Abstract
The UV-damaged DNA-binding activity protein (UV-DDB) consists of two subunits, DDB1 and DDB2, and functions in DNA repair and cell cycle regulation. The DDB1 subunit is a target for the hepatitis B virus X protein (HBx). Binding of HBx to DDB1 interferes with cell growth and viability in culture and has been implicated in the establishment of viral infection. DDB1 also interacts with the V proteins encoded by several paramyxoviruses including simian virus 5 (SV5), which prevent interferon signaling by targeting either STAT1 or STAT2 proteins for proteolysis. The role of V binding to DDB1, however, remains unclear. Here we show that the V protein of SV5 (SV5-V) and HBx exhibit strikingly similar DDB1 binding properties. Thus, SV5-V and HBx bind to DDB1 in a mutually exclusive manner, and SV5-V shares with HBx the ability to enhance the steady-state levels of DDB1 and to inhibit its association with DDB2. Yet only HBx induces cell death, and SV5-V can prevent HBx from doing so by blocking its interaction with DDB1. Binding of SV5-V to DDB1 may serve another function, since SV5-V shows a decreased ability to induce STAT1 degradation in cells expressing reduced amounts of DDB1. These findings demonstrate that HBx performs a unique function through its association with DDB1 for which SV5-V cannot substitute and suggest that SV5-V and HBx have evolved to bind DDB1 to achieve distinct functions, both by a mechanism that does not involve DDB2.
DDB1 is a 127-kDa protein that associates with DDB2, a UV-inducible 48-kDa nuclear protein that transports DDB1 from the cytoplasm to the nucleus (27, 36). In the nucleus DDB1 and DDB2 form the UV-DDB complex, which exhibits high binding affinity for UV-damaged DNA (1, 10, 14, 15, 17, 28). UV-DDB activity has been directly implicated previously in nucleotide excision repair (30, 35, 43, 45) and is absent in some cancer-prone xeroderma pigmentosum group E patients due to mutations of the DDB2 gene (10, 15, 28). No mutations in DDB1 have been reported. A role for UV-DDB other than in DNA repair has also been suggested. UV-DDB functionally interacts with the cell cycle transcription factor E2F1 to stimulate transcription of E2F1-regulated genes, suggesting that it plays a role in the cell cycle (36). The DDB2 subunit itself is a cell cycle-regulated protein whose level peaks at the G1/S boundary and decreases in S phase (24, 25). This regulation may involve cullin-4A (24), a member of the cullin family of proteins, which are components of E3 ubiquitin ligases believed to be involved in selecting specific targets for ubiquitination (reviewed in reference 33). Cullin-4A associates with UV-DDB and when overexpressed stimulates degradation of DDB2 through the ubiquitin-proteasome pathway (9, 24, 37). Cullin-4A can form a complex with DDB1 in the absence of DDB2, but the levels of DDB1 are only modestly affected by cullin-4A overexpression (9). Although the two known activities implicating DDB1, damaged-DNA binding and stimulation of E2F1-activated transcription, both require its association with DDB2, the high evolutionary conservation of DDB1, but not of DDB2, suggests that DDB1 carries out important functions in the cell independently of DDB2 (42, 46).
The DDB1 subunit of UV-DDB is a cellular target for the X protein of hepatitis B virus (HBx). HBx is a small regulatory protein that is well conserved among the mammalian members of the family Hepadnaviridae, which also includes the hepatitis viruses of the woodchuck and ground squirrel. HBx is essential for establishing natural viral infection (8, 47) and has been specifically implicated elsewhere in the development of liver cancer associated with chronic infection (reviewed in reference 3). Binding to DDB1 is a conserved feature among the mammalian X proteins (41), and evidence has been presented that this interaction is critical for efficient hepatitis virus infection in the woodchuck (39). In cell culture, HBx exhibits pleiotropic activities affecting transcription, DNA repair, cell growth, and apoptotic cell death (for reviews, see references 3 and 23). Binding of HBx to DDB1 is essential for HBx to activate transcription (40) and to induce cell death (22, 40). Recent work has indicated that HBx exerts deleterious activities by forming a complex with DDB1 in the nuclear compartment (6). How exactly HBx translocates into the nucleus remains uncertain, however, since both DDB1 (40) and DDB2 (25) have been implicated in the nuclear localization of HBx, whereas our studies are more consistent with HBx reaching the nucleus independently of UV-DDB (6). Unexpectedly, once in the nucleus HBx competes with DDB2 for binding to DDB1. As a result, increased levels of DDB2 can relieve HBx-mediated cell death (6), indicating that HBx will exert its activities in association with DDB1 depending on the relative concentrations of DDB1 and DDB2 in the cell.
The DDB1 subunit of UV-DDB is also a potential target for another viral accessory protein that shows no similarity to HBx. DDB1 interacts with the V protein encoded by several members of the Paramyxoviridae family of negative-strand RNA viruses including simian virus 5 (SV5), human parainfluenza virus 2 (hPIV2), mumps virus, and measles virus (21). The role of the V protein in the viral life cycle remains to be fully established, but evidence exists that the protein is essential for pathogenicity of these viruses in the natural host (31, 44). The V protein is expressed from the viral P gene, which also codes for the P protein. The V and P proteins have the same amino-terminal sequence but differ in their carboxy termini (for a review of paramyxoviruses, see reference 19). The unique C-terminal domain of the V protein is highly conserved among the paramyxoviruses and is critical for interaction of the V protein with DDB1 (21). This region is also essential for the well-recognized ability of the V proteins of SV5, mumps virus, and hPIV2 to block interferon signaling, and thereby induction of an antiviral state, by specifically targeting STAT1 or STAT2 proteins for proteasome-mediated degradation (2, 11, 18, 29, 32). However, a role for DDB1 in STAT protein degradation by the V proteins has not yet been demonstrated. The only effect of V protein binding to DDB1 documented to date concerns the ability of the V protein of SV5 (SV5-V) to slow the progression of the cell cycle when produced in large amounts (2, 20).
We report here that SV5-V, which shows strong interaction with DDB1 (21), exhibits DDB1 binding properties that are strikingly similar to those of HBx. Yet in contrast to HBx the SV5-V protein lacks cytotoxic activity, which we show requires HBx and DDB1 to be interacting through their natural binding regions. Most significantly, SV5-V can prevent HBx from inducing cell death by displacing it from DDB1. The interaction of SV5-V with DDB1 appears to mediate another activity, since DDB1 is essential for SV5-V to induce STAT1 degradation. These findings demonstrate that HBx performs a unique function through its interaction with DDB1 for which SV5-V cannot substitute. They also point to an important role of DDB1 in the cell that does not involve DDB2 and suggest that SV5-V and HBx may bind to DDB1 to assist in the viral life cycle by serving distinct functions.
MATERIALS AND METHODS
Expression constructs.
All recombinant DNA work was carried out according to standard procedures. Details of the plasmid constructions are available upon request.
The mammalian expression vectors pSRαS, EBS-PL, and KEBOB-PL used in this study have been described elsewhere. Plasmid pSRαS is a modified version of pCI-neo (Promega) in which the original cytomegalovirus promoter was replaced by an SRα promoter (6). The episomal Epstein-Barr virus-based vector EBS-PL carries a hygromycin resistance gene and permits expression from the strong SRα promoter (22). The episomal vector KEBOB-PL contains a blasticidin resistance marker and a simian virus 40 early promoter (6). Green fluorescent protein (GFP) produced either from pEGFP-C1 (Clontech) or from the GFP open reading frame of pEGFP-N1 (Clontech) cloned into pSRαS was used to assess transfection efficiencies.
The regions encoding full-length SV5-V or the unique carboxyl Vu segment of SV5-V extending from amino acids 164 to 222 were amplified by PCR with a cDNA clone derived from SV5 (strain W3a)-infected cells (34) (kindly provided by Joe Curran, University of Geneva Medical School, Geneva, Switzerland) and primers that introduced convenient restriction endonuclease sites to allow direct subcloning. The PCR fragments were cloned into pBluescript (Stratagene, La Jolla, Calif.), sequenced, and used to construct the various Saccharomyces cerevisiae and mammalian expression vectors. myc-HBx and myc-SV5-V are similar to myc-DDB2 (6) and carry a triple myc epitope at the amino terminus. They were constructed by inserting three copies of a double-stranded oligonucleotide encoding peptide MEQKLISEEDLHMH (myc epitope tag in boldface) in front of the HBx and SV5-V open reading frames. GFP-SV5-V was generated by fusing in frame the GFP coding region excised out of pEGFP-C1 (Clontech) to the N terminus of the SV5-V coding region.
The DDB1 insertion mutants were generated as follow. The DDB1 cDNA cloned into pBluescript was cleaved at the following naturally occurring unique restriction endonuclease sites: EcoRI, 601; SphI, 1194; ClaI, 2110; and XhoI, 2837 (the numbering starts with the A of the ATG initiator codon). The linearized plasmids were made blunt ended with the Klenow fragment or T4 DNA polymerase and then religated in the presence of an excess of a double-stranded linker oligonucleotide of appropriate length to maintain the reading frame. The integrity of the open reading frame at the insertion site was confirmed by sequencing. The inserted sequences introduce a unique AscI restriction site and result in DDB1 protein mutants that contain two or four extra amino acids at the positions indicated in Fig. 1C.
FIG. 1.
Binding of SV5-V and HBx to DDB1 is mutually exclusive. (A) Schematic diagram of the SV5-V protein. The carboxyl segment unique to SV5-V (Vu) extending from amino acids 164 to 222 is indicated in black. Full-length SV5-V, the Vu domain, and HBx fused carboxyl-terminally to the transcriptionally inactive human RFX-binding protein were individually tested for interaction with DDB1 linked to the VP16 activation domain in a yeast two-hybrid assay (22). The black bars represent the relative activities of an integrated lacZ reporter gene bearing a single RFX-binding site in strains expressing the indicated RFX fusion proteins alone (−) or together (+) with VP16-DDB1. Basal levels of lacZ activity were detected in cells expressing VP16-DDB1 alone (data not shown). The data are representative of four independent experiments. (B) SV5-V and HBx expressed in their native form at high levels were tested for their ability to competitively inhibit activation caused by interaction between RFX-HBx and VP16-DDB1 (top panel) or RFX-SV5-V and VP16-DDB1 (middle panel) or for their effects on activation by VP16-RFX to assess specificity of inhibition (bottom panel). Results from one of three independent experiments are shown. (C) DDB1 mutants bearing single two- or four-residue insertions introduced at the indicated positions along the 1,140-amino-acid protein were fused to the VP16 activation domain and tested for interaction with RFX-HBx (upper panel) or RFX-SV5-V (lower panel) in the yeast two-hybrid system. Immunoblotting with antibodies against DDB1 detected comparable levels of the wild-type and mutant proteins (data not shown). The data are representative of three independent experiments.
HBx, GFP-HBx, and the HBx(R96E) point mutant expressed as a native protein or as an amino-terminal fusion to wild-type DDB1 [HBx(R96E)-DDB1 in Fig. 4C] have been previously described (22). The HBx(R96E)-DDB1(i947) fusion construct was obtained by replacing the sequence encoding wild-type DDB1 downstream of HBx(R96E) by the sequence encoding the DDB1 insertion mutant. All the other mammalian expression plasmids are described in reference 6.
FIG. 4.
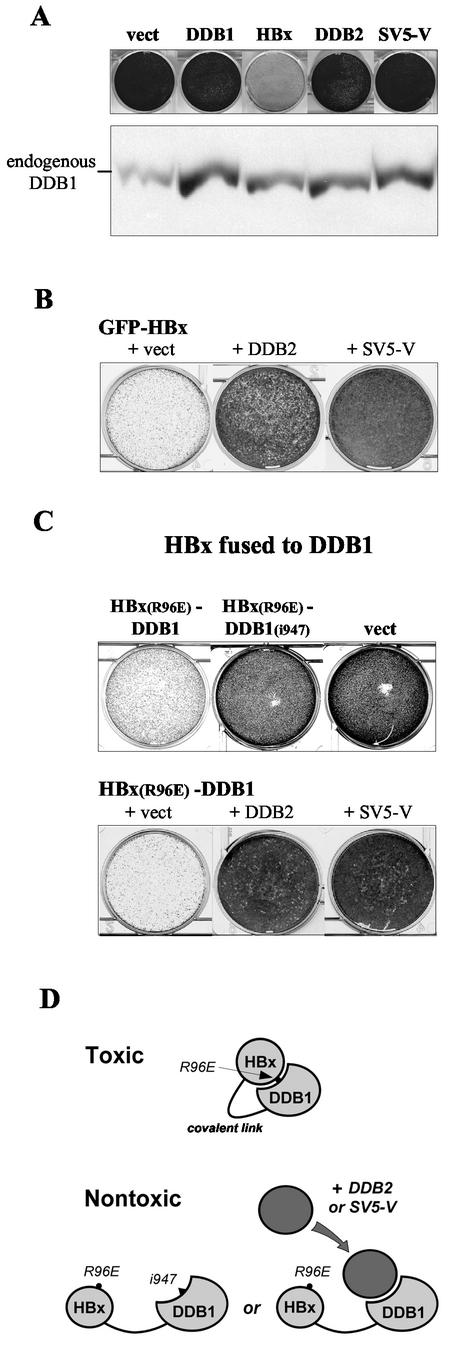
SV5-V lacks cytotoxic activities and can prevent HBx from inducing cell death. (A) HeLa cells transfected with constructs expressing the indicated proteins from the episomal vector EBS-PL, which carries a hygromycin resistance-conferring gene, or with empty vector (vect), were selected with hygromycin for resistant colonies. A GFP gene was cotransfected to assess for comparable transfection efficiencies by FACS analysis (data not shown). Drug-resistant colonies were fixed and stained with crystal violet 8 days after transfection (upper panel). The steady-state levels of endogenous DDB1 in these cells were examined 72 h after transfection by Western blot analysis with anti-DDB1 antibodies. The data are representative of three independent transfection experiments. (B) HeLa cells were cotransfected with GFP-HBx expressed from KEBOB-PL, which carries a blasticidin resistance marker, and equal amounts of empty vector EBS-PL (+ vect) or derivatives expressing DDB2 or SV5-V. The transfected cells were cultured in medium containing blasticidin and hygromycin to select for both plasmids. Drug-resistant cells were fixed and stained with crystal violet 8 days after transfection. Results from one of four independent transfection experiments are shown. (C) The DDB1-binding-defective HBx(R96E) point mutant was examined for its ability to suppress clonal outgrowth in HeLa cells when expressed as a fusion to wild-type DDB1 [HBx(R96E)-DDB1] or to the DDB1(i947) insertion mutant that lacks HBx-binding activity [HBx(R96E)-DDB1(i947)] (upper panel). The effect of DDB2 or SV5-V on cell death induced by the HBx(R96E)-DDB1 fusion protein was assessed by cotransfecting equal amounts of control vector (+ vect) or plasmids expressing the relevant proteins (lower panel). All the constructs were expressed in EBS-PL. Transfection efficiencies and hygromycin-resistant colony formation were assessed as described for panel A. The data are representative of three independent transfection experiments. (D) A covalent link between HBx(R96E), which cannot interact with the endogenous DDB1 protein, and DDB1 is proposed to act as a clamp forcing interaction between the two protein moieties, thereby restoring cytotoxic activity to the HBx mutant. The i947 mutation in DDB1 or ectopic expression of DDB2 or SV5-V prevents this from occurring by further compromising this interaction.
The DDB1-specific small interfering RNA (siRNA)-like transcript used for long-term inhibition of DDB1 gene expression was produced from the episomal vector EBOB-PL, which is identical to KEBOB-PL except that it carries a beta-lactamase gene in place of the kanamycin resistance gene for selection in bacteria. A 635-bp NaeI-KpnI fragment containing the RNA polymerase III H1-RNA promoter and T5 termination signal was excised from pSUPER (7) (a generous gift from René Bernards, The Netherlands Cancer Institute, Amsterdam, The Netherlands) and inserted in the polylinker region of EBOB-PL between BstEII blunt ended with Klenow enzyme and KpnI, yielding plasmid EBB-SUP. A DDB1 targeting sequence directed against nucleotides 3242 to 3260 of the DDB1 coding region was inserted into the resulting vector in the form of a double-stranded oligonucleotide (top strand, 5′-GATCCCCGACAGAACCAGCCACAGGTTTCAAGAGAACCTGTGGCTGGTTCTGTCTTTTTGGAAA-3′; DDB1 sequences in boldface) according to the author's recommendations (7).
Yeast two-hybrid and β-galactosidase assay.
All the proteins are encoded by single-copy plasmids marked with the TRP1, URA3 (38), or ADE2 (12) gene. VP16-DDB1, VP16-RFX, and RFX-HBx have been described previously (22). RFX-SV5-V(full-length) and RFX-SV5-V(Vu) were constructed by joining full-length SV5-V or the Vu domain unique to SV5-V in frame to the carboxyl terminus of RFX, respectively. All the constructs are expressed from the TBP promoter with the exception of the VP16-DDB1 constructs, which are expressed under control of the galactose-inducible GAL1,10 regulatory sequences, and native SV5-V and HBx proteins that were overproduced in the experiment shown in Fig. 1B by placing their genes under the control of the strong DED1 promoter.
The β-galactosidase assays were performed in a yeast strain carrying an RFX-dependent lacZ reporter gene integrated at the HIS3 locus and bearing a single RFX-binding site upstream of the CYC1 core promoter. Transformed yeast cells were grown to early log phase in selective medium and assayed for β-galactosidase activity as described elsewhere (12).
Cell culture, transfection, and colony-forming assay.
HeLa cells were grown at 37°C in the presence of 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 100 U of penicillin/ml, 100 μg of streptomycin/ml, 2 mM l-glutamine, and 10% (vol/vol) fetal calf serum (Chemie Brunschwig). The cells were transfected by using the FuGENE 6 reagent (Roche) according to the manufacturer's instructions. For the immunoprecipitation experiments presented in Fig. 2, about 4 × 105 HeLa cells were seeded in an 85-mm-diameter plate and transfected with 2.7 μg of myc-SV5-V, 0.6 μg of myc-DDB2, and 0.3 μg of HA-DDB1 constructs in pSRαS, or combinations thereof. The total amount of DNA was kept constant at 4 μg in all transfections by supplementation with vector DNA. For the pulse-labeling and chase experiments presented in Fig. 3B, about 2 × 105 HeLa cells were seeded in a 50-mm-diameter plate and transfected with 0.45 μg of myc-HBx or myc-SV5-V construct and 1.35 μg of the indicated constructs, all proteins being expressed from EBS-PL. When not made as a fusion protein, an expression plasmid for GFP was cotransfected (10% of total DNA). At 24 h posttransfection, cells were trypsinized and a fraction (usually one-third) was scanned by fluorescence-activated cell sorting (FACS) for GFP fluorescence to assess for transfection efficiencies; transfection efficiencies were generally 50 to 80% with variations of less than 10% within any single experiment. The colony-forming assays presented in Fig. 4 were performed exactly as described previously (6).
FIG. 2.
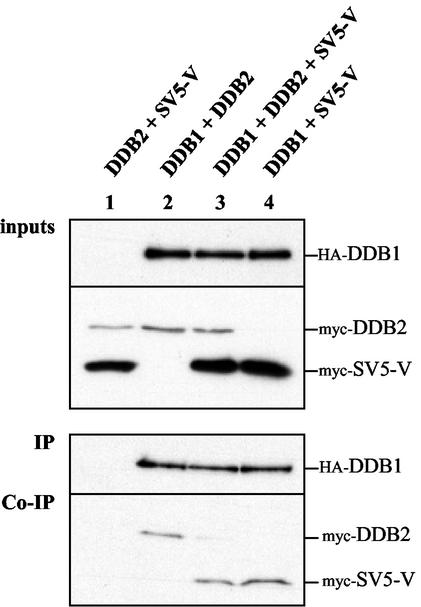
SV5-V interferes with UV-DDB complex formation. Shown are results of coimmunoprecipitation experiments with extracts from transiently transfected HeLa cells. N-terminally epitope-tagged HA-DDB1, myc-DDB2, and myc-SV5-V produced from the expression vector pSRαS were transfected at a 1:2:9 DNA ratio in pairwise combinations or all three together. Total plasmid DNA was kept equal by adding empty plasmid DNA. Whole-cell extracts (100 μg) were prepared 1 day after transfection and subjected to immunoprecipitation with a MAb against the HA epitope. The immunoprecipitates were separated by SDS-PAGE and analyzed by Western blot assays for the presence of HA-DDB1 with the anti-HA antibody (IP). The presence of coimmunoprecipitated myc-DDB2 and myc-SV5-V was detected with an anti-myc MAb (Co-IP). The upper panel (inputs) shows 1/25 of the cell extract used in the immunoprecipitations to assess for comparable protein levels. The data are representative of two independent transfection experiments.
FIG. 3.
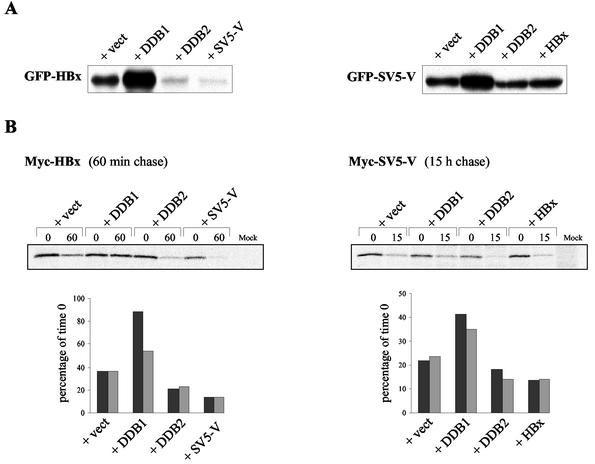
HBx and SV5-V are stabilized by their interaction with DDB1. (A) Steady-state protein levels of HBx and SV5-V in cells expressing higher levels of DDB1 or DDB2. HeLa cells were cotransfected with GFP-HBx (left panel) or GFP-SV5-V (right panel) made from the expression vector KEBOB-PL and equal amounts of either control vector EBS-PL (+ vect) or derivatives thereof expressing the indicated proteins under the control of the strong SRα promoter. Transfection efficiencies were comparable as assessed by FACS analysis (data not shown). Cells lysates were prepared 72 h after transfection, and equal protein amounts per sample were resolved by SDS-PAGE. The accumulation of the GFP fusion proteins was examined by Western blot analysis with a MAb directed against GFP. Results from one of three independent transfection experiments are shown. (B) Pulse-labeling and chase experiments. HeLa cells were cotransfected with myc-HBx (left panel) or myc-SV5-V (right panel) and a threefold excess of either empty vector (+ vect) or constructs expressing the indicated proteins. All the proteins were produced from EBS-PL. Two days after transfection, myc-HBx and myc-SV5-V were immunoprecipitated with myc antibody from extracts prepared at the indicated chase period after metabolic pulse-labeling, separated by SDS gel electrophoresis, and either visualized by autoradiography (upper panels) or quantitated by phosphorimager analysis (lower panels). A chase period of 60 min for myc-HBx and 15 h for SV5-V was chosen based on the half-life of the proteins calculated from linear regression lines fitted to multiple chase time points (data not shown). The lower panels represent quantitative data from two independent transfection experiments, one of which corresponds to the autoradiography presented in the upper panels. The labeled protein levels are expressed as percentages of their values at time zero.
Immunoprecipitation, cell extracts, and Western blotting.
The coimmunoprecipitation experiments shown in Fig. 2 were performed with whole-cell extracts prepared 24 h after transfection from 4 × 105 HeLa cells lysed on the plate in 1.0 ml of NP-40 lysis buffer as described in reference 6. HA-DDB1 was immunoprecipitated from 100 μg of whole-cell extracts by incubation with 50 μl of antihemagglutinin (anti-HA) affinity matrix (Roche) in a final volume adjusted to 500 μl with lysis buffer. After incubation for 2 h at 4°C with constant rotation on a rocker, the beads were washed twice in lysis buffer and then resuspended and boiled in Laemmli buffer. One-half of the supernatant was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 9% polyacrylamide). After separation, the proteins on the gels were transferred to Immobilon P membranes (Millipore) and subjected to immunoblotting as described previously (22). Membranes were probed with anti-myc monoclonal antibody (MAb) 9E10 or anti-HA MAb 16B12 (BAbCo). Binding of primary antibody was detected with anti-mouse immunoglobulin, horseradish peroxidase-linked whole antibody (Amersham Pharmacia Biotech). Blotted proteins were visualized with Lumi-Light or Lumi-LightPLUS blotting reagents (Roche).
The Western blot analyses presented in Fig. 3A, 4A, and 5 were performed with cell extracts prepared as described previously (22). Membranes were probed with anti-GFP MAb (mixture of clones 7.1 and 13.1 from Roche), anti-p127 (DDB1) polyclonal antibody (a generous gift from Vesna Rapić Otrin, University of Pittsburgh, Pittsburgh, Pa.), or anti-N-terminal STAT1 MAb (Transduction Laboratories).
FIG. 5.
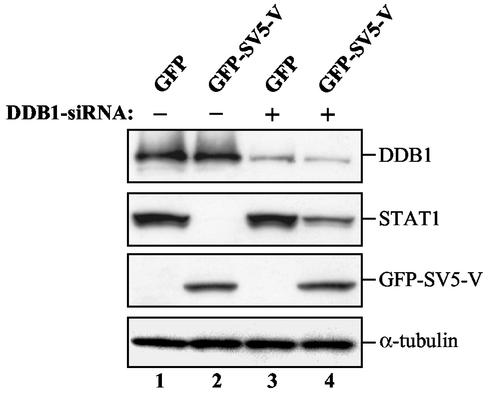
SV5-V induces STAT1 degradation less efficiently in cells expressing reduced amounts of DDB1. HeLa cells were cotransfected with GFP or with GFP-SV5-V and equal amounts of either control vector EBB-SUP (−) or a construct (+) that directs the synthesis of a DDB1-specific siRNA from the RNA polymerase III H1-RNA gene promoter (7). Transfection efficiencies were comparable as assessed by FACS analysis (data not shown). Whole-cell extracts prepared 8 days after transfection and growth in selective medium were separated on SDS-polyacrylamide gels and immunoblotted to detect DDB1, STAT1, and GFP-SV5-V proteins. Quantitation by serial dilution (data not shown) showed that a 10- to 20-fold-higher amount of STAT1 can be detected in lane 4 than in lane 2, which corresponds to 20 to 30% of the amount of STAT1 present in extracts from cells expressing no SV5-V (lanes 1 and 3). In these experiments, GFP-SV5-V was produced from the expression vector KEBOB-PL at reduced levels compared to those in Fig. 4A, which may explain why no SV5-V-mediated increase in endogenous DDB1 protein levels is observed. Results from one of two independent experiments are shown.
Metabolic labeling and chase experiments.
Transfected HeLa cells were metabolically labeled 48 h after transfection. The cells were starved for 45 min in methionine- and cysteine-free minimal essential medium (Sigma) and then pulse-labeled for 45 min at 37°C in fresh medium supplemented with a [35S]methionine-[35S]cysteine mix (Hartmann Analytic) to a final concentration of 0.15 mCi/ml. After labeling, the cells were rinsed twice in minimal essential medium containing 2% fetal calf serum, 10 mM methionine, and 10 mM cysteine (Sigma) and either harvested immediately (time zero) or further incubated in the same medium. At each time point, cells were harvested and lysed as described previously (6). The lysates were cleared by centrifugation for 15 min at 4°C in an Eppendorf microcentrifuge. myc-HBx and myc-SV5-V were immunoprecipitated with anti-myc MAb 9E10 from 200 μg of whole-cell extracts precleared with protein A-Sepharose beads (CL-4B; Amersham Pharmacia Biotech) for 90 min at 4°C. After an overnight incubation at 4°C, protein A-Sepharose beads were added, and the mixture was rotated at 4°C for 2 h. Beads were subsequently washed twice in 1% β-mercaptoethanol-0.5 M NaCl and then twice in radioimmunoprecipitation assay buffer containing 0.1% SDS, and finally resuspended and boiled in Laemmli buffer. One-half of the supernatant was loaded on SDS-polyacrylamide gels. The labeled protein bands were quantitated with a Bio-Rad phosphorimager and Quantity One software. The subtracted background intensity for each band was determined from a rectangle of equal size drawn immediately above the rectangle for each protein band.
RESULTS
SV5-V and HBx interact with DDB1 at overlapping binding sites.
The SV5-V protein has been shown elsewhere to interact with DDB1 via its unique carboxyl-terminal Vu domain (21). We examined whether SV5-V and DDB1 would also interact in the yeast transcription-based two-hybrid protein interaction assay that allowed us to identify DDB1 as a cellular partner of HBx (22). As bait, we used either the full-length SV5-V protein or only the Vu domain fused carboxy terminal to RFX, a human DNA-binding protein with no transcriptional activity in yeast. Figure 1A shows that both hybrid proteins exhibit transactivation activities compared to the transcriptionally inactive RFX-HBx variant in a yeast strain carrying an RFX-dependent lacZ reporter gene. This indicates that the fusion proteins are stably expressed. HBx, as expected, and the full-length SV5-V fusions show increased activity when tested in combination with a VP16 activation domain-tagged DDB1 derivative (Fig. 1A). By contrast, no increase in lacZ activity is observed when the Vu domain of SV5-V is used as bait. These results indicate that SV5-V and DDB1 can form a complex in yeast, and they raise the possibility that sequences outside the Vu domain of SV5-V contribute to interaction between the two proteins.
To determine whether SV5-V and HBx recognize distinct or overlapping surfaces of the DDB1 protein, we tested the effect of overexpressing native SV5-V on transcription activation caused by interaction between RFX-HBx and VP16-DDB1 in the yeast two-hybrid system. If the two viral proteins bind DDB1 through nonoverlapping sites, SV5-V should form a ternary complex with RFX-HBx and VP16-DDB1 and thereby increase RFX-dependent lacZ activity due to its intrinsic transactivation property (Fig. 1A). By contrast, if SV5-V and HBx were to bind DDB1 through overlapping sites and thus in a mutually exclusive fashion, high-level expression of SV5-V should competitively inhibit the interaction between RFX-HBx and VP16-DDB1 and therefore interfere with activation. The results presented in Fig. 1B fully support the latter prediction. Indeed, overexpressed SV5-V reduces activation mediated by RFX-HBx and VP16-DDB1 to nearly uninduced levels (top panel in Fig. 1B, compare first and third lanes). Under these conditions SV5-V also abolishes its own ability to function as bait (middle panel in Fig. 1B). Overexpression of HBx causes a similar but less pronounced reduction in lacZ activity in a two-hybrid assay with RFX-HBx while having no effect when RFX-SV5-V is used as bait (top and middle panels in Fig. 1B, compare first and second lanes to fourth lane). The specificity of inhibition is demonstrated by a lack of effect of the viral proteins on transcription stimulated by VP16-RFX (bottom panel in Fig. 1B). These results indicate that SV5-V and HBx compete for interaction with DDB1, and they suggest that SV5-V may bind to DDB1 with higher affinity than that of HBx.
The DDB1 protein is particularly sensitive both to terminal (22) and to internal (data not shown) deletions. As a first step toward identifying regions of DDB1 important for interaction with SV5-V and HBx, we generated a series of DDB1 mutants containing 2- or 4-amino-acid insertions at various positions along the protein. The mutants were tested as fusions to VP16 for interaction with RFX-HBx and RFX-SV5-V, respectively. Figure 1C shows that all the DDB1 mutants retain at least partial binding activities in both assays, with the exception of mutant DDB1(i947), which contains the most carboxyl-terminal insertion. This mutant is completely defective for interaction with HBx while retaining normal SV5-V-binding ability (Fig. 1C). The mutant also remains capable of interacting with DDB2 (data not shown). We conclude from these results that SV5-V and HBx bind to DDB1 in a distinct yet mutually exclusive manner, most likely by recognizing overlapping determinants on the DDB1 protein.
SV5V shares with HBx the property of disrupting the UV-DDB complex.
Our previous work has shown that the binding of HBx to DDB1 blocks the binding of DDB2 to DDB1 (6). The finding that SV5-V and HBx interact with DDB1 in a similar fashion prompted us to examine whether SV5-V might also interfere with UV-DDB complex formation. We therefore performed coimmunoprecipitation experiments using extracts from transfected cells to assess whether an excess of SV5-V protein would interfere with the binding of DDB2 to limiting amounts of DDB1. Plasmids expressing N-terminally epitope-tagged HA-DDB1, myc-DDB2, and myc-SV5-V proteins were transiently transfected into HeLa cells, either in pairwise combinations or all three together. In these experiments we used low amounts of HA-DDB1 plasmid and an excess of myc-SV5-V over myc-DDB2. Whole-cell extracts were prepared, HA-DDB1 was immunoprecipitated with an anti-HA antibody, and proteins present in the extract before or after immunoprecipitation were detected by Western blot analysis. As shown in Fig. 2, each protein was expressed at comparable levels in all the transfected cells, and myc-SV5-V was clearly overproduced relative to myc-DDB2 (“inputs” panel in Fig. 2). Yet comparable amounts of myc-DDB2 and myc-SV5-V are recovered in the immunoprecipitates from extracts of cells expressing either one of the two proteins together with HA-DDB1 (Co-IP panel in Fig. 2, compare lanes 2 and 4). This suggests that the amount of HA-DDB1 available to interact with myc-DDB2 or myc-SV5-V is limiting. Under these conditions, the excess of myc-SV5-V strongly reduces the amount of myc-DDB2 that coimmunoprecipitates with HA-DDB1 (Co-IP panel in Fig. 2, compare lanes 2 and 3). By contrast, myc-SV5-V coimmunoprecipitates with similar efficiencies in the absence and in the presence of myc-DDB2 (Co-IP panel in Fig. 2, compare lanes 3 and 4). These results demonstrate that SV5-V exhibits the same striking ability as does HBx to displace DDB2 from DDB1.
Binding to DDB1 enhances the stability of SV5-V and HBx.
HBx is a short-lived protein that is strongly stabilized through its physical interaction with DDB1 (5a, 6). As a result, the cellular levels of HBx largely depend on the absolute amount of DDB1 and on the relative concentrations of DDB1 and DDB2 in the cell (5a, 6). To examine whether SV5-V also accumulates in a DDB1-dependent fashion, we compared the effects of expressing DDB1 or DDB2 in excess over the endogenous proteins on the levels of HBx and SV5-V variants bearing an amino-terminal GFP. As reported previously (6), cotransfection of DDB1 leads to an accumulation of GFP-HBx whereas DDB2 reduces the amount of GFP-HBx, most likely by displacing it from endogenous DDB1 (left panel in Fig. 3A). Overexpression of DDB1 and DDB2 similarly affects GFP-SV5-V protein levels, although the reduction in the amount of GFP-SV5-V by DDB2 is much more modest (right panel in Fig. 3A). We also examined whether SV5-V and HBx would influence each other's steady-state levels by mutually exclusive binding when expressed in the same cell. The left panel in Fig. 3A shows that SV5-V markedly decreases GFP-HBx protein levels, thus confirming that the abundance of HBx is indeed regulated by its interaction with endogenous DDB1 (6). By contrast, HBx shows no obvious effect on GFP-SV5-V (right panel in Fig. 3A), consistent with the possibility that SV5-V exhibits higher affinity for DDB1 than does HBx.
To test the effect of DDB1 binding on the half-life of HBx and SV5-V, we performed pulse-labeling and chase experiments with cells expressing myc epitope-tagged versions of HBx and SV5-V. The half-life of myc-HBx and myc-SV5-V was first estimated by determining the fraction of the labeled proteins remaining at various chase periods after metabolic pulse-labeling (data not shown). The half-life of HBx during the chase was calculated to be ∼40 min, which is consistent with previous studies (13b). SV5-V proved to be much more stable, with an estimated half-life of nearly 8 h. This latter estimate largely exceeds the 45-min half-life reported elsewhere for a C-terminal truncation mutant of SV5-V that cannot bind DDB1 (13a), suggesting that native SV5-V may be strongly stabilized by its interaction with endogenous DDB1. The effect of DDB1 and DDB2 on myc-HBx and myc-SV5-V stability was then examined in cotransfection experiments by determining the percentage of pulse-labeled proteins remaining in the cell following an appropriate chase period. Figure 3B shows that cotransfection of DDB1 resulted in an increase of the half-life of both HBx and SV5-V proteins, whereas a modest but reproducible reduction in their half-life was observed upon cotransfection of DDB2 or the other viral protein. Taken together, these results indicate that the cellular levels of SV5-V and HBx are determined, at least in part, by stabilization of the proteins through their interaction with DDB1.
SV5-V can prevent HBx from inducing cell death through its interaction with DDB1.
HBx affects the growth and survival of both primary and immortalized cells. This activity requires interaction of HBx with DDB1 and explains the failure of HBx-expressing cells to grow in a colony formation assay (22, 40). We examined whether expression of the SV5-V protein in HeLa cells, which has been documented previously to slow their progression through the cell cycle (20), would inhibit their ability to form colonies under conditions where HBx exhibits strong growth suppression activities. Figure 4A reveals that this is not the case. Interestingly, SV5-V shares with HBx and DDB2 the property of inducing an accumulation of endogenous DDB1 (Fig. 4A). These results indicate that HBx-mediated cell death is not simply due to HBx causing a harmful accumulation of DDB1 in the cell. Consistent with this view, HeLa cells overexpressing DDB1 to comparable levels in the absence of HBx proliferate normally (Fig. 4A).
The mutually exclusive nature of SV5-V and HBx binding to DDB1, together with the finding that SV5-V lacks cytotoxic activities, predicts that high levels of SV5-V should prevent HBx from inducing cell death by displacing it from endogenous DDB1. To test this hypothesis, we transfected HeLa cells with the GFP-HBx gene alone or together with constructs expressing DDB2 or SV5-V from a strong promoter. As reported previously (6), overexpression of DDB2 partially relieves suppression of colony formation by HBx (Fig. 4B). Remarkably, SV5-V exhibits the same ability to overcome HBx-dependent cell death and is actually more effective than DDB2 in doing so (Fig. 4B).
The finding that DDB2 and SV5-V can block the interaction between HBx and DDB1 gave us an opportunity to examine whether this interaction simply serves a tethering function or whether it plays a more critical role in HBx activity. Previous work has shown that the functionally defective HBx(R96E) point mutant, which cannot interact with the endogenous DDB1 protein, regains cytotoxic properties when covalently fused to DDB1 (22). One mechanism whereby a covalent link between HBx(R96E) and DDB1 may alleviate the defective phenotype of the HBx mutant is by acting as a “clamp” forcing the two protein moieties into their natural interaction (Fig. 4D) (see Discussion). If so, one would predict that further compromising this interaction by mutagenesis or by overexpression of DDB2 or SV5-V should interfere with the ability of the HBx(R96E)-DDB1 fusion protein to exert deleterious activities (Fig. 4D). The experiments presented in Fig. 4C fully confirmed this prediction. Fusion of the DDB1(i947) variant that specifically lacks HBx-binding abilities (Fig. 1C) fails to restore activity to the HBx(R96E) mutant (upper panel in Fig. 4C), and overexpression of DDB2 or SV5-V largely overcomes the inhibitory effect of the HBx(R96E)-DDB1 fusion protein in the colony formation assay (lower panel in Fig. 4C). Hence, a covalent link between HBx and DDB1 does not bypass the need for an interaction between the two proteins, indicating that this interaction plays a more critical role than simply bringing the two proteins together (see Discussion).
DDB1 is essential for SV5-V to induce degradation of STAT1.
The best-characterized activity of the V protein of SV5 is its ability to block interferon signaling by specifically targeting STAT1 protein for proteasome-mediated degradation (2, 11). We tested whether HBx would exhibit similar properties and found that this is not the case (data not shown). To determine whether DDB1 plays a role in SV5-V activity, we examined the ability of SV5-V to induce STAT1 degradation in cells expressing reduced levels of DDB1 protein. For this purpose, we constructed an episomal plasmid producing an siRNA designed to inhibit DDB1 gene expression (7). As shown in Fig. 5, HeLa cells transfected with this construct showed reduced DDB1 protein expression, while STAT1 protein levels were not affected (Fig. 5, compare lanes 1 and 3). These results suggest that the steady-state level of STAT1 in the cell does not depend on DDB1. Remarkably, however, SV5-V is 10- to 20-fold less competent in inducing degradation of STAT1 in the DDB1 knockdown cells (Fig. 5, compare lanes 2 and 4). Hence, DDB1 is essential for SV5-V-induced STAT1 degradation, suggesting that SV5-V mediates this activity through its association with DDB1. Given these data, we propose that, despite exhibiting a number of common properties with respect to DDB1 binding, the SV5-V and HBx viral proteins mediate distinct activities through this interaction.
DISCUSSION
The DDB1 subunit of UV-DDB is a cellular target of at least two unrelated viral regulatory proteins, the X proteins encoded by the mammalian hepatitis B viruses, among which the HBx protein of human hepatitis B virus is the prototype, and the V proteins of several members of the Paramyxoviridae family, including SV5. In the present study, we show that HBx and SV5-V exhibit very similar properties with regard to DDB1 binding. Firstly, HBx and SV5-V compete for association with DDB1, most likely as a result of recognizing overlapping surfaces on the DDB1 protein. Secondly, both HBx and SV5-V exert a stabilizing effect on cellular DDB1 upon binding, and conversely, both viral proteins are stabilized by DDB1. Lastly, SV5-V shares with HBx the striking ability to displace DDB2 from DDB1. Despite these common features, however, only HBx exhibits the distinctive property of inducing cell death. Most significantly, HBx cytotoxic activity can be relieved by coexpression of SV5-V. These findings exclude the possibility that HBx interferes with cell viability by sequestering DDB1 and thereby preventing it from performing its normal activities in association with DDB2. They also argue against the possibility that HBx induces a harmful accumulation of DDB1 in the cell. Instead, they indicate that HBx performs a unique function through its association with DDB1 for which SV5-V cannot substitute.
As yet we do not know by what mechanism HBx triggers cell death. However, our approach of testing HBx cytotoxic activity when ectopically fused to DDB1 might provide an insight into how the native HBx protein functions in association with cellular DDB1 and what role the interaction between HBx and DDB1 plays in this activity. We envision two possible scenarios. First, HBx and DDB1 may perform independent functions that become deleterious when physically associated. In this instance, interaction between the two proteins would simply serve a tethering function that is bypassed by covalently linking the DDB1-binding-defective HBx(R96E) mutant to DDB1. The alternative hypothesis is that cell death involves an activity that requires HBx and DDB1 to be interacting through their natural binding sites. In this scenario, a covalent link between HBx(R96E) and DDB1 would alleviate the defective phenotype of the HBx mutant by acting as a clamp forcing the two protein moieties into their natural interaction (Fig. 4D). Our results strongly argue in favor of the second hypothesis. Indeed, the simplest explanation for the observation that DDB2 or SV5-V can overcome the cytotoxic effect of the HBx(R96E)-DDB1 fusion protein (Fig. 4C) is that they act by precluding the HBx(R96E) and DDB1 moieties from interacting with one another (Fig. 4D). According to this model, the DDB1(i947) variant that is specifically compromised for HBx binding would fail to rescue the HBx(R96E) mutant because the mutation in DDB1 further compromises interaction between the two proteins (Fig. 4C and D). This leads us to propose that HBx acquires deleterious activities through its association with DDB1 in a way that requires the two proteins to be interacting via their natural binding regions.
What molecular mechanism, then, might lie behind the need for HBx to interact normally with DDB1 to exhibit cytotoxic properties? One possibility is that upon forming a complex HBx and DDB1 create a new surface for interaction with another cellular factor to form a ternary complex, with each of the two proteins contributing to the interaction. This would readily explain why SV5-V cannot substitute for HBx to induce cell death. Intriguingly, however, of the 12 HBx charge reversal point mutants that we have analyzed, the two mutants that fail to induce cell death also lack DDB1-binding activities (22; data not shown). This raises the possibility that HBx may act solely through its association with DDB1 to somehow confer deleterious activities on DDB1, perhaps by inducing a conformational change within the protein. Studies aimed at further addressing this issue are under way.
DDB1 is a large protein of 1,140 amino acids that has been proposed elsewhere to consist mainly of WD40-related repeats predicted to fold into β-propeller domains (26). These are likely to present multiple surfaces for interactions with other proteins. Our finding that HBx, SV5-V, and cellular DDB2 all associate with DDB1 in a mutually exclusive manner is therefore unexpected. One possibility is that DDB1 functions in a complex with other cellular components such that it offers only a limited surface for interaction with additional proteins. Alternatively, HBx and SV5-V may have evolved to inhibit UV-DDB cellular function by preventing DDB2 from binding to DDB1. An interaction between the two UV-DDB subunits is required for the function of UV-DDB in stimulation of E2F1-activated transcription (36) and in DNA repair (28, 30, 35, 43). The viral proteins are therefore expected to interfere with cell cycle regulation and DNA repair when produced in large amounts. That this is indeed the case for HBx has been well documented (4, 5, 13, 16), and evidence has been recently presented that SV5-V can slow progression of the cell cycle through its binding to DDB1 when expressed at high levels (20). Whether the viral proteins are expressed in sufficiently high amounts to exhibit such activities in the course of natural infection and whether this is of significance in terms of viral pathogenesis remain to be determined.
Despite strikingly similar DDB1-binding properties, HBx and SV5-V perform apparently unrelated activities in the cell. Whereas HBx has the potential to interfere with cell viability in culture, we provide evidence here that under the same experimental conditions SV5-V does not. Conversely, SV5-V and the V protein of hPIV2, which also binds DDB1 (21), exhibit the ability to block interferon signaling by specifically targeting STAT1 (2, 11) and STAT2 (2, 32), respectively, for proteasome-mediated degradation, whereas HBx lacks this activity (data not shown). What common function then might these unrelated viral proteins carry out through their association with DDB1 and independently of DDB2? The DDB1 subunit, unlike DDB2, is highly conserved among species and is expressed among all mammalian tissues (42, 46). DDB1 is therefore likely to perform important, yet to be discovered functions in the cell that do not involve DDB2. DDB1 binds cullin-4A, a member of a family of proteins that possess ubiquitin ligase activity, and cullin-4A stimulates degradation of DDB2 through the ubiquitin-proteasome pathway (9, 24, 37). DDB1 may thus have a specific role in proteasome-mediated degradation of DDB2 and other cellular proteins. It is conceivable, therefore, that the V proteins of SV5 and hPIV2 function as adapters, through their association with DDB1, to target the STAT proteins for proteolysis. Our finding that DDB1 is essential for STAT1 degradation induced by the V protein of SV5 is fully consistent with this hypothesis. This raises the possibility that HBx may interfere with cell viability through its binding to DDB1 by targeting important cellular control proteins for proteolysis. To our knowledge, however, no HBx-interacting protein has yet been described that shows decreased stability in the presence of HBx. Thus, the existence and identity of such a protein(s) remain to be established.
Acknowledgments
We are most grateful to René Bernards for the pSUPER vector, to Joe Curran for the SV5-V plasmid, and to Vesna Rapić Otrin for anti-p127 DDB1 antibodies. Special thanks go to Dominique Garcin and Dan Kolakofsky for sharing their expertise on interferon signaling assays, to Rachel Imoberdorf for yeast technical support, and to Géraldine Silvano Gargano for expert technical assistance. We are also very grateful to Joe Curran, Dan Kolakofsky, and Walter Reith for their careful and critical reading of the manuscript and helpful discussions.
O.L. was a recipient of an Ernst and Lucie Schmidheiny Foundation fellowship. S.B. was supported by the Geneva Cancer League. This work was supported by a grant from the Swiss National Science Foundation to M.S. (no. 31-49′792.96).
REFERENCES
- 1.Abramic, M., A. S. Levine, and M. Protic. 1991. Purification of an ultraviolet-inducible, damage-specific DNA-binding protein from primate cells. J. Biol. Chem. 266:22493-22500. [PubMed] [Google Scholar]
- 2.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbuthnot, P., A. Capovilla, and M. Kew. 2000. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J. Gastroenterol. Hepatol. 15:357-368. [DOI] [PubMed] [Google Scholar]
- 4.Becker, S. A., T. H. Lee, J. S. Butel, and B. L. Slagle. 1998. Hepatitis B virus X protein interferes with cellular DNA repair. J. Virol. 72:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benn, J., and R. J. Schneider. 1995. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci. USA 92:11215-11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Bergametti, F., D. Sitterlin, and C. Transy. 2002. Turnover of hepatitis B virus X protein is regulated by damaged DNA-binding complex. J. Virol. 76:6495-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bontron, S., N. Lin-Marq, and M. Strubin. 2002. HBx associated with UV-DDB1 induces cell death in the nucleus and is functionally antagonized by UV-DDB2. J. Biol. Chem. 277:38847-38854. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H. S., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276:48175-48182. [DOI] [PubMed] [Google Scholar]
- 10.Chu, G., and E. Chang. 1988. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242:564-567. [DOI] [PubMed] [Google Scholar]
- 11.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Couto, E., N. Klages, and M. Strubin. 1997. Synergistic and promoter-selective activation of transcription by recruitment of transcription factors TFIID and TFIIB. Proc. Natl. Acad. Sci. USA 94:8036-8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groisman, I. J., R. Koshy, F. Henkler, J. D. Groopman, and M. A. Alaoui-Jamali. 1999. Downregulation of DNA excision repair by the hepatitis B virus-X protein occurs in p53-proficient and p53-deficient cells. Carcinogenesis 20:479-483. [DOI] [PubMed] [Google Scholar]
- 13a.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 13b.Hu, Z., Z. Zhang, E. Doo, O. Coux, A. L. Goldberg, and T. J. Liang. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 73:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang, B. J., and G. Chu. 1993. Purification and characterization of a human protein that binds to damaged DNA. Biochemistry 32:1657-1666. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, B. J., S. Toering, U. Francke, and G. Chu. 1998. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol. Cell. Biol. 18:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia, L., X. W. Wang, and C. C. Harris. 1999. Hepatitis B virus X protein inhibits nucleotide excision repair. Int. J. Cancer 80:875-879. [DOI] [PubMed] [Google Scholar]
- 17.Keeney, S., G. J. Chang, and S. Linn. 1993. Characterization of a human DNA damage binding protein implicated in xeroderma pigmentosum E. J. Biol. Chem. 268:21293-21300. [PubMed] [Google Scholar]
- 18.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal CYS-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 19.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 20.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 22.Lin-Marq, N., S. Bontron, O. Leupin, and M. Strubin. 2001. Hepatitis B virus X protein interferes with cell viability through interaction with the p127-kDa UV-damaged DNA-binding protein. Virology 287:266-274. [DOI] [PubMed] [Google Scholar]
- 23.Murakami, S. 2001. Hepatitis B virus X protein: a multifunctional viral regulator. J. Gastroenterol. 36:651-660. [DOI] [PubMed] [Google Scholar]
- 24.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nag, A., A. Datta, K. Yoo, D. Bhattacharyya, A. Chakrabortty, X. Wang, B. L. Slagle, R. H. Costa, and P. Raychaudhuri. 2001. DDB2 induces nuclear accumulation of the hepatitis B virus X protein independently of binding to DDB1. J. Virol. 75:10383-10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuwald, A. F., and A. Poleksic. 2000. PSI-BLAST searches using hidden Markov models of structural repeats: prediction of an unusual sliding DNA clamp and of beta-propellers in UV-damaged DNA-binding protein. Nucleic Acids Res. 28:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols, A. F., T. Itoh, J. A. Graham, W. Liu, M. Yamaizumi, and S. Linn. 2000. Human damage-specific DNA-binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem. 275:21422-21428. [DOI] [PubMed] [Google Scholar]
- 28.Nichols, A. F., P. Ong, and S. Linn. 1996. Mutations specific to the xeroderma pigmentosum group E Ddb-phenotype. J. Biol. Chem. 271:24317-24320. [DOI] [PubMed] [Google Scholar]
- 29.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otrin, V. R., M. McLenigan, M. Takao, A. S. Levine, and M. Protic. 1997. Translocation of a UV-damaged DNA binding protein into a tight association with chromatin after treatment of mammalian cells with UV light. J. Cell Sci. 110:1159-1168. [DOI] [PubMed] [Google Scholar]
- 31.Parisien, J. P., J. F. Lau, and C. M. Horvath. 2002. STAT2 acts as a host range determinant for species-specific paramyxovirus interferon antagonism and simian virus 5 replication. J. Virol. 76:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 33.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 34.Precious, B., D. F. Young, A. Bermingham, R. Fearns, M. Ryan, and R. E. Randall. 1995. Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of NP-P and NP-V interactions. J. Virol. 69:8001-8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapiæ Otrin, V., I. Kuraoka, T. Nardo, M. McLenigan, A. P. Eker, M. Stefanini, A. S. Levine, and R. D. Wood. 1998. Relationship of the xeroderma pigmentosum group E DNA repair defect to the chromatin and DNA binding proteins UV-DDB and replication protein A. Mol. Cell. Biol. 18:3182-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiyanov, P., S. A. Hayes, M. Donepudi, A. F. Nichols, S. Linn, B. L. Slagle, and P. Raychaudhuri. 1999. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 19:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiyanov, P., A. Nag, and P. Raychaudhuri. 1999. Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274:35309-35312. [DOI] [PubMed] [Google Scholar]
- 38.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sitterlin, D., F. Bergametti, P. Tiollais, B. C. Tennant, and C. Transy. 2000. Correct binding of viral X protein to UVDDB-p127 cellular protein is critical for efficient infection by hepatitis B viruses. Oncogene 19:4427-4431. [DOI] [PubMed] [Google Scholar]
- 40.Sitterlin, D., F. Bergametti, and C. Transy. 2000. UVDDB p127-binding modulates activities and intracellular distribution of hepatitis B virus X protein. Oncogene 19:4417-4426. [DOI] [PubMed] [Google Scholar]
- 41.Sitterlin, D., T. H. Lee, S. Prigent, P. Tiollais, J. S. Butel, and C. Transy. 1997. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J. Virol. 71:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takao, M., M. Abramic, M. Moos, Jr., V. R. Otrin, J. C. Wootton, M. McLenigan, A. S. Levine, and M. Protic. 1993. A 127 kDa component of a UV-damaged DNA-binding complex, which is defective in some xeroderma pigmentosum group E patients, is homologous to a slime mold protein. Nucleic Acids Res. 21:4111-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang, J. Y., B. J. Hwang, J. M. Ford, P. C. Hanawalt, and G. Chu. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 5:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wakasugi, M., A. Kawashima, H. Morioka, S. Linn, A. Sancar, T. Mori, O. Nikaido, and T. Matsunaga. 2002. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 277:1637-1640. [DOI] [PubMed] [Google Scholar]
- 46.Zolezzi, F., and S. Linn. 2000. Studies of the murine DDB1 and DDB2 genes. Gene 245:151-159. [DOI] [PubMed] [Google Scholar]
- 47.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]