Abstract
The complete DNA sequence of herpes B virus (Cercopithecine herpesvirus 1) strain E2490, isolated from a rhesus macaque, was determined. The total genome length is 156,789 bp, with 74.5% G+C composition and overall genome organization characteristic of alphaherpesviruses. The first and last residues of the genome were defined by sequencing the cloned genomic termini. There were six origins of DNA replication in the genome due to tandem duplication of both oriL and oriS regions. Seventy-four genes were identified, and sequence homology to proteins known in herpes simplex viruses (HSVs) was observed in all cases but one. The degree of amino acid identity between B virus and HSV proteins ranged from 26.6% (US5) to 87.7% (US15). Unexpectedly, B virus lacked a homolog of the HSV γ134.5 gene, which encodes a neurovirulence factor. Absence of this gene was verified in two low-passage clinical isolates derived from a rhesus macaque and a zoonotically infected human. This finding suggests that B virus most likely utilizes mechanisms distinct from those of HSV to sustain efficient replication in neuronal cells. Despite the considerable differences in G+C content of the macaque and B virus genes (51% and 74.2%, respectively), codons used by B virus are optimal for the tRNA population of macaque cells. Complete sequence of the B virus genome will certainly facilitate identification of the genetic basis and possible molecular mechanisms of enhanced B virus neurovirulence in humans, which results in an 80% mortality rate following zoonotic infection.
Comparative genome analyses of closely related viruses offer insight into the protein coding capacities of viral genomes (18, 69), a means to biologically classify viruses (2, 19) and identify viral genes involved in virulence and pathogenicity (1, 37, 67). The number of completely sequenced viral genomes has been increasing rapidly and now exceeds 1,000, including 29 herpesvirus genomes (GenBank data, http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/viruses.html).
B virus (Cercopithecine herpesvirus 1, monkey B virus) is a member of the subfamily Alphaherpesvirinae, which together with human herpes simplex virus types 1 and 2 (HSV-1 and HSV-2) constitutes the genus Simplexvirus. B virus generally causes only mild localized or asymptomatic infections in its natural hosts, Asian monkeys of the genus Macaca (33, 34, 74). In contrast, B virus infections in foreign hosts, humans or monkey species other than macaques, often result in encephalitis, encephalomyelitis, and death (53, 73, 74).
The genome organization of B virus is similar to that of HSV-1 and HSV-2: the unique long (UL) and unique short (US) segments flanked by inverted long (RL) and short (RS) repeat sequences are covalently joined in four possible isomeric configurations (26). Sequence analysis of the partial US regions of B virus and simian agent 8 virus demonstrated that human and primate viruses are colinear in this genomic segment (51). However, only a limited number of B virus gene sequences from the UL region have been published (3, 36, 62, 63), and nothing has been reported about the structure and the gene content of the repeat genomic elements.
The size of the B virus genome was estimated previously as 165 kb (26) and 162.5 kb (40), which is significantly larger than the HSV-1 and HSV-2 genomes (152 and 155 kb, respectively). Extra DNA might contain additional genes that are not present in human viruses and may provide insight about B virus pathogenicity in foreign hosts.
In this study, the complete genomic sequence of the B virus reference strain E2490, isolated from a rhesus macaque (Macaca mulatta), was determined, and viral DNA termini were located. The genomic structure and gene content were analyzed and compared to those of human herpesviruses HSV-1 and HSV-2.
MATERIALS AND METHODS
Viruses, cells, and media.
B virus laboratory strain E2490 and two B virus clinical isolates were obtained from the National B Virus Resource Laboratory (Atlanta, Ga.). Viruses were propagated in Vero cells maintained in minimal essential medium (MEM) supplemented with 2% fetal bovine serum (Gibco). Viral DNA was purified from the infected-cell lysate by ultracentrifugation in an NaI density gradient as described previously (70). Escherichia coli Top10 (Invitrogen) cells were used for propagation of plasmids.
DNA cloning and sequencing.
The BamHI, KpnI, SalI, PstI, and XhoI restriction fragments of B virus genomic DNA were cloned into the pUC19 vector digested with the same enzyme. The following three techniques were employed to generate full sequences of the clones: primer walking, unidirectional deletions, and random transposon insertions (Epicentre Technologies).
To obtain the sequence of the IRL-IRS regions, two shotgun libraries of random overlapping clones were prepared from the adjacent PstI fragments P13 and P1, covering this region (Fig. 1). DNA fragments of 13 kb (P13) and 4.8 kb (P1) were isolated after separation of PstI-digested genomic DNA on a 0.8% agarose gel. Each DNA fragment was then treated with DNase I from a DNaseI Shotgun kit (Novagen) according to the manufacturer's suggested protocol. The DNase I-treated fragments were fractionated on a 1.4% agarose gel, and then 300- to 1,000-bp fragments were eluted from the gel, blunt ended with T4 DNA polymerase, dA-tailed with Tth polymerase and a dA tailing kit (Novagen), and ligated into the pScreen T vector (Novagen). The ligation mixtures were transformed into E. coli NovaBlue cells (Novagen).
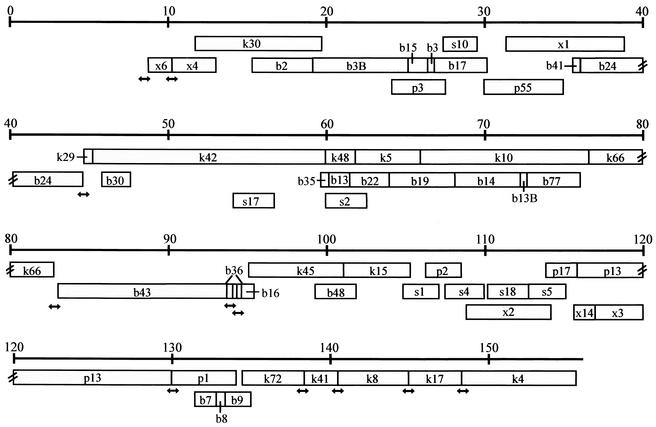
FIG. 1.
Molecular cloning of B virus genome. The first letter in the clone designations indicates the cloning site: B, BamHI; K, KpnI; P, PstI, S, SalI; X, XhoI. The number in the clone designations reflects the order of clone isolation. Double-sided arrows indicate PCR fragments used for verification of clone junctions. The map scale is in kilobases.
DNA sequencing was performed with BigDye Terminator (PE Applied Biosystems) sequencing chemistry on an automatic DNA sequencer ABI 377 (Applied Biosystems). Sequencing reactions were performed in the presence of Sequence Enhancer A (Gibco) to resolve sequence compressions due to the high G+C content of templates. A 3:1 mixture of regular and dGTP BigDye sequencing kits (Applied Biosystems) was used in order to overcome premature termination of sequencing reactions in G-rich regions. DNAStar SeqMan program (DNAStar Inc.) was used to assemble the complete genome sequence from the sequences of the overlapping plasmid clones. For adjacent fragments with no overlap, areas across the junction were amplified with B virus genomic DNA as the template, and the resulting PCR fragments were sequenced.
Cloning of genomic DNA termini.
B virus genomic DNA (≈0.5 μg) was blunt ended by treatment with 5 U of T4 DNA polymerase (Novagen) as suggested by the manufacturer. After purification on a Qiaquick spin column (Qiagen), the blunt-ended DNA was digested overnight with restriction enzyme SphI and ligated into SmaI- and SphI-digested vector pUC19. E. coli NovaBlue (Novagen) competent cells were transformed with the ligation mixture.
Southern blots.
SphI-digested genomic DNA was transferred from a 0.8% agarose gel onto a nylon membrane (Roche) by alkali blotting according to the instructions supplied with the membranes. Plasmid DNA was labeled with digoxigenin with a HighPrime labeling kit (Roche). Prehybridization, hybridization, and washing steps were carried out according to standard protocols (59). Chemiluminescence detection of the bound probe was performed with an ECL detection kit (Amersham Pharmacia Biotech).
DNA sequence analysis.
Identification of open reading frames (ORFs), repeats, and DNA regulatory sequences was performed with DNAStar suite of programs. GenBank database searches were carried out with BlastN, BlastP, and BlastX with default settings. Multiple alignments between B virus, HSV-1, and HSV-2 genes and proteins were performed with and analyzed by DNAStar MegAlign program (version 4.0.3) with the PAM250 amino acid substitution matrix and Joltun Hein method (30) and the following parameters: gap opening penalty = 11 and gap extension penalty = 3. Identity values were determined from the generated alignments.
The GeneMark and GeneMark.hmm gene-finding programs were used to refine the procedure of gene identification (4, 41). The parameters of the statistical models were defined by training on the set of B virus ORFs encoding protein products homologous to known HSV-1 and HSV-2 proteins. These statistical methods are able to identify frameshifts and unusual gene starts in addition to detecting genes not found by similarity search. To prove that a homolog of the HSV neurovirulence factor is absent in the B virus genome, BlastP and PSI-Blast searches with ICP34.5 protein as the query were used against the whole B virus genome translated in six frames. In addition, a Hidden Markov Model profile was created from the conservative domain of the ICP34.5 protein and used to scan the translated B virus genome.
In a search for unique genes, the PSI-Blast and RPS-Blast programs were used to search for possible conserved domains in proteins predicted by statistical methods. Predicted proteins shorter than 20 residues were excluded from the analysis. Analyses of protein structures were performed with the DNAStar Protean program (version 4.0.3), signal peptide prediction program SignalP version 2.0.b2 (http://www.cbs.dtu.dk/services/SignalP-2.0/), and transmembrane prediction programs TMHMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), DAS (http://www.sbc.su.se/≈miklos/DAS/), and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html).
GenBank accession numbers.
The complete sequence of the B virus genome (GenBank accession number AF533768) and the L-terminal sequences of two clinical B virus isolates (GenBank accession numbers AY230747 and AY230748) have been deposited in the NCBI database.
RESULTS AND DISCUSSION
Cloning and sequencing of B virus genome.
The relative positions of all fragments used for B virus genome sequencing are shown in Fig. 1. The UL and US genomic sequences were derived from the nucleotide sequences of the plasmid-cloned BamHI, KpnI, SalI, PstI, and XhoI overlapping genomic fragments. The sequences of the internal copies of the large and small repeat regions (IRL and IRS, respectively) were determined from the nucleotide sequences of short overlapping clones derived from shotgun libraries prepared from the adjacent P13 and P1 genomic fragments. Terminal copies of repeat elements (TRL and TRS) were assumed to be identical to the internal copies, as in the HSV-1 and HSV-2 genomes (18, 43, 44). An assembly of 1,389 single readings resulted in generation of the UL-IRL-IRS-US genomic sequence comprising 139,269 bp, with an average redundancy of 3.9. Exact UL-TRL, UL-IRL, US-IRS, and US-TRS boundaries were determined by sequence comparisons of PCR fragments amplified across these junctions.
Location of B virus genomic termini.
To determine the first and last nucleotides in the B virus genome, genomic termini were defined. Initially, the locations of the internal a sequence and the joint between the B virus long and short internal repeat regions (L/S joint) were estimated after aligning the HSV-1 and B virus IRL-IRS sequences (not shown). Two SphI restriction sites were located approximately 1.8 kb (in the IRL region) and 1.5 kb (in the IRS region) from the predicted internal a sequence. Since the RL and RS internal and terminal copies in alphaherpesvirus genomes are identical, SphI restriction sites were expected to be located ≈1.8 kb and ≈1.5 kb from the B virus L and S genomic termini, respectively, and therefore could be used for cloning the B virus genomic ends.
Blunt-ended B virus genomic DNA was then digested with SphI and ligated into SmaI- and SphI-digested vector pUC19. All internal genomic fragments had SphI sticky ends on both sides, and consequently only terminal genomic fragments, which had one blunt end and another SphI end, were compatible with the prepared vector and successfully cloned by this procedure. As predicted, two sets of recombinant clones were generated with inserts of the estimated sizes. To confirm that the clones isolated included viral DNA ends, plasmid DNA from clones containing 1.5-kb and 1.8-kb inserts was used to probe SphI-digested B virus genomic DNA on Southern blots. Theoretically, if these clones indeed contain genomic ends, they will hybridize to the two SphI genomic fragments of 1.5 kb or 1.8 kb (terminal) and 3.1 kb (internal from the L/S junction). These fragments and two additional 2.1-kb and 3.3-kb fragments of lower intensity were detected (Fig. 2A). The observed cross-hybridization patterns indicated that the two genomic ends had common sequences. The additional fragments are most likely L-terminal (2.1 kb) and junction (3.3 kb) fragments containing an extra copy of a sequence. The variable number of a sequence repeats has also been described at the L terminus and L/S junction in HSV-1 (57).
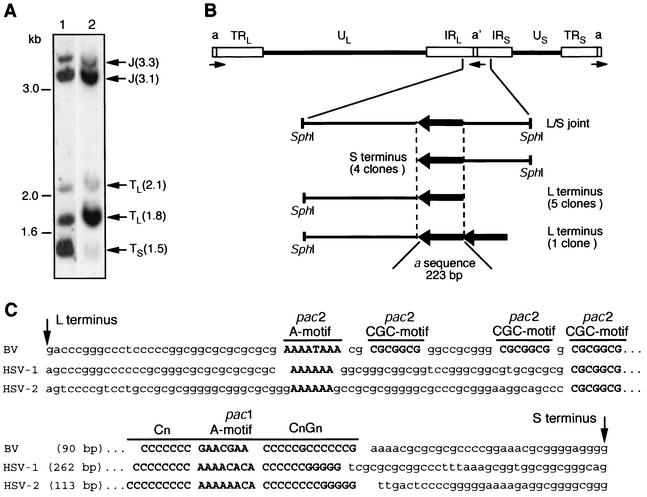
FIG. 2.
Identification of B virus genomic termini. (A) Southern blot of SphI-digested genomic DNA hybridized with digoxigenin-labeled plasmids containing 1.6-kb (lane 1) and 1.8-kb (lane 2) terminal fragments. An autoradiograph of the membranes is shown. The positions of DNA size markers are shown on the left. Arrows indicate terminal small (TS), terminal large (TL), and junction (J) fragments. (B) Cloned genomic fragments containing B virus termini. The structural organization of the B virus genome is shown, with the UL and US regions represented by solid lines and the TRL, IRL, TRS, and IRS regions represented by open boxes. Terminal a sequences and the oppositely oriented internal a′ sequence are indicated. Below the genome diagram, a schematic alignment of the isolated terminal fragments is shown. Arrows denote the locations of a sequence copies and their orientations in the genome. The numbers in parentheses indicate the number of clones sequenced. (C) Alignments of the a sequences of B virus (BV), HSV-1, and HSV-2. Arrows indicate genomic ends. The conserved motifs of pac1 and pac2 signals are shown in bold.
Nucleotide sequences adjacent to the vector cloning site SmaI (genomic termini) were derived from the inserts of 10 selected clones: six clones originated from the large terminal repeat (L-terminal clones) and four originated from the small terminal repeat (S-terminal clones) (Fig. 2B). All contained identical sequences (223-bp long) present in direct orientation on both genomic ends and in reverse orientation at the L/S joint in either one (nine clones) or two (one clone) directly repeated copies. The B virus 223-bp terminal sequence is a homolog of the HSV-1 a sequence that is also present in other herpesvirus genomes. Two cis-acting elements, pac1 and pac2, that are required for cleavage and packaging of herpesvirus genomes exist in the a sequence (17, 68).
Although overall sequence homology between the a sequences of B virus, HSV-1, and HSV-2 was low, the sequence composition of the cleavage and packaging signals was very well conserved (Fig. 2C). An A-rich motif followed by three CGCGGCG motifs formed the B virus pac2 signal. Interestingly, most herpesviruses have only one copy of the CGCGGCG motif, which contributes significantly to the efficiency of genome cleavage and packaging (45). The B virus pac1 contained three conserved motifs, an A-rich region flanked by two stretches of seven C residues and 13 C+G residues. Both pac1 and pac2 are located at conserved distances from the genomic ends (17), 31 bp from the L terminus and 34 bp from the S terminus of B virus DNA. There were no DR2 repeats between the pac1 and pac2 signals in the B virus a sequence, which were thus noticeably shorter than the HSV-1 a sequence.
Overall genome organization.
The complete B virus genomic sequence was assembled according to the HSV-1 prototype genome structure (57). The genome of B virus is 156,789 bp in length, considerably shorter than earlier predictions based on the constructed B virus physical map (26) or direct measurements by electron microscopy (40). The 74.5% G+C base composition of the genome is very similar to earlier estimates of 75% determined by DNA buoyant density centrifugation (40). Table 1 presents a comparison of the length and G+C content of B virus, HSV-1, and HSV-2 by genomic region. The G+C composition was elevated in all B virus regions compared to that in HSV types 1 and 2, with unique regions demonstrating the highest G+C content among known herpesviruses. The sizes of genomic segments were comparable among the viruses with the exception of RS, which in the B virus genome was ≈1.5 kb longer.
TABLE 1.
Comparison of B virus, HSV-1, and HSV-2 genomic regions
| Region | Virus | Length (bp) | % G+C |
|---|---|---|---|
| UL | B virus | 107,815 | 72.9 |
| HSV-1 | 107,947 | 66.9 | |
| HSV-2 | 109,689 | 68.9 | |
| US | B virus | 14,687 | 73.2 |
| HSV-1 | 12,980 | 64.3 | |
| HSV-2 | 14,329 | 66.2 | |
| TRL | B virus | 9,021 | 79.4 |
| HSV-1 | 9,912 | 71.6 | |
| HSV-2 | 9,297 | 74.4 | |
| TRS | B virus | 8,234 | 80.4 |
| HSV-1 | 6,677 | 79.5 | |
| HSV-2 | 6,711 | 80.1 | |
| Whole genome | B virus | 156,789 | 74.5 |
| HSV-1 | 152,261 | 68.3 | |
| HSV-2 | 154,746 | 70.4 |
Ten sets of tandem reiterations were identified in the B virus genome. Genomic locations of sets, numbers of repeat units in each set, sizes, and sequences are given in Table 2. Most reiterations were present at locations compatible with the locations of HSV-1 reiterations. Six sets were present in two copies in the genome due to location in the terminal and internal copies of the RS and RL segments. Only two reiteration sets were located in protein-coding regions, the UL36 and US4 (gG) genes.
TABLE 2.
Sets of reiterated sequences in the B virus genome
| Set | Genomic region | Location of set
|
Sequence of repeat unit | Unit size (bp) | No. of units | Comments | |
|---|---|---|---|---|---|---|---|
| First copy | Second copya | ||||||
| 1 | RL | 824-1022 | 124836-125034 | GGGGGTCCTGGGGGTCCGGGGTCGCC | 26 | 8 | |
| 2 | RL | 2244-2315 | 123543-123614 | GGGGGTCTC | 9 | 8 | Located in ICP0 intron |
| 3 | RL | 3658-3729 | 122129-122200 | GCCCGGCGCCCAAGTCCC | 18 | 4 | |
| 4 | RL | 5164-5244 | 120614-120694 | CCAGAAGCAGAGAGGGGCGGGGGCTCC | 27 | 3 | |
| 5 | UL | 43039-43123 | GGGGGTGCGGGGGCGGT | 17 | 5 | Located between UL21 and UL22 | |
| 6 | UL | 71559-71756 | GCAGGGGCA | 9 | 22 | Encodes PAA repeat in UL36 | |
| 7 | UL | 115984-116238 | CCCCCCTCCCCTCCCCCGCG | 20 | 13 | ||
| 8 | RS | 131851-132461 | 149963-150573 | CCCTTCCCCCCTT | 13 | 47 | Flanks oriS |
| 9 | RS | 133448-133785 | 148639-148976 | CCCCCGCGCACCCCTCGCCCTCCCCT | 26 | 13 | Flanks oriS |
| 10 | US | 139337-139444 | CCCGCCCCCACCACCACC | 18 | 6 | Encodes PAPTTT repeat in gG | |
A location of the second copy of a set is given for reiteration sets from the RS and RL genomic regions.
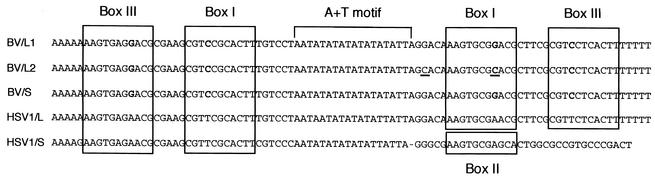
Both origins of B virus replication, oriL and oriS, were tandemly duplicated and present in locations corresponding to HSV oriL and oriS locations. Thus, six origins of DNA replication exist: two copies of oriL (oriL1 and oriL2) in the UL region and two copies each of oriS (oriS1 and oriS2) in the terminal and internal RS regions. Duplications of either the oriL or oriS sequence were also found in HSV-1 strain ANG and HSV-2 strain HG52, respectively (18, 24), but concurrent duplication of both origins has not been described previously for herpes simplex viruses. All B virus origins share the core element of a 94-bp perfect palindrome (Fig. 3) containing two predicted binding sites for the origin-binding protein (OBP), box I, and box III (22, 27, 52). Furthermore, the nucleotide sequences of the B virus oriS and oriL core elements were extremely conserved and almost identical to HSV oriL but differed from HSV oriS (25, 42). Whether the existence of six nearly identical origins of replication in the B virus genome has any functional significance or is just an artifact due to repeated passage of the virus in cell culture currently remains unknown.
FIG. 3.
Comparison of B virus and HSV-1 origins of replication. The DNA sequences of the oriL and oriS core elements are shown. BV/L1, B virus oriL1; BV/L2, B virus oriL2; BV/S, B virus oriS1 and oriS2; HSV1/L, HSV-1 oriL; HSV1/S, HSV-1 oriS. The OBP-binding sites (box I, box II, and box III) are boxed. Residue substitutions in all B virus origins relative to HSV-1 oriL are shown in bold. Residue substitutions in B virus oriL2 relative to oriL1 are underlined.
Gene characterization.
Both major approaches to gene identification, extrinsic and intrinsic, were used to identify and characterize B virus genes. Extrinsic methods, such as Blast, identify protein-coding genes by detecting similarity of translated protein sequences to the primary structure of a known protein. The intrinsic approach, an ab initio statistical method such as GeneMark, identifies protein-coding regions by detecting specific frequency patterns in nucleotide order, including the codon usage pattern. These two types of methods have complementary strengths in terms of sensitivity and specificity. Extrinsic, similarity search methods have high specificity but may miss some genes, up to 30 to 40%, which could be recovered by the intrinsic, statistical method. The B virus genomic sequence can be efficiently analyzed by similarity search methods due to extensive knowledge of the closely related viral species HSV-1 and HSV-2. Still, unique virus-specific genes could be missed. Therefore, in addition to the similarity search, we used ab initio methods, GeneMark and GeneMark.hmm, trained on the set of genes confirmed by similarity search. These methods were modified to allow a noncanonical translation initiation codon, GTG (46), that could be used in this highly G+C-rich genome.
(i) HSV gene homologs.
All but one B virus gene were identified on the basis of sequence homology to HSV-1 and HSV-2 genes and named correspondingly. Table 3 summarizes their locations in the genome, predicted sizes, and percent identities with the corresponding HSV-1 and HSV-2 gene products. Seventy-two genes existed as a single copy within unique genomic regions, whereas two genes, ICP0 and ICP4, appeared twice due to duplication of the large and small repeat regions where they reside. Two ORFs (RL2 and UL15) are predicted to have introns matching ones in the corresponding HSV genes.
TABLE 3.
ORFs and other features of the B virus genome
| ORF or feature | Location
|
Strand | Length (codons) | Identity (%)
|
Characteristics and predicted function(s)a (reference) | ||
|---|---|---|---|---|---|---|---|
| Start | Stop | HSV-1 | HSV-2 | ||||
| a sequence | 1 | 223 | Terminal direct repeat | ||||
| TRL | 1 | 9021 | Terminal copy of large repeat region | ||||
| RL2 | + | 701 | 43.5 | 40.1 | Immediate-early protein ICP0; multifunctional regulatory protein | ||
| Exon 1 | 2194 | 2241 | + | 16 | |||
| Exon 2 | 2461 | 3003 | + | 181 | |||
| Exon 3 | 3125 | 4636 | + | 504 | |||
| UL | 9022 | 116837 | Unique large region | ||||
| UL1 | 9072 | 9746 | + | 225 | 65.5 | 53.4 | Virion membrane glycoprotein L; in complex with gH; membrane fusion |
| UL2 | 9814 | 10575 | + | 254 | 71.3 | 68.9 | Uracil-DNA glycosylase; DNA repair |
| UL3 | 10711 | 11394 | + | 228 | 61.8 | 61.8 | Colocolization with ICP22 and UL4 in small, dense nuclear bodies |
| UL4 | 12120 | 11509 | − | 204 | 56.1 | 56.5 | Colocolization with ICP22 and UL3 in small, dense nuclear bodies |
| UL5 | 14820 | 12172 | − | 883 | 82.7 | 83.0 | Component of helicase-primase complex |
| UL6 | 14819 | 16870 | + | 684 | 69.5 | 71.6 | Capsid protein; DNA cleavage/packaging |
| UL7 | 16821 | 17711 | + | 297 | 65.2 | 65.2 | Capsid protein; DNA cleavage/packaging |
| UL8 | 20171 | 17886 | − | 762 | 58.2 | 60.4 | Component of helicase-primase complex |
| UL9 | 22847 | 20223 | − | 875 | 78.3 | 78.8 | ori binding protein; helicase activity |
| UL10 | 22714 | 24132 | + | 473 | 65.2 | 65.1 | Virion membrane glycoprotein M; proposed role in capsid envelopment |
| UL11 | 24733 | 24437 | − | 99 | 50.0 | 53.1 | Myristylated tegument protein; capsid envelopment |
| UL12 | 26553 | 24679 | − | 625 | 67.8 | 69.1 | DNase; endoexonuclease; processing of DNA replication intermediates |
| UL13 | 28097 | 26553 | − | 515 | 66.9 | 68.8 | Virion protein kinase |
| UL14 | 28513 | 27869 | − | 215 | 65.6 | 66.5 | Minor tegument protein |
| UL15 | + | 739 | 87.5 | 87.5 | DNA cleavage/packaging; transiently associated with maturing capsids | ||
| Exon 1 | 28599 | 29624 | + | 342 | |||
| Exon 2 | 33199 | 34389 | + | 397 | |||
| UL16 | 30898 | 29795 | − | 368 | 64.2 | 66.7 | Capsid associated; DNA cleavage/packaging; located in UL15 intron |
| UL17 | 33015 | 30919 | − | 699 | 66.8 | 69.5 | Tegument protein; DNA cleavage/packaging |
| UL18 | 35574 | 34618 | − | 319 | 82.8 | 82.4 | Capsid protein VP23; forms triplexes with VP19C that connect pentons and hexons in capsids |
| UL19 | 39918 | 35785 | − | 1,378 | 86.9 | 85.5 | Major capsid protein VP5; forms pentons and hexons of capsid shell |
| UL20 | 40783 | 40112 | − | 224 | 64.6 | 66.4 | Virion membrane protein; virion egress; syn5 locus |
| UL21 | 41384 | 42964 | + | 527 | 66.2 | 68.3 | Nucleotidylylated phosphoprotein; interacts with microtubules and facilitates intracellular transport of the virus (65) |
| UL22 | 45736 | 43193 | − | 848 | 59.1 | 60.9 | Virion membrane glycoprotein H; in complex with gL; membrane fusion, entry, cell-to-cell spread |
| UL23 | 47107 | 45995 | − | 371 | 59.8 | 60.7 | Thymidine kinase |
| UL24 | 47042 | 47842 | + | 267 | 66.4 | 66.4 | Nonglycosylated membrane-associated protein; syn5 locus |
| UL25 | 48036 | 49775 | + | 580 | 79.8 | 79.8 | Minor capsid protein; DNA packaging; possible role in DNA anchoring (50) |
| UL26 | 49937 | 51763 | + | 609 | 65.7 | 65.4 | Capsid maturation protease |
| UL26.5 | 50834 | 51763 | + | 310 | 58.8 | 57.0 | Scaffolding protein |
| UL27 | 54815 | 52137 | − | 893 | 79.9 | 80.4 | Virion membrane glycoprotein B; cell entry; contains syn3 locus |
| UL28 | 57192 | 54835 | − | 786 | 84.0 | 84.8 | DNA cleavage/packaging; transiently associated with maturing capsids |
| UL29 | 61257 | 57667 | − | 1,197 | 82.7 | 82.2 | Single-strand DNA-binding protein; key role in assembly of DNA replication proteins |
| oriL1 | 61592 | 61789 | Center of replication origin oriL1 | ||||
| oriL2 | 61795 | 61992 | Center of replication origin oriL2 | ||||
| UL30 | 62173 | 65916 | + | 1,248 | 79.6 | 80.0 | DNA polymerase catalytic subunit; complexes with UL42 |
| UL31 | 66766 | 65861 | − | 302 | 80.9 | 81.3 | Nuclear phosphoprotein; interacts with UL34; capsid egress from nucleus |
| UL32 | 68531 | 66759 | − | 591 | 74.2 | 74.5 | DNA packaging; not associated with capsids |
| UL33 | 68530 | 68928 | + | 133 | 72.1 | 72.9 | DNA packaging; not associated with capsids |
| UL34 | 68988 | 69803 | + | 272 | 66.8 | 71.3 | Type II nuclear membrane-associated phosphoprotein; interacts with UL31; capsid egress from nucleus |
| UL35 | 69939 | 70283 | + | 115 | 51.8 | 47.3 | Basic phosphorylated capsid protein VP26 |
| UL36 | 80356 | 70490 | − | 3,289 | 61.2 | 62.2 | Very large tegument protein; interacts with UL19 and UL37 |
| UL37 | 84145 | 80624 | − | 1,174 | 69.4 | 68.7 | Minor tegument protein |
| UL38 | 84598 | 85974 | + | 459 | 70.6 | 69.1 | Capsid protein VP19C, forms triplexes with VP23 that connect pentons and hexons in capsids |
| UL39 | 86392 | 89385 | + | 998 | 66.7 | 66.0 | Large subunit of ribonucleotide reductase |
| UL40 | 89434 | 90450 | + | 339 | 79.7 | 80.4 | Small subunit of ribonucleotide reductase |
| UL41 | 92073 | 90619 | − | 485 | 72.3 | 73.0 | Tegument phosphoprotein; virion-associated host shutoff (vhs) protein |
| UL42 | 92575 | 94002 | + | 476 | 48.6 | 48.8 | Double-stranded DNA-binding protein, DNA polymerase subunit |
| UL43 | 94131 | 95270 | + | 380 | 44.4 | 49.1 | Predicted membrane-associated protein |
| UL44 | 95527 | 96930 | + | 468 | 49.9 | 51.5 | Virion membrane glycoprotein C; cell attachment; blocking host immune response |
| UL45 | 97166 | 97690 | + | 175 | 63.6 | 59.0 | Type II membrane protein; possible role in cell fusion |
| UL46 | 100140 | 97978 | − | 721 | 57.7 | 58.9 | Tegument phosphoprotein VP11/12; modulates alpha trans-inducing factor activity |
| UL47 | 102327 | 100267 | − | 687 | 59.9 | 58.5 | Tegument phosphoprotein VP13/14; O-glycosylated; modulate α-TIF activity; RNA binding (60) |
| UL48 | 104246 | 102783 | − | 488 | 69.3 | 69.2 | Major tegument protein VP16 (α-TIF); trans-activator of α genes |
| UL49 | 105449 | 104580 | − | 290 | 45.9 | 45.0 | Major tegument protein VP22; binds RNA; carrier of mRNA from infected to uninfected cells (60) |
| UL49A | 106092 | 105853 | − | 80 | 40.5 | 43.0 | Envelope protein |
| UL50 | 106107 | 107216 | + | 370 | 55.9 | 54.3 | Deoxyuridine triphosphatase |
| UL51 | 108064 | 107381 | − | 228 | 67.5 | 69.3 | Capsid/tegument-associated phosphoprotein (15) |
| UL52 | 108126 | 111302 | + | 1,059 | 73.0 | 73.1 | Component of helicase-primase complex |
| UL53 | 111254 | 112267 | + | 338 | 66.0 | 68.6 | Membrane glycoprotein K; virion egress; contains syn1 locus |
| UL53A | − | 300 | Hypothetical protein predicted by GeneMark and GeneMark.hmm | ||||
| Exon 2 | 112186 | 112495 | − | 197 | |||
| Exon 1 | 112755 | 113344 | − | 103 | |||
| UL54 | 112644 | 114116 | + | 491 | 59.1 | 60.8 | Immediate-early protein ICP27; regulates some early and all late gene expression |
| UL55 | 114429 | 115001 | + | 191 | 62.0 | 64.3 | Nuclear matrix-associated protein |
| UL56 | 115834 | 115154 | − | 227 | 39.7 | 42.9 | Type II membrane protein (38); involved in virus pathogenicity (35) |
| IRL | 116837 | 125634 | Internal copy of large repeat region | ||||
| RL2 | − | 701 | 43.5 | 40.1 | Immediate-early protein ICP0; multifunctional regulatory protein | ||
| Exon 3 | 122733 | 121222 | − | 504 | |||
| Exon 2 | 123397 | 122855 | − | 181 | |||
| Exon 1 | 123664 | 123617 | − | 16 | |||
| a′ sequence | 125635 | 125857 | Inverted copy of a sequence | ||||
| IRS | 125858 | 133868 | Internal copy of small repeat region | ||||
| RS1 | 131284 | 127724 | − | 1,187 | 65.0 | 66.8 | Immediate-early protein ICP4; regulator of gene expression |
| Oris1 | 132795 | 132796 | Center of replication origin oriS1 | ||||
| Oris2 | 132998 | 132999 | Center of replication origin oriS2 | ||||
| Us | 133869 | 148554 | Unique small region | ||||
| US1 | 133900 | 135252 | + | 451 | 41.1 | 43.5 | Immediate-early protein ICP22; required for optimal ICP0 expression |
| US2 | 136386 | 135478 | − | 303 | 56.1 | 54.0 | Tegument protein |
| US3 | 136708 | 138084 | + | 459 | 60.8 | 61.1 | Protein kinase; antiapoptotic activity |
| US4 | 138221 | 140242 | + | 674 | 29.2 | 39.2 | Virion membrane glycoprotein G; entry into polarized cells |
| US5 | 140465 | 140830 | + | 122 | 34.0 | 26.6 | Glycoprotein J; block apoptosis |
| US6 | 141296 | 142480 | + | 395 | 57.0 | 59.0 | Virion membrane glycoprotein D; cell entry; interacts with cellular receptors |
| US7 | 142680 | 143885 | + | 402 | 46.1 | 51.0 | Virion membrane glycoprotein I; in complex with gE; basolateral viral spread |
| US8 | 144253 | 145872 | + | 540 | 46.0 | 48.0 | Virion membrane glycoprotein E; in complex with gI; basolateral viral spread |
| US8.5 | 145817 | 146185 | + | 123 | 42.3 | 45.9 | Nucleolar phosphoprotein |
| US9 | 146309 | 146581 | + | 91 | 58.9 | 57.3 | Tegument protein |
| US10 | 148139 | 147204 | − | 312 | 43.7 | 45.9 | Tegument protein |
| US11 | 148290 | 147847 | − | 148 | 45.2 | 46.7 | RNA-binding tegument protein; interacts with protein kinase R |
| US12 | 148555 | 148310 | − | 82 | 26.8 | 26.8 | Immediate-early protein ICP47; inhibits antigen presentation |
| TRs | 148556 | 156566 | Terminal copy of small repeat region | ||||
| oriS2 | 149425 | 149426 | Center of replication origin oriS2 | ||||
| oriS1 | 149628 | 149629 | Center of replication origin oriS1 | ||||
| RS1 | 151140 | 154700 | + | 1187 | 65.0 | 66.8 | Immediate-early protein ICP4; regulator of gene expression |
| a sequence | 156567 | 156789 | Terminal direct repeat | ||||
It was inferred that the B virus UL1 and UL2 genes use the GTG codon as a start codon at genomic sequence positions 9072 and 9814, respectively. This prediction was made statistically and was corroborated by the presence of conserved regions upstream of the canonical ATG. Both predicted translational initiation GTG codons aligned with the ATGs of the HSV UL1 and UL2 genes. Conservation of the UL1 GTG start codon was confirmed by sequence analysis of the 5′ region of the UL1 gene from two B virus clinical isolates (data not shown).
The extent of amino acid identity between B virus and HSV polypeptides varied from 26.6% (US5) to 87.7% (UL15). The conservation of specific protein domains observed in HSV-1 and HSV-2 was mirrored when HSV-1 and B virus were compared. The following proteins were significantly conserved in B virus: DNA cleavage and packaging proteins, i.e., UL15, UL28, UL32, and UL33; capsid proteins, i.e., UL18, UL19, and UL38; proteins involved in DNA replication, i.e., UL2, UL5, UL29, and UL30; and glycoprotein B. The three least-conserved proteins in B virus were US4, US5, and US12. Similar levels of conservation have been described for homologous proteins in many other mammalian herpesviruses (58).
The B virus proteins could be divided into three groups based on the degree of similarity to HSV-1 and HSV-2 proteins. The largest group (46 proteins) showed greater similarity to HSV-2 proteins, e.g., DNA cleavage and packaging proteins. The second group (20 proteins), with capsid proteins among others, showed higher levels of similarity to HSV-1 proteins. In seven proteins, differences in similarity to HSV-1 or HSV-2 homologs were marginal, and these proteins formed the third group. The proteins that enable HSV-1 to replicate and reactivate more efficiently at orofacial sites and HSV-2 at genital sites are unknown, but the fact that B virus replicates and is reactivated with similar efficiencies at both sites may be explained by the similarities of selected B virus proteins with either HSV type 1 or 2 proteins. These hybrid properties in B virus raise many questions about alphaherpesvirus evolution.
(ii) Gene homologs absent.
In recent years, the existence of additional HSV-1-specific genes has been proposed and substantiated by experimental data. UL20.5 is located between UL20 and UL21, ORF P and ORF O map antisense to the 34.5 gene, and UL43.5 and UL27.5 map antisense to UL43 and UL27, respectively (8, 55, 56, 71, 72). Like HSV-2 (18), B virus has no equivalents to these proposed genes.
One well-established gene, the HSV γ134.5 (RL1) homolog, was observed to be absent in B virus. This conclusion was reached after repeated searches with state-of-the-art computational genome analysis tools (see Materials and Methods). To determine that this observation was not limited to the laboratory strain of B virus, we cloned and sequenced L-terminal SphI restriction fragments from two low-passage clinical isolates, one of which was derived from a rhesus macaque (54) and the other post mortem from a zoonotically infected human patient (16). Sequence comparison of these fragments with the corresponding fragment of the laboratory strain did not reveal any significant differences: only single nucleotide substitutions and variations in the number of copies of short reiterations were found in the clinical isolates relative to the laboratory strain. Since the absence of the γ134.5 gene homolog was verified in three independent B virus strains, we concluded that it was a genuine feature of the B virus genome.
The protein product of the HSV-1 γ134.5 gene, infected-cell protein 34.5 (ICP34.5), is a neurovirulence factor with at least two known, distinct functional activities (9, 10, 12-14, 64, 66). One function, encoded by the carboxyl-terminal domain, negates the antiviral effect of induced protein kinase R by redirecting the host protein phosphatase 1α to dephosphorylate translation initiation factor eIF2α, preventing protein synthesis shutoff in infected cells (9, 11, 12, 29, 39). Another function, mapped to both amino-terminal and carboxyl-terminal domains, somehow enables the virus to replicate in the peripheral and central nervous systems of experimentally infected animals (10, 64). Deletion of γ134.5 leads to complete neuroattenuation of highly neurovirulent HSV-1 strains. In the absence of γ134.5, other genes may supply these functions, given the striking similarities between the replication characteristics of B virus and the human simplex viruses.
We predict that B virus currently uses compensatory strategies to block host responses to infection, similar to those described for HSV-1. For example, the HSV-1 US11 protein can inhibit protein kinase R activation and compensate for the absence of the ICP34.5 function in deletion mutants if expressed early in infection (5, 6, 28, 48). It was proposed that the US11 gene encodes an alternative mechanism to preclude the shutoff of protein synthesis that is currently inactive in HSV-1 (5). In addition, B virus might have evolved unique mechanisms to prevent termination of protein synthesis and elude the DNA replication blocks imposed by neuronal cells. To examine this possibility, a search for B virus-specific genes was performed.
(iii) Unique genes in B virus genome.
Two major sequence differences between the B virus and HSV-1 and HSV-2 genomes were detected. The B virus RS region contains an additional ≈1.5 kb of sequence between the S terminus and the ICP4 gene homolog, while the RL region is shorter in B virus than in the human viruses, with no sequence homology to the HSV ICP0 flanking regions. However, no potential genes were identified in these regions despite rigorous analyses.
A putative two-exon gene (UL53A) was identified by statistical analysis in the B virus UL region on a complementary strand (positions 112186 to 112495 and 112755 to 113344). This putative gene had a codon usage pattern compatible with codons present in established B virus genes. The UL53A protein is a hydrophilic basic protein (pI 11.9) with an amino acid composition biased to Pro and Arg residues. The first 21 residues were predicted to be a signal peptide by the SignalP program, and residues 222 to 240 were predicted to be a transmembrane domain by two out of three programs applied (TMpred and DAS), but probability scores were not high enough to classify this protein as a membrane protein. PSI-Blast analysis detected sequence similarity to domains in three neuronal proteins encoded in mammalian genomes: neural cell adhesion molecule NCAM-180 (GenBank accession number P13595, E value of 0.16), calcineurin inhibitor cabin 1 (GenBank accession number AAD40846, E value of 0.014), and brain calcium channel α 1A subunit (GenBank accession number AAB64179, E value of 0.007). However, the similarity was insufficient to confidently characterize the gene at this time, and pending experimental evidence will be critical to determine whether this gene, which is missing in HSV-1 and HSV-2, provides B virus with a unique mechanism to attack neural cells.
B virus genome high G+C content and optimization of codon usage.
The G+C content of the protein-coding regions in B virus, 74.4%, is only slightly lower than the G+C content of the noncoding regions, 75.4%. These data indicate a strong mutation pressure toward G and C nucleotides in B virus DNA evolution. Obviously, the genetic drift toward the abundance of G's and C's has been compensated for by positive selection for conservative A's and T's in the first (26% A and T) and second (44% A and T) positions of a codon in protein-coding regions and by positive selection for A's and T's in a few evolutionarily conserved regions (promoters, repeats, and other regulatory sites) in noncoding DNA. The G+C content of the third position of codons in B virus genes is extremely high, 93.1%. This bias, created by mutation pressure, seems to be a driving force in the formation of the codon usage pattern in B virus.
Genomes of primates tend to have low G+C content, and the logical question arises whether the highly G+C-biased codon usage in B virus matches the proportions of the isoacceptor tRNA in host cells. Unfortunately, no experimental data are currently available regarding the tRNA pool in macaque cells. Still, relative amounts of the isoacceptor tRNAs could be predicted based on the frequencies of codon use in rhesus macaque genes because a strong direct correlation between these parameters has been demonstrated in a number of organisms (23, 32, 49).
A codon usage catalog of rhesus macaque genes was generated based on 288 protein-coding sequences from the GenBank database and compared with the codon usage in B virus genes (Table 4). As expected, the overall codon usage patterns of B virus and the macaque hosts differed substantially. However, remarkably, the favored codons of the B virus genes mirrored the most frequently used codons in the rhesus macaque genes (except Ser and Arg, both encoded by six codons). Moreover, these most frequent codons in B virus were almost the only codons used for each amino acid. This observation correlates with intriguing data from a number of unicellular and multicellular organisms that highly expressed genes utilize preferable codons in each synonymous group, the optimal codons (20, 21, 23, 61). Thus, the combined effects of directional mutation pressure and selection for preferential codons put the B virus genome in a favorable state to efficiently employ the host cell translation machinery for expression of viral proteins.
TABLE 4.
Codon usage in rhesus macaque and B virus genesa
| Amino acid | Codon |
Macaca mulatta
|
B virus
|
||
|---|---|---|---|---|---|
| No. of occurrences | % of occurrences | No. of occurrences | % of occurrences | ||
| His | CAU | 972 | 43 | 31 | 3 |
| CAC | 1,284 | 57 | 870 | 97 | |
| Tyr | UAU | 1,229 | 43 | 60 | 6 |
| UAC | 1,602 | 57 | 887 | 94 | |
| Gln | CAA | 1,161 | 30 | 55 | 6 |
| CAG | 2,756 | 70 | 929 | 94 | |
| Asn | AAU | 1,625 | 46 | 28 | 4 |
| AAC | 1,902 | 54 | 692 | 96 | |
| Lys | AAA | 2,094 | 45 | 50 | 10 |
| AAG | 2,561 | 55 | 455 | 90 | |
| Asp | GAU | 1,633 | 44 | 144 | 7 |
| GAC | 2,050 | 56 | 1,978 | 93 | |
| Glu | GAA | 2,141 | 42 | 157 | 7 |
| GAG | 3,018 | 58 | 1,986 | 93 | |
| Phe | UUU | 1,696 | 44 | 268 | 21 |
| UUC | 2,187 | 56 | 1,028 | 79 | |
| Cys | UGU | 1,141 | 47 | 83 | 12 |
| UGC | 1,284 | 53 | 601 | 88 | |
| Ile | AUU | 1,475 | 32 | 52 | 6 |
| AUC | 2,303 | 50 | 808 | 89 | |
| AUA | 809 | 18 | 47 | 5 | |
| Val | GUU | 1,056 | 19 | 141 | 5 |
| GUC | 1,440 | 25 | 1,139 | 41 | |
| GUA | 516 | 09 | 46 | 2 | |
| GUG | 2,686 | 47 | 1,426 | 52 | |
| Pro | CCU | 1,366 | 28 | 155 | 4 |
| CCC | 1,476 | 31 | 2,014 | 53 | |
| CCA | 1,457 | 30 | 120 | 3 | |
| CCG | 518 | 11 | 1,527 | 40 | |
| Thr | ACU | 1,300 | 24 | 31 | 2 |
| ACC | 2,076 | 37 | 992 | 49 | |
| ACA | 1,528 | 28 | 58 | 3 | |
| ACG | 577 | 11 | 935 | 46 | |
| Gly | GGU | 926 | 17 | 127 | 4 |
| GGC | 1,666 | 30 | 1,724 | 54 | |
| GGA | 1,641 | 29 | 209 | 6 | |
| GGG | 1,352 | 24 | 1,180 | 36 | |
| Ala | GCU | 1,548 | 27 | 144 | 2 |
| GCC | 2,192 | 39 | 3,471 | 55 | |
| GCA | 1,298 | 23 | 127 | 2 | |
| GCG | 635 | 11 | 2,599 | 41 | |
| Arg | CGU | 398 | 9 | 82 | 2 |
| CGC* | 727 | 17 | 2,109 | 56 | |
| CGA | 368 | 8 | 208 | 5 | |
| CGG | 749 | 17 | 1,200 | 31 | |
| AGA* | 1,077 | 26 | 47 | 1 | |
| AGG | 1,014 | 23 | 180 | 5 | |
| Leu | UUG | 1,109 | 12 | 80 | 2 |
| UUA | 665 | 7 | 14 | 0 | |
| CUU | 1,261 | 14 | 55 | 1 | |
| CUC | 2,930 | 21 | 1,373 | 36 | |
| CUA | 562 | 6 | 58 | 2 | |
| CUG | 3,465 | 40 | 2,256 | 59 | |
| Ser | UCU | 1,428 | 20 | 93 | 4 |
| UCC* | 1,731 | 23 | 648 | 29 | |
| UCA | 1,122 | 16 | 37 | 2 | |
| UCG* | 326 | 5 | 778 | 35 | |
| AGU | 1,080 | 15 | 41 | 2 | |
| AGC | 1,505 | 21 | 620 | 28 | |
| Overall G+C | 51.03 | 74.36 | |||
| First position G or C | 52.55 | 73.73 | |||
| Second position G or C | 42.12 | 56.22 | |||
| Third position G or C | 58.42 | 93.11 | |||
The sequence of the M. mulatta genes were obtained from the GenBank database. The most frequently observed codons are in boldface. Codons for which discrepant results were obtained are marked with an asterisk. Nondegenerate amino acids (Met and Trp) are not included.
Conclusions.
The complete genomic sequence of the B virus E2490 laboratory strain demonstrates that the genome organization and gene repertoire are similar but not identical to those of the HSV-1 and HSV-2 genomes. Interestingly, B virus lacks the gene homolog encoding the neurovirulence factor ICP34.5. We postulate that B virus utilizes different mechanisms to block host antivirus responses and facilitate replication in neurons. The enhanced neurovirulence of B virus in human hosts may reflect the inability of human cells to mount efficient antiviral cellular responses against these divergent strategies. The other implication of our findings is that because B virus does not possess the γ134.5 gene homolog, acquisition of this gene by human viruses is an even more recent evolutionary event than was suggested previously (6, 7).
Comparative analysis of the B virus and HSV genomes is essential to understand the mechanisms of B virus pathogenesis in humans. The complete sequence of the B virus reference strain also supplies much-needed information to determine the genetic basis of phenotypic and pathogenic differences among B virus isolates derived from different macaque subspecies. With this information, new antiviral and vaccine strategies can be designed to target critical viral components in order to control this deadly zoonotic agent.
Acknowledgments
This work was supported by Public Health Service grants R01 RR03162 and P40 RR05162 from the NIH's National Center for Research Resources and in part by grant GM-47853 from the National Institutes of Health.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahr, U., and G. Darai. 2001. Analysis and characterization of the complete genome of tupaia (tree shrew) herpesvirus. J. Virol. 75:4854-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, A. M., L. Harrington, and D. C. Kelly. 1992. Nucleotide sequence analysis of genes encoding glycoproteins D and J in simian herpes B virus. J. Gen. Virol. 73:2963-2967. [DOI] [PubMed] [Google Scholar]
- 4.Borodovsky, M., and J. McIninch. 1993. Recognition of genes in DNA sequence with ambiguities. Biosystems 30:161-171. [DOI] [PubMed] [Google Scholar]
- 5.Cassady, K. A., M. Gross, G. Y. Gillespie, and B. Roizman. 2002. Second-site mutation outside of the US10-12 domain of Δγ134.5 herpes simplex virus 1 recombinant blocks the shutoff of protein synthesis induced by activated protein kinase R and partially restores neurovirulence. J. Virol. 76:942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the alpha subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassady, K. A., M. Gross, and B. Roizman. 1998. The second-site mutation in herpes simplex virus recombinants lacking the γ134.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF-2α. J. Virol. 72:7005-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. E., L. Menotti, F. Filatov, G. Campadelli-Fiume, and B. Roizman. 1998. UL27.5 is a novel γ2 gene antisense to the herpes simplex virus 1 gene encoding glycoprotein B. J. Virol. 72:6056-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of a Mr 90,000 phosphoprotein with protein kinase protein kinase R in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 11.Chou, J., A. P. Poon, J. Johnson, and B. Roizman. 1994. Differential response of human cells to deletions and stop codons in the γ134.5 gene of herpes simplex virus. J. Virol. 68:8304-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, J., and B. Roizman. 1992. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou, J., and B. Roizman. 1994. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc. Natl. Acad. Sci. USA 91:5247-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou, J., and B. Roizman. 1990. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J. Virol. 64:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daikoku, T., K. Ikenoya, H. Yamada, F. Goshima, and Y. Nishiyama. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J. Gen. Virol. 79:3027-3031. [DOI] [PubMed] [Google Scholar]
- 16.Davenport, D. S., D. R. Johnson, G. P. Holmes, D. A. Jewett, S. C. Ross, and J. K. Hilliard. 1994. Diagnosis and management of human B virus (Herpesvirus simiae) infections in Michigan. Clin. Infect. Dis. 19:33-41. [DOI] [PubMed] [Google Scholar]
- 17.Deiss, L. P., J. Chou, and N. Frenkel. 1986. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J. Virol. 59:605-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duret, L. 2000. tRNA gene number and codon usage in the C. elegans genome are coadapted for optimal translation of highly expressed genes. Trends Genet. 16:287-289. [DOI] [PubMed] [Google Scholar]
- 21.Duret, L., and D. Mouchiroud. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 96:4482-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elias, P., C. M. Gustafsson, and O. Hammarsten. 1990. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication oris. J. Biol. Chem. 265:17167-17173. [PubMed] [Google Scholar]
- 23.Gouy, M., and C. Gautier. 1982. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10:7055-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray, C. P., and H. C. Kaerner. 1984. Sequence of the putative origin of replication in the UL region of herpes simplex virus type 1 ANG DNA. J. Gen. Virol. 65:2109-2119. [DOI] [PubMed] [Google Scholar]
- 25.Hardwicke, M. A., and P. A. Schaffer. 1995. Cloning and characterization of herpes simplex virus type 1 oriL: comparison of replication and protein-DNA complex formation by oriL and oriS. J. Virol. 69:1377-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington, L., L. V. Wall, and D. C. Kelly. 1992. Molecular cloning and physical mapping of the genome of simian herpes B virus and comparison of genome organization with that of herpes simplex virus type 1. J. Gen. Virol. 73:1217-1226. [DOI] [PubMed] [Google Scholar]
- 27.Hazuda, D. J., H. C. Perry, A. M. Naylor, and W. L. McClements. 1991. Characterization of the herpes simplex virus origin binding protein interaction with OriS. J. Biol. Chem. 266:24621-24626. [PubMed] [Google Scholar]
- 28.He, B., J. Chou, R. Brandimarti, I. Mohr, Y. Gluzman, and B. Roizman. 1997. Suppression of the phenotype of γ134.5− herpes simplex virus 1: failure of activated RNA-dependent protein kinase to shut off protein synthesis is associated with a deletion in the domain of the α47 gene. J. Virol. 71:6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hein, J. 1990. Unified approach to alignment and phylogenies. Methods Enzymol. 183:626-645. [DOI] [PubMed] [Google Scholar]
- 31.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 32.Ikemura, T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2:13-34. [DOI] [PubMed] [Google Scholar]
- 33.Keeble, S. A. 1960. B virus infection in monkeys. Ann. N.Y. Acad. Sci. 85:960-969. [DOI] [PubMed] [Google Scholar]
- 34.Keeble, S. A., G. J. Christofinis, and W. Wood. 1958. Natural B virus infection in rhesus monkeys. J. Pathol. Bacteriol. 76:189-199. [DOI] [PubMed] [Google Scholar]
- 35.Kehm, R., A. Rosen-Wolff, and G. Darai. 1996. Restitution of the UL56 gene expression of HSV-1 HFEM led to restoration of virulent phenotype; deletion of the amino acids 217 to 234 of the UL56 protein abrogates the virulent phenotype. Virus Res. 40:17-31. [DOI] [PubMed] [Google Scholar]
- 36.Killeen, A. M., L. Harrington, L. V. Wall, and D. C. Kelly. 1992. Nucleotide sequence analysis of a homologue of herpes simplex virus type 1 gene US9 found in the genome of simian herpes B virus. J. Gen. Virol. 73:195-199. [DOI] [PubMed] [Google Scholar]
- 37.Kingham, B. F., V. Zelnik, J. Kopacek, V. Majerciak, E. Ney, and C. J. Schmidt. 2001. The genome of herpesvirus of turkeys: comparative analysis with Marek's disease viruses. J. Gen. Virol. 82:1123-1135. [DOI] [PubMed] [Google Scholar]
- 38.Koshizuka, T., F. Goshima, H. Takakuwa, N. Nozawa, T. Daikoku, O. Koiwai, and Y. Nishiyama. 2002. Identification and characterization of the UL56 gene product of herpes simplex virus type 2. J. Virol. 76:6718-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leib, D. A., M. A. Machalek, B. R. G. Williams, R. H. Silverman, and H. W. Virgin. 2000. From the cover: specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig, H., G. Pauli, H. R. Gelderblom, G. Darai, H.-G. Koch, R. M. Flugel, B. Norrild, and M. D. Daniel. 1983. B virus (Herpesvirus simiae), p. 385-428. In B. Roizman (ed.), The herpesviruses, vol. 2. Plenum Press, New York, N.Y.
- 41.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, D. W., S. P. Deb, J. S. Klauer, and S. Deb. 1991. Analysis of the herpes simplex virus type 1 OriS sequence: mapping of functional domains. J. Virol. 65:4359-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGeoch, D. J., C. Cunningham, G. McIntyre, and A. Dolan. 1991. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J. Gen. Virol. 72:3057-3075. [DOI] [PubMed] [Google Scholar]
- 44.McGeoch, D. J., A. Dolan, S. Donald, and D. H. Brauer. 1986. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 14:1727-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McVoy, M. A., D. E. Nixon, S. P. Adler, and E. S. Mocarski. 1998. Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J. Virol. 72:48-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehdi, H., E. Ono, and K. C. Gupta. 1990. Initiation of translation at CUG, GUG, and ACG codons in mammalian cells. Gene 91:173-178. [DOI] [PubMed] [Google Scholar]
- 47.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohr, I., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 49.Moriyama, E. N., and J. R. Powell. 1997. Codon usage bias and tRNA abundance in Drosophila. J. Mol. Evol. 45:514-523. [DOI] [PubMed] [Google Scholar]
- 50.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohsawa, K., D. H. Black, H. Sato, and R. Eberle. 2002. Sequence and genetic arrangement of the US region of the monkey B virus (cercopithecine herpesvirus 1) genome and comparison with the US regions of other primate herpesviruses. J. Virol. 76:1516-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olivo, P. D., N. J. Nelson, and M. D. Challberg. 1988. Herpes simplex virus DNA replication: the UL9 gene encodes an origin-binding protein. Proc. Natl. Acad. Sci. USA 85:5414-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palmer, A. E. 1987. B virus, herpesvirus simiae: historical perspective. J. Med. Primatol. 16:99-130. [PubMed] [Google Scholar]
- 54.Perelygina, L., I. Patrusheva, N. Manes, M. J. Wildes, P. Krug, and J. K. Hilliard. 2003. Quantitative real-time PCR for detection of monkey B virus (cercopithecine herpesvirus 1) in clinical samples. J. Virol. Methods 109:245-251. [DOI] [PubMed] [Google Scholar]
- 55.Randall, G., M. Lagunoff, and B. Roizman. 1997. The product of ORF O located within the domain of herpes simplex virus 1 genome transcribed during latent infection binds to and inhibits in vitro binding of infected cell protein 4 to its cognate DNA site. Proc. Natl. Acad. Sci. USA 94:10379-10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Randall, G., and B. Roizman. 1997. Transcription of the derepressed open reading frame P of herpes simplex virus 1 precludes the expression of the antisense γ134.5 gene and may account for the attenuation of the mutant virus. J. Virol. 71:7750-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 58.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Sciortino, M. T., B. Taddeo, A. P. W. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 99:8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharp, P. M., T. M. Tuohy, and K. R. Mosurski. 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14:5125-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slomka, M. J., L. Harrington, C. Arnold, J. P. Norcott, and D. W. Brown. 1995. Complete nucleotide sequence of the herpesvirus simiae glycoprotein G gene and its expression as an immunogenic fusion protein in bacteria. J. Gen. Virol. 76:2161-2168. [DOI] [PubMed] [Google Scholar]
- 63.Smith, A. L., D. H. Black, and R. Eberle. 1998. Molecular evidence for distinct genotypes of monkey B virus (herpesvirus simiae) which are related to the macaque host species. J. Virol. 72:9224-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taha, M. Y., G. B. Clements, and S. M. Brown. 1989. A variant of herpes simplex virus type 2 strain HG52 with a 1.5 kb deletion in RL between 0 to 0.02 and 0.81 to 0.83 map units is non-neurovirulent for mice. J. Gen. Virol. 70:705-716. [DOI] [PubMed] [Google Scholar]
- 65.Takakuwa, H., F. Goshima, T. Koshizuka, T. Murata, T. Daikoku, and Y. Nishiyama. 2001. Herpes simplex virus encodes a virion-associated protein which promotes long cellular processes in overexpressing cells. Genes Cells 6:955-966. [DOI] [PubMed] [Google Scholar]
- 66.Thompson, R. L., S. K. Rogers, and M. A. Zerhusen. 1989. Herpes simplex virus neurovirulence and productive infection of neural cells is associated with a function which maps between 0.82 and 0.832 map units on the HSV genome. Virology 172:435-450. [DOI] [PubMed] [Google Scholar]
- 67.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 74:7980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varmuza, S. L., and J. R. Smiley. 1985. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell 41:793-802. [DOI] [PubMed] [Google Scholar]
- 69.Virgin, H. W. t., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walboomers, J. M., and J. T. Schegget. 1976. A new method for the isolation of herpes simplex virus type 2 DNA. Virology 74:256-258. [DOI] [PubMed] [Google Scholar]
- 71.Ward, P. L., D. E. Barker, and B. Roizman. 1996. A novel herpes simplex virus 1 gene, UL43.5, maps antisense to the UL43 gene and encodes a protein which colocalizes in nuclear structures with capsid proteins. J. Virol. 70:2684-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward, P. L., B. Taddeo, N. S. Markovitz, and B. Roizman. 2000. Identification of a novel expressed open reading frame situated between genes UL20 and UL21 of the herpes simplex virus 1 genome. Virology 266:275-285. [DOI] [PubMed] [Google Scholar]
- 73.Weigler, B. J. 1992. Biology of B virus in macaque and human hosts: a review. Clin. Infect. Dis. 14:555-567. [DOI] [PubMed] [Google Scholar]
- 74.Whitley, R. J., and J. K. Hilliard. 2001. Cercopithecine herpesvirus (B virus), p. 2835-2848. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.