Abstract
NADPH oxidase is implicated in the pathogenesis of various cardiovascular disorders. In vascular smooth muscle cells (VSMC), expression of NOX1 (NADPH oxidase 1), a catalytic subunit of NADPH oxidase, is low and is induced upon stimulation by vasoactive factors, while it is abundantly expressed in colon epithelial cells. To clarify the regulatory mechanisms underlying such cell-specific expression, the upstream regions directing transcription of the NOX1 gene were explored. In P53LMACO1 cells, a cell line originated from mouse VSMCs, two novel Nox1 mRNA species, the c- and f-type, were isolated. These transcripts contained 5′-untranslated regions that differed from the colon type mRNA (a-type) and encoded an additional N-terminal peptide of 28 amino acids. When these transcripts were fused to the c-myc tag and expressed in human embryonic kidney 293 cells, a fraction of translated proteins demonstrated the size containing the additional peptide. Proteins encoded by the c- and f-type mRNAs exhibited superoxide-producing activities equivalent to the activity of the a-type form. The a-type mRNA was expressed in the colon and in the intact aorta, whereas the c-type mRNA was detected in the primary cultured VSMCs migrated from aortic explants, in vascular tissue of a wire-injury model and in the thoracic aorta of mice infused with angiotensin II. The promoter region of the c-type mRNA exhibited transcriptional activity in P53LMACO1 cells, but not in MCE301 cells, a mouse colon epithelial cell line. These results suggest that expression of the Nox1 gene is regulated by alternative promoters and that the novel c-type transcript is induced under phenotypic modulation of VSMCs.
Keywords: alternative promoters, colon epithelial cell (CEC), NADPH oxidase, NOX1, phenotypic modulation, vascular smooth muscle cell (VSMC)
Abbreviations: ATF-1, activating transcription factor 1; CEC, colon epithelial cell; DMEM, Dulbecco's modified Eagle's medium; FBS, foetal bovine serum; HEK-293, human embryonic kidney 293; NOX1, NADPH oxiadase 1; 5′-RACE, 5′-rapid amplification of c-DNA ends; RT-PCR, reverse transcriptase-PCR; 5′-UTR, 5′-untranslated region; VSMC, vascular smooth muscle cell
INTRODUCTION
In cardiovascular tissues, ROS (reactive oxygen species) such as superoxide anion (O2−) and hydrogen peroxide act as signalling molecules; they are involved in phenomena ranging from vasorelaxation to vascular remodelling. NADPH oxidase is recognized as a main source of ROS in the vascular system. The enzyme, initially characterized in phagocytes, is composed of multiple subunits including a membrane-spanning catalytic subunit, named NOX1 (NADPH oxiadase 1) [1,2]. NADPH oxidase in phagocytes consists of the catalytic subunit NOX2 (gp91phox), with the regulatory subunits p22phox, p47phox, p40phox, p67phox and the small GTPase Rac [3]. Six homologues of phagocyte-type NOX2 have been reported so far. Among them, NOX1/NADPH oxidase has been implicated in the proliferation and hypertrophy of cultured VSMCs (vascular smooth muscle cells). Although the basal level of NOX1 expressed in VSMC is relatively low, it is increased upon stimulation by numerous vasoactive factors, including angiotensin II and platelet-derived growth factor [1,4]. We also observed that prostaglandin F2α, one of the primary prostanoids generated in vascular tissue, causes hypertrophy of VSMCs by induction of NOX1 and a subsequent increase in O2− generation [5]. Thus up-regulation of NOX1 may play a key role in the development of vascular remodelling associated with various metabolic and cardiovascular disorders [6].
However the transcriptional regulation of the NOX1 gene has not been elucidated fully. We reported previously the involvement of ATF-1 (activating transcription factor 1), a transcription factor of the cAMP-response-element-binding protein/ATF family, in induction of Nox1 in rat VSMCs [7]. Recently, an interferon-γ-responsive element was identified in the upstream region of the human NOX1 gene [8]. In colon epithelial Caco-2 cells, maximal promoter activity was reported to be dependent on a GATA-binding site located in the proximal promoter region of the human NOX1 gene [9]. In contrast with the abundant expression of NOX1 in CECs (colon epithelial cells) and in colon-carcinoma cell lines [1,10], low expression of NOX1 has been documented in vascular tissue. Accordingly, the NOX1 gene may have distinct mechanisms of transcriptional control between VSMCs and CECs. To clarify the regulatory mechanisms underlying the cell-specific expression of NOX1, we characterized the upstream regions directing transcription of the NOX1 gene. We here report the presence of novel NOX1 transcripts induced under phenotypic modulation of VSMCs. Expression of these transcripts was regulated by alternative promoters directing the cell-specific expression of the gene.
EXPERIMENTAL
RNA preparation
Total RNA was extracted from mouse tissues or cultured cells using Sepasol reagent (Nacalai Tesque, Kyoto, Japan) and purified further with RNeasy columns (Qiagen, Hilden, Germany), including DNaseI treatment according to the manufacturer's instructions. The cell line, P53LMACO1, originating from mouse VSMCs was obtained from the Japan Health Science Foundation (Osaka, Japan). P53LMACO1 cells were cultured in DMEM (Dulbecco's modified Eagle's medium) containing 10% (v/v) FBS (foetal bovine serum) as described previously [11]. MCE301 cells, a mouse cell line originating from CECs, were cultured on collagen type I-coated dishes in DMEM/Hams-F12 (1:1, v/v) medium containing 3% FBS, insulin-transferrin-selenium supplement and 10 ng/ml epidermal growth factor as described previously [12]. The thoracic aorta and colon were dissected from 8–12 week-old male C57BL/6J mice. To obtain dedifferentiated VSMCs, primary culture was performed using the migration method described previously [13].
Animal models
Transluminal arterial injury surgery was carried out on the left legs of 6-week-old male C57BL/6J mice as described previously [14]. At 14 days after surgery, the mice were killed and the femoral arteries were dissected. For angiotensin II infusion, 8-week-old male mice were anaesthetized with sodium pento-barbital (80 mg/kg) and osmotic minipumps were implanted as described previously [15]. Angiotensin II was delivered for 7 days at a rate of 0.7 mg/kg per day. Animal studies were carried out following the guidelines for animal care with the approval of the Committee for Animal Research at Kyoto Prefectural University of Medicine (Kyoto, Japan).
5′-RACE (5′-Rapid amplification of c-DNA ends) analysis
5′-RACE was carried out using a 3′/5′-RACE kit (Roche, Indianapolis, IN, U.S.A.). Total RNA isolated from either P53LMACO1 cells or the colon was reverse transcribed with the primer NR1, which is complementary to the nucleotides 80–104 of mouse Nox1 mRNA (Genbank AF539799, Figure 1A). The cDNAs were tailed with dATP using terminal deoxytransferase and then the first round of amplification was carried out using the oligo dT-anchor primer and the second primer NR2, which is complementary to the nucleotides 55–79 of the mouse Nox1 mRNA. The resulting PCR products were used as templates for a subsequent nested amplification of cDNAs specific for Nox1 with the anchor primer and the primer NR3, which is complementary to the region covering the start codon (Figure 1A). In another set of 5′-RACE experiments for the a-type mRNA, total RNA from the colon was reverse transcribed with the primer NR3. The product was amplified with the primer NR4 and then with the primer NR5. For the 5′-end of the c-type mRNA, total RNA from P53LMACO1 cells was reverse transcribed with the primer 1c-ant00. The product was amplified with the primer 1c-ant01 and then with the primer 1c-ant02. For the f-type mRNA, total RNA from P53LMACO1 cells was reverse transcribed with the primer 1f-ant00. The product was amplified with the primer 1f-ant01 and then with the primer 1f-ant02. On the basis of sequencing analyses, 5′-ends of the longest cDNA clones were regarded as the transcriptional start sites and denoted as the +1 position.
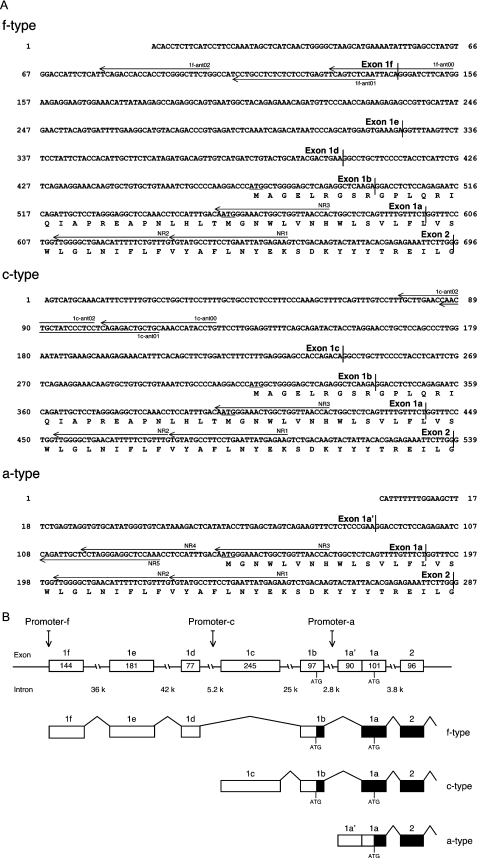
Figure 1. Structure of the novel transcript forms of the Nox1 gene.
(A) 5′-Nucleotide sequences of the f-, c- and a-type Nox1 cDNA and deduced amino acid sequences at the beginning of open reading frames. Translational start sites are underlined. Primers used for 5′-RACE are indicated with arrows. (B) The exon/intron structure of the Nox1 gene and its alternative splicing pathways. Open boxes indicate exons. Numbers in the box denote the size of each exon (bp). The size of the intron is indicated under the broken lines (bp). Closed boxes show the open reading frames of the transcript.
Detection of Nox1 proteins by Western blotting
To examine the integrity of the c-, f- and a-type Nox1 transcripts, cDNAs including the initial exon and exon 13, containing the stop codon, were amplified by PCR. Restriction enzyme mapping of the PCR products revealed that the downstream constituents of the exon 1a were identical in all Nox1 transcript forms. A shorter spliced variant devoid of the exon 11 [16,17] was not detected in mouse tissues or cultured cells (results not shown).
To detect NOX1 proteins, an oligonucleotide encoding the c-myc tag sequence (EQKLISEEDL) was introduced at the stop codon of Nox1 cDNA by PCR. Isolated cDNAs were cloned into pcDNA3 (Invitrogen, Carlsbad, CA). These plasmids (3 μg) were transfected to HEK-293 (human embryonic kidney 293) cells using FuGENE™ 6 transfection reagent (Roche). The whole cell extracts were prepared 24 h after transfection and Western blotting was carried out essentially as described previously [18]. The c-myc-tagged NOX1 proteins were detected with the mouse monoclonal antibody 9B11 (Cell Signaling) and horseradish-peroxidase-linked secondary antibody.
Measurement of superoxide production
Full-length cDNAs of NOXA1 and NOXO1 were isolated from human colon epithelia. HEK-293 cells were seeded on 24-well plates, and pcDNA3 plasmids containing Nox1 (without the c-myc tag), NOXA1 and NOXO1 cDNAs (0.25 μg each/well) were transfected to the cells using Lipofectamine 2000 (Invitrogen). After 24 h, the cells were trypsinized and the superoxide-producing activities were measured using lucigenin in the presence of 10 μM diphenyleneiodonium chloride or without (Sigma) as described previously [19].
RT-PCR (reverse transcriptase-PCR) analysis
To detect expression of each transcript, total RNA was reverse transcribed using Superscript III (Invitrogen) with the primer 5-R, which is complementary to the nucleotides 456–483 (GTTTGGAGACTGGATGGGATTTAGCCAA) of the mouse Nox1 mRNA. Different sets of the primers specific for the respective 5′-untranslated region (5′-UTR) of Nox1 transcripts (see Supplementary Table 1 at http://www.BiochemJ.org/bj/398/bj3980303add.htm) were next used to analyse the expression of each transcript form. PCR was performed, with 41 cycles required to detect the c-type and a-type forms and the common region, while 48 cycles were required to detect the f-type form. The identity of the PCR products was verified by sequencing or restriction-enzyme mapping analyses.
Reporter constructs and luciferase assay
The 5′-flanking regions of the corresponding exons for the identified transcripts were isolated from a mouse genomic library (Lambda FIXII Mouse 129/SVJ, Stratagene, La Jolla, CA, U.S.A.), and cloned into a pGL3-basic vector (Promega). The 1.5 kb 5′-flanking region of the f-type transcript was cloned into the SacI/SmaI site, the 3.0 kb region upstream of the c-type into the HindIII site and the 2.0 kb region upstream of the a-type into the HindIII/NcoI site of pGL3-basic respectively. A series of 5′-deletion constructs were made by cleavage with restriction enzymes or PCR. All constructs were subjected to sequence analyses to verify the orientations and fidelity of the insert.
Luciferase plasmids (0.75 μg/well) and a pSV-β-galactosidase control vector (0.25 μg/well; Promega) were co-transfected into the cells. P53LMACO1 and MCE301 cells were cultured for a further 24 or 48 h respectively, after which time luciferase activities in cell lysates were determined and normalized to β-galactosidase activity.
RESULTS
5′-UTRs of the novel transcripts and corresponding exons in the NOX1 gene
The human NOX1 mRNA expressed in the colon consists of 13 exons with the translational start codon located in the first exon [1,2]. The structure of the Nox1 gene is highly conserved in the mouse (Genbank AF539799). Several PCR products were depicted by the 5′-RACE analysis of the colon RNA. The longest 5′-end of the colon products was 146 bases upstream of the translational start site (Figure 1A, a-type).
In P53LMACO1 cells, a major band of approx. 450 bp and a minor band of 600 bp were detected in the first 5′-RACE analysis. Sequence analyses of these two products and another set of 5′-RACE experiments verified two splice isoforms containing different 5′-UTRs of the mouse Nox1 gene. The 5′-ends of these forms were different from those isolated from the colon. On the basis of genomic structure, we designated these novel Nox1 transcripts as the c-type and f-type mRNA. The conventional transcript isolated from the colon was tentatively designated as the a-type mRNA.
Both c- and f-type mRNA contained the same sequence as the a-type up to 56 bases upstream of the translational initiation codon, whereas the sequence further upstream was divergent. Accordingly, the first exon previously assigned to the colon Nox1 transcript form was divided into a common region (exon 1a) and its contiguous region (exon 1a'). As shown in Figure 1(B), the c-type form was initiated from a novel exon 1c and contained another exon 1b, both of which were placed on chromosome X (Genbank AL671915). Exon 1c was located approx. 30 kb upstream of exon 1a. On the other hand, the f-type form was not an extended form of the c-type since it did not contain exon 1c. It was initiated from exon 1f and contained additional exons 1e, 1d and 1b, each localized on chromosome X (Genbank AL672215 and AL671915). Exon 1f was placed approximately 110 kb upstream of exon 1a. All of these newly identified exons were spliced according to the GT-AG rule (see Supplementary Table 2 at http://www.BiochemJ.org/bj/398/bj3980303add.htm).
Characterization of NOX1 proteins encoded by the c- and f-type Nox1 mRNAs
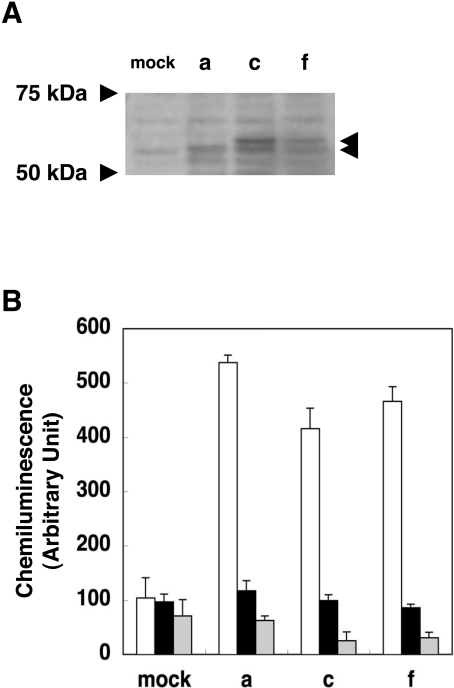
The a-type mRNA has an in-frame translational start site in exon 1a, indicating that the open reading frame starts from exon 1a. On the other hand, the f- and c-type mRNAs contain a putative translation start site with a perfect Kozak consensus sequence, CCATGG, in exon 1b located further upstream of exon 1a. To test whether the translation of the f- and c-type mRNAs is initiated at this site to yield an additional N-terminal peptide of 28 amino acids (Figure 1A), the c-myc tag was introduced into the C-terminus of Nox1. When these fusion proteins were expressed in HEK-293 cells, the antibody against the c-myc tag detected a single band at approx. 57 kDa in the cells transfected with the plasmid encoding the a-type fusion protein (Figure 2A, lane a). Whereas, doublet bands were detected in c- or f-type transfected cells. The faster migrating band was detected at 57 kDa, while the slower one was at approx. 60 kDa (Figure 2A, lanes c and f). These findings suggest that translation of the c- and f-type Nox1 mRNA is initiated at both the AUG in exon 1b and the start site in exon 1a.
Figure 2. Characterization of NOX1 proteins encoded by the novel transcript forms.
(A) NOX1–c-myc fusion proteins expressed in HEK-293 cells. Cells were transfected with vectors containing the f-, c- or a-type Nox1–c-myc tag fusion cDNA. Whole cell lysates (10 μg) were separated by SDS/PAGE (8.4% gels) and the c-myc-tagged proteins were detected with the mouse monoclonal antibody as described in the Experimental section. (B) Superoxide-producing activities of NOX1 proteins encoded by the novel transcripts. HEK-293 cells transfected with pcDNA3 containing Nox1, NOXA1 and NOXO1 cDNAs were incubated for 5 min in Krebs/Hepes buffer containing 5 μM lucigenin and 100 μM NADPH in the presence or absence of 10 μM diphenyleneiodonium chloride (DPI). The chemiluminescence was measured by a luminometer and expressed as means±SE (n=4). Open bar, cells transfected with NOX1, NOXA1 and NOXO1; closed bar, NOX1 and NOXA1 and NOXO1 and DPI; grey bar, NOX1 only.
Functional analysis of proteins encoded by the c- and f-type Nox1 mRNAs was next carried out in HEK-293 cells co-transfected with plasmids containing NOXA1 and NOXO1. As shown in Figure 2(B), NOX1 proteins encoded by the c- and f-type mRNAs exhibited superoxide-producing activities equivalent to the activity observed in the a-type form.
Cell specific expression of Nox1 transcripts
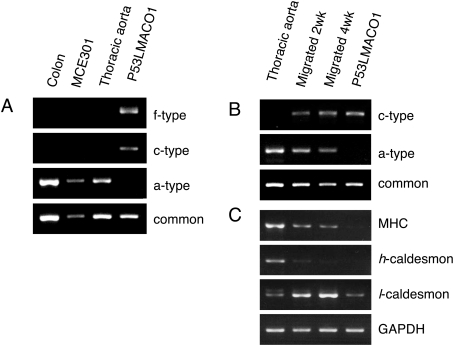
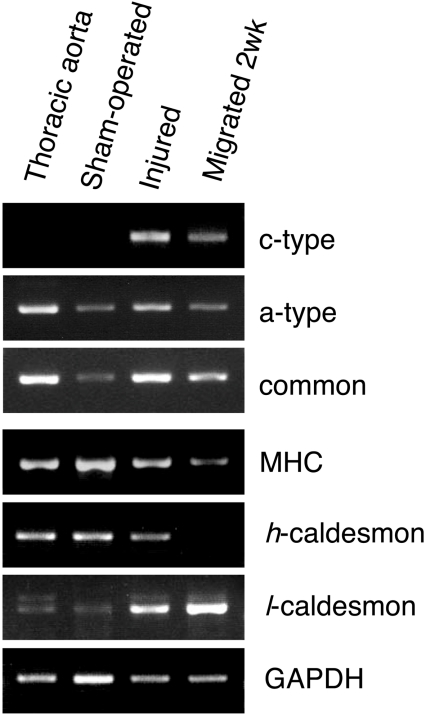
As shown in Figure 3(A), only the a-type Nox1 transcript was detected in the colon and MCE301 cells, a mouse colon epithelial cell line. Similarly, only the a-type mRNA was detected in the intact thoracic aorta. On the other hand, the f- and c-type mRNAs were detected in P53LMACO1 cells, but not the a-type. VSMCs within mature and intact aorta are in a differentiated state, while the dedifferentiated cells lose contractility and display phenotypic modulation, which results in cell proliferation, migration and changes in the expression of smooth muscle-specific molecular markers [20]. Accordingly, the expression of the f- and c-type transcripts might be associated with phenotypic modulation during cell culture. To confirm this, we analysed the 5′-UTR of the Nox1 mRNA in migrated VSMCs isolated from the thoracic aorta (Figure 3B). In these cells, expression of the c-type mRNA was observed while the level of the a-type decreased as the cells were grown in culture over a longer term. The f-type mRNA was undetectable in the migrated cells (results not shown). Among smooth muscle-specific molecular markers, the mRNA level of the myosin heavy chain was attenuated, while the expression of h-caldesmon was converted to l-caldesmon (Figure 3C). As the down-regulation of the myosin heavy chain and isoform conversion of caldesmon are known to take place during dedifferentiation [20], these results suggest that the conversion of Nox1 mRNA species from the a-type to the c-type is associated with dedifferentiation of VSMCs.
Figure 3. Cell-specific expression of the Nox1 transcripts.
(A) Expression of the f-, c-, and a-type Nox1 transcripts in mouse tissues and cell lines. Nox1 transcripts containing different 5′-UTRs or the common sequence (common) were amplified by RT-PCR with total RNA from indicated tissues and cells, and separated on 2.5% agarose gel. (B) Expression of the c-type transcript in migrated VSMCs. VSMCs migrated from aortic explants were cultured for 2- or 4-weeks, and expression of each transcript form was analysed by RT-PCR. (C) Expression of smooth muscle-specific molecular markers. Myosin heavy chain (MHC), h-, l-caldesmon and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNAs were detected in the total RNA used in (B). Figures are representative of at least three independent experiments.
Functional analysis of the Nox1 promoters
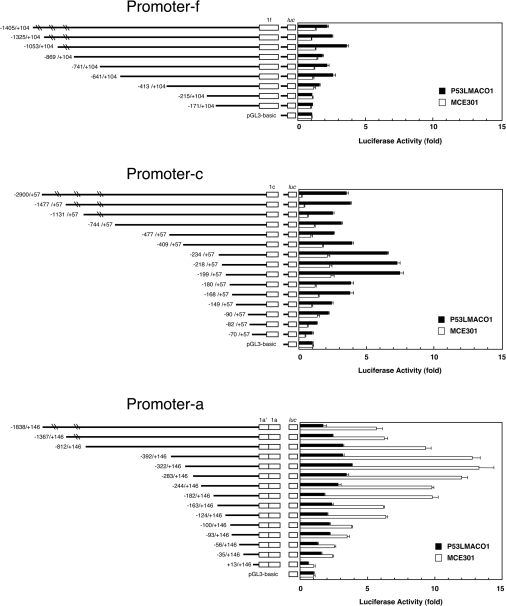
We determined the sequences of the 5′-flanking regions of the three Nox1 exons, 1f, 1c and 1a. These regions were designated as Promoter-f, -c and -a respectively (Figure 1B and Supplementary Figures IA–C at http://www.BiochemJ.org/bj/398/bj3980303add.htm). Nox1 promoter activities were determined using luciferase reporter constructs containing Promoter-f, -c and -a. As shown in Figure 4, Promoter-f showed luciferase activity in only the P53LMACO1 cells, but the activity itself was very low. In MCE301 cells, all of the 5′-deletion constructs were inert. These findings were consistent with the RT-PCR data, which showed the low expression of the f-type transcript in P53LMACO1 cells, while the f-type PCR product was absent in MCE301 cells. The deletion constructs of Promoter-c (between −2900 and −149 bp relative to the transcription start site) demonstrated between 2.5- and 17-fold higher activities in P53LMACO1 cells compared with MCE301 cells. By contrast, Promoter-a constructs showed preferential reporter activities in MCE301 cells. The region between −1838 and −124 bp relative to the transcription start site, showed between 2.5- and 4-fold higher activities in MCE301 cells compared with P53LMACO1 cells. These results were also consistent with the RT-PCR data, demonstrating the absence of the c-type transcript in MCE301 cells and of the a-type in P53LMACO1 cells.
Figure 4. Analyses of the Nox1 promoters in different cell lines.
(A) Schematic diagram of the promoter-luciferase fusion plasmids is shown on the left, where the 5′/3′ ends of the construct relative to the transcription initiation site are indicated. Each construct was transiently transfected into P53LMACO1 (closed bar) or MCE301 cells (opened bar). The β-galactosidase-expression vector was co-transfected as an internal control. The relative luciferase activity was denoted as the ratio to the activity of the pGL3-basic vector. Bars represent means ±SE of at least three experiments.
In Promoter-c, the region upstream of −1477 bp relative to the transcription start site seemed to augment the preferential expression in P53LMACO1 cells, because the constructs containing this region showed 10-fold higher reporter activities in P53LMACO1 cells compared with MCE301 cells. On the other hand, the sequence between −409 and −234 bp relative to the transcription start site appeared to suppress promoter activity. Deletion of the two regions, −199 to −180 bp and −168 to −149 bp relative to the transcription start site elicited a marked decrease in the activities of Promoter-c, which suggests that these regions may contain elements that are essential for the basal expression of the c-type mRNA. In Promoter-a, regions −182 to −163 and −124 to −100 relative to the transcription start site appeared to be pivotal for the expression of the a-type mRNA in MCE301 cells.
Expression of the c-type Nox1 mRNA in an injured artery
VSMCs in the vessels of adult animals dedifferentiate in response to injury to allow for remodelling and repair. To elucidate whether expression of the c-type Nox1 mRNA takes place in vivo and if it is associated with vascular remodelling, we analysed the transcripts isolated from the femoral artery of a wire-injury model. The c-type mRNA was expressed at 14 days after the injury with concomitant down-regulation of the myosin heavy chain and isoform conversion of caldesmon (Figure 5). Accordingly, expression of the c-type mRNA was demonstrated not only in in vitro cultured cells, but also in in vivo injured arteries. A close relationship between the expression of the c-type Nox1 mRNA and dedifferentiation of VSMCs was suggested. The f-type mRNA was not detected in the injured artery (results not shown).
Figure 5. Expression of the c-type Nox1 transcript in injured arteries.
Nox1 mRNA containing different 5′-UTRs, the common sequence (common), smooth muscle-specific molecular markers and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were detected by RT-PCR. A wire-injury was inflicted in the femoral artery of the left leg of each mouse. At 14 days after the surgery, total RNA was extracted from pooled arteries from the sham-operated right legs or the injured left legs of three mice. The thoracic aorta and migrated VSMCs cultured for 2 weeks were used as controls.
Expression of the c-type Nox1 mRNA in the aorta of mouse infused with angiotensin II
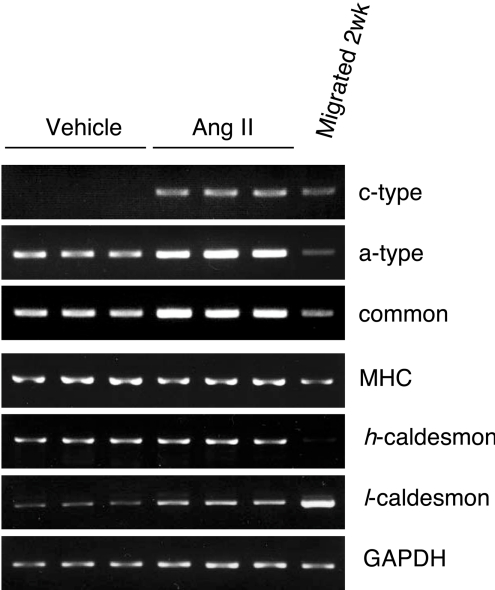
Angiotensin II is known to induce expression of the Nox1 gene in cultured VSMCs [4] as well as in the mouse aorta [15]. We therefore investigated which type of Nox1 mRNA was expressed in the thoracic aorta of mice infused with angiotensin II. After the infusion of a pressor dose of angiotensin II for 7 days, expression of the c-type Nox1 mRNA was clearly detected with concomitant up-regulation of the a-type mRNA (Figure 6). Quantitative real-time PCR in mice infused with angiotensin II for 14 days revealed that expression level of the c-type was approx. 15% of the a-type transcript (results not shown). The mRNA levels of the myosin heavy chain and h-caldesmon were relatively unchanged, whereas the level of l-caldesmon was increased in angiotensin II infused-mice. These results suggest that expression of the c-type Nox1 mRNA is up-regulated in angiotensin II-induced hypertrophy.
Figure 6. Expression of the c-type Nox1 transcript in the thoracic aorta of mice infused with angiotensin II.
Nox1 mRNA containing different 5′-UTRs, the common sequence (common), smooth muscle-specific molecular markers and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were detected by RT-PCR. Total RNA was isolated from three mice infused with angiotensin II or vehicle for 7 days.
DISCUSSION
The present study demonstrates the presence of novel Nox1 transcripts in the vascular cell lineage. These transcripts, designated as c-type and f-type mRNA, were initially identified in P53LMACO1 cells, an established cell line originated from mouse VSMCs.
Expression of the c-type mRNA was detected in the primary cultured VSMCs migrated from aortic explants, in vascular tissue of a wire-injury model and in the thoracic aorta of mice infused with angiotensin II. In migrated VSMCs, down-regulation of the myosin heavy chain and conversion of the caldesmon isoform were observed, while the f-type form was totally undetectable. The f-type mRNA appears to be expressed in fully dedifferentiated VSMCs after a long period of cultivation, or may be specific to P53LMACO1 cells, which were established from p53-deficient mice [11]. In the colon, where abundant expression of Nox1 is noted, only the conventional transcript form was detected. This transcript form, termed the a-type mRNA, was structurally similar to the form described previously in the human colon [1,2]. Predominant expression of the a-type mRNA was demonstrated in the intact aorta, while the c-type form was undetectable by RT-PCR in the unharmed aortic tissue. These results suggest that expression of the new c-type Nox1 mRNA was linked to the phenotypic modulation of VSMCs.
Expression of these Nox1 gene transcripts was governed by alternative promoters in a cell-specific fashion. Promoter reporter analyses revealed regions essential for expression of each Nox1 transcript form. In Promoter-c, these regions were located between −199 and −180 bp, and between −168 and −149 bp relative to the transcription start site respectively. In Promoter-a, the regions between −182 and −163 bp, and between −124 and −100 bp relative to the transcription start site respectively, were instrumental in the promoter activity in MCE301 cells. Putative binding sites for such transcription factors as HFH-2, HNF-3β, Oct-1, C/EBP, Cdx-1 and c-Ets-1 were found in these regions (see Supplementary Figures IB and IC at http://www.BiochemJ.org/bj/398/bj3980303add.htm). Binding of such transcription factors to the different promoters of the Nox1 gene may determine the cell-specific expression of the respective transcripts. Recently, it was reported that transcription of the human NOX1 gene in Caco-2 cells is governed by GATA-binding sites located in the proximal promoter region, one of which is conserved in the mouse Promoter-a (−199 to −194 bp relative to the transcription start site) [9]. Our reporter assay demonstrated, however, that this site is not essential for transcription in MCE301 cells. We also carried out luciferase assays using human Caco-2 cells. In these cells, the activity of a mouse Promoter-a construct (−392 to +146) was less than 3-fold higher than the basic vector, whereas that of a human NOX1 promoter construct (−349 to +221) was approx. 35-fold higher. Conversely, the human construct did not show any transcriptional activity in mouse MCE301 cells (results not shown). Accordingly, transcription of the NOX1 gene in CEC appears to be highly species-specific.
Nox1 proteins encoded by the f- and c-type mRNAs demonstrated superoxide-producing activities equivalent to the activity observed in the a-type form. While the translational start codon of the a-type transcript exists in exon 1a, translation of the f- and c-type mRNAs is initiated at the AUG in exon 1b as well as at the codon in exon 1a. We recently identified the expression of the c-type-like mRNA in A7r5, a rat vascular smooth muscle cell line. This transcript, however, did not encode an additional N-terminal peptide demonstrated in the mouse mRNAs (results not shown). In human chromosome X, a region homologous to exon 1c was found upstream of exon 1a. Yet, the c-type-like mRNA has not been identified in the human aorta or in a vascular smooth muscle cell line T/G HA-VSMC (results not shown). It should be noted that another catalytic subunit of NADPH oxidase NOX5 is expressed in human VSMCs [21], but not in rodents. Thus there seems to be considerable species-specific differences in the regulation of the NOX gene expression. Identification of novel transcripts of the Nox1 gene in the present study highlights the diversity of the catalytic subunit of NADPH oxidase expressed in the vascular lineage.
Online data
Acknowledgments
This work was supported in part by Grant-in-Aid for Young Scientists (B) 14770036 and 17790173 from The Ministry of Education, Culture, Sports, Science and Technology of Japan. We are grateful to Dr C. Sakakura (Kyoto Prefectural University of Medicine, Kyoto, Japan), for providing us with human colon epithelia. We also thank Dr T. Nishinaka (Kyoto Prefectural University of Medicine, Kyoto, Japan) and Dr S. Tsuchiya (Kyoto University, Kyoto, Japan) for valuable discussions and advice.
References
- 1.Suh Y. A., Arnold R. S., Lassègue B., Shi J., Xu X., Sorescu D., Chung A. B., Griendling K. K., Lambeth J. D. Cell transformation by the superoxide-generating oxidase Mox1. Nature (London) 1999;401:79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- 2.Bánfi B., Maturana A., Jaconi S., Arnaudeau S., Laforge T., Sinha B., Ligeti E., Demaurex N., Krause K. H. A mammalian H+ channel generated through alternative splicing of the NADPH oxidase homolog NOH-1. Science. 2000;287:138–142. doi: 10.1126/science.287.5450.138. [DOI] [PubMed] [Google Scholar]
- 3.Lassègue B., Clempus R. E. Vascular NAD(P)H oxidases: specific features, expression, and regulation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;285:R277–R297. doi: 10.1152/ajpregu.00758.2002. [DOI] [PubMed] [Google Scholar]
- 4.Lassègue B., Sorescu D., Szöcs K., Yin Q., Akers M., Zhang Y., Grant S. L., Lambeth J. D., Griendling K. K. Novel gp91phox homologues in vascular smooth muscle cells: nox1 mediates angiotensin II-induced superoxide formation and redox-sensitive signaling pathways. Circ. Res. 2001;88:888–894. doi: 10.1161/hh0901.090299. [DOI] [PubMed] [Google Scholar]
- 5.Katsuyama M., Fan C., Yabe-Nishimura C. NADPH oxidase is involved in prostaglandin F2α-induced hypertrophy of vascular smooth muscle cells: induction of NOX1 by PGF2α. J. Biol. Chem. 2002;277:13438–13442. doi: 10.1074/jbc.M111634200. [DOI] [PubMed] [Google Scholar]
- 6.Guzik T. J., West N. E., Black E., McDonald D., Ratnatunga C., Pillai R., Channon K. M. Vascular superoxide production by NAD(P)H oxidase: association with endothelial dysfunction and clinical risk factors. Circ. Res. 2000;86:E85–E90. doi: 10.1161/01.res.86.9.e85. [DOI] [PubMed] [Google Scholar]
- 7.Katsuyama M., Fan C., Arakawa N., Nishinaka T., Miyagishi M., Taira K., Yabe-Nishimura C. Essential role of ATF-1 in induction of NOX1, a catalytic subunit of NADPH oxidase: involvement of mitochondrial respiratory chain. Biochem. J. 2005;386:255–261. doi: 10.1042/BJ20041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuwano Y., Kawahara T., Yamamoto H., Teshima-Kondo S., Tominaga K., Masuda K., Kishi K., Morita K., Rokutan K. Interferon-γ activates transcription of NADPH oxidase 1 gene and upregulates production of superoxide anion by human large intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2006;290:C433–C443. doi: 10.1152/ajpcell.00135.2005. [DOI] [PubMed] [Google Scholar]
- 9.Brewer A. C., Sparks E. C., Shah A. M. Transcriptional regulation of the NADPH oxidase isoform, Nox1, in colon epithelial cells: role of GATA-binding factor(s) Free Radical Biol. Med. 2006;40:260–274. doi: 10.1016/j.freeradbiomed.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 10.Geiszt M., Lekstrom K., Brenner S., Hewitt S. M., Dana R., Malech H. L., Leto T. L. NAD(P)H oxidase 1, a product of differentiated colon epithelial cells, can partially replace glycoprotein 91phox in the regulated production of superoxide by phagocytes. J. Immunol. 2003;171:299–306. doi: 10.4049/jimmunol.171.1.299. [DOI] [PubMed] [Google Scholar]
- 11.Ohmi K., Masuda T., Yamaguchi H., Sakurai T., Kudo Y., Katsuki M., Nonomura Y. A novel aortic smooth muscle cell line obtained from p53 knock out mice expresses several differentiation characteristics. Biochem. Biophys. Res. Commun. 1997;238:154–158. doi: 10.1006/bbrc.1997.7218. [DOI] [PubMed] [Google Scholar]
- 12.Tabuchi Y., Ohta S., Arai Y., Kawahara M., Ishibashi K., Sugiyama N., Horiuchi T., Furusawa M., Obinata M., Fuse H., et al. Establishment and characterization of a colonic epithelial cell line MCE301 from transgenic mice harboring temperature-sensitive simian virus 40 large T-antigen gene. Cell Struct. Funct. 2000;25:297–307. doi: 10.1247/csf.25.297. [DOI] [PubMed] [Google Scholar]
- 13.Qin H., Ishiwata T., Wang R., Kudo M., Yokoyama M., Naito Z., Asano G. Effects of extracellular matrix on phenotype modulation and MAPK transduction of rat aortic smooth muscle cells in vitro. Exp. Mol. Pathol. 2000;69:79–90. doi: 10.1006/exmp.2000.2321. [DOI] [PubMed] [Google Scholar]
- 14.Sata M., Maejima Y., Adachi F., Fukino K., Saiura A., Sugiura S., Aoyagi T., Imai Y., Kurihara H., Kimura K., et al. A mouse model of vascular injury that induces rapid onset of medial cell apoptosis followed by reproducible neointimal hyperplasia. J. Mol. Cell. Cardiol. 2000;32:2097–2104. doi: 10.1006/jmcc.2000.1238. [DOI] [PubMed] [Google Scholar]
- 15.Matsuno K., Yamada H., Iwata K., Jin D., Katsuyama M., Matsuki M., Takai S., Yamanishi K., Miyazaki M., Matsubara H., Yabe-Nishimura C. Nox1 is involved in angiotensin II-mediated hypertension: a study in Nox1-deficient mice. Circulation. 2005;112:2677–2685. doi: 10.1161/CIRCULATIONAHA.105.573709. [DOI] [PubMed] [Google Scholar]
- 16.Geiszt M., Lekstrom K., Leto T. L. Analysis of mRNA transcripts from the NAD(P)H oxidase 1 (Nox1) gene. Evidence against production of the NADPH oxidase homolog-1 short (NOH-1S) transcript variant. J. Biol. Chem. 2004;279:51661–51668. doi: 10.1074/jbc.M409325200. [DOI] [PubMed] [Google Scholar]
- 17.Harper R. W., Xu C., Soucek K., Setiadi H., Eiserich J. P. A reappraisal of the genomic organization of human Nox1 and its splice variants. Arch. Biochem. Biophys. 2005;435:323–330. doi: 10.1016/j.abb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 18.Fan C., Katsuyama M., Nishinaka T., Yabe-Nishimura C. Transactivation of the EGF receptor and a PI3 kinase-ATF-1 pathway is involved in the upregulation of NOX1, a catalytic subunit of NADPH oxidase. FEBS Lett. 2005;579:1301–1305. doi: 10.1016/j.febslet.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Ueno N., Takeya R., Miyano K., Kikuchi H., Sumimoto H. The NADPH oxidase Nox3 constitutively produces superoxide in a p22phox-dependent manner: its regulation by oxidase organizers and activators. J. Biol. Chem. 2005;280:23328–23339. doi: 10.1074/jbc.M414548200. [DOI] [PubMed] [Google Scholar]
- 20.Owens G. K., Kumar M. S., Wamhoff B. R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 21.Bánfi B., Molnár G., Maturana A., Steger K., Hegedûs B., Demaurex N., Krause K. H. A Ca2+-activated NADPH oxidase in testis, spleen, and lymph nodes. J. Biol. Chem. 2001;276:37594–37601. doi: 10.1074/jbc.M103034200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.