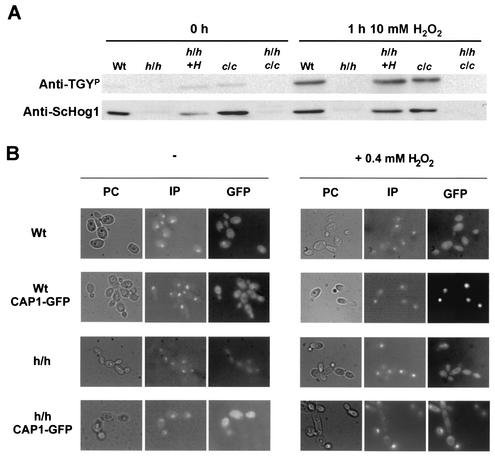
FIG. 5.
Upon oxidative stress, Cap1 did not affect the phosphorylation of Hog1, and Hog1 did not affect the nuclear localization of Cap1. (A) Hog1 is phosphorylated in the presence of hydrogen peroxide independently of Cap1. A Western blot shows the Cap1-independent activation of Hog1 after 1 h in the presence of 10 mM H2O2. The same blot was assayed with a monoclonal antibody raised against phospho-p38 (Anti-TGYP) and a polyclonal antibody raised against S. cerevisiae Hog1 (Anti-ScHog1) (see Materials and Methods). (B) Cap1 localization is not affected in a hog1 mutant upon oxidative stress. Cap1 localizes to the nucleus in the presence of oxidative stress (0.4 mM H2O2, 15 min) irrespective of the genetic background. Cells were observed by phase-contrast microscopy (PC), and nuclei were observed by using propidium iodide (IP) to ascertain the localization of the Cap1-GFP fusion (GFP). More than 300 cells were observed, and representative images of different strains were chosen.
StyI controls the localization of Pap1 to the nucleus in S. pombe (65). We tested the possibility that Hog1 could influence the nuclear localization of Cap1. For that purpose, we integrated a CAP1-GFP construct into the CAP1 locus in either a wild-type or a hog1 background to generate strain RCG1 or HCG1, respectively. The correct integration was confirmed by PCR (data not shown), and the expression of the full-length fusion was detected by using anti-GFP antibodies (data not shown; see Materials and Methods for details). Cap1 localized to the nucleus in the presence of H2O2 in both wild-type and hog1 backgrounds (Fig. 5B), showing that in the absence of Hog1, Cap1 is still able to enter the nucleus. All of these data confirm that Hog1 and Cap1 act in parallel—and independently—in sensing and responding to oxidative stress damage in C. albicans.
hog1 mutants are defective in chlamydospore formation, an oxygen-dependent morphogenetic program.
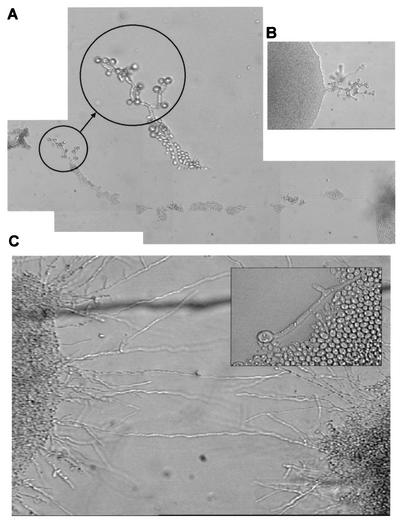
Alonso-Monge et al. previously showed that C. albicans Hog1 functions as a repressor of morphological transitions in both S. cerevisiae and C. albicans (3), a result in close agreement with the situation for S. cerevisiae Hog1 (50). In view of the involvement of this MAP kinase in both morphogenesis and oxidative stress responses, a specific phenotype for chlamydospore formation was anticipated for Hog1-deficient strains. Chlamydospores are thick-walled structures that develop under certain experimental conditions (low temperature, defined nutritional media and, probably related to oxidative stress, the absence of light and the presence of a microaerophilic environment). Wild-type cells (SC5314) were able to form chlamydospores after 4 to 5 days of growth at 24°C on cornmeal agar (Fig. 6). These structures appeared at the tips of growing hyphae (either long [Fig. 6A] or short [Fig. 6B]) under microaerophilic conditions. We observed the formation of abundant chlamydospores, which were round and had thick walls, in wild-type cells. In contrast, in hog1 mutants, chlamydospore formation was completely abolished. The colonies displayed the hyperfilamentous phenotype characteristic of hog1 mutants and, very infrequently, some structures at some hyphal tips slightly resembled chlamydospores and might have represented immature chlamydospores (Fig. 6C, inset). cap1Δ mutants were able to form chlamydospores, but hog1 cap1Δ cells were not. Since the only gene that has been described to date to play a role in this process is EFG1 (62), which encodes a transcription factor also shown to be involved in morphogenesis (64), it is feasible that the activation of EFG1 is HOG1 dependent. The overexpression of EFG1 under the control of the regulated PCK1 promoter resulted in enhanced filamentous growth, as expected, but did not suppress the defect in chlamydospore formation (data not shown). These results support a role for HOG1 in chlamydospore formation in C. albicans through a mechanism independent from EFG1.
FIG. 6.
hog1 mutants show defects in chlamydospore formation. Images show the wild-type strain (A and B) and hog1 mutants (C) on corn meal agar. No formation of chlamydospores can be observed at the end of the hog1 mutant hyphae, in contrast to those of the wild-type strain.
DISCUSSION
Novel mechanism for oxidative stress resistance in a pathogenic fungus.
In this work, we have shown for the first time that the Hog1 MAP kinase plays a role in the responses to oxidants of the pathogenic fungus C. albicans. This finding is particularly relevant for pathogenesis, since adaptation to the human body requires the pathogen to cope with host natural defenses, such as macrophages or neutrophils, whose capacity for oxidative killing of the invader is essential to controlling the outcome of fungal infections (66, 70). Other responses of pathogenic fungi to natural oxidative stress are the blocking of the macrophage oxidative burst in Histoplasma capsulatum (29); the production of polyols, such as mannitol, to scavenge reactive oxygen intermediates in Cryptococcus neoformans (7); and the activation of a TEY-MAP kinase-mediated pathway in Pneumocystis carinii (20). Our results show a novel way (activation of a “stress-activated” MAP kinase route [38]) in which a pathogenic fungus can escape from oxidative stress. An analysis of the killing of Candida hog1 mutants by macrophages and/or neutrophils could explain the reduced virulence of hog1 mutants (3) as being due to the reduced ability of these mutants to respond to oxidative stress, as shown by the higher sensitivity of hog1 mutants to peroxide- or superoxide-producing compounds and UV light.
Cells of the immune system produce oxidants on a continuous basis, thus maintaining constant concentrations (up to 0.5 mM ONOO− per min [67] or 0.5 mM H2O2 [56]) in the phagolysosomes of macrophages. Our experiments in which a small amount of H2O2 was maintained could mimic this phenomenon in vivo. In other experiments, we used larger amounts of oxidants (10 to 100 mM) to study the acute response of the cells to a single challenge of oxidants, which is probably exhausted over the time course of the experiments. In both situations, hog1 strains showed increased sensitivity to oxidants, indicating that Hog1 function is needed for both acute and adaptive responses to oxidants. Pretreatment of hog1 cells with a sublethal amount of oxidant (10 mM hydrogen peroxide for 1 h) resulted in enhanced survival when the cells were challenged with higher doses of oxidants (100 mM). The viability of hog1 cells increased from 52% to 83% when the cells were pretreated with 10 mM hydrogen peroxide for 1 h. On the contrary, the viability of hog1 cap1Δ cells was not increased by the same treatment (data not shown). Adaptation to oxidative stress does not simply imply a Hog1-dependent transcriptional adaptive mechanism but probably implies underlying altered levels of antioxidant defenses. However, our evidence clearly shows the specificity of the Hog1-mediated response to oxidative stress, because Hog1 was activated by phosphorylation in the presence of H2O2 (Fig. 2). We also showed that an increase in external osmolarity is signaled by Hog1 phosphorylation. It should be noted that this is the first time that stimuli previously presumed to activate a MAP kinase route in C. albicans have been demonstrated biochemically. Most of the data concerning the involvement of MAP kinase routes in different functions in this microorganism have been deduced from epistasis experiments and extrapolation of data obtained for model yeasts, such as S. cerevisiae (46).
Relationship of Hog1 to other proteins involved in the oxidative stress response.
Cap1 and Cat1 are the only proteins that had been associated with the oxidative stress response in C. albicans (1, 2, 70, 71). Our results from epistasis analyses also demonstrated that Hog1 and Cap1 play separate roles in oxidative stress defense and operate through different mechanisms. There are several arguments in favor of these conclusions. First, the patterns of susceptibility to the same oxidant agent (either hydrogen peroxide or MD) are different in cap1Δ and hog1 mutants, and the phenotype of the double mutant is aggravated (Fig. 3 and 4). Second, Hog1 and Cap1 respond—and maintain—separate responses to different stimuli, indicating that they do indeed participate in different physiological processes, such as heavy metal resistance, the osmotic response (Fig. 4A), or chlamydospore formation (Fig. 6), a morphological process related to oxygen availability and in which only the EFG1 regulator has been shown to play a role (62). hog1 and hog1 cap1Δ mutants are defective in chlamydospore formation or growth in high-osmolarity media, defects that are not observed in cap1Δ mutants. Third, in cap1Δ mutants, oxidative stress-induced Hog1 activation is not substantially altered in response to the stimulus (Fig. 5A). Fourth, Cap1 nuclear localization is not affected in hog1 mutants (Fig. 5B). All of these data suggest distinct and complementary roles of Hog1 and Cap1 in resistance to oxidative stress, as recently described for the homologous proteins in S. pombe (54).
With regard to catalase, we found that the activity of this enzyme was increased in both hog1 and cap1Δ mutants (Fig. 4C), although both mutants were still sensitive to H2O2. These results suggest that this activity is independent of both proteins and that there are alternative ways to sense and respond to oxidative stress that could induce the activity of catalase independently of Hog1 or Cap1. Such a mechanism would operate as an emergency mechanism for cell rescue in the absence of the capacity (which the HOG pathway seems to provide) to sense and respond to oxidative stress in C. albicans. The higher basal levels and the greater increase in the levels of catalase activity in hog1 and hog1 cap1Δ mutants (Fig. 4C) could suggest this explanation. Although catalase activity seems to be important for resistance to oxidative stress (70), the results shown in Fig. 4C indicate that in C. albicans, mechanisms other than catalase activity are important for overcoming oxidative stress challenge, since hog1 and cap1Δ strains were very sensitive, despite their ability to increase their catalase activity.
A molecular model for the oxidative stress response in C. albicans.
Several data indicate that the response of C. albicans to oxidative stress has several specificities compared to those of the classical species used as yeast model systems. For example, C. albicans hog1 mutants display intermediate sensitivity to UV light (Fig. 1). Whereas S. cerevisiae hog1 mutants are not sensitive to UV irradiation (61), S. pombe sty1− mutants are very sensitive, StyI being phosphorylated in wild-type strains in the presence of UV light (12). Moreover, cap1Δ mutants can increase their catalase activity in the presence of H2O2 (Fig. 4C), while S. pombe pap1− mutants cannot (although the expression of ctt1+ increases) (45). In addition, pap1− mutants show intermediate sensitivity to acute oxidative stress similar to that of cap1Δ mutants (Fig. 3B); both types of mutants are less sensitive than sty1− or hog1 strains (Fig. 3B) (54). Thus, the mechanism of the oxidative stress response in C. albicans is different from those in S. cerevisiae or S. pombe.
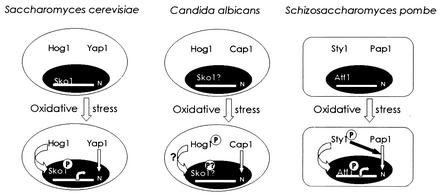
Models of the oxidative stress responses in S. cerevisiae and S. pombe are depicted in Fig. 7. In S. cerevisiae, Hog1 is not phosphorylated upon oxidative stress (data not shown) (61), although hog1 mutants are sensitive to H2O2 (55). A bZIP transcription factor, Sko1, has been shown to be a repressor of the transcription of some oxidative stress response genes. Interestingly, they are also downstream targets of Yap1 (17), and their expression—even in sko1Δ mutants—is dependent on Yap1. The phosphorylation of Sko1 by Hog1 disturbs the Ssn6-Tup1-Sko1 repressor complex (53), probably allowing the transcription of these genes (55). S. cerevisiae Sko1 is the homologue of S. pombe Atf1 (Fig. 7). In S. pombe, StyI plays two roles: it activates through phosphorylation the transcription factor Atf1 (54, 60, 69), and it influences the nuclear localization of Pap1 during oxidative stress (Fig. 7) (65). In addition, sty1− mutants show high sensitivity to H2O2, which is higher than that of the atf1− or pap1− single mutants but very similar to that of the atf1− pap1− mutants, indicating that StyI controls both mechanisms of the oxidative stress response in S. pombe (54).
FIG. 7.
C. albicans seems to respond to oxidative stress in a manner different from those of S. cerevisiae and S. pombe. The scheme shows the different ways in which C. albicans, S. cerevisiae, and S. pombe respond to oxidative stress. N, nucleus. The white bar inside the nucleus represents DNA. See Discussion for a detailed explanation.
The phenotypes of C. albicans hog1 mutants share some characteristics with counterparts in the two model systems, so that a mixed scheme can be envisioned for C. albicans on the basis of important distinctive features: Hog1 is phosphorylated in the presence of oxidative stress, and no relationship that mediates different responses to oxidative stress seems to exist between Hog1 and Cap1 (Fig. 7). Moreover, the nuclear localization of Cap1 upon oxidative stress is not affected in hog1 mutants, and neither is the basal phosphorylation status of Hog1 in cap1Δ mutants, thus reinforcing our hypothesis of Cap1 and Hog1 sensing and responding to oxidative stress in independent ways. The existence in the genome of C. albicans of an Sko1 homologue whose structure is more similar to that of Atf1 than to that of S. cerevisiae Sko1 could suggest a mechanism of transmission of the signal to the nucleus more like that of S. pombe, but regulated by C. albicans Hog1. C. albicans Sko1 has a region which is similar to the Hog1 phosphorylation region in S. cerevisiae Sko1 (53). We are currently examining this possibility through the cloning and disruption of SKO1 in different backgrounds.
Given the fact that these three species live in different environments, it is not strange that they have developed different strategies to respond to oxidative stress, using the same elements but in distinct ways. In any case, our data indicate that this pathway is essential in a pathogenic fungus for resistance to oxidative stress, a result with potential implications in the field of antifungal therapy.
Acknowledgments
R. Alonso-Monge and F. Navarro-García contributed equally to this work.
R. M. Pérez-Díaz and H. Martín are acknowledged for excellent technical assistance during some parts of this work and for helpful suggestions regarding the manuscript, respectively. We thank W. S. Moye-Rowley and D. Sanglard for the CAP1-GFP construct.
R. Alonso-Monge is a fellow of the Ramón y Cajal Program of the Ministerio de Ciencia y Tecnología and was supported initially by a postdoctoral fellowship from Comunidad Autónoma de Madrid (Proyecto Estratégico). B. Eisman and E. Román hold fellowships from Universidad Complutense de Madrid and Ministerio de Ciencia y Tecnología, respectively. This work was supported by grant BIO2000-0729 and by Proyecto Estratégico de la Comunidad Autónoma de Madrid.
REFERENCES
- 1.Alarco, A. M., I. Balan, D. Talibi, N. Mainville, and M. Raymond. 1997. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272:19304-19313. [DOI] [PubMed]
- 2.Alarco, A. M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso-Monge, R., F. Navarro-García, G. Molero, R. Diez-Orejas, M. Gustin, J. Pla, M. Sánchez, and C. Nombela. 1999. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossier, P., L. Fernandes, D. Rocha, and C. Rodrigues-Pousada. 1993. Overexpression of YAP2, coding for a new yAP protein, and YAP1 in Saccharomyces cerevisiae alleviates growth inhibition caused by 1,10-phenanthroline. J. Biol. Chem. 268:23640-23645. [PubMed] [Google Scholar]
- 5.Brewster, J. L., T. de Valoir, N. D. Dwyer, E. Winter, and M. C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760-1763. [DOI] [PubMed] [Google Scholar]
- 6.Cano, E., and L. C. Mahadevan. 1995. Parallel signal processing among mammalian MAPKs. Trends Biochem. Sci. 20:117-122. [DOI] [PubMed] [Google Scholar]
- 7.Chaturvedi, V., B. Wong, and S. L. Newman. 1996. Oxidative killing of Cryptococcus neoformans by human neutrophils. Evidence that fungal mannitol protects by scavenging reactive oxygen intermediates. J. Immunol. 156:3836-3840. [PubMed] [Google Scholar]
- 8.Chiou, T. J., and W. F. Tzeng. 2000. The roles of glutathione and antioxidant enzymes in menadione-induced oxidative stress. Toxicology 154:75-84. [DOI] [PubMed] [Google Scholar]
- 9.Christman, M. F., G. Storz, and B. N. Ames. 1989. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. USA 86:3484-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, S. T., E. A. Epping, S. M. Steggerda, and W. S. Moye-Rowley. 1999. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 19:8302-8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Backer, M. D., P. T. Magee, and J. Pla. 2000. Recent developments in molecular genetics of Candida albicans. Annu. Rev. Microbiol. 54:463-498. [DOI] [PubMed] [Google Scholar]
- 12.Degols, G., and P. Russell. 1997. Discrete roles of the Spc1 kinase and the Atf1 transcription factor in the UV response of Schizosaccharomyces pombe. Mol. Cell. Biol. 17:3356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degols, G., K. Shiozaki, and P. Russell. 1996. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 16:2870-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Fuente, J. M., A. Alvarez, C. Nombela, and M. Sánchez. 1992. Flow cytometric analysis of Saccharomyces cerevisiae autolytic mutants and protoplasts. Yeast 8:39-45. [DOI] [PubMed] [Google Scholar]
- 15.Delaunay, A., A. D. Isnard, and M. B. Toledano. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demple, B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53-57. [DOI] [PubMed] [Google Scholar]
- 17.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 18.Estruch, F., and M. Carlson. 1993. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox, D., and A. G. Smulian. 1999. Mitogen-activated protein kinase Mkp1 of Pneumocystis carinii complements the slt2Δ defect in the cell integrity pathway of Saccharomyces cerevisiae. Mol. Microbiol. 34:451-462. [DOI] [PubMed] [Google Scholar]
- 21.Fox, J. L. 1993. Fungal infection rates are increasing. ASM News 10:515-518. [Google Scholar]
- 22.Galcheva-Gargova, Z., B. Dérijard, I.-H. Wu, and R. J. Davis. 1994. An osmosensing signal transduction pathway in mammalian cells. Science 265:806-808. [DOI] [PubMed] [Google Scholar]
- 23.Gillum, A. M., E. Y. H. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 24.Gounalaki, N., and G. Thireos. 1994. Yap1p, a yeast transcriptional activator that mediates multidrug resistance, regulates the metabolic stress response. EMBO J. 13:4036-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, J., J.-D. Lee, L. Bibbs, and R. J. Ulevitch. 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265:808-811. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan, D. 1988. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning. IRL Press, Oxford, England.
- 27.Harshman, K. D., W. S. Moye-Rowley, and C. S. Parker. 1988. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell 53:321-330. [DOI] [PubMed] [Google Scholar]
- 28.Hauptmann, N., and E. Cadenas. 1997. The oxygen paradox: biochemistry of active oxygen, p. 1-20. In J. G. Scandalios (ed.), Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Ikeda, T., and J. R. Little. 1995. Deactivation of macrophage oxidative burst in vitro by different strains of Histoplasma capsulatum. Mycopathologia 132:133-141. [DOI] [PubMed] [Google Scholar]
- 30.Jamieson, D. J. 1992. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J. Bacteriol. 174:6678-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14:1511-1527. [DOI] [PubMed] [Google Scholar]
- 32.Jamieson, D. J., D. W. Stephen, and E. C. Terriere. 1996. Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol. Lett. 138:83-88. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi, N., and K. McEntee. 1993. Identification of cis and trans components of a novel heat shock stress regulatory pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:248-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Köhler, G. A., T. C. White, and N. Agabian. 1997. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J. Bacteriol. 179:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Köhler, J., and G. R. Fink. 1996. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc. Natl. Acad. Sci. USA 93:13223-13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krems, B., C. Charizanis, and K. D. Entian. 1996. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet. 29:327-334. [DOI] [PubMed] [Google Scholar]
- 37.Kuge, S., N. Jones, and A. Nomoto. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kultz, D., and M. Burg. 1998. Evolution of osmotic stress signaling via MAP kinase cascades. J. Exp. Biol. 201:3015-3021. [DOI] [PubMed] [Google Scholar]
- 39.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 40.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 41.Martin, H., J. M. Rodriguez-Pachon, C. Ruiz, C. Nombela, and M. Molina. 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:1511-1519. [DOI] [PubMed] [Google Scholar]
- 42.Moradas-Ferreira, P., and V. Costa. 2000. Adaptive response of the yeast Saccharomyces cerevisiae to reactive oxygen species: defences, damage and death. Redox Rep. 5:277-285. [DOI] [PubMed] [Google Scholar]
- 43.Moradas-Ferreira, P., V. Costa, P. Piper, and W. Mager. 1996. The molecular defences against reactive oxygen species in yeast. Mol. Microbiol. 19:651-658. [DOI] [PubMed] [Google Scholar]
- 44.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakagawa, C. W., K. Yamada, and N. Mutoh. 2000. Role of Atf1 and Pap1 in the induction of the catalase gene of fission yeast Schizosaccharomyces pombe. J. Biochem.(Tokyo) 127:233-238. [DOI] [PubMed] [Google Scholar]
- 46.Navarro-García, F., B. Eisman, E. Román, C. Nombela, and J. Pla. 2001. Signal transduction pathways and cell-wall construction in Candida albicans. Med. Mycol. 39:87-100. [PubMed] [Google Scholar]
- 47.Navarro-García, F., M. Sánchez, J. Pla, and C. Nombela. 1995. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol. Cell. Biol. 15:2197-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Negredo, A., L. Monteoliva, C. Gil, J. Pla, and C. Nombela. 1997. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143:297-302. [DOI] [PubMed] [Google Scholar]
- 49.Odds, F. C. 1988. Candida and candidosis. Baillière Tindall, London, England.
- 50.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pla, J., C. Gil, L. Monteoliva, F. Navarro-García, M. Sánchez, and C. Nombela. 1996. Understanding Candida albicans at the molecular level. Yeast 12:1677-1702. [DOI] [PubMed] [Google Scholar]
- 52.Pla, J., R. M. Pérez-Díaz, F. Navarro-García, M. Sánchez, and C. Nombela. 1995. Cloning of the Candida albicans HIS1 gene by direct complementation of a C. albicans histidine auxotroph using an improved double-ARS shuttle vector. Gene 165:115-120. [DOI] [PubMed] [Google Scholar]
- 53.Proft, M., A. Pascual-Ahuir, E. de Nadal, J. Arino, R. Serrano, and F. Posas. 2001. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 20:1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quinn, J., V. J. Findlay, K. Dawson, J. B. Millar, N. Jones, B. A. Morgan, and W. M. Toone. 2002. Distinct regulatory proteins control the graded transcriptional response to increasing H2O2 levels in the fission yeast S. pombe. Mol. Biol. Cell 13:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rep, M., M. Proft, F. Remize, M. Tamas, R. Serrano, J. M. Thevelein, and S. Hohmann. 2001. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 40:1067-1083. [DOI] [PubMed] [Google Scholar]
- 56.Root, R. K., J. Metcalf, N. Oshino, and B. Chance. 1975. H2O2 release from human granulocytes during phagocytosis. I. Documentation, quantitation, and some regulating factors. J. Clin. Investig. 55:945-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samejima, I., S. Mackie, and P. A. Fantes. 1997. Multiple modes of activation of the stress-responsive MAP kinase pathway in fission yeast. EMBO J. 16:6162-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.San José, C., R. Alonso-Monge, R. M. Pérez-Díaz, J. Pla, and C. Nombela. 1996. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J. Bacteriol. 178:5850-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schnell, N., B. Krems, and K. D. Entian. 1992. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr. Genet. 21:269-273. [DOI] [PubMed] [Google Scholar]
- 60.Shiozaki, K., and P. Russell. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10:2276-2288. [DOI] [PubMed] [Google Scholar]
- 61.Singh, K. K. 2000. The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic. Biol. Med. 29:1043-1050. [DOI] [PubMed] [Google Scholar]
- 62.Sonneborn, A., D. P. Bockmuhl, and J. F. Ernst. 1999. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67:5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stephen, D. W., S. L. Rivers, and D. J. Jamieson. 1995. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol. Microbiol. 16:415-423. [DOI] [PubMed] [Google Scholar]
- 64.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toone, W. M., S. Kuge, M. Samuels, B. A. Morgan, T. Toda, and N. Jones. 1998. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase Sty1/Spc1. Genes Dev. 12:1453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vazquez-Torres, A., and E. Balish. 1997. Macrophages in resistance to candidiasis. Microbiol. Mol. Biol. Rev. 61:170-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vazquez-Torres, A., J. Jones-Carson, and E. Balish. 1996. Peroxynitrite contributes to the candidacidal activity of nitric oxide-producing macrophages. Infect. Immun. 64:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wallace, S. S. 1997. Oxidative damage to DNA and its repair, p. 49-90. In J. G. Scandalios (ed.), Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 69.Wilkinson, M. G., M. Samuels, T. Takeda, W. M. Toone, J. C. Shieh, T. Toda, J. B. Millar, and N. Jones. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10:2289-2301. [DOI] [PubMed] [Google Scholar]
- 70.Wysong, D. R., L. Christin, A. M. Sugar, P. W. Robbins, and R. D. Diamond. 1998. Cloning and sequencing of a Candida albicans catalase gene and effects of disruption of this gene. Infect. Immun. 66:1953-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, X., M. De Micheli, S. T. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36:618-629. [DOI] [PubMed] [Google Scholar]
- 72.Zitomer, R. S., and C. V. Lowry. 1992. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 56:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]