Abstract
Human immunodeficiency virus (HIV) type 1 subtype B sequences (whole envelope and the p17 region of gag) were obtained from peripheral blood mononuclear cell samples collected in 1981 from seven HIV-infected U.S. individuals and in 1982 from one infected Canadian resident. Phylogenetic and nucleotide distance analyses were performed by using database sequences representing North American strains collected from 1978 to 1995. The estimated phylogeny was starlike, with early strains represented on different lineages. When sequences were grouped by years of collection, nucleotide distance comparisons demonstrated an increase in diversity over time and indicated that contemporary strains are more closely related to early epidemic strains than to each other. Using a recently developed likelihood ratio reduction procedure, the date of origin of the U.S. epidemic was estimated to be 1968 ± 1.4 years. A coalescent approach was also used to estimate the population history of the U.S. subtype B epidemic. Our analyses provide new information that implies an exponential growth rate from the beginning of the U.S. HIV epidemic. The dating results suggest a U.S. introduction date (or date of divergence from the most recent common ancestor) that precedes the date of the earliest known AIDS cases in the late 1970s. Furthermore, the estimated epidemic growth curve shows a period of exponential growth that preceded most of the early documented cases and also indicates a leveling of prevalence rates in the recent past.
The main group of human immunodeficiency virus type 1 (HIV-1), the strains responsible for the pandemic, has been classified into nine subtypes and at least 14 circulating recombinant forms (20), with most clustering by geographic region. In North America, subtype B is the predominant strain (20). Although non-B subtype infections have been identified in the United States in recent years (2, 12), most of these infections have occurred in African-born individuals (38). The large proportion of subtype B infections presumably reflects founder effects of early strains in North America (7, 25). In the United States, the AIDS epidemic was first recognized in June 1981 with the report of an outbreak of Pneumocystis pneumonia among homosexual men (3). Subsequent retrospective studies identified AIDS cases among U.S. residents and Haitian immigrants during 1978 to 1981 (22, 24, 26, 36), with the earliest evidence of HIV in North America probably occurring in 1977 (40).
Since the genetic variability of HIV is known to increase over time within epidemics (11), we obtained samples collected in 1981-1982 from seven U.S. (13) and one Canadian (1) HIV-infected individual(s) and used the subsequent HIV sequences to evaluate the evolution of the virus over the course of the U.S. epidemic. We used nucleotide distance comparisons and phylogenetic reconstruction to analyze the early strains in conjunction with contemporary North American HIV sequences. Phylogeny-based methods have previously been used to estimate dates for HIV-1 common ancestors (16, 34, 37, 44). Here, we use an improved (P. Lemey, M. Salemi, B. Wang, N. K. Saksena, W. H. Hall, N. Duffy, and A.-M. Vandamme, submitted for publication) site stripping for clock detection (SSCD) procedure (34) to estimate the date of origin of U.S. HIV subtype B. Finally, phylogenetic methods incorporating coalescent theory (14) have been used previously to infer demographic histories from representative HIV-infected (9, 28, 43) as well as hepatitis C virus-infected (29) populations. We used an updated algorithm (30) that can analyze noncontemporaneously sampled sequences to estimate the epidemic history of HIV in the United States.
MATERIALS AND METHODS
Specimens, PCR, and DNA sequencing.
Blood specimens from homosexual men with AIDS were collected (September to November of 1981) from seven individuals residing in Georgia, New York, New Jersey, and California and during 1982 from one Canadian resident. Peripheral blood mononuclear cell processing from the blood specimens and PCR amplification with primers specific for the gene coding for the p17 protein of the gag region were as previously described (35). The Expand Long Template PCR Kit (Boehringer Mannheim, Indianapolis, Ind.) was used for nested amplification of the env gene (gp160) with primary amplification primers KR1173 (5′-ATGGAGCCAGTAGATCCTAGACTAG) and KR1174 (5′-CCTGAGGTGTGACTGGAAAAC) and secondary (nested) primers KR1175 (5′-GCAGCATTAGTAGTAGCAATAATA) and KR1176 (5′-GTGCTTCTAGCCAGGCACAAG). Primary thermocycling conditions were 1 cycle at 94°C for 2 min; 30 cycles at 94°C for 10 s, 55°C for 30 s, and 68°C for 4 min; and 1 cycle at 72°C for 30 min. Secondary PCR conditions were 5 μl of the primary PCR product (100 μl) and the thermocycling conditions of the primary PCR, with the exception of the annealing temperature of 60°C. The PCR-amplified products were purified by Qiagen PCR purification kits (Qiagen Inc., Chatsworth, Calif.).
Purified DNA was sequenced by using the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, Calif.). Sequencing primers for the p17 coding region of gag were as previously described (35), while primers used for gp160 sequencing included the secondary PCR primers (KR1175 and KR1176), as well as LK1 and LK2 (17), ED5 and ED12 (5), and various env primers (35). The lengths (nucleotides) of PCR fragments sequenced were 396 for the p17 region of gag and approximately 2,500 for the entire env.
Distance and phylogenetic analyses.
Maximum-likelihood tree reconstructions were performed on alignments of the p17 or gp160 sequences, combined with U.S. and Haitian sequences from the HIV database (Los Alamos National Laboratories [LANL], Los Alamos, N.Mex.). Manual alignment was performed with the Se-Al v2 program (Se-Al; A. Rambaut, distributed by the author at http://evolve.zoo.ox.ac.uk/software/), with sites containing any gaps subsequently removed. Sequences of strains from the database were selected for p17 and gp160 sequences from the same individual where possible, as well as similar numbers of sequences within the year of collection. For gp160 nucleotide distance comparisons, additional sequences from the HIV database were added for incorporation into groups by year of collection (1981-1982, n = 8; 1983-1984, n = 6; 1985-1986, n = 16; 1988-1989, n = 6; 1991-1992, n = 11; and 1994-1995, n = 19). Subsequently, the V3V5 region (∼600 bp) was extracted and used in an alignment with four published V3V5 sequences collected in 1978-1979 (n = 4) (7). As fewer p17 sequences were available for distance comparisons, p17 groups were assembled as follows: 1981-1982, n = 8; 1983-1984, n = 5; 1985-1986, n = 4; and 1994-1995, n = 6. Intergroup and intragroup calculations (means and standard errors of the means) were performed with the MEGA program (S. Kumar, K. Tamura, I. Jakobsen, and M. Nei, MEGA: molecular evolutionary genetic analysis; distributed by the authors at www.megasoftware.net/). Statistical significance of the differences between the mean distances of groups was measured by the t test. For both tree construction and distance comparisons, the Modeltest program (27) was used to select and evaluate the DNA substitution models used. Maximum-likelihood trees were constructed with PAUP 4.0 (39), using heuristic searches incorporating tree bisection-reconnection (TBR) branch swapping.
Estimation of the origin date of U.S. HIV subtype B.
To provide tree nodes outside the subtype B cluster, subtype A and D reference sequences were added to the previous gp160 alignment used for tree construction. Alignment was performed in DAMBE (41), and sites containing gaps were excluded. The Modeltest program (27) was applied to determine the appropriate DNA substitution model, and PAUP 4.0 was used to construct a maximum-likelihood tree. The alignment and the maximum-likelihood tree were used in a recently described SSCD procedure (Lemay et al., submitted), which removes positions in an alignment that interfere with clocklike behavior. The Baseml program of PAML (42) was used to estimate site likelihoods under the DR (different rate) and SRDT (single rate-dated tip) (31) models, using the general time reversible substitution model (21) with gamma rate heterogeneity. The difference in log-likelihood scores for the two models for each site was ordered according to their contribution to the overall log-likelihood difference under the two models. This order was subsequently used in stripping sites from the alignment (five by five, in a progressive fashion). These separate data sets were combined in a single file and rerun in Baseml, which provided mutation rates and internal dates for each progressively stripped alignment. The 95% confidence level for the likelihood ratio test (LRT) statistic necessary for rejection of a molecular clock was established by likelihood ratio testing. In addition, a 99% level of confidence for determining the number of sites needing stripping for clock detection was established by data set simulation through the Seq-Gen program (32) and subsequent input of the replicates into PAML for site-specific likelihood calculation.
Estimation of population history.
An additional 40 gp160 sequences with known year of collection were gathered from the LANL database and included with the sequences used in the dating analysis. This sequence set was manually aligned in Se-Al (Se-Al; A. Rambaut, distributed by the author at http://evolve.zoo.ox.ac.uk/software/), after removal of sites containing gaps. No evidence of hypermutation was found in the subtype B strains in this alignment when ConB95 was used as the reference strain in the HYPERMUT program (http://hiv-web.lanl.gov/content/hiv-db/HYPER/hypermut.html) (33). The Modeltest program (27) was used to estimate the model of DNA substitution and gamma rate heterogeneity. PAUP 4.0 (39) was used to build a neighbor-joining tree, which served as input for a heuristic search with TBR branch swapping to construct a maximum-likelihood tree. The tree was edited with the TreeEdit program (TreeEdit; A. Rambaut and M. Charleston, 2001, phylogenetic tree editor; distributed by the authors at http://evolve.zoo.ox.ac.uk/software/) to remove the subtype A and D and Haitian subtype B sequences and to root the tree using the Z2 strain (a subtype D strain collected in 1985), which was also subsequently removed. This tree, along with the sequence alignment, served as input into TipDate (31) to generate a tree based on the SRDT model, which transformed the noncontemporaneous sequences into branch lengths representing time. A maximum-likelihood rate of molecular evolution (nucleotide substitution rate) was also estimated. The resulting tree was analyzed with the program GENIE (30), which uses a coalescent approach to infer viral population history from a given phylogeny. Both parametric and nonparametric estimates of effective population size were obtained. An LRT was used to determine the parametric demographic with the best fit. Demographic parameter estimates with approximate 95% confidence intervals were also obtained from GENIE.
Nucleotide sequence accession numbers.
The gp160 nucleotide sequences from this study have been assigned GenBank accession no. AY247218 to AY247225, and the p17 nucleotide sequences have accession no. AY247226 to AY247233.
RESULTS
Phylogenetic tree analysis.
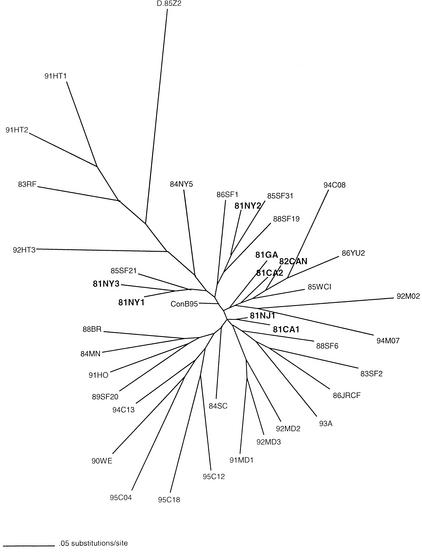
HIV-1 sequences with known year of sampling were extracted from the LANL HIV database in similar proportions to represent temporal groups of the North American subtype B epidemic, including the 1981-1982 strains determined in this study. The maximum-likelihood phylogeny estimated from the gp160 sequences is depicted in Fig. 1. The earliest sampled strains display short branches, typical of low-diversity strains in the beginning stages of an epidemic (15, 18, 19), while the consensus subtype B sequence (LANL, 1995) appears closest to the root of the tree. Four 1981 sequences grouped in two separate clusters (81NY1 and 81NY3 and 81NJ1 and 81CA1), which were supported by >70% bootstrap values in a separate neighbor-joining bootstrap tree (data not shown). These clusters and the other early sequences are, however, scattered in different lineages of the tree. Construction of a maximum-likelihood tree from p17 sequences resulted in similar short branch lengths and scattered lineages for the early strains but showed no supported clustering within the 1981-1982 strains (data not shown).
FIG. 1.
Maximum-likelihood tree of gp160 sequences, depicting the relationship of the early U.S. sequences (bold) to North American subtype B strains collected in later years. The numbers preceding the sequence names refer to the collection year. Strain D.85Z2 (subtype D) is the outgroup, and ConB95 is the consensus sequence for subtype B (HIV database, LANL).
Genetic distance comparisons.
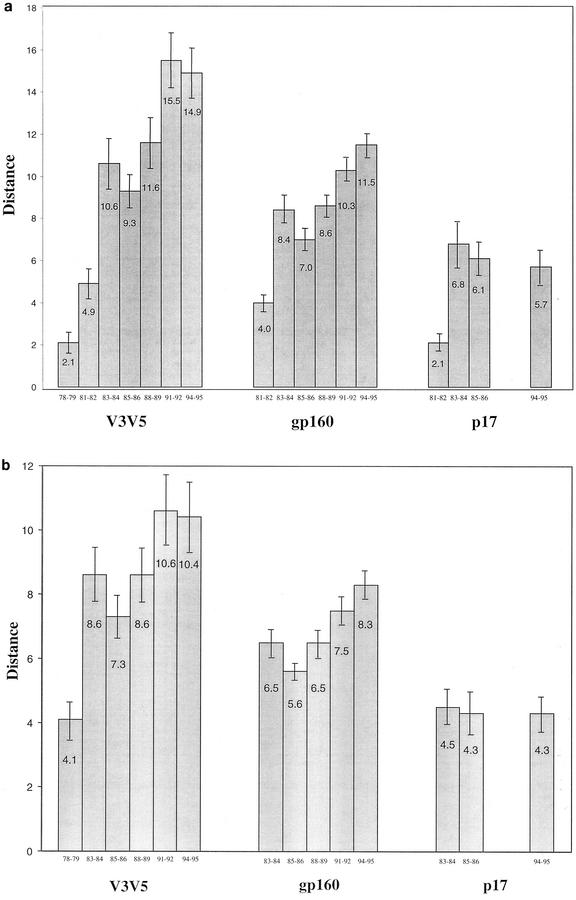
The within-group (years of collection) nucleotide distances depicted in Fig. 2a show an increasing diversity over time, across all the gene regions, with a leveling-off seen in the strains from the 1990s. The dip in the 1985-1986 group distances, seen especially in V3V5 and gp160, could be a sampling bias from the limited number of U.S. HIV sequences available from different time periods in the epidemic or alternatively could be indicative of a nonlinear relationship of diversity versus time in the epidemic or time of infection in individuals. The V3V5 region comparison displays the largest group distances, and the differences between groups through 1992 are significant (P < 0.05), with the exception of nonsignificant differences within the three groups from the 1980s (1983-1984, 1985-1986, and 1988-1989). There is a substantial difference between even the earliest strains (1978-1979 versus 1981-1982), although the sampling size is small (see Materials and Methods). The increasing distances within gp160 groups through 1992 are also statistically supported (P < 0.05), again with the exception of the three groups from the 1980s. By contrast, the p17 distances do not significantly differ from 1983-1984 through 1994-1995, following a significant (P < 0.001) increase from 1981-1982 to 1983-1984. Between-group (1981-1982 versus other groups) V3V5 and gp160 distances (Fig. 2b) also show an increasing diversity through time, with the exception of the 1981-1982 to 1985-1986 comparison, and a flattening of distances in the sequence groups from the 1990s. However, p17 between-group distances were very similar (Fig. 2b). In all gene regions, between-group distances were less than (in some cases not significantly) the corresponding later group's within-group distance.
FIG. 2.
Nucleotide distance comparisons within sequences grouped by year of collection, by gene region (a). (b) Between-group (1981-1982 group versus other groups) distances are shown. Numbers within columns are mean distances, and error bars are ± standard errors of the mean.
Estimated date of origin for U.S. HIV subtype B.
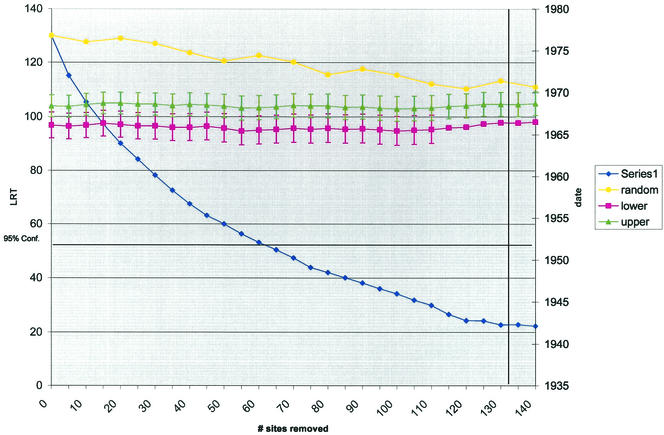
Stripping of sites from the gp160 sequence alignment, based on their contribution to the overall LRT statistic, produced a data set behaving in a molecular clock-like manner (Fig. 3). The 95% confidence limit according to a chi-square (χ2) distribution, for which higher LRT values signify a significant rejection of the molecular clock, is 56, which would indicate clock behavior of this data set after approximately 60 of the total 2,389 sites have been removed (Fig. 3). Using a more stringent data simulation and replicate likelihood distribution (see Materials and Methods) to establish a 99% site-specific cutoff (0.421), 133 sites in the alignment must be stripped to obtain a molecular clock (vertical bar in Fig. 3). Therefore, the dating results obtained from the alignment with 135 and 140 sites removed (approximately 5% of the total 2,389 sites) are the most rigorous for this data set. The date of the node, including Haiti and U.S. strains (Fig. 3, lower series), is 1966 ± 1.4, and the date of the node which includes only U.S. strains (Fig. 3, upper series) is 1968 ± 1.4. A smooth, consistently decreasing LRT value versus an increased site-stripping tracing was observed (Fig. 3, blue trace). The leveling in the later stages of stripping suggests that a relative “diminishing returns” is approached as higher numbers of sites are removed or that the LRT changes are no longer representative for site stripping. The power of the test is not diminished substantially at these levels of site removal, however, since the random stripping of variable sites (Fig. 3, yellow trace) does not dip appreciably during the procedure of stripping up to 140 sites. Also of note, the dating results were not significantly affected (overlapping of confidence intervals depicted in Fig. 3, both lower and upper series) by the amount of stripping, suggesting no time bias in the nonclock data.
FIG. 3.
Chart depiction of data from the SSCD analysis, showing estimated dates for the two internal gp160 tree nodes encompassing U.S. strains only (upper) and both U.S. and Haiti strains (lower). Each symbol within the lines represents the respective calculation performed on alignments with a progressive number of sites stripped (x axis). The blue line (series 1) represents the LRT test statistic 2 X (DR-SRDT) calculated for the alignments with sites removed based on their contribution to the overall LRT; the yellow line represents the LRT statistic calculated for alignments where variable sites were progressively removed at random (testing the power of the procedure). The 95% confidence limit according to a χ2 distribution for which higher LRT values signify a significant rejection of the molecular clock is drawn as a horizontal line. The more rigorous 99% site-specific cutoff, signifying the number of sites needing stripping (133) for molecular clock rejection, is shown as a vertical line. The chart simultaneously plots the LRT values (blue and yellow lines) versus the number of sites removed, together with the date of the internal node (upper and lower) corresponding to the number of sites removed.
Population history of the U.S. HIV epidemic.
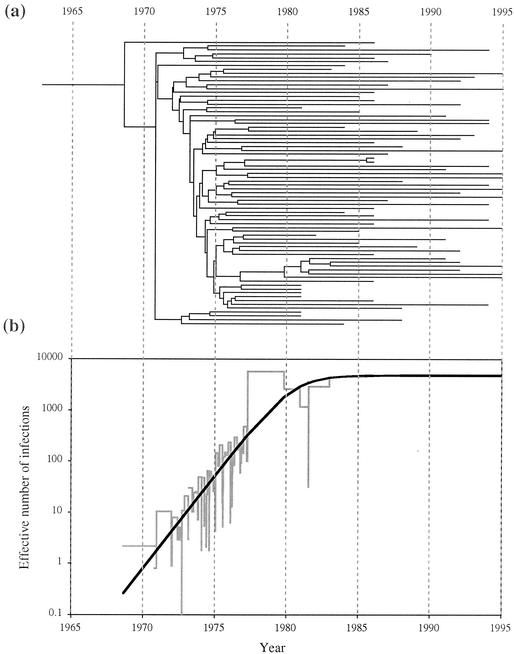
Coalescent analysis of the gp160 U.S. subtype B sequences involved three steps: (i) estimating the phylogeny with the TipDate program, assuming a constant rate of substitution (molecular clock), while accommodating the different collection dates (stripping of sites was not utilized to obtain a clocklike data set); (ii) obtaining a nonparametric estimate of viral population history (skyline plot); and (iii) determining parametric maximum-likelihood estimates of the viral population history under two models—exponential growth and logistic growth. Figure 4 depicts the maximum-likelihood TipDate tree on the same time scale as the parametric and nonparametric estimates of demographic history. The molecular rate of evolution used to scale the graph to years was estimated by TipDate to be 0.00473 substitution/site/year. By comparison, the substitution rate estimated by Korber et al. (16) for a large data set of gp160 sequences from multiple HIV-1 subtypes was 0.0024, while the substitution rate used by Leitner et al. (23) for a small set of V3 sequences was 0.0067. These differences may be related to the exact gene region, the alignment, the model of substitution, and/or the degree of homogeneity in the data set used for estimation. The stepwise nonparametric estimate of effective population size (skyline plot) is followed cleanly by the maximum-likelihood (parametric) estimate (Fig. 4b). The logistic growth model was a significantly better fit than the exponential growth model. Approximate 95% confidence intervals of parameter estimates for the logistic model were as follows: current effective population size (theta) = 4,830 (1995, 26,750); exponential growth rate (rho) = 0.834 (0.72, 0.945); and logistic shape parameter (c) = 5.30 E-06 (3.7 E-07; 1.29 E-04). The lack of coalescent events between roughly 1985 and 1995 (Fig. 4a) indicates that little information is available about changes in effective population size during that time period. The flat line, seen between the early 1980s and 1995 in Fig. 4b, cannot therefore be unambiguously interpreted as constant growth, but it does reflect the harmonic mean of effective population size during that time period. The demographic signal preceding the earliest sampled sequences in this data set (1981), however, is strong. The rooting of the TipDate tree is consistent with the analysis estimate represented in Fig. 3 (approximately 1968).
FIG. 4.
Population growth history analysis of U.S. subtype B gp160 sequences using the TipDate and Genie programs. (a) TipDate-generated maximum-likelihood tree (SRDT model). (b) Genie analysis depicting coalescent estimates of effective population size (number of infections). The stepwise plot (shaded line) is the nonparametric estimate. The smooth plot (bold curve) is the maximum-likelihood parametric estimate obtained under the logistic growth model.
DISCUSSION
Using the sequences of HIV-1 strains isolated early in the U.S. epidemic in conjunction with HIV database sequences, we conducted a detailed analysis to evaluate both the evolution of the subtype B viruses and the dynamics of the epidemic in North America. The early epidemic strains were collected in the same year (1981) that AIDS cases were first reported (3) and were part of a case control study (13) involving patients from three U.S. regions (New York, California, and Georgia). Given the presumed early time point in the U.S. epidemic when these strains were circulating, the expectation exists for a close phylogenetic association among strains collected in later years, if a very limited number of viruses were responsible for the start of the epidemic. The observation of eight strains along four different lineages would reject the hypothesis of a single founder virus responsible for the U.S. subtype B epidemic. Alternatively, HIV may have been introduced in the late 1960s, allowing these strains sufficient time to diverge. Comparing nucleotide genetic distances between and among sequences grouped by year of collection also provided useful evolutionary information. While the increasing HIV-1 diversity seen over the course of roughly two decades was expected (15, 18), the leveling off of later year intragroup and intergroup distances could be due to stochastic (random) effects or to the fact that the relationship between diversity and time is not linear throughout this epidemic. In a previous study of V3V5 sequences collected from San Francisco in 1978-1979 (7), distances of later year sequences were shown to be closer to those of earlier strains than to each other, a trend also observed in our study for both V3V5 and gp160 sequences. This observation, along with the fact that the subtype B gp160 consensus sequence (LANL, 1995) lies closest to the tree root and a consensus of 1978-1981 V3 loop sequences is an exact match to the V3 loop of the gp160 consensus sequence, implies that a consensus of early strains may be a reasonable vaccine candidate.
Estimating the date of origin of HIV subtype B, which spread into the U.S. population, is important for understanding the dynamics of the epidemic in North America. Using the recently developed clock detection tool (SSCD based on likelihood ratio reduction), we achieved a data set that behaves in a clocklike manner. Even without sites being stripped in this sequence set (the molecular clock theoretically not in effect), the dating was remarkably similar to estimates from stripped alignments, suggesting that the rate variability did not bias the overall estimations of the date and evolutionary rate in this homogeneously sampled data set. Given that the time course for HIV infection to AIDS onset can vary substantially in individuals, potentially confounding the relationship of year of collection specimens accurately reflecting the respective year of the epidemic, one might have expected more rate variation (and subsequent increased variability in date estimates) than was actually observed in our data. A linear regression analysis of the U.S. strains we used, plotted by sampling date versus divergence (substitutions/site) from the root, resulted in tight clustering of the samples to the regression line (data not shown), which also would indicate that the sample collection dates were representative of the epidemic year. Additionally, our time estimates were similar to those in another study (16) that modeled for the uncertainty of viral sequence year of origin into their analysis. The finding that estimates for date of origin of Haitian sequences closely coincide with those for the U.S. strains is consistent with commonly held assumptions of an epidemiological association between Haitian and U.S. homosexual contacts, which led to HIV spread between the countries (19). Our estimate of 1968 as the date of origin of U.S. HIV subtype B, roughly a decade before the earliest documented U.S. infection in 1977 (40), is similar to findings of other investigators (16). Since the median clinical latency from HIV infection is typically 10 years (6), the time interval of the introduction of HIV to the first recognition of AIDS in 1981 is consistent with the long latency period for the virus. Ten years is only the median latency period, and thus, there is a presumption of the existence of individuals with a shorter latency; thereby, AIDS cases in the early to mid 1970s likely existed. AIDS cases are difficult to confirm retrospectively, yet there are a few possible and probable cases from Haiti and the United States in the early 1970s (10). Also, assuming all 12 of the U.S. AIDS cases previously identified in 1978 (36) were not fortuitously rapid progressors (HIV infection progression to AIDS in 2 to 3 years or less), there must have been at least some HIV infections in the early 1970s. The date of origin of the founder virus(es) represents the most recent common ancestor (8, 16) of the U.S. subtype B strains, in essence the time period of divergence apart from the group M HIV-1 viruses. Whether this diversification began before, during, or after HIV-infected individuals arrived in the United States cannot be answered by this analysis.
Last, the gp160 data set was expanded to include additional HIV database sequences collected from throughout the U.S. epidemic. This larger data set was used in a coalescent theory framework (30) to estimate the population growth dynamics of the U.S. HIV epidemic. The theoretical problem, discussed above, of some sample collection dates potentially not reflecting the times of infection should not appreciably interfere with this analysis, since a random sampling of strains from throughout the population is an important assumption of coalescent analysis. Additionally, the crucial relationship of branch lengths being approximately proportional to time is not affected by lengths of infection. As anticipated, the estimated current effective population size (for the year 1995) of 4,830 (95% confidence intervals of 1995 to 26,750) is smaller than the number of U.S. AIDS cases in 1995 (approximately 200,000) (4); effective population sizes are routinely smaller than true population sizes because they are a function of several parameters, including the variance in reproductive success among infections and times between transmission events. Although these parameters may influence estimates of the effective numbers of infections, they do not affect the shape of the epidemic growth curve or the estimate of exponential growth rate. Our results are most useful in providing information about the increase in HIV infections in the time predating HIV specimen collection (before 1981, for the gp160 sequences). The slope of the epidemic curve, detailing the increase in effective number of HIV infections, implies rapid, exponential growth from the introduction of the strain(s) into the susceptible population. The best-fitting model of logistic growth is consistent with epidemiological evidence of a decline in new U.S. HIV infections from the mid-1980s to the mid-1990s, as a result of behavioral interventions and other HIV prevention strategies (4). The estimated exponential growth rate for the epidemic is 0.834/year, corresponding to a population doubling time of roughly 1 year. By comparison, similar analysis of growth rate estimates for the globally distributed hepatitis C virus subtype 1a was 0.098 (29) and for HIV-1 infections in the Democratic Republic of Congo was 0.1676 (43). Additionally, the growth estimates in effective infections show an increase of almost four orders of magnitude in a little more than a decade. Taken together, these results suggest no lag time in the spread of HIV upon introduction into the United States, but rather, an immediate, exponentially growing proliferation into the susceptible population. Discernible demographic information for the time period 1980-1995 was not obtainable (a flatline characteristic in the genealogical plots) for the currently available U.S. gp160 sequences, presumably due to the paucity of coalescent events estimated from this data set. As more sequences from the U.S. subtype B epidemic become available, it would be interesting to determine if demographic information later in the epidemic is obtainable with this methodology.
Acknowledgments
We thank Charles Schable at the Centers for Disease Control and Prevention for help in obtaining the early samples.
P.L. was supported by the Flemish Institute for Scientific-Technological Research in Industry (IWT); M.S. is a postdoctoral fellow with the Fonds voor Wetenschappelijk Onderzoek (FWO). In part, this work was supported by the Flemish Fonds voor Wetenschappelijk Onderzoek (FWO G.0288.01).
REFERENCES
- 1.Auerbach, D. M., W. W. Darrow, H. W. Jaffe, and J. W. Curran. 1984. Cluster of cases of the acquired immune deficiency syndrome. Am. J. Med. 76:487-492. [DOI] [PubMed] [Google Scholar]
- 2.Brodin, S. K., J. R. Mascola, P. J. Weiss, S. I. Ito, K. R. Porter, A. W. Artenstein, F. C. Garland, F. E. McCutchan, and D. S. Burke. 1995. Detection of diverse subtypes in the USA. Lancet 346:1198-1199. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1981. Pneumocystis pneumonia—Los Angeles. Morb. Mortal. Wkly. Rep. 30:1-3. [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2001. HIV/AIDS—United States, 1981-2000. Morb. Mortal. Wkly. Rep. 50:430-433. [PubMed] [Google Scholar]
- 5.Delwart, E. L., E. G. Shpaer, F. E. McCutchan, J. Louwagie, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a heteroduplex mobility assay: analysis of HIV env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 6.Evans, L. A., and J. A. Levy. 1993. The heterogeneity and pathogenicity of HIV, p. 29-73. In W. J. W. Morrow and N. L. Haigwood (ed.), HIV molecular organization, pathogenicity and treatment. Elsevier, Amserdam, The Netherlands.
- 7.Foley, B., H. Pan, S. Buchbinder, and E. L. Delwart. 2000. Apparent founder effect during the early years of the San Francisco HIV type 1 epidemic (1978-1979). AIDS Res. Hum. Retrovir. 16:1463-1469. [DOI] [PubMed] [Google Scholar]
- 8.Hillis, D. M. 2000. Origins of HIV. Science 288:1757-1759. [DOI] [PubMed] [Google Scholar]
- 9.Holmes, E. C., O. G. Pybus, and P. H. Harvey. 1999. The molecular population dynamics of HIV-1, p. 177-207. In K. A. Crandall (ed.), The evolution of HIV. The Johns Hopkins University Press, Baltimore, Md.
- 10.Hooper, E. 1999. In The river, p. 77-82; 440-443. Little, Brown & Co., Boston, Mass.
- 11.Hu, D. J., T. J. Dondero, M. A. Rayfield, J. R. George, G. Schochetman, H. W. Jaffe, C.-C. Luo, M. L. Kalish, B. G. Weniger, C.-P. Pau, C. A. Schable, and J. W. Curran. 1996. The emerging genetic diversity of HIV. JAMA 275:210-216. [PubMed] [Google Scholar]
- 12.Irwin, K. L., C.-P. Pau, D. Lupo, D. Pienazek, C.-C. Luo, N. Olivo, M. Rayfield, D. J. Hu, J. T. Weber, R. A. Respess, R. Janssen, P. Minor, and J. Ernst. 1997. Presence of human immunodeficiency virus (HIV) type 1 subtype A infection in a New York community with high HIV prevalence. J. Infect. Dis. 176:1629-1633. [DOI] [PubMed] [Google Scholar]
- 13.Jaffe, H. W., K. Choi, P. A. Thomas, H. W. Haverkos, D. M. Auerbach, M. E. Guinan, M. F. Rogers, T. J. Spira, W. W. Darrow, M. A. Kramer, S. M. Friedman, J. M. Monroe, A. E. Friedman-Kien, L. J. Laubenstein, M. Marmor, B. Safai, S. K. Dritz, S. J. Criopi, S. L. Fannin, J. P. Orkwis, A. Kelter, W. R. Rushing, S. B. Thacker, and J. W. Curran. 1983. National case-control study of Kaposi's sarcoma and Pneumocystis carinii pneumonia in homosexual men: part 1, epidemiologic results. Ann. Int. Med. 99:145-151. [DOI] [PubMed] [Google Scholar]
- 14.Kingman, J. F. C. 1982. The coalescent. Stochastic Processes Their Appl. 13:235-248. [Google Scholar]
- 15.Korber, B., J. Theiler, and S. Wolinsky. 1998. Limitations of a molecular clock applied to considerations of the origin of HIV-1. Science 280:1868-1871. [DOI] [PubMed] [Google Scholar]
- 16.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 17.Kostrikis, L. G., S. Shin, and D. H. Ho. 1998. Genotyping HIV-1 and HCV strains by a combinatorial DNA melting assay (COMA). Mol. Med. 4:443-453. [PMC free article] [PubMed] [Google Scholar]
- 18.Kuiken, C. L., G. Zwart, E. Baan, R. A. Coutinho, J. A. R. van den Hoek, and J. Goudsmit. 1993. Increasing antigenic and genetic diversity of the V3 variable domain of the human immunodeficiency virus envelope protein in the course of the AIDS epidemic. Proc. Natl. Acad. Sci. USA 90:9061-9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuiken, C., R. Thakallapalli, A. Eskild, and A. de Ronde. 2000. Genetic analysis reveals epidemiologic patterns in the spread of human immunodeficiency virus. Am. J. Epidemiol. 152:814-822. [DOI] [PubMed] [Google Scholar]
- 20.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinsky (ed). 2000. Human retroviruses and AIDS 2000: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 21.Lanave, C., G. Preparata, C. Saccone, and G. Serio. 1984. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20:86-93. [DOI] [PubMed] [Google Scholar]
- 22.Laverdiere, M., J. Tremblay, R. Lavallee, Y. Bonny, M. Lacombe, J. Boileau, J. Lachapelle, and C. Lamoureux. 1983. AIDS in Haitian immigrants and in a Caucasian woman closely associated with Haitians. Can. Med. Assoc. J. 129:1209-1212. [PMC free article] [PubMed] [Google Scholar]
- 23.Leitner, T., D. Ecanilla, C. Franzen, M. Uhlen, and J. Albert. 1996. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc. Natl. Acad. Sci. USA 93:10864-10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masur, H., M. A. Michelis, J. B. Greene, I. Onorato, R. A. Stouwe, R. S. Holzman, G. Wormser, L. Brettman, M. Lang, H. W. Murray, and S. Cunningham-Rundles. 1981. An outbreak of community-acquired Pneumocystis carinii pneumonia: initial manifestation of cellular immune dysfunction. N. Engl. J. Med. 305:1431-1438. [DOI] [PubMed] [Google Scholar]
- 25.Myers, G. 1994. Tenth anniversary perspectives on AIDS. HIV: between past and future. AIDS Res. Hum. Retrovir. 10:1317-1324. [DOI] [PubMed] [Google Scholar]
- 26.Noel, G. E. 1988. Another case of AIDS in the pre-AIDS era. Rev. Infect. Dis. 10:688-689. [DOI] [PubMed] [Google Scholar]
- 27.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 28.Pybus, O. G., A. Rambaut, and P. H. Harvey. 2000. An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics 155:1429-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pybus, O. G., M. A. Charleston, S. Gupta, A. Rambaut, E. C. Holmes, and P. H. Harvey. 2001. The epidemic behavior of the hepatitis C virus. Science 292:2323-2325. [DOI] [PubMed] [Google Scholar]
- 30.Pybus, O. G., and A. Rambaut. 2002. GENIE: estimating demographic history from molecular phylogenies. Bioinformatics 18:1404-1405. [DOI] [PubMed] [Google Scholar]
- 31.Rambaut, A. 2000. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics 16:395-399. [DOI] [PubMed] [Google Scholar]
- 32.Rambaut, A., and N. C. Grassly. 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput. Appl. Biosci. 13:235-238. [DOI] [PubMed] [Google Scholar]
- 33.Rose, P. P., and B. T. Korber. 2000. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics 16:400-401. [DOI] [PubMed] [Google Scholar]
- 34.Salemi, M., K. Strimmer, W. W. Hall, M. Duffy, E. Delaporte, S. Mboup, M. Peters, and A.-M. Vandamme. 2001. Dating the common ancestor of SIVcpz and HIV-1 group M and the origin of HIV-1 subtypes by using a new method to uncover clock-like molecular evolution. FASEB J. 15:276-278. [DOI] [PubMed] [Google Scholar]
- 35.Schochetman, G. S., S. Subbarao, and M. L. Kalish. 1996. Methods for studying genetic variation of the human immunodeficiency virus, 25-41. In K. A. Adolph (ed.), Viral genome methods. CRC Press Inc., Boca Raton, Fla.
- 36.Selik, R. M., H. W. Haverkos, and J. W. Curran. 1984. Aquired immune deficiency syndrome (AIDS) trends in the United States, 1978-1982. Am. J. Med. 76:491-500. [DOI] [PubMed] [Google Scholar]
- 37.Sharp, P. M., D. L. Robertson, F. Gao, and B. H. Hahn. 1994. Origins and diversity of human immunodeficiency viruses. AIDS 8:S27-S43. [Google Scholar]
- 38.Sullivan, P. S., A. N. Do, D. Ellenberger, C.-P. Pau, S. Paul, K. Robbins, M. Kalish, C. Storck, C. A. Schable, H. Wise, C. Tetteh, J. I. Jones, J. McFarland, C. Yang, R. B. Lal, and J. W. Ward. 2000. Human immunodeficiency virus (HIV) subtype surveillance of African-born persons at risk for group O and group N HIV infections in the United States. J. Infect. Dis. 181:463-469. [DOI] [PubMed] [Google Scholar]
- 39.Swofford, D. L. 1998. PAUP*, phylogenetic analysis using parsimony (* and other methods). Sinauer & Assoc., Sunderland, Mass.
- 40.Thomas, P., R. O'Donnel, R. Williams, and M. A. Chiasson. 1988. HIV infection in hetero-sexual female intravenous drug users in New York City, 1977-1980. N. Engl. J. Med. 319:374.. [PubMed] [Google Scholar]
- 41.Xia, X. 2000. DAMBE (data analysis in molecular biology and evolution). Kluwer Academic Publishers, Boston, Mass.
- 42.Yang, Z. 1997. PAML: a program for phylogenetic analysis by maximum likelihood. Comput. Appl. Biol. Sci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 43.Yusim, K., M. Peeters, O. G. Pybus, T. Bhattacharya, E. Delaporte, C. Mulanga, M. Muldoon, J. Theiler, and B. Korber. 2001. Using human immunodeficiency virus type 1 sequences to infer historical features of the acquired immune deficiency syndrome epidemic and human immunodeficiency virus evolution. Philos. Trans. R. Soc. Lond. B 356:855-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, T., B. T. Korber, A. J. Nahmias, E. Hooper, P. M. Sharp, and D. D. Ho. 1998. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature 391:594-597. [DOI] [PubMed] [Google Scholar]