Abstract
The herpes simplex virus type 1 (HSV-1) UL6, UL15, and UL28 proteins are essential for cleavage of replicated concatemeric viral DNA into unit length genomes and their packaging into a preformed icosahedral capsid known as the procapsid. The capsid-associated UL6 DNA-packaging protein is located at a single vertex and is thought to form the portal through which the genome enters the procapsid. The UL15 protein interacts with the UL28 protein, and both are strong candidates for subunits of the viral terminase, a key component of the molecular motor that drives the DNA into the capsid. To investigate the association of the UL6 protein with the UL15 and UL28 proteins, the three proteins were produced in large amounts in insect cells with the baculovirus expression system. Interactions between UL6 and UL28 and between UL6 and UL15 were identified by an immunoprecipitation assay. These results were confirmed by transiently expressing wild-type and mutant proteins in mammalian cells and monitoring their distribution by immunofluorescence. In cells expressing the single proteins, UL6 and UL15 were concentrated in the nuclei whereas UL28 was found in the cytoplasm. When the UL6 and UL28 proteins were coexpressed, UL28 was redistributed to the nuclei, where it colocalized with UL6. In cells producing either of two cytoplasmic UL6 mutant proteins and a functional epitope-tagged form of UL15, the UL15 protein was concentrated with the mutant UL6 protein in the cytoplasm. These observed interactions of UL6 with UL15 and UL28 are likely to be of major importance in establishing a functional DNA-packaging complex at the portal vertex of the HSV-1 capsid.
Herpes simplex virus type 1 (HSV-1) DNA is packaged into a preassembled icosahedral capsid, referred to as the procapsid, within the cell nucleus (28, 29, 31). During this process, the replicated concatemeric viral DNA is cleaved into unit length genomes, the internal viral scaffolding proteins occupying the cavity of the procapsid are removed, and the procapsid undergoes a rapid transition into a stable, more angularized capsid (35, 38, 46). Seven HSV-1 DNA-packaging proteins, encoded by the UL6, UL15, UL17, UL25, UL28, UL32, and UL33 genes, have been identified through genetic analysis (2, 3, 5-7, 12, 22, 26, 33, 34, 40, 43, 45). Of these proteins, only the UL6, UL17, and UL25 products are readily detectable in the DNA-containing capsid and are also constituents of the mature virion (15, 26, 32, 42). Recent results have revealed that the purified UL6 product forms a dodecameric ring that is remarkably similar in appearance to the complexes of the portal or connector protein from double-stranded DNA bacteriophage such as φ29, T4, and P22 (30, 47). These connectors all have radial projections around a central channel through which the viral DNA is inserted into the capsid. Like the bacteriophage connectors, the UL6 protein is localized at a single vertex on the capsid. These similarities with the bacteriophage connectors strongly suggest that the UL6 protein forms the portal vertex on the procapsid, which is the docking site for the other components of the DNA-packaging machinery and the viral genome.
In contrast to the UL6 product, the UL15 and UL28 packaging proteins are only transiently associated with the capsid and are found in reduced amounts in DNA-containing capsids in comparison with the levels in procapsids (42, 50). The incorporation of UL15 in angularized scaffold-containing (B) capsids, which are dead-end products, is dependent on the presence of UL28, suggesting that these two proteins interact (50). In addition, indirect evidence of an interaction between homologues of UL15 and UL28 was obtained from studies on inhibitors of human or murine cytomegalovirus DNA packaging and cleavage, which found that drug resistance mutations occurred in either UL15 or UL28 homologues (10, 20). The findings that UL15 copurified with UL28 from virus-infected cells, redirected the normally cytoplasmic UL28 protein to the nuclei in cells transiently expressing both proteins, and was efficiently coimmunoprecipitated with UL28 confirmed that the two proteins form a complex (1, 18, 19). Evidence is accumulating that this complex is probably analogous to the terminase of double-stranded DNA bacteriophage. This enzyme, which usually consists of two subunits, binds to specific sequences on the bacteriophage genome and interacts with the connector, delivering the DNA to the site of packaging. The terminase initiates the translocation of DNA into the capsid by cleaving replicated concatemeric viral DNA at a specific site and provides energy for the packaging process through the hydrolysis of ATP. It terminates the process by a second cleavage event. The terminase and the connector are the key components of a very powerful molecular motor for pumping the viral genome into the capsid (11, 14). The first clue that UL15 might be a component of the terminase came from protein homology searches that identified a relationship between the channel catfish herpesvirus homologue of UL15 protein and bacteriophage T4 terminase subunit gp17 (13). The homology was highest in regions of gp17 that bound ATP, and mutation of one of the binding sites inactivated the function of UL15 (49). Although no ATPase activity has been reported for UL15 or its homologues, a recent report claimed that the human cytomegalovirus (HCMV) UL28 homologue is an ATPase whose activity is enhanced by the UL15 homologue (16). The finding that purified UL28 binds to the pacI motif within the HSV-1 packaging and cleavage signals of the “a” sequence provides further evidence that this protein is a terminase subunit (4). The HCMV homologue, UL56, has also been shown to interact with the corresponding HCMV DNA sequence (9). Recently, purified HCMV UL56 and the UL15 homologue have been reported to have nonspecific nuclease activity in vitro that is enhanced when both proteins are present (41).
In the present report, we provide further evidence in support of the idea that UL15 and UL28 are terminase subunits by showing that these proteins interact with the UL6 portal protein.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Spodoptera frugiperda strain IPLB-SF-21 (Sf) cells (17) were cultured in TC100 medium supplemented with 5% fetal calf serum and the above antibiotics. The HSV-1 UL6 null mutant lacZ-UL6− was propagated on a transformed Vero cell line, G33, and the UL28 null mutant gCB was grown in Vero cell-derived C1 cells (33, 45). The recombinant baculoviruses AcUL6 and AcUL6AU1, expressing the UL6 product and AU1 epitope-tagged UL6, respectively, were produced by recombination of the baculovirus transfer vector containing the HSV-1 gene with Bsu36I-cleaved baculovirus AcPAK6 DNA essentially as described by Kitts et al. (17). The construction of the AcUL28 and AcUL15pp65 baculoviruses expressing the UL28 protein and an epitope-tagged form of the UL15 product, respectively, was described by Abbotts et al. (1).
Plasmids.
The HSV-1 DNA-packaging genes were placed under the control of the major HCMV immediate-early promoter in plasmid pCMV10 for transient expression in mammalian cells (44). The construction of pAS30, containing full-length UL6 in pCMV10, has been described previously by Patel et al. (33). The UL6 open reading frame (ORF) was removed from pAS30 in two pieces, a BamHI-SstI fragment and an SstI-PstI fragment, and ligated to baculovirus transfer vector pAcCL29 cleaved with BamHI and PstI (23). The AU1 epitope-tagged UL6 mammalian expression construct was generated in two stages. Plasmid pAS30 was digested with BamHI and HindIII, and the pCMV10 fragment was purified. This vector was ligated to the BamHI-SalI fragment from the 5′ end of the UL6 ORF and to a short SalI-HindIII double-stranded oligonucleotide. This oligonucleotide contained the codons for the AU1 epitope (TYRYI) inserted between codons 379 and 380 of UL6 and the KpnI sequence spanning codons 380 and 381. The full-length UL6 ORF was regenerated by inserting the truncated mutated UL6 gene as a KpnI fragment into pAS30 cleaved with KpnI to produce plasmid pUL6in379E (Table 1). A recombinant baculovirus transfer vector containing the AU1 epitope-tagged UL6 gene was constructed by partially digesting pAS30AU1 with HindIII, treating the linear partials with T4 polymerase in the presence of deoxyribonucleotides, and ligating the blunt-ended DNA to a BamHI oligonucleotide linker. The UL6 gene was removed from this plasmid by digestion with BamHI and inserted into BamHI-digested pAcCL29.1. The mutant proteins UL6in161, UL6in269, UL6in368, and UL6in378 (Table 1) were constructed by ligating a self-annealing oligonucleotide, 5′ CGCGAGATCTCG 3′, containing an internal BglII site to linear partials of pAS30 cleaved with TaqI (UL6in161, UL6in269, and UL6in378) or MspI (UL6in368). The pCMV10 derivative containing UL28 (pUL28) has been previously described (1). Plasmid pUL28-c-myc was derived from pUL28Δ3 (1), which was cleaved at the unique XmaI site, treated with calf intestinal phosphatase, and ligated to an annealed phosphorylated oligonucleotide specifying the c-myc epitope and containing an internal BglII site (refer to Table 2). In the resulting protein, amino acids 466 to 476 of UL28 are replaced by the sequence EQKLISEEDL. The UL15 protein linked to the HCMV pp65 tag (ERKTPRVTGG) at its C terminus was expressed from plasmid pJM19 in mammalian cells and by the recombinant baculovirus AcUL15pp65 as described previously (1).
TABLE 1.
Complementation of UL6 null mutant by HSV-1 UL6 insertional mutants
| Plasmid | UL6 amino acid change (from/to) | Localizationb | Virus titer (PFU/ml)c |
|---|---|---|---|
| pCMV10 | <20 | ||
| pAS30 | Nuclear | 1.0 × 106 | |
| pUL6in161 | 161I-162D/I-ARSR-D | Cytoplasmic | <10 |
| pUL6in269 | 269F-D270/F-ARSR-D | Cytoplasmic | <10 |
| pUL6in368 | 368A-369G/A-ARSR-G | Nuclear | <10 |
| pUL6in378 | 378V-379D/DV-ARSR-D | Nuclear | 8.6 × 106 |
| pUL6in379Ea | 379D-380G/D-TYRYI-G | Nuclear | 8.8 × 106 |
This plasmid contains the coding sequences for the AU1 epitope (amino acids TYRYI) inserted into the UL6 ORF.
The distribution of the transiently expressed protein in Vero cells was determined by using an indirect immunofluorescence assay.
Vero cells were transfected with plasmid DNA and subsequently infected with the UL6 null mutant. The virus yield was determined on the complementing cell line.
TABLE 2.
Complementation of the UL28 null mutant by HSV-1 UL28c-myc
| Plasmid | UL28 amino acid change (from/to) | Virus titer (PFU/ml)a |
|---|---|---|
| pCMV10 | <10 | |
| pUL28 | 3.0 × 104 | |
| pUL28-c-myc | 465G-GGGEDEDRRRG-P/ G-EQKLISEEDL-Pb | 1.5 × 104 |
Vero cells were transfected with plasmid DNA and subsequently infected with the UL28 null mutant. The virus yield was determined on the complementing cell line.
Amino acids 465 to 477 were replaced by 10 amino acids specifying c-myc epitope.
Antibodies.
Two rabbit polyclonal antibodies, YE583 and R123, which are specific for the UL6 and UL28 proteins, respectively, have been described previously (1, 33). Commercial purified mouse monoclonal antibodies reactive to the AU1 epitope tag (BABCO), the c-myc tag (Calbiochem), the HCMV pp65 epitope tag (Capricorn), or the six-His tag (Sigma) were also used.
Immunoprecipitation assays.
Immunoprecipitation assays were carried out essentially as described by McLean et al. (25). Sf cell monolayers (1.2 × 106 cells per 22-mm-diameter tissue culture well) were infected in duplicate with recombinant baculoviruses at 5 PFU per cell, and one set was labeled with l-[35S]methionine at 24 h postinfection. Soluble extracts (150 μl per sample) were prepared at 42 h postinfection with ice-cold EZ buffer (100 mM Tris HCl [pH 8], 100 mM KCl, 1% NP-40, 0.5% sodium deoxycholate, 10 μM zinc acetate, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, 0.001 mM leupeptin, 0.001 mM pepstatin A) and clarified by centrifugation at 44,000 × g for 20 min at 4°C. A sample of supernatant (135 μl) was incubated with the relevant antibodies for 1.5 h at 4°C. The complexes were collected on protein A-Sepharose beads and washed with ice-cold EZ buffer containing 1% Tween 20, and the proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Gels containing [35S]methionine-labeled proteins were dried and analyzed with a phosphorimager. To detect unlabeled proteins, Western blot analysis was performed by the enhanced chemiluminescence method based on the luminol detection system (Amersham).
Immunofluorescence assays.
Vero cells were seeded onto glass coverslips in Linbro wells at 0.4 × 105 cells per coverslip (13-mm diameter). Approximately 200 ng of plasmid DNA was transfected into cells with Lipofectamine reagent (GIBCO-Invitrogen Corporation). At 12 to 16 h posttransfection, the cells were fixed with 5% formaldehyde in phosphate-buffered saline (PBS) containing 2% sucrose and subsequently permeabilized with 0.5% NP-40 in PBS containing 10% sucrose. Primary antibodies were diluted in PBS containing 1% fetal calf serum (PBSF). After 1 h of incubation at room temperature, the coverslips were washed three to six times with PBSF prior to treatment with fluorescein isothiocyanate (FITC)-conjugated sheep anti-rabbit immunoglobulin G (IgG; Sigma) and Cy5-conjugated goat anti-mouse IgG (Amersham), diluted 1/200 and 1/500, respectively. Following a 45-min incubation at room temperature, the coverslips were washed with PBSF and mounted with Mowiol 4-88 (Hoechst) or AF1 (Citifluor). The coverslips were examined with a Zeiss LSM 510 confocal microscope system with a Zeiss Axioplan 63× oil immersion objective lens and two lasers with excitation wavelengths of 488 and 633 nm. The two channels were scanned individually with the same settings maintained throughout. Captured images were exported and compiled with Photoshop version 5.5.
Transient-complementation yield assay.
BHK cell monolayers were transfected with UL28 constructs by the calcium phosphate procedure as described by Abbotts et al. (1) or, in the case of the UL6 plasmids, transfected with Lipofectin reagent essentially as outlined by Preston et al. (36). The cells were infected with 5 PFU of gCB virus per cell at 6 h posttransfection (for the UL28 constructs) or with 1 to 2 PFU of lacZ-UL6− per cell at 24 h posttransfection (for the UL6 constructs). After 1 h of incubation at 37°C, unabsorbed virus was inactivated with an acid glycine wash (39) and the cells were overlaid with tissue culture medium. Following incubation at 37°C for 18 to 24 h, the cells were harvested and the virus yield was determined on complementing cell lines G33 (for UL6 null mutant infections) and C1 (for UL28 null mutant infections).
RESULTS
Transient-complementation studies.
To study the interaction of UL6 with the putative terminase subunits UL15 and UL28, a variety of constructs were used, including epitope-tagged forms of UL6, UL15, and UL28 and three other insertional mutant forms of UL6. The latter UL6 mutant proteins were selected from a panel of mutant proteins generated by linker-scanning mutagenesis in which four amino acids were inserted in frame at commonly occurring restriction enzyme sites. Upon initial characterization of these mutant proteins by immunofluorescence, two UL6 proteins, one carrying the insertion at amino acid 161 (UL6in161) and the other carrying the insertion at amino acid 269 (UL6in269), had a cytoplasmic distribution when transiently expressed in cells, in contrast to the UL6 wild-type (wt) product, which localized to the nuclei (33). However, like wt UL6, two mutant proteins, UL6in368 and UL6in378, that had insertions at amino acids 368 and 378, respectively, were concentrated in the cell nuclei.
The UL6 mutant proteins were screened for the ability to complement the UL6 null mutant, lacZ-UL6−, in a transient-complementation assay. Nonpermissive Vero cells were transfected with the mammalian expression plasmid encoding the wt or mutated UL6 gene and subsequently infected with lacZ-UL6−. Following incubation for 18 to 24 h, the virus yield was determined on permissive G33 cells. As expected, the wt UL6 protein consistently complemented the mutant virus whereas mutant proteins UL6in161, UL6in269, and UL6in368 failed to do so. The results of a representative experiment are given in Table 1. Interestingly, the UL6in378 mutant virus efficiently complemented the null mutant. For this reason, we inserted an oligonucleotide encoding five amino acids (TYRYI) that specifies the epitope recognized by the AU1 monoclonal antibody in frame at position D379. This epitope-tagged UL6 protein also complemented the lacZ-UL6− virus to wt levels in the transient-complementation assay, indicating that the insertion at amino acid 379 does not impair the function of the UL6 protein (Table 1).
Previous work had shown that epitope-tagged UL15 protein UL15pp65 complemented the UL15 null mutant as efficiently as the wt product in a transient-complementation assay (1). A similar experiment was carried out to ensure that the epitope-tagged version of the UL28 protein was also functional. Positive complementation values were consistently obtained, and the results of a representative experiment are shown in Table 2. These findings demonstrate that the c-myc tag in the UL28 protein (plasmid pUL28-c-myc) did not have a significant effect on the activity of the protein. The levels of virus revertants in the experiments shown in Tables 1 and 2 were negligible.
Interaction of UL6 with the putative terminase subunits.
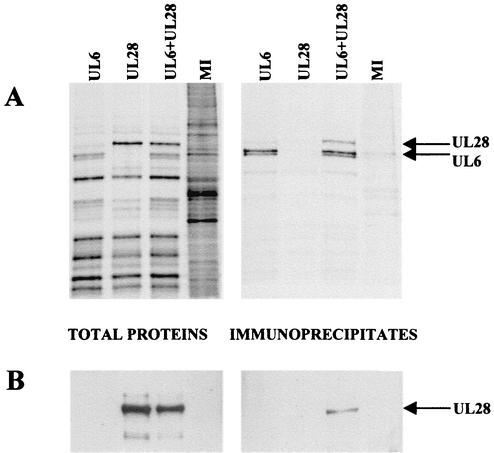
Large amounts of the UL6, UL15, and UL28 proteins were produced in insect cells infected with baculoviruses containing the HSV genes under the control of the polyhedrin promoter. In each case, the major recombinant protein product had an apparent molecular weight similar to that predicted on the basis of its ORF (24). In contrast to the UL15 and UL28 abundant products, the UL6 protein was consistently resolved into two bands on an SDS-polyacrylamide gel (Fig. 1, lane 1). Since a significant portion of each of the HSV-1 proteins was soluble, the baculovirus expression system was used to determine whether there was an interaction between UL6 and UL28 and similarly between UL6 and UL15 in an immunoprecipitation assay. Initially, soluble radiolabeled cell extracts were prepared from Sf cells singly infected with AcUL6 and AcUL28 or coinfected with both viruses. The extracts were incubated with anti-UL6 antibody YE583, and immune precipitates were purified. Total cell protein and immune precipitates were analyzed by SDS-PAGE. Figure 1A shows that UL6 was precipitated from the extract of cells infected with AcUL6 and the extract infected with both AcUL6 and AcUL28, while a protein band of the size of the UL28 product was only detectable in the extract containing both UL6 and UL28. The identity of the UL28 protein band was confirmed by Western blotting with anti-UL28 antibody R123 (Fig. 1B). Thus, coprecipitation of UL28 by anti-UL6 antibody was dependent on the presence of UL6.
FIG. 1.
Coimmunoprecipitation of UL6 and UL28. Sf cells were infected with AcUL6, AcUL28, or the two viruses together, and the virus-infected cell polypeptides were labeled with [35S]methionine. Cell extracts were incubated with anti-UL6 antibody (YE583), and the immunoprecipitates, along with samples of total cell proteins, were analyzed by SDS-PAGE. The radioactive protein bands were detected in the dried gel by phosphorimager analysis (A). The positions of UL6 and UL28 are indicated. In a separate polyacrylamide gel, the proteins were transferred to a nitrocellulose membrane and the UL28 protein was detected with antibody R123 by Western blot analysis (B). MI, mock-infected cell extract.
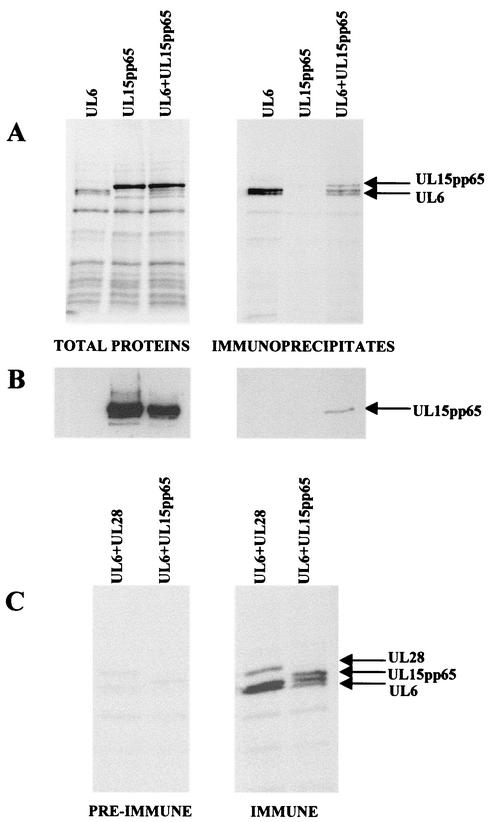
Similar experiments were done with extracts prepared from recombinant baculovirus-infected cells expressing UL6, UL15pp65, or both proteins. A functional epitope-tagged version of UL15 was expressed instead of the wt protein because of the lack of a suitable UL15-specific polyclonal antibody. The extracts were incubated with anti-UL6 antibody YE583, and the immune precipitates were analyzed by SDS-PAGE and subsequent Western blotting with anti-pp65 antibody. Figure 2A shows that UL6 was precipitated from the AcUL6-infected cell extract and the extract of cells infected with both AcUL6 and AcUL15pp65, whereas a band of the size of UL15 was only precipitated from cells coinfected with both viruses. The identity of this putative UL15 protein band was confirmed by Western blotting analysis (Fig. 2B).
FIG. 2.
Coimmunoprecipitation of UL6 and UL15. Sf cells were infected with AcUL6, AcUL15pp65, or the two viruses together, and the virus-infected cell polypeptides were labeled with [35S]methionine. The cell extracts were incubated with anti-UL6 antibody (YE583), and the immunoprecipitates, along with the total extracts, were analyzed by SDS-PAGE. The radioactive protein bands were detected in the dried gel with a phosphorimager (A). In a separate gel, the proteins were transferred to a nitrocellulose membrane and the UL15pp65 protein was detected with anti-pp65 antibody by Western blot analysis (B). The complexes precipitated from extracts of cells coinfected with AcUL6 and AcUL15pp65 or AcUL6 and AcUL28, as indicated, with UL6 immune or preimmune serum are shown in panel C.
Preimmune serum from the animal used to generate the anti-UL6 serum, YE583, was similarly tested for the ability to precipitate complexes from cells coexpressing UL6 and UL15 or UL6 and UL28. Figure 2C shows that, in comparison to YE583, only very small amounts of the three proteins were detected in the proteins precipitated by the preimmune serum. In addition, it can be seen that other background bands were present at similar levels with the two sera, suggesting that nonspecific trapping of proteins in the UL6-containing complexes does not occur to any significant extent. Similar experiments were also performed with cells coinfected with an AU1 epitope-tagged form of UL6, in this case with a mouse monoclonal antibody to the AU1 epitope. Again, both UL15 and UL28 coprecipitated with UL6, indicating that the results are not due to artifacts associated with the YE583 serum (results not shown).
To exclude the possibility that the interaction of UL6 with UL15 and UL28 might occur indirectly through binding to DNA or RNA, the effects of DNase I, RNase, and ethidium bromide on the immunoprecipitated complexes formed with YE583 were examined. Complexes collected on protein A Sepharose beads were resuspended in 10 mM Tris-HCl (pH 7.5)-10 mM KCl-1.5 mM MgCl2 and incubated for 15 min at 37°C in 50 U of DNase I per ml-5 μg of RNase A per ml plus 50 U of RNase T1 per ml or 50 μg of ethidium bromide per ml (21). The beads were spun down and washed twice with EZ buffer prior to gel electrophoresis. None of the treatments reduced the amounts of UL15 or UL28 coprecipitated with UL6 (data not shown), suggesting that DNA or RNA molecules are unlikely to act as bridges between protein molecules.
Taken together, the results of the immunoprecipitation experiments demonstrate that, in the absence of other HSV-1 proteins, UL6 can establish direct and specific protein-protein interactions with both UL15 and UL28.
Colocalization of UL6 with UL28.
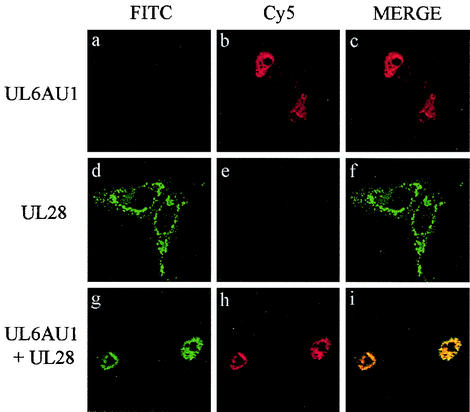
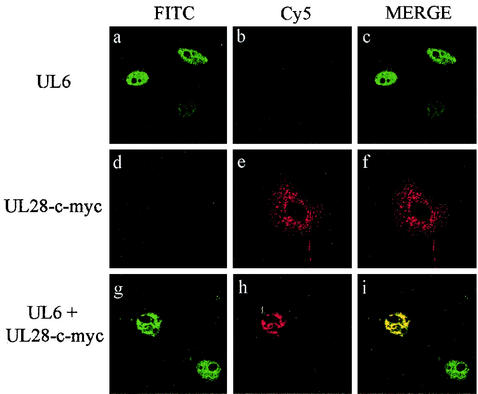
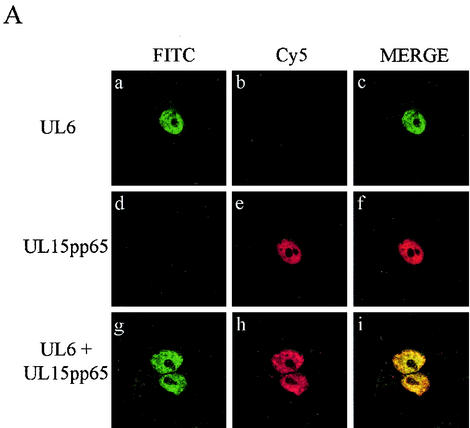
The immunoprecipitation results strongly suggested that both UL15 and UL28 interacted with UL6. To extend these findings, the HSV-1 proteins were transiently expressed in mammalian cells and their intracellular distribution was determined with an indirect immunofluorescence assay. In order to identify two different proteins by this method, the primary antibodies have to be derived from different species. Since only rabbit polyclonal antibodies, specific for the packaging proteins, were available, epitope tags (for which commercial mouse monoclonal antibodies were obtainable) were inserted into UL15 and UL28. Vero cells were transfected with pAS30AU1 (encoding an AU1 epitope-tagged form of UL6) and pUL28 either separately or in combination. The cells were fixed and stained with a mixture of anti-AU1 and anti-UL28 (R123) antibodies, followed by a mixture of Cy5- and FITC-conjugated secondary antibodies. Figure 3 shows that UL6AU1 was concentrated in the nuclei when expressed alone (Fig. 3a to c) whereas UL28 was localized in the cytoplasm (Fig. 3d to f). In cotransfected cells, both proteins were present in the nuclei, where they exhibited extensive colocalization (Fig. 3g to i). Similar findings were obtained when the experiment was repeated with wt UL6 protein and an epitope-tagged form of UL28, UL28-c-myc (Fig. 4). Cells expressing the UL28-c-myc protein on its own, like those containing the wt protein, had speckles and larger areas of immunofluorescence throughout the cytoplasm (Fig. 4d to f). By contrast, in the presence of UL6, UL28-c-myc was almost exclusively concentrated in the nuclei along with UL6 (Fig. 4g to i).
FIG. 3.
Intracellular colocalization of UL6AU1 and UL28. Vero cells were transfected with pAS30AU1 expressing UL6AU1 (panels a to c), pUL28 expressing UL28 (panels d to f), or both plasmids together (panels g to i). The proteins were detected by an indirect immunofluorescence assay with the AU1 mouse monoclonal antibody, specific for UL6AU1, and the R123 polyclonal rabbit antibody, specific for UL28, as primary antibodies and Cy5-conjugated anti-mouse IgG and FITC-conjugated anti-rabbit IgG as secondary antibodies. The cells were examined by confocal microscopy. Each set of three digital confocal images (from left to right) shows the same field of cells, with FITC fluorescence in green, Cy5 fluorescence in red, and a merged image of the two.
FIG. 4.
Intracellular colocalization of UL6 and UL28-c-myc. The digital confocal images show transfected Vero cells expressing UL6 (panels a to c), UL28-c-myc (panels d to f), and UL6 and UL28-c-myc together (panels g to i). The HSV-1 proteins were detected by treating cells with YE583 polyclonal rabbit antibody (specific for UL6) and c-myc mouse monoclonal antibody (specific for UL28-c-myc) and then incubating them with FITC-conjugated anti-rabbit IgG and Cy5-conjugated anti-mouse IgG. Each set of three panels shows FITC staining of UL6 (left), Cy5 staining of UL28-c-myc (middle), and a merged image of the two for the same field of transfected cells (right). Note that in panels g to i, only one of the two UL6-expressing cells contains UL28-c-myc.
Intracellular distribution of UL6 insertional mutant proteins and their association with UL15.
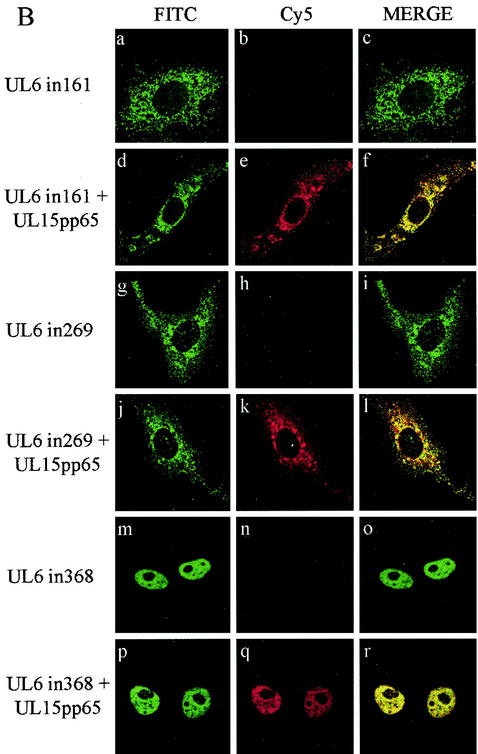
To examine the localization of UL6 and UL15pp65, the proteins were transiently expressed in Vero cells separately or together. The cells were fixed, stained with anti-UL6 rabbit antibodies (YE583) and HCMV pp65 mouse monoclonal antibody, and subsequently treated with a mixture of Cy5- and FITC-conjugated secondary antibodies. Both UL6 and UL15pp65 were concentrated in the nuclei of cells when expressed on their own as described previously (1, 33) (Fig. 5A, a to f). Since these products also had the same nuclear distribution when they were coexpressed in cells, it was difficult to establish whether a true interaction was occurring between the proteins (Fig. 5A, g to i).
FIG. 5.
Intracellular distribution of wt UL6, mutant UL6, and UL15pp65. (A). Digital images of transfected Vero cells expressing wt UL6 (a to c), UL15pp65 (d to f), or a mixture of the two proteins (g to i). (B). Images of transfected Vero cells expressing mutant protein UL6in161 alone (a to c) or with UL15pp65 (d to f), UL6in269 alone (g to i) or with UL15pp65 (j to l), and UL6in368 alone (m to o) or withUL15pp65 (p to r). The expressed proteins were detected by confocal microscopy with polyclonal rabbit antibody YE583 (specific for UL6) and mouse monoclonal antibody pp65 (specific for UL15pp65) as primary antibodies, followed by FITC-conjugated anti-rabbit IgG and Cy5-conjugated anti-mouse IgG. The sets of three panels show FITC staining of wt or mutant UL6 protein, Cy5 staining of UL15pp65, and a merged image of the same field of cells.
To investigate the interaction of UL6 with the UL15 protein by immunofluorescence, two cytoplasmic UL6 mutant proteins, UL6in161 and UL6in269, were examined. Another nonfunctional insertional mutant, UL6in368, which has a nuclear localization, was included as a control in addition to the wt protein. In transfected cells, UL6in161 had a pattern of distribution similar to that of UL6in269 when expressed alone, with large aggregates of protein accumulating in the cytoplasm (Fig. 5B, a to c and g to i). When the cells were transfected with either of the cytoplasmic UL6 mutant proteins together with UL15pp65, the intracellular location of UL15pp65 became cytoplasmic (Fig. 5B, d to f and j to l). The patterns of immunofluorescence were very similar for UL15pp65 and UL6 in these cells, with UL6 mutant protein and UL15pp65 colocalizing in distinct cytoplasmic aggregates, often concentrated around the nuclei. The behavior of the UL6 insertional mutant form UL6in368, when coexpressed with UL15pp65, was similar to that of the wt UL6 protein, and both proteins had a nuclear distribution (Fig. 5B, p to r). The ability of UL6in161 and UL6in269 to alter the intracellular localization of UL15pp65 therefore supports the immunoprecipitation data indicating that UL6 and UL15 interact.
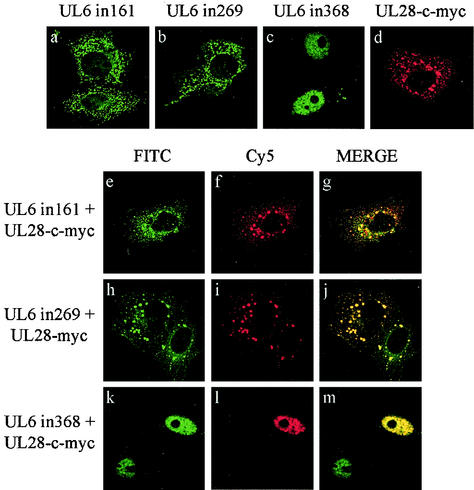
Colocalization of UL28 with UL6 insertional mutant proteins.
Immunoprecipitation and immunofluorescence experiments suggested that UL6 interacted with UL28. Additional work was carried out to determine whether the three nonfunctional UL6 insertional mutant proteins described above affected the distribution of UL28 in transfected cells. Figure 6 indicates that mutant proteins UL6in161 and UL6in269 formed distinct aggregates in the cytoplasm that showed a high degree of colocalization with those of UL28 (Fig. 6e to g and h to j). The other UL6 mutant form, UL6in368, retained the ability to redistribute UL28 to the nucleus and in this respect behaved similarly to the wt protein (Fig. 6k to m). Thus, the insertion of four amino acids between amino acids 368 and 369 did not appear to disrupt the ability of UL6 to interact with UL28. In addition, the strong colocalization of the two cytoplasmic UL6 mutant proteins with UL28 suggests that UL6 containing an insertion of four amino acids after either amino acid residue 161 or 269 probably retains its ability to associate with UL28.
FIG. 6.
Intracellular localization of UL6 mutant proteins and UL28. Digital confocal images of transfected Vero cells expressing UL6in161, UL6in269, or UL6in368 alone or with UL28-c-myc. The HSV proteins were detected by confocal microscopy with YE583, which is specific for UL6, and anti-c-myc, which is specific for UL28-c-myc, as primary antibodies and FITC-conjugated anti-rabbit IgG and Cy5-conjugated anti-mouse IgG as secondary antibodies. The top row (a to d) shows merged FITC and Cy5 fluorescent images of the HSV protein expressed on its own. The remainder of the figure shows individual FITC or Cy5 staining and a merged image for UL6in161 and UL28-c-myc (e to g), UL6in269 and UL28-c-myc (h to j), and UL6in368 and UL28-c-myc (k to m). Note that in panels k to m, only one of the UL6-expressing cells contains UL28-c-myc.
DISCUSSION
This paper presents the first description of an interaction between UL6 and the putative terminase subunits UL15 and UL28. Two independent approaches were used to investigate the association of these proteins in the absence of other HSV-1 polypeptides. In the immunoprecipitation assay, the interaction between soluble HSV proteins in recombinant baculovirus-infected cell extracts was examined, and in the immunofluorescence assay, the localization of HSV proteins within the transfected cell was determined. The results of these assays provided compelling evidence that UL6 can interact with both UL15 and UL28. In the immunofluorescence assay, we found that UL6 directly affected the localization of UL28. Our results suggest that UL6 may have a role in transporting UL28 to the nucleus. Previous studies have shown that, in transfected cells, UL15 also has the ability to relocate UL28 to the nucleus (1, 18, 19). The observation that UL28 is present in B capsids purified from cells infected with the HSV-1 UL15 or UL6 null mutant is consistent with idea that there is more than one pathway for the transport of UL28 into the nucleus (50). It is also possible that a complex of all three proteins normally forms prior to capsid assembly and that the portal is incorporated into the capsid with other components of the packaging machinery present. In this context, it is interesting that angularized procapsids with cleaved internal scaffolds retain the UL15 and UL28 proteins even though these capsids are incapable of packaging viral DNA (50).
In contrast to our data, a recent report stated that UL6 was not detectable in immunoprecipitates of UL15 and UL28 from HSV-1-infected cells (8). Although it is difficult to make direct comparisons of the experimental systems used, a possible explanation for the apparent discrepancy is that such a complex of UL6, UL15, and UL28 might be rapidly incorporated into the capsid and hence be unavailable for precipitation from HSV-1-infected cells. Alternatively, the relative affinities of the UL6, UL15, and UL28 proteins could be altered in the presence of replicated viral DNA containing packaging signals and other components of the packaging machinery, or relevant epitopes could become masked.
Although we were unable to demonstrate that wt UL6 interacted with UL15 by using the immunofluorescence assay because both proteins were nuclear, we observed in cell nuclei that contained localized aggregates of wt UL6 that UL15 was also concentrated in these regions. The ability of the two cytoplasmic UL6 mutant proteins to prevent UL15 from migrating to the nucleus, however, provided confirmation of the findings from the immunoprecipitation experiments that the two proteins interact. Recent findings by Newcomb and Brown support this conclusion (27). They found that the inhibitor WAY-150138 reduced the incorporation of both UL6 and UL15 into capsids in virus-infected cells but concluded that the drug probably directly affected UL6 rather than UL15 since only UL6 drug-resistant mutant proteins had been isolated (27, 48). It would be interesting to investigate whether the inhibitor also affected the amount of UL28 in capsids. The finding that UL15 interacts with UL6 is also consistent with an earlier report by Yu and Weller (50) that UL15 is present in reduced amounts in UL6-negative B capsids in comparison to the levels in wt capsids. Interestingly, the levels of UL28 were similar in wt, UL6-negative, and UL15-negative capsids (50). It is unclear whether the presence of UL28 in UL6-negative capsids is due to an interaction with another DNA-packaging protein with a capsid shell protein or is a nonspecific association with the capsid. The observation that UL28 bound to UL6-negative capsids was resistant to 2 M guanidine hydrochloride treatment favors the possibility of a specific interaction (50).
The interaction of UL15 and UL28 with UL6 provides additional evidence consistent with the idea that UL15 and UL28 are terminase subunits. The form of UL6 precipitated with UL15 and UL28 in our experiments remains to be determined. The centrifugation procedure used to produce the soluble extracts would not be expected to pellet dodecameric UL6 ring structures, and therefore, it is possible that UL15 and UL28 interact with either UL6 monomers or higher structures. Future experiments will aim to characterize these complexes in further detail and also to determine whether the three proteins can assemble into a single structure. Recently, the HCMV homologues of UL15 and UL28 have been reported to form ring-like monomers (41). It would also be interesting to determine (i) whether UL15 and UL28 are required to form ring-like structures in order to interact with UL6, (ii) the subunit composition of any such assemblies, and (iii) whether UL6 can affect the conformation adopted by these proteins. The UL33 protein has recently been found to interact with UL28 and UL15 but is not associated with the capsid (8, 37), suggesting that other factors are also important in the assembly of a functional packaging complex at the portal vertex.
Acknowledgments
We are grateful to Fred Homa for supplying the UL28 null mutant and the complementing cell line. We thank Duncan McGeoch for critically reading the manuscript and Michayla Williams for her valuable contribution to the early stages of the project.
REFERENCES
- 1.Abbotts, A. P., V. G. Preston, M. Hughes, A. H. Patel, and N. D. Stow. 2000. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J. Gen. Virol. 81:2999-3009. [DOI] [PubMed] [Google Scholar]
- 2.Addison, C., F. J. Rixon, J. W. Palfreyman, M. Ohara, and V. G. Preston. 1984. Characterization of a herpes simplex virus type-1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246-259. [DOI] [PubMed] [Google Scholar]
- 3.Addison, C., F. J. Rixon, and V. G. Preston. 1990. Herpes simplex virus type-1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 71:2377-2384. [DOI] [PubMed] [Google Scholar]
- 4.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene-product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 6.Baines, J. D., C. Cunningham, D. Nalwanga, and A. Davison. 1997. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 product. J. Virol. 71:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines, J. D., A. P. W. Poon, J. Rovnak, and B. Roizman. 1994. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol. 68:8118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex type 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buerger, I., J. Reefschlaeger, W. Bender, P. Eckenberg, A. Popp, O. Weber, S. Graeper, H.-D. Klenk, H. Ruebsamen-Waigmann, and S. Hallenberger. 2001. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via UL89 and UL56 gene products. J. Virol. 75:9077-9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham, C., and A. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 13.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 14.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2:537-545. [DOI] [PubMed] [Google Scholar]
- 15.Goshima, F., D. Watanabe, H. Takakuwa, K. Wada, T. Daikoku, M. Yamada, and Y. Nishiyama. 2000. Herpes simplex virus UL17 protein is associated with B capsids and colocalizes with ICP35 and VP5 in infected cells. Arch. Virol. 145:417-426. [DOI] [PubMed] [Google Scholar]
- 16.Hwang, J.-S., and E. Bogner. 2002. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J. Biol. Chem. 277:6943-6948. [DOI] [PubMed] [Google Scholar]
- 17.Kitts, P. A., and R. D. Possee. 1993. A method for producing recombinant baculovirus expression vectors at high frequency. BioTechniques 14:810-817. [PubMed] [Google Scholar]
- 18.Koslowski, K. M., P. R. Shaver, J. T. Casey II, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koslowski, K. M., P. R. Shaver, X.-Y. Wang, D. J. Tenney, and N. E. Pederson. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71:9118-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W.-H. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai, J.-S., and W. Herr. 1992. Ethidium bromide provides a simple test for identifying genuine DNA-dependent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livingstone, C., and I. Jones. 1989. Baculovirus expression vectors with single strand capability. Nucleic Acids Res. 17:2366.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 25.McLean, G. W., A. P. Abbotts, M. E. Parry, H. S. Marsden, and N. D. Stow. 1994. The herpes simplex virus type 1 origin binding protein interacts specifically with the viral UL8 protein. J. Gen. Virol. 75:2699-2706. [DOI] [PubMed] [Google Scholar]
- 26.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newcomb, W. W., and J. C. Brown. 2002. Inhibition of herpes simplex virus replication by WAY-150138: assembly of capsids depleted of the portal and terminase proteins involved in DNA encapsidation. J. Virol. 76:10084-10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 29.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Cheng, A. Steven, F. Booy, and J. C. Brown. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newcomb, W. W., B. L. Trus, N. Cheng, A. C. Steven, A. K. Sheaffer, D. J. Tenney, S. K. Weller, and J. C. Brown. 2000. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J. Virol. 74:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel, A. H., and J. B. Maclean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206:465-478. [DOI] [PubMed] [Google Scholar]
- 33.Patel, A. H., F. J. Rixon, C. Cunningham, and A. J. Davison. 1996. Isolation and characterization of herpes simplex virus type 1 mutants defective in the UL6 gene. Virology 217:111-123. [DOI] [PubMed] [Google Scholar]
- 34.Poon, A. P. W., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preston, V. G., J. A. V. Coates, and F. J. Rixon. 1983. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J. Virol. 45:1056-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preston, V. G., J. Kennard, F. J. Rixon, A. J. Logan, R. W. Mansfield, and I. M. McDougall. 1997. Efficient herpes simplex virus type 1 (HSV-1) capsid formation directed by the varicella-zoster virus scaffolding protein requires the carboxy-terminal sequences from the HSV-1 homologue. J. Gen. Virol. 78:1633-1646. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds, A. E., Y. Fan, and J. D. Baines. 2000. Characterization of the UL33 gene product of herpes simplex virus 1. Virology 266:310-318. [DOI] [PubMed] [Google Scholar]
- 38.Rixon, F. J., and D. McNab. 1999. Packaging-competent capsids of a herpes simplex virus temperature-sensitive mutant have properties similar to those of in vitro-assembled procapsids. J. Virol. 73:5714-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenthal, K. S., M. D. Leuther, and B. G. Banisas. 1984. Herpes simplex virus binding and entry modulate cell surface protein mobility. J. Virol. 49:980-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheffczik, H., C. G. W. Savva, A. Holzenburg, L. Kolesnikova, and E. Bogner. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 30:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stow, N. D., O. Hammarsten, M. I. Arbuckle, and P. Elias. 1993. Inhibition of herpes simplex virus type 1 DNA replication by mutant proteins of the origin-binding protein. Virology 196:413-418. [DOI] [PubMed] [Google Scholar]
- 45.Tengelsen, L. A., N. E. Pederson, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263:447-462. [DOI] [PubMed] [Google Scholar]
- 47.Valpuesta, J. M., and J. L. Carrascosa. 1994. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys. 27:107-155. [DOI] [PubMed] [Google Scholar]
- 48.van Zeijl, M., J. Fairhurst, T. R. Jones, S. K. Vernon, J. Morin, J. LaRocque, B. Feld, B. O'Hara, J. D. Bloom, and S. V. Johann. 2000. Novel class of thiourea compounds that inhibit herpes simplex virus type 1 DNA cleavage and encapsidation: resistance maps to the UL6 gene. J. Virol. 74:9054-9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32-44. [DOI] [PubMed] [Google Scholar]
- 50.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]