Abstract
Several human neurodegenerative diseases result from expansion of CTG/CAG or CGG/CCG triplet repeats. The finding that single-stranded CNG repeats form hairpin-like structures in vitro has led to the hypothesis that DNA secondary structure formation is an important component of the expansion mechanism. We show that single-stranded DNA loops containing 10 CTG/CAG or CGG/CCG repeats are inefficiently repaired during meiotic recombination in Saccharomyces cerevisiae. Comparisons of the repair of DNA loops with palindromic and nonpalindromic sequences suggest that this inefficient repair reflects the ability of these sequences to form hairpin structures in vivo.
Expansion of repetitive tracts of trinucleotide repeats in the human genome is associated with about 12 diseases including Fragile X syndrome and myotonic dystrophy (1, 2). All but one of these diseases, Friedreich’s ataxia (3), result from expansion of a CTG/CAG or a CCG/CGG tract.
One model to explain expansions of trinucleotide repeat tracts is DNA polymerase slippage (ref. 4; Fig. 1a). In this model, transient dissociation of the primer and template strand within a repetitive DNA tract during DNA replication is followed by a misaligned reassociation of the two strands generating an unpaired loop. If this loop is not repaired, it will result in an addition (if the loop occurred in the primer strand, as shown in Fig. 1a), or deletion (if the loop occurred in the template strand). In a second model (Fig. 1b), expansions occur during displacement synthesis of an Okazaki fragment (5–7). Although a displaced DNA strand would be expected to be a target for the FEN1 endonuclease, hairpin formation within this strand could make the strand resistant to FEN1 (8), resulting in an elevated frequency of expansion (6). Secondary structures formed within mispaired loops (Fig. 1a) or displaced single strands (Fig. 1b) might increase repeat tract instability and, in addition, strand-specific tendencies to form such structures could lead to a bias for expansion or contraction, depending on the orientation of the tract relative to the DNA replication origin (9).
Figure 1.
Two models for expansions of trinucleotide repeat tracts. (a) Expansion as a consequence of DNA polymerase slippage (4). During replication of a trinucleotide repeat tract (repeats indicated by rectangles), the primer and template strands transiently dissociate (step 1). Reassociation occurs with a DNA loop formed in the primer strand (step 2). Continued synthesis with no repair of the loop (step 3) would result in an addition of repeats. We show the loop stabilized by formation of a hairpin structure. (b) Expansion of a trinucleotide repeat tract as a consequence of displacement of an Okazaki fragment (5–7). In this model, synthesis displaces the 5′ end (indicated by ∗) of the neighboring Okazaki fragment (step 1′). The displaced strand folds back on itself to form a hairpin (step 2′) and is resistant to processing by FEN1 endonuclease (8). Continued synthesis and ligation (step 3′) results in an expanded trinucleotide repeat tract.
Central to both of these models is the formation of secondary structures unique to certain trinucleotide repeat tracts that result in instability. Several observations were made that supported the idea that these structures could form in vivo. First, oligonucleotides containing trinucleotide repeats involved in expansion diseases (CTG/CAG and CCG/CGG) are capable of folding into hairpin structures in vitro (10–13). Second, long CNG tracts in both Escherichia coli (14, 15) and Saccharomyces cerevisiae (16–18), undergo orientation-dependent expansions and large deletions suggestive of palindromic behavior. Third, by using an in vivo assay based on the observation that palindromic DNA sequences in the bacteriophage λ result in small plaque size, Darlow and Leach (19) obtained evidence suggesting that as few as two triplet repeats were capable of forming stable base pairs at the tip of a long hairpin structure.
We examined trinucleotide repeat secondary structure formation in vivo by constructing yeast strains in which heteroduplex formation associated with meiotic recombination resulted in the generation of single-stranded DNA loops. From our analysis of such strains, we demonstrate that DNA loops with CNG repeats are inefficiently repaired in meiosis, AGT/ACT repeats result in intermediate levels of repair, and loops containing AAG/CTT and CAA/TTG repeats are readily repaired. From these results and our previous demonstration that loops with palindromic DNA sequences are inefficiently repaired (20), we conclude that single-stranded trinucleotide repeats of CTG, CAG, CCG, or CGG (but not AAG or CTT) are capable of forming hairpin structures in vivo.
MATERIALS AND METHODS
Plasmids.
Plasmids used to construct mutant his4 alleles with various insertions were pDN9 derivatives (20). This plasmid contains a 1.6-kb XhoI–BglII fragment containing the 5′ end of HIS4 ligated into SalI- and BamHI-treated YIp5 (21). To create mutant alleles of HIS4 with various insertions, we annealed 36-bp complementary oligonucleotides to generate double-stranded DNA molecules with ends compatible with SalI. These molecules were inserted into the unique SalI site of pDN9, located within the HIS4 coding sequence, 463 bp from the initiating codon; the sequence of these insertions is shown in Table 1. Plasmid constructions were confirmed by DNA sequence analysis.
Table 1.
Names of plasmids, mutant alleles and yeast strains used in the study and the sequence of mutant insertions
| Plasmid | Mutant allele | Sequence of insertion | Haploid strain | Diploid strain |
|---|---|---|---|---|
| pNH1 | his4-CAG10 | TCGAC(CAG)10G | PG78 | PG84 |
| pNH2 | his4-CTG10 | TCGAC(CTG)10G | HMY20 | HMY21 |
| pNH3 | his4-CGG10 | TCGAC(CGG)10G | HMY25 | HMY26 |
| pNH4 | his4-CCG10 | TCGAC(CCG)10G | HMY29 | HMY32 |
| pNH5 | his4-AAG10 | TCGAC(AAG)10G | HMY30 | HMY33 |
| pNH6 | his4-AGT10 | TCGAC(AGT)10G | HMY12 | HMY16 |
| pNH7 | his4-CAA10 | TCGAC(CAA)10G | HMY14 | HMY18 |
| pNH8 | his4-R | TCGACCCCTGTTGCTGCC | PG86 | PG88 |
| GGCTTGGCCGCGTCTTTG | ||||
| pNH9 | his4-CAGd | TCGACCAGACT(CAG)8G | HMY36 | HMY38 |
| pNH10 | his4-Bi | TCGAC(CGG)5(AGT)5G | HMY40 | HMY42 |
| pNH11 | his4-Pal | TCGACCTCGTCCTGCTCG | HMY41 | HMY43 |
| TGCTCGTGCTGGTCGTGG |
All plasmids were derived from pDN9 (as described in Materials and Methods) and contain a 36-bp insertion within a SalI site in the HIS4 coding sequence. The sequence of the insertion is depicted 5′ to 3′ as it is arranged in the nontranscribed strand; boldface type indicates the 30 bp that differ between insertions. The plasmids were used in two-step transplacement transformations of the haploid AS13 (22), and the resulting transformants were mated to AS4 to generate the diploid strains. All strains are isogenic except for the sequence of the insertions.
Strains.
The haploid yeast strains were derivatives of AS13 (a leu2-Bst ura3 ade6) or AS4 (α trp1–1 arg4–17 tyr7–1 ura3 ade6) (22). Haploid derivatives of AS13 with mutant his4 genes with various 36-bp insertions (Table 1) were created by two-step transplacement (23). The plasmids with the mutant allele were treated with SnaBI before transformation. Ura+ transformants were selected, purified, and then grown on plates containing 0.1% 5-fluoro-orotate to select for Ura− derivatives (24). Ura− strains were screened for those that were also His−. Constructions were confirmed by Southern analysis and PCR. Diploids were obtained by mating AS13 derivatives with AS4. All strains were isogenic except for the changes introduced by transformation.
Genetic Techniques.
Standard methods and media were used except where noted (25). Because sporulation at low temperatures elevates HIS4 meiotic recombination in our strain background (26), diploid strains were sporulated at 18°C on solid medium [1% potassium acetate, 0.1% yeast extract, 0.05% dextrose, 2% agar supplemented with adenine (6 μg/ml)]. For most experiments, the sporulated cells were dissected on rich growth medium (yeast extract/peptone/dextrose). After spore colonies had formed, they were replica-plated to various omission media to score the segregating markers. We examined spore colonies for sectored growth patterns by microscopy to detect postmeiotic segregation (PMS) events.
Experiments to determine whether unrepaired DNA loops within heteroduplexes at the HIS4 locus involved the transcribed or nontranscribed strand were done as described (27). In brief, sporulated cells were dissected onto plates lacking histidine and incubated for 8–9 hr at 30°C. The dissected spores then were examined microscopically and scored as His+ (more than three cells) or His− (one cell). Histidine then was added to the medium by placing the agar slabs with the spores onto yeast extract/peptone/dextrose plates overlaid with 1 ml of a 1% histidine solution. Spore colonies were grown for an additional 3 days at 30°C before being replica-plated to omission media. A comparison of the spore phenotype on the dissection plate lacking histidine with the spore colony sectoring phenotype allows one to determine whether the transcribed or nontranscribed strand contained the unrepaired loop of triplet repeat sequences.
PCR Analysis.
We used PCR to analyze the sizes of the trinucleotide repeat insertions for some yeast spore colonies. DNA was isolated and PCR performed as described (28) except, for this study, we used 0.2 μM of each primer and 2.5 mM MgCl2. The products of PCR were examined on DNA sequencing gels. The primers (fHIS419 [5′ TTGGTGAAGTACGTACAGACCG] and rHIS587 [5′ TTGATCCAGATTTCATTCCTAG]) were within the HIS4 gene flanking the insertion and yielded an amplified product of about 200 bp.
Statistical Analysis.
Comparisons were made by using the Fisher’s exact test with two-tailed P values. Results were considered statistically significant if P < 0.05. When comparing levels of PMS between strains, the total number of tetrads with one or more PMS events was compared with the number of tetrads with no PMS events.
RESULTS
Experimental Rationale.
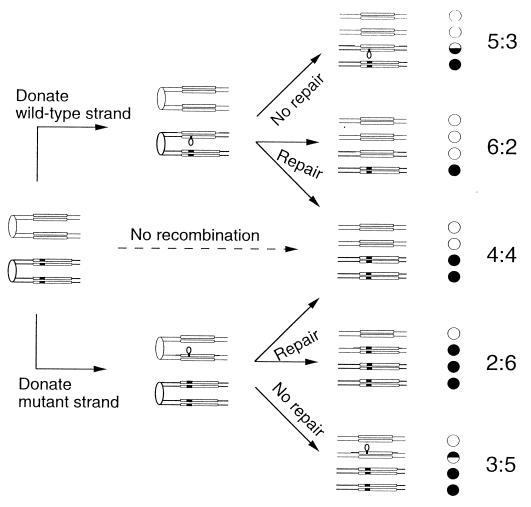
Our assay of the behavior of trinucleotide repeats within single-stranded DNA loops is based on an analysis of patterns of meiotic segregation. If a yeast diploid heterozygous for a mutation in gene A (wild-type allele A and mutant allele a) is sporulated and the resulting tetrads are dissected, in the absence of recombination near the marker, one will observe two A spore colonies and two a spore colonies (4:4 segregation). If recombination is initiated near the heterozygous marker, DNA strand transfer may generate a heteroduplex involving the marker, with one strand derived from the A allele and one derived from the a allele (29). The heteroduplex will contain a DNA loop if the wild-type and mutant genes differ by an insertion as shown in Fig. 2. There are several possible fates for this loop. Repair of the loop will either restore normal Mendelian segregation or generate gene conversion events (6A:2a or 2A:6a; Fig. 2). Failure to repair the loop will result in PMS, the segregation of the two alleles at the first mitotic division of the spore (5A:3a or 3A:5a; Fig. 2). Such events are detected by production of a spore colony with a sectored A/a genotype.
Figure 2.
Expected patterns of meiotic segregation in a yeast strain heterozygous for a mutant insertion in HIS4. We show the chromatids as double-stranded DNA molecules with sister chromatids held together at the centromeres (shown by the ovals). The HIS4 genes are depicted as long rectangles; the mutant insertion is shown as a black marking. His+ spore colonies are shown as white circles, and His− spore colonies are shown as black circles.
The HIS4 locus has a very high rate of meiotic recombination (50% of tetrads with a detectable event) associated with a high level of heteroduplex formation (20, 30). Most importantly, strains heterozygous for palindromic insertions within HIS4 have high levels of PMS and low levels of gene conversion, indicating that DNA loops capable of hairpin structures are inefficiently repaired (20). In contrast, most loops incapable of hairpin formation are efficiently repaired (20, 31).
We constructed diploid strains heterozygous for his4 mutant alleles generated by 30-bp insertions of trinucleotide repeats. After sporulation and tetrad dissection, we examined colonies for the segregation of the his4 mutation. If the trinucleotide repeats formed hairpin structures when located in heteroduplexes, we would expect high frequencies of PMS tetrads relative to gene conversion tetrads. Failure to form such structures would result in high levels of gene conversion tetrads compared with PMS tetrads.
DNA Loops Containing CNG Repeats Are Inefficiently Repaired.
The strains PG84 and HMY21 were heterozygous for his4 mutant alleles caused by insertion of 10 repeats of CAG/CTG (Table 1). In PG84, the CAG sequence was in the nontranscribed strand of HIS4, whereas, in HMY21, the same insertion was in the opposite orientation. In both strains, high levels of PMS tetrads were observed (28–33%), and more than three-quarters of the aberrant segregation events at HIS4 were PMS (Table 2), indicating that DNA loops containing CAG/CTG repeats are inefficiently corrected by the DNA repair machinery. The frequency of PMS tetrads and the fraction of aberrant segregants that were PMS were very similar to those observed previously for 26-bp perfect palindromic insertions (20). Strains HMY26 and HMY32 (two orientations of CGG/CCG repeats, Table 1) had patterns of aberrant segregation similar to those observed for PG84 and HMY21 (Table 2). Thus, DNA loops with CGG/CCG repeats also are inefficiently repaired.
Table 2.
Meiotic segregation patterns in strains heterozygous for small palindromic or nonpalindromic insertions at the HIS4 locus
| Strain | Mutant allele | 4:4 | 6:2 | 2:6 | 5:3 | 3:5 | Other* | Total tetrads | Aberrant segregation, % of total | PMS, % of total | PMS, % of Ab. seg. | HIS4/LEU2 distance, cM† |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PG84 | his4-CAG10 | 343 | 29 | 20 | 82 | 52 | 32 | 558 | 39 | 28 | 76 | 31 |
| HMY21 | his4-CTG10 | 304 | 20 | 20 | 79 | 74 | 19 | 516 | 41 | 33 | 80 | 37 |
| HMY26 | his4-CGG10 | 433 | 47 | 16 | 102 | 120 | 46 | 764 | 43 | 33 | 79 | 36 |
| HMY32 | his4-CCG10 | 219 | 20 | 17 | 48 | 22 | 14 | 339 | 36 | 23 | 69 | 33 |
| HMY33 | his4-AAG10 | 451 | 30 | 47 | 9 | 12 | 4 | 553 | 18 | 4 | 21 | 35 |
| HMY18 | his4-CAA10 | 385 | 65 | 54 | 14 | 6 | 6 | 530 | 27 | 4 | 15 | 33 |
| PG88 | his4-R | 388 | 32 | 33 | 18 | 6 | 1 | 478 | 19 | 5 | 27 | 35 |
| HMY16 | his4-AGT10 | 349 | 31 | 45 | 16 | 33 | 8 | 482 | 28 | 11 | 40 | 33 |
| HMY43 | his4-Pal | 211 | 14 | 8 | 55 | 43 | 16 | 347 | 39 | 32 | 84 | 40 |
| HMY38 | his4-CAGd | 122 | 13 | 12 | 32 | 30 | 4 | 213 | 43 | 31 | 72 | 33 |
| HMY42 | his4-Bi | 259 | 41 | 61 | 25 | 8 | 6 | 400 | 35 | 9 | 24 | 31 |
In contrast, strains with the AAG/CTT (HMY33) or CAA/TTG (HMY18) repeats had low levels of PMS (4%) and a small percentage (15–21%) of the aberrant segregants were PMS tetrads. PMS levels in these strains were not significantly different (P = 0.6 and P = 0.7, respectively) from that observed in strain PG88 (5% PMS and 27% of the aberrant segregants were PMS) that was heterozygous for a nonpalindromic insertion of the same size. The results argue that DNA loops with either AAG/CTT or CAA/TTG (as well as loops with “random” DNA sequences) are efficiently corrected. The strain HMY16, heterozygous for an insertion of AGT/ACT, had a PMS frequency of 11% (40% of the aberrant segregants were PMS), which is intermediate to, but significantly (P < 0.01 for both comparisons) different from, the values observed for the other two classes, indicating an intermediate level of repair.
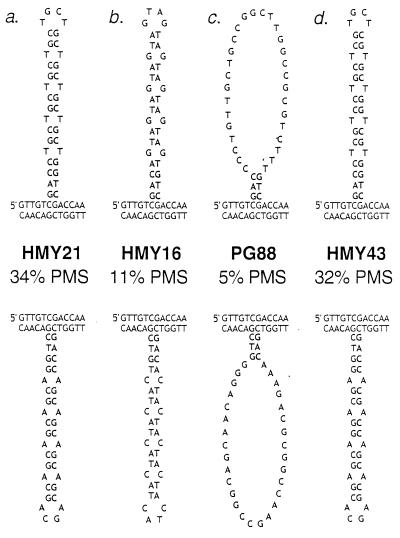
There are two possible interpretations of these data. We favor the idea that the efficiency of correction of the DNA loops is inversely related to the stability of hairpin-like structures in the loop. DNA loops with CNG repeats could form pseudo-hairpins (Fig. 3a) in which there are two GC bp and one mismatch per repeat. The AGT/ACT repeats could form less stable structures with two AT bp and one mismatch per repeat (Fig. 3b). The loops with AAG/CTT, CAA/TTG, and random sequences (Fig. 3c) would not be expected to form hairpin structures.
Figure 3.
Expected secondary structures in DNA loops containing various insertions within HIS4. As discussed in the text, heteroduplex formation between a his4 gene with a mutant insertion and a wild-type HIS4 gene will produce a DNA loop. The upper strand in each duplex is the nontranscribed strand. (Upper) The loops result from a heteroduplex in which the nontranscribed strand is mutant and the transcribed strand is wild type. (Lower) The nontranscribed strand is wild type and the transcribed strand is mutant. The inserted sequence is shown in bold. Strain names and the percentages of the unselected tetrads representing PMS tetrads are shown in the middle. A high percentage of PMS reflects inefficient repair of the DNA loop. The relevant genotypes are: (a) HMY21 (heterozygous for his4-CTG10); (b) HMY16 (heterozygous for his4-AGT10); (c) PG88 (heterozygous for his4-R), his4-R represents an insertion of 36 bp with a randomized DNA sequence that should be incapable of forming stable secondary structures, and (d) HMY43 (heterozygous for his4-Pal), his4-Pal is a 36-bp, nontriplet repeat sequence that should have similar base-pairing properties to his4-CTG10.
An alternative interpretation of the data is that certain DNA loops are inefficiently corrected because of the primary sequence of the insertion, rather than as a consequence of the formation of a secondary structure. To distinguish between these two interpretations, we constructed several additional strains. The insertion in HMY43 was designed to mimic the secondary structure of the CAG/CTG repeats without the same primary sequence (Fig. 3d). Although this insertion did not have triplet repeats, it had the same placement of CG bp and mismatches as the CAG/CTG repeats. No significant differences (P > 0.3 for all comparisons) were found in the levels of PMS tetrads for HMY43 and the CAG/CTG insertions (strains PG84 and HMY21; Table 2), suggesting that it is the base-pairing properties of this loop and not the DNA sequence that is responsible for its inefficient repair.
We also tested a strain, HMY42 with an insertion [(CGG)5(AGT)5], that would form a loop with one arm composed of CGG repeats and the other composed of AGT repeats. Our expectation was that, if primary sequence was responsible for inefficient DNA repair, this strain would have a level of PMS intermediate to the levels seen for his4-AGT10 and his4-CGG10 (about 20%). Alternatively, if secondary structure was responsible for inefficient DNA loop repair, this construct would result in a low level of PMS. Although the observed level of PMS for HMY42 was low (9%), it is significantly greater (P < 0.03 for all comparisons) than the levels observed in strains with nonpalindromic insertions (about 5%). One explanation of this result is that each half of the insertion can form a small, relatively unstable hairpin. Very small numbers of trinucleotide repeats, as few as two or three, still manifest secondary structure in vitro (12, 13), and 14-bp palindromic sequences form stable hairpins in vivo (33).
Finally, we tried to perturb the secondary structure by making an interruption of the stem (strain HMY38, [(CAG)ACT(CAG)8]). PMS frequencies were not significantly reduced compared with the (CTG/CAG)10 strains (P > 0.6). This result is not unexpected because Nag and Kurst (34) showed that single interruptions located within the stem of a palindromic insertion do not result in efficient repair of the insertion.
Strand Specificity of DNA Loop Repair.
Each strain used in our analysis has the potential for producing DNA loops of two types. Both 5:3 (donation of wild-type strand to an insertion mutant gene) and 3:5 (donation of an insertion mutant strand to a wild-type gene) tetrads can be formed by transferring either the nontranscribed or the transcribed strand to the heteroduplex. Which strand is transferred determines which sequence will be present in the DNA loop (27). For example, the PMS spore in a 5:3 tetrad of HMY21 in which the transcribed strand was donated from a wild-type gene to a mutant gene would have an unrepaired DNA loop of (CTG)10 (Fig. 3a, Upper). By using an altered protocol for tetrad dissection (described in Materials and Methods), we were able to directly test whether the unrepaired loop was composed of transcribed or nontranscribed sequences. As shown in Table 3, there were no significant differences in the number of unrepaired loops found in either strand for all strains tested (all P values >0.09). This finding indicates that DNA loops with the four CNG sequences evade repair to approximately the same extent. We suggest, therefore, that all four CNG sequences form hairpins with similar in vivo stabilities.
Table 3.
Analysis of the DNA sequences in unrepaired DNA loops by strand-transfer experiments
| Strain | Strand containing loop | Sequence of loop | No. of spores with PMS event | Total number of tetrads |
|---|---|---|---|---|
| PG84 | T | (CTG)10 | 39 | 204 |
| NT | (CAG)10 | 32 | ||
| HMY21 | T | (CAG)10 | 26 | 184 |
| NT | (CTG)10 | 36 | ||
| HMY26 | T | (CCG)10 | 38 | 260 |
| NT | (CGG)10 | 63 | ||
| HMY16 | T | (ACT)10 | 10 | 197 |
| NT | (AGT)10 | 14 | ||
| HMY18 | T | (TTG)10 | 7 | 197 |
| NT | (CAA)10 | 2 | ||
| HMY33 | T | (CTT)10 | 3 | 157 |
| NT | (AAG)10 | 4 |
As described in Materials and Methods, a modification of the standard tetrad dissection protocol allowed us to determine whether a sectored His+/His− colony contained mutant information in the transcribed or nontranscribed DNA strand of the heteroduplex. From our knowledge of the DNA sequence of the mutant insertions in each strain, we inferred the sequence of the unrepaired DNA loop that resulted in the PMS event (reflected by the sectored colony). T, transcribed strand; NT, nontranscribed strand.
Other Parameters of Meiotic Recombination Involving Trinucleotide Repeats.
A mutant insertion associated with the ability to initiate meiotic recombination (hotspot activity) is expected to have a greater sum of 6:2 and 5:3 classes than 2:6 and 3:5 classes because the initiating chromosome is the recipient in strand exchange (ref. 29; Fig. 2). Although marginally significant differences of this type were found for two of the strains in this study, PG84 and HMY32 (P = 0.05), other data indicate that these strains do not have strong hotspot activity. First, HIS4-LEU2 recombination distances were not elevated in these strains compared with the other strains (Table 2). In addition, in a rad50S derivative of PD84, we detected a meiosis-specific double-strand DNA break (DSB) in the upstream regulatory region of HIS4, as we previously have observed (26), but no DSB associated with the trinucleotide repeat insertion (data not shown); all recombination hotspots thus far characterized are associated with DSBs (35). We conclude that none of these insertions have strong hotspot activity.
To determine whether repair of triplet repeat loops during gene conversion often was associated with additions or deletions of trinucleotide repeat units, we examined the size of the insertions in all four spores of 2:6 tetrads by PCR analysis (details in Materials and Methods). In 12 of 12 such tetrads (four PG84 and eight HMY21), the original tract size was observed in all His− spores and the His+ spore lacked any insertion. We conclude that the repair events involving DNA loops with small trinucleotide repeat tracts usually are accurate.
DISCUSSION
We used an in vivo DNA repair assay to assess the potential secondary structure of single-stranded DNA loops generated during meiotic recombination in yeast. We previously showed that DNA loops containing palindromic insertions were inefficiently repaired (20, 33), in contrast to the efficient repair of most DNA loops containing nonpalindromic insertions (29). In this study, we find that DNA loops containing CNG repeats (predicted to form hairpin structures by in vitro studies; refs. 10–13) were as inefficiently repaired as perfect hairpin structures. DNA loops containing AAG/CTT and CAA/GTT triplet repeats or other sequences not likely to be capable of hairpin formation were efficiently repaired. This correlation between predicted secondary structures and the properties of in vivo repair strongly indicates that DNA loops with CNG repeats form stable hairpin structures in vivo. One caveat to this conclusion is that one nonpalindromic DNA insertion results in an inefficiently repaired DNA loop (29). Because of this exception, we cannot completely rule out a role of the primary DNA sequence in DNA loop repair.
In most in vitro studies, single-stranded oligonucleotides containing CTG repeats appear to form more stable hairpin structures than those with CAG repeats (12, 36–40) and CGG repeats result in more stable structures than CCG repeats (41, 42). Although we found that CTG and CGG loops were repaired less efficiently than their CAG and CCG counterparts (Table 3), these differences were not statistically significant, suggesting that all four CNG sequences form hairpins with similar in vivo stabilities. Experimental differences between the in vivo and in vitro experiments (ion concentrations, length of CNG tract, the presence of cellular proteins, etc.) or the inability of the in vivo assay to detect subtle variations in repair efficiency may account for this discrepancy.
In some, but not all (17), studies of the stability of CNG trinucleotide repeat tracts in E. coli (14, 15) and yeast (16, 18), large deletions are observed when CTG (or CGG) is in the lagging strand, whereas fewer deletions and occasional additions are found when CAG (or CCG) is in the lagging strand. These results are consistent with the slippage model shown in Fig. 1a, assuming that DNA sequences on the lagging strand have more opportunity to form secondary structures and the CTG and CGG repeats yield more stable hairpin structures than the CAG and CCG repeats (as predicted from the in vitro studies). Given our findings, factors other than the in vivo stability of single-strand CNG loops may help to explain these observations. For example, sequence-dependent slowing of the rate of DNA replication may provide greater opportunity for the formation of secondary structures when the repeat tract is in one orientation.
The formation of secondary structure within trinucleotide repeat tracts could destabilize these tracts in different ways. First, as described above, hairpin formation might potentiate DNA polymerase slippage by formation of DNA loops (Fig. 1a). Alternatively, hairpin-like structures at the 5′ end of a displaced Okazaki fragment (Fig. 1b) might be inefficiently removed by the FEN1 endonuclease (8, 43), leading to tract expansions (6). Finally, formation of secondary structures could cause DNA polymerase pausing (44, 45), increasing the probability of DNA polymerase slippage or sister-strand recombination.
In humans, long tracts of trinucleotide repeats exhibit somatic instability (1). Are CNG loops created during replication slippage or other mechanisms poor substrates for mitotic repair? Although it is clear that 14-bp palindromic DNA loops escape correction in vegetative yeast (46), the correction of nonpalindromic loops of comparable size has not been examined. In mammalian cells, both palindromic and nonpalindromic DNA loops, in certain artificial recombination substrates, can be corrected (47, 48), although a fraction of palindromic loops escapes correction (49). No direct comparison of the relative repair efficiencies of palindromic and nonpalindromic DNA loops of the same size and in the same context has been carried out in mammalian cells.
In yeast cells with large trinucleotide repeat tracts, deletion events outnumber expansions (16–18) whereas, in humans, expansions are the primary mutagenic event. This difference could reflect fundamentally different modes of DNA replication, DNA repair, and/or recombination in yeast and human cells. A simpler possibility, however, is that a difference in the activity of a single enzyme is responsible. For example, mutations in RAD27 (encoding the yeast homologue of FEN1) result in expansions of trinucleotide repeat tracts (50, 51). If the human FEN1 endonuclease is less active or more sensitive to DNA secondary structures than Rad27p, expansions of trinucleotide repeat tracts would be more common in human cells than in yeast. It is likely, therefore, that the DNA secondary structures that we have detected in yeast will be relevant to understanding the trinucleotide repeat tract expansions observed in human cells.
In summary, we present evidence that DNA loops with two of the most common types of trinucleotide repeats (CTG/CAG, CCG/CGG) associated with trinucleotide repeat tract expansions in several human diseases are inefficiently corrected when located in heteroduplexes formed during yeast meiotic recombination. This inefficient correction is likely to reflect in vivo formation of hairpin secondary structures within the loop. The third class of triplet repeats associated with expansions (AAG/CTT) does not share this property. Although all triplet repeat expansions may involve DNA secondary structures, expansions of AAG/CTT tracts are thought to reflect formation of triplex DNA rather than hairpin structures (13, 52); triplex formation would not be expected in 30-bp single-stranded loops.
Acknowledgments
We thank N. Kleckner and D. Nag for plasmids used in the study, and D. Leach for comments on the manuscript. This research was supported by National Institutes of Health grants to T.D.P. (GM24110) and N.A. (R37 GM36745).
ABBREVIATION
- PMS
postmeiotic segregation
References
- 1.Ashley C T, Warren S T. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- 2.Richards R I, Sutherland G R. Trends Biochem Sci. 1997;22:432–436. doi: 10.1016/s0968-0004(97)01108-0. [DOI] [PubMed] [Google Scholar]
- 3.Campuzano V, Montermini L, Molto M D, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 4.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Cold Spring Harbor Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Richards R I, Sutherland G R. Nat Genet. 1994;6:114–116. doi: 10.1038/ng0294-114. [DOI] [PubMed] [Google Scholar]
- 6.Gordenin D A, Kunkel T A, Resnick M A. Nat Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 7.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bambara R A, Murante R S, Henricksen L A. J Biol Chem. 1997;272:4647–4650. doi: 10.1074/jbc.272.8.4647. [DOI] [PubMed] [Google Scholar]
- 9.Sinden R R, Wells R D. Curr Opin Biotechnol. 1992;3:612–622. doi: 10.1016/0958-1669(92)90005-4. [DOI] [PubMed] [Google Scholar]
- 10.Wells R D. J Biol Chem. 1996;271:2875–2878. doi: 10.1074/jbc.271.6.2875. [DOI] [PubMed] [Google Scholar]
- 11.Mitas M. Nucleic Acids Res. 1997;25:2245–2253. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao X, Huang X, Smith G K, Zheng M. In: Genetic Instabilities and Hereditary Neurological Disorders. Wells R D, Warren S T, editors. San Diego: Academic; 1998. pp. 623–646. [Google Scholar]
- 13.Mariappan S V S, Chen X, Catasti P, Bradbury E M, Gupta G. In: Genetic Instabilities and Hereditary Neurological Disorders. Wells R D, Warren S T, editors. San Diego: Academic; 1998. pp. 647–676. [Google Scholar]
- 14.Kang S, Jaworski A, Ohshima K, Wells R D. Nat Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 15.Shimizu M, Gellibolian R, Oostra B A, Wells R D. J Mol Biol. 1996;258:614–626. doi: 10.1006/jmbi.1996.0273. [DOI] [PubMed] [Google Scholar]
- 16.Maurer D J, O’Callaghan B L, Livingston D M. Mol Cell Biol. 1996;16:6617–6622. doi: 10.1128/mcb.16.12.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miret J J, Pessoa-Brandao L, Lahue R S. Mol Cell Biol. 1997;17:3382–3387. doi: 10.1128/mcb.17.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freudenreich C H, Stavenhagen J B, Zakian V A. Mol Cell Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darlow J M, Leach D R F. Genetics. 1995;141:825–832. doi: 10.1093/genetics/141.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nag D K, White M A, Petes T D. Nature (London) 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 21.Struhl K, Stinchomb D T, Scherer S, Davis R W. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stapleton A, Petes T D. Genetics. 1991;127:39–51. doi: 10.1093/genetics/127.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scherer S, Davis R W. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeke J D, Lacroute F, Fink G R. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 25.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. [Google Scholar]
- 26.Fan Q-Q, Xu F, Petes T D. Mol Cell Biol. 1995;5:1679–1688. doi: 10.1128/mcb.15.3.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nag D K, Petes T D. Genetics. 1990;125:753–761. doi: 10.1093/genetics/125.4.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wierdl M, Greene C N, Datta A, Jinks-Robertson S, Petes T D. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petes T D, Malone R E, Symington L S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Broach J R, Jones E W, Pringle J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 407–521. [Google Scholar]
- 30.Detloff P, White M A, Petes T D. Genetics. 1992;132:113–123. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirkpatrick D T, Petes T D. Nature (London) 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 32.Perkins D D. Genetics. 1949;34:607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nag D K, Petes T D. Genetics. 1991;129:669–673. doi: 10.1093/genetics/129.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nag D K, Kurst A. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roeder G S. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 36.Gacy A M, Goellner G, Juranic N, Macura S, McMurray C T. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 37.Yu A, Dill J, Wirth S S, Huang G, Lee V H, Haworth I S, Mitas M. Nucleic Acids Res. 1995;23:2706–2714. doi: 10.1093/nar/23.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu A, Dill J, Mitas M. Nucleic Acids Res. 1995;23:4055–4057. doi: 10.1093/nar/23.20.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petruska J, Arnheim N, Goodman M F. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson C E, Sinden R R. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 41.Mitas M, Yu A, Dill J, Kamp T J, Chambers E J, Haworth I S. Nucleic Acids Res. 1995;23:1050–1059. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu A, Barron M D, Romero R M, Christy M, Gold B, Dai J, Gray D M, Haworth I S, Mitas M. Biochemistry. 1997;36:3687–3699. doi: 10.1021/bi9625410. [DOI] [PubMed] [Google Scholar]
- 43.Lieber M R. BioEssays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 44.Ohshima K, Wells R D. J Biol Chem. 1997;272:16798–16806. doi: 10.1074/jbc.272.27.16798. [DOI] [PubMed] [Google Scholar]
- 45.Samadashwily G M, Raca G, Mirkin S M. Nat Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 46.Weng Y, Nickoloff J A. Genetics. 1998;148:59–70. doi: 10.1093/genetics/148.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss U, Wilson J H. Proc Natl Acad Sci USA. 1987;84:1619–1623. doi: 10.1073/pnas.84.6.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taghian D G, Hough H, Nickoloff J A. Genetics. 1998;148:1257–1268. doi: 10.1093/genetics/148.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bollag R J, Elwood D R, Tobin E D, Godwin A R, Liskay R M. Mol Cell Biol. 1992;12:1546–1552. doi: 10.1128/mcb.12.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schweitzer J K, Livingston D M. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 51.Freudenreich C H, Kantrow S M, Zakian V A. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 52.Gacy A M, Goellner G M, Spiro C, Chen X, Gupta G, Bradbury E M, Dyer R B, Mikesell M J, Yao J Z, Johnson A J, et al. Mol Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]