Abstract
Background
The Tat pathway transports folded proteins across the cytoplasmic membrane of bacteria and the thylakoid membrane of plants. In Eschericha coli, Tat transport requires the integral membrane proteins TatA, TatB and TatC. In this study we have tested the ability of tat genes from the eubacterial species Pseudomonas syringae, Streptomyces coelicolor and Aquifex aeolicus, to compensate for the absence of the cognate E. coli tat gene, and thus to form functional Tat translocases with E. coli Tat components.
Results
All three subunits of the Tat system from the Gram positive organism Streptomyces coelicolor were able to form heterologous translocases with substantive Tat transport activity. However, only the TatA and TatB proteins of Pseudomonas syringae were able to functionally interact with the E. coli Tat system even though the two organisms are closely related. Of the Tat components from the phylogenetically distant hyperthermophillic bacterium Aquifex aeolicus only the TatA proteins showed any detectable level of heterologous functionality. The heterologously expressed TatA proteins of S. coelicolor and A. aeolicus were found exclusively in the membrane fraction.
Conclusion
Our results show that of the three Tat proteins, TatA is most likely to show cross-species complementation. By contrast, TatB and TatC do not always show cross-complementation, probably because they must recognise heterologous signal peptides. Since heterologously-expressed S. coelicolor TatA protein was functional and found only in the membrane fraction, it suggests that soluble forms of Streptomyces TatA reported by others do not play a role in protein export.
Background
There are two general pathways by which proteins are translocated across the cytoplasmic membranes of bacteria. The Sec pathway, which is ubiquitous, uses a threading mechanism to transport unfolded polypeptides across the membrane, driven by the energy of ATP hydrolysis and the transmembrane proton gradient [1]. By contrast, the Tat pathway, which is encoded in about half of bacterial genomes sequenced so far, exports only pre-folded proteins. Substrate proteins are targeted to the Tat machinery by N-terminal signal peptides that harbour an almost invariant pair of arginine residues, which are critical for transport by the Tat pathway [reviewed in [2,3]]. Tat transport is driven solely by the protonmotive force [4].
Much of our understanding of protein transport by the Tat pathway has come from dual studies of the bacterial Tat pathway and the homologous ΔpH/Tat pathway in plant thylakoids. The Tat system of Escherichia coli is comprised of the three major components, TatA, TatB and TatC, along with the minor component TatE which is a poorly expressed TatA orthologue [5-9]. Protein purification and cross-linking studies have identified two major types of Tat protein complexes in the membranes of E. coli, and analogous complexes have also been identified in thylakoid membranes. An equimolar complex of TatB and TatC, which contains multiple copies of each component, acts as the receptor for Tat substrates [10-12]. Site-specific cross-linking studies have implicated TatC as the component that recognizes the twin arginine motif of the substrate signal peptide [12]. The TatA protein forms a separate, highly heterogeneous complex, which varies in size because it contains different numbers of TatA protomers [13-16]. Examination of purified TatA complexes by negative stain electron microscopy reveal that it forms channel complexes with internal diameters large enough to accommodate folded substrate proteins [16]. Cross-linking studies suggest that TatA transiently associates with the substrate-bound TatBC complex during active protein translocation [12,17,18].
The Tat systems of some Gram positive bacteria, exemplified by Bacillus subtilis, and some Archaea show a slightly different organization in that they lack TatB and therefore have translocases that are comprised solely of TatA and TatC [19]. The structural arrangement of subunits in these minimal Tat systems is currently unknown. However, a number of reports have indicated that at least some of the TatA protein of Haloferax volcanii, B. subtilis, and of the TatA and TatB proteins of Streptomyces lividans exists in a soluble form in the cytoplasmic fractions of these organisms [20-22]. These findings are significant, because they imply that the Tat systems in these prokaryotes may operate by a somewhat different mechanism to the canonical Tat systems of E. coli and plant thylakoids.
Previous studies looking at heterologous interactions during Tat transport have generally focused on the ability of Tat systems to recognize and transport foreign Tat substrates. Thus tat genes from different bacterial sources have been expressed in an E. coli strain deleted for all Tat components [23], which inform on the capacity of foreign translocases to recognize and transport E. coli Tat substrate proteins. Conversely, foreign Tat substrates have also been expressed in E. coli, to test the capacity of the system to recognize non-native signal peptides and passenger proteins [e.g. [24-28]]. However, very few studies have looked at the ability of individual Tat subunits to substitute for the absence of the cognate E. coli Tat component. It was reported that Helicobacter pylori tatA could partially complement the Tat defect of an E. coli ΔtatAΔtatE strain, but that H. pylori tatB could not substitute for E. coli tatB [8]. A very recent study suggested that the P. syringae pv tomato DC3000 tatC gene could also complement the E. coli tatC deletion strain [29].
In this work, we have systematically examined the ability of tat genes from three different bacterial species to compensate for the absence of the cognate E. coli tat gene. The organisms we selected for this study are Aquifex aeolicus, a thermophilic bacterium which forms the deepest branch in bacterial phylogeny, Streptomyces coelicolor, a Gram positive actinomycete and Pseudomonas syringae pv maculicola ES4326, which, like E. coli, is a gamma Proteobacterium. E. coli diverged from A. aeolicus approximately 4 billion years ago, from S. coelicolor approximately 3.2 billion years ago and from P. syringae approximately 1.3 billion years ago [30]. The percentage identities of the Tat proteins from these organisms with the paralogous E. coli Tat proteins is shown in Table 1. Our results indicate that TatA proteins from any of these organisms are able to, at least partially, restore Tat activity to a strain lacking E. coli tatA and tatE. The P. syringae tatB and S. coelicolor tatB and tatC genes were also able to complement the cognate E. coli tat deletion strains. Cell fractionation experiments demonstrate that the heterologously expressed TatA proteins of S. coelicolor and A. aeolicus are found exclusively in the membrane.
Table 1.
Percentage identities between E. coli Tat proteins and those from other eubacteria analysed in this study.
| Protein | Length (amino acids) | Percentage identity to E. coli homologue |
| E. coli TatA | 89 | |
| P. syringae TatA | 91 | 60% over 48 residue overlap |
| S. coelicolor TatA | 95 | 36% over 40 residue overlap |
| A. aeolicus TatA1 | 59 | 41% over 46 residue overlap |
| A. aeolicus TatA2 | 77 | 38% over 55 residue overlap |
| E. coli TatB | 171 | |
| P. syringae TatB | 153 | 40% over 71 residue overlap |
| S. coelicolor TatB | 161 | 33% over 55 residue overlap |
| A. aeolicus TatB | 117 | 23% over 102 residue overlap |
| E. coli TatC | 258 | |
| P. syringae TatC | 266 | 62% over 245 residue overlap |
| S. coelicolor TatC | 301 | 27% over 247 residue overlap |
| A. aeolicus TatC | 240 | 41% over 234 residue overlap |
Results and discussion
Experimental design
Throughout these experiments we have used three different tests to assess functionality of the Tat system, each of which depends upon the transport of one or more native E. coli Tat substrates. E. coli tat mutants show a pleiotropic cell envelope defect due to an inability to export two Tat-dependent periplasmic amidases, AmiA and AmiC, which are involved in cell wall remodelling [31,32]. Strains with an inactive Tat system are unable to grow on solid media in the presence of 2% SDS and therefore the ability to grow in the presence of this detergent is a qualitative indication of Tat function [33]. Likewise, E. coli is able to grow anaerobically using trimethylamine-N-oxide (TMAO) as an electron acceptor due to the Tat-dependent export of two enzymes, TMAO reductase (TorA, which is a soluble periplasmic protein) and dimethylsulphoxide (DMSO) reductase (DmsABC, which is membrane-bound, with its active site facing the periplasm [34]). Therefore the ability of strains to grow anaerobically on minimal media with glycerol as a carbon source and TMAO as sole electron acceptor is also a qualitative indicator of Tat functionality. Finally we have also assayed the activity of TMAO reductase in periplasmic extracts as a more quantitative assessment of the level of Tat functionality.
We have observed previously that the stoichiometry of Tat subunit expression is critical for activity of the Tat translocase. In particular, high level expression of TatB relative to TatA and TatC has a severe inactivating effect on the E. coli Tat system [8]. Thus in the following experiments we have routinely used pcnB derivatives of each of the tat mutant strains, which drastically lowers the copy number of plasmids with the ColE1 replicon, typically to less than 5 copies per cell [35,36].
Heterologous expression of TatA proteins
As shown in Fig 1, expression of the S. coelicolor or P. syringae TatA proteins in the E. coli tatA/tatE mutant strain resulted in significant restoration of Tat system function. Thus the tatA/tatE strain expressing either of these constructs showed robust growth in the presence of SDS (Fig 1A), with TMAO as sole terminal electron acceptor (Fig 1B) and had significant TMAO reductase (TorA) activity in the periplasmic fraction (Fig 1C). A. aeolicus is unusual in that it has a tandem duplication of tatA genes, designated tatA1 and tatA2, which share 68% identity with each other over a 50 amino acid overlap. Expression of either or both of these tatA genes in the E. coli tatA/tatE strain was sufficient to support growth in the presence of SDS or with TMAO as sole electron acceptor (Fig 1A and 1B). However, the level of periplasmic TMAO reductase activity supported by these strains was not significantly above background (Fig 1C) indicating that the rate of TorA export was very low. None-the-less, these results suggest that TatA proteins from all of the three species tested are capable of some level of heterologous interaction with the E. coli TatBC proteins.
Figure 1.
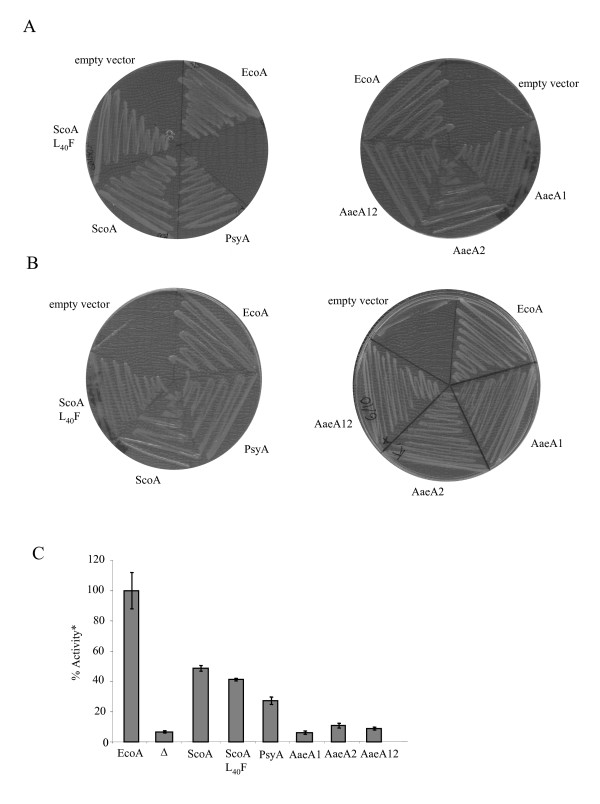
Complementation of the E. coli ΔtatAΔtatE, pcnB strain, JARV16-P, with tatA genes from other bacteria. Strain JARV16-P was transformed with either: E. coli tatA encoded on pFAT415 (EcoA), P. syringae tatA on plasmid pUniprom-PA (PsyA), S. coelicolor tatA from plasmid pUniprom-SA (ScoA), or the L40F derivative (ScoA L40F), A. aeolicus tatA1 (AaeA1), tatA2 (Aae2) or tatA12 (Aae12) from pQEAQ1, pQEAQ2 and pQEAQ12, respectively, or pBluescript (empty vector; marked as Δ in panel C). A. Growth of strains on LB medium containing 2% SDS. B. Growth of strains anaerobically on minimal glycerol TMAO medium. C. TMAO reductase activities from periplasmic fractions. *100% activity is taken as that determined from the periplasmic fraction of JARV16-P carrying pFAT415 and corresponds to 1.3μmol benzyl viologen oxidised/min/mg protein. Error bars represent the standard error of the mean (n = 3).
We have previously demonstrated that residue F39 of E. coli TatA is critical for TatA function and that mutation to anything other than tyrosine or tryptophan not only inactivates TatA function but also shows a dominant phenotype such that in co-expression studies it grossly affects the function of wild type TatA [37,38]. Surprisingly, S. coelicolor is one of a very few organisms that does not have phenylalanine at that position. Instead the amino acid at residue 40, which is the equivalent position in S. coelicolor TatA, is leucine. Although a leucine substitution of E. coli F39 is barely tolerated [38], the S. coelicolor TatA protein shows a significant level of functionality in E. coli. We tested whether the activity of S. coelicolor TatA could be enhanced by mutagenesis of L40 to phenylalanine. However, as shown in Fig 1C, this substitution did not significantly alter the functionality of S. coelicolor TatA in the E. coli system. We conclude that other residues in S. coelicolor TatA compensate for the presence of a leucine at this position.
Heterologous expression of TatB
The TatB protein, where present, is an essential component of the Tat system. It forms an equimolar complex with TatC and site-specific cross-linking analysis has indicated that it contacts Tat signal peptides close to the twin arginine motif, but also within the hydrophobic core [10,12]. In addition TatB has also been shown to co-purify with TatA, with which it shares some sequence homology [39]. It has been suggested to function as the adaptor between the TatBC complex and the TatA channel complex and therefore high level expression of TatB may disrupt Tat function by interfering with the co-assembly of the two individual complexes [8]. Since it interacts with each of the other Tat components and with Tat signal peptides, cross-complementation with TatB might be expected to be less efficient than with other Tat proteins. As shown in Fig 2A and 2B, the TatB proteins from either S. coelicolor or P. syringae permitted significant growth of the E. coli tatB strain, BØD-P, on selective media containing either SDS or TMAO. By contrast A. aeolicus TatB (co-expressed on a construct that also contains tatC) did not allow any significant growth of the E. coli tatB mutant on either medium. It should, however, be noted that we were not able to demonstrate expression of the A. aeolicus TatB protein in E. coli (data not shown) so it is possible that A. aeolicus TatB does not complement the E. coli tatB mutant because the protein is not produced. Fractionation of the tatB strain harbouring the different tatB genes and assay for periplasmic TMAO reductase activity, shown in Fig 2C, demonstrated that, as seen for TatA, the S. coelicolor TatB homologue gave the highest level of cross-species complementation.
Figure 2.

Complementation of the E. coli ΔtatB, pcnB strain, BØD-P, with tatB genes from other bacteria. Strain BØD-P was transformed with either: E. coli tatB encoded on pFAT416 (EcoB), P. syringae tatB on plasmid pUniprom-PB (PsyB), S. coelicolor tatB from plasmid pUniprom-SB (ScoB), A. aeolicus tatBC from plasmid pQEAQBC (AaeBC) or pBluescript (empty vector; marked as Δ in panel C). A. Growth of strains on LB medium containing 2% SDS. B. Growth of strains anaerobically on minimal glycerol TMAO medium. C. TMAO reductase activities from periplasmic fractions. *100% activity is taken as that determined from the periplasmic fraction of BØD-P carrying pFAT416 and corresponds to 0.83μmol benzyl viologen oxidised/min/mg protein. Error bars represent the standard error of the mean (n = 3).
It has been reported previously that the TatA and TatB proteins of S. lividans, an extremely close relative of S. coelicolor have partially overlapping activities [40]. We therefore tested the ability of the S. coelicolor tatB gene to complement the E. coli tatA/E mutant strain, and likewise the S. coelicolor tatA gene to complement the E. coli tatB mutant. In each case we saw no detectable complementation (results not shown).
Heterologous expression of TatC
The TatC protein is the largest and most conserved Tat component. It has been implicated as a specificity determinant for Tat-dependent secretion, most likely through recognition of Tat signal peptides [19,41]. Moreover, it has also been shown to cross-link with Tat signals when the site-specific cross-linker was incorporated adjacent to the twin arginines, suggesting that it probably recognizes the twin arginine motif of the signal peptide [12]. As shown in Fig 3A, B and 3C, only the TatC protein of S. coelicolor restored any detectable level of Tat activity to the E. coli tatC, pcnB mutant strain. The tatC genes of P. syringae and A. aeolicus (the latter co-expressed with tatB) completely failed to complement the tatC strain, even for growth on SDS-containing medium, which is the most sensitive test for native Tat substrate export [42]. As shown in Fig 3D, despite its lack of functionality when expressed in the E. coli tatC strain, the A. aeolicus his-tagged TatC protein encoded on our construct was clearly produced in E. coli and moreover was localized to the membrane fraction. We were not able to demonstrate whether the P. syringae TatC protein was expressed in these experiments because we lack a native antibody to the protein. However in the section below we demonstrate clearly that it is expressed from an analogous clone that also encodes P. syringae tatA and tatB. Taken together our results suggest that S. coelicolor Tat subunits show the greatest level of cross-complementation, even though this organism is more distantly related to E. coli than is P. syringae.
Figure 3.
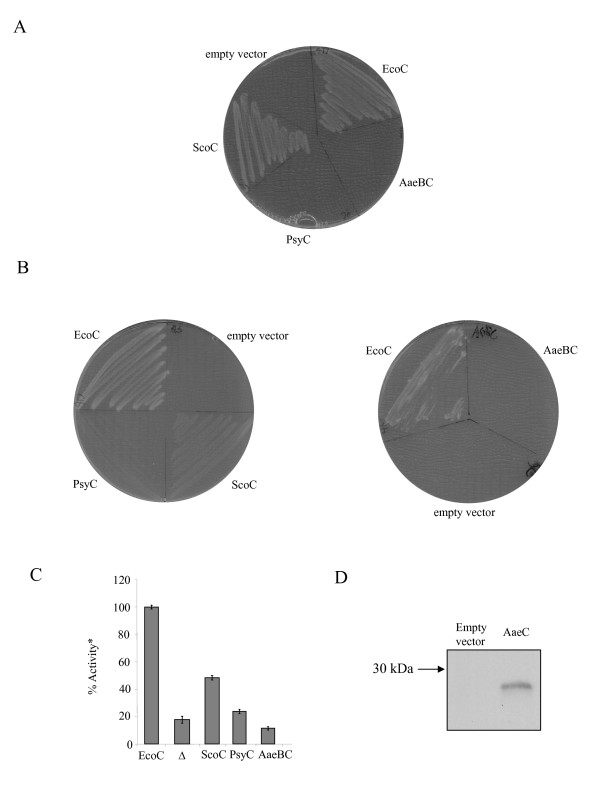
Complementation of the E. coli ΔtatC, pcnB strain, B1LK0-P, with tatC genes from other bacteria. Strain B1LK0-P was transformed with either: E. coli tatC encoded on pFAT417 (EcoC), P. syringae tatC on plasmid pUniprom-PC (PsyC), S. coelicolor tatC from plasmid pUniprom-SC (ScoC), A. aeolicus tatBC from plasmid pQEAQBC (AaeBC) or pBluescript (empty vector; marked as Δ in panel C). A. Growth of strains on LB medium containing 2% SDS. B. Growth of strains anaerobically on minimal glycerol TMAO medium. C. TMAO reductase activities from periplasmic fractions. *100% activity is taken as that determined from the periplasmic fraction of B1LK0-P carrying pFAT417 and corresponds to 0.79μmol benzyl viologen oxidised/min/mg protein. Error bars represent the standard error of the mean (n = 3). D. The A. aeolicus TatC protein is produced from clone pQEAQBC. Strain M15 [pREP4] harboring either pQE60 (empty vector) or pQEAQBC (AaeC) was cultured in LB medium until OD600 of 0.4 was reached, after which production of the TatChis protein was induced by addition of 1 mM isopropyl-β-D-galactopyranoside (IPTG) for a further 2 hours. Membrane fractions were prepared, proteins (50μg of total membrane protein), separated by SDS-PAGE, blotted onto nitrocellulose and developed using anti-penta-His antiserum.
Heterologous expression of tatABC genes in a strain of E. coli devoid of all Tat components
The experiments described above where individual Tat subunits are expressed in E. coli examine the ability of that particular subunit to interact with E. coli Tat components, and, in addition, to recognize E. coli Tat signal peptides and substrate proteins. Under these circumstances, it is apparent that whilst all of the TatA proteins displayed some level of heterologous function, the A. aeolicus tatB and tatC genes, and P. syringae tatC, failed to complement the cognate E. coli tat mutant strains. In order to determine whether this was because of an inability to form functional complexes with E. coli Tat components rather than an inability to interact with the test E. coli substrates, we examined whether transport activity was observed when the full set of heterologous Tat proteins were co-expressed. As shown in Fig 4, the P. syringae tatABC operon showed a low but detectable level of Tat function, indicating that the P. syringae TatC protein alone was not functional in the E. coli tatC strain most likely because it could not either interact with E. coli TatB and/or TatA, or because it was expressed at an inappropriate ratio with these proteins. An alternative explanation is that P. syringae tatC in vector pUniprom-PC fails to express, although we note that it clearly does express from the same vector in the presence of P. syringae tatA and tatB. Conversely, the A. aeolicus tatA1, tatA2, tatB and tatC genes when expressed together from an artificial operon could not complement the E. coli total tat deletion strain. With the proviso that all of the A. aeolicus tatA, tatB and tatC genes express in E. coli, this result suggests that the A. aeolicus Tat proteins are unable to recognize any of the AmiA, AmiC, TorA or DmsA signal peptides.
Figure 4.
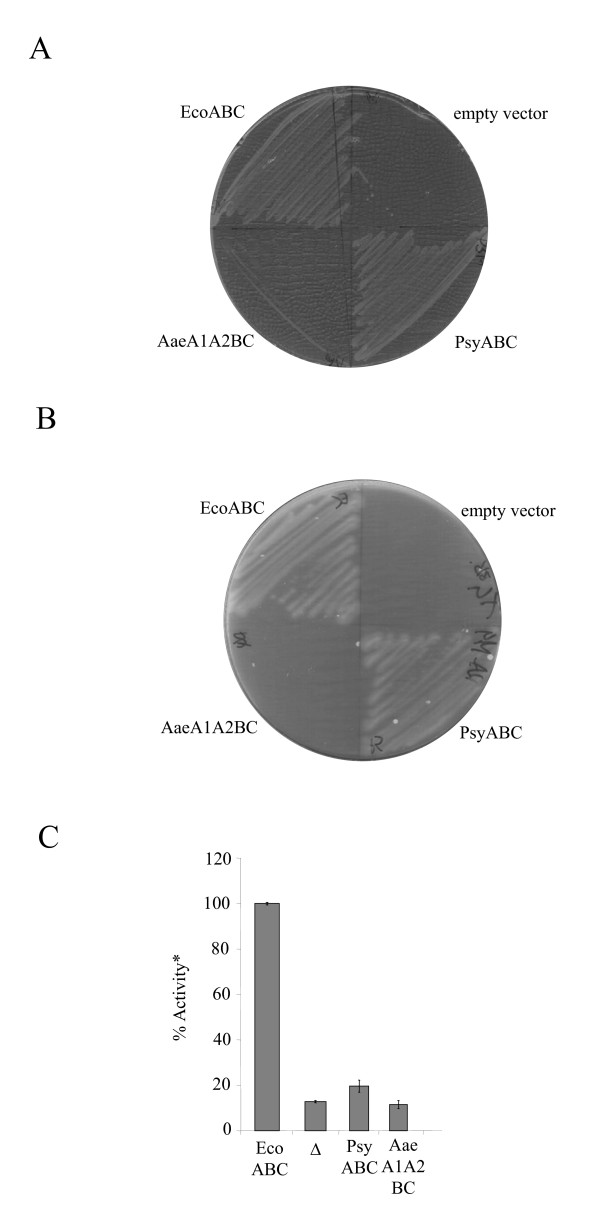
Complementation of the E. coli ΔtatABCDΔtatE, pcnB strain, DADE-P, with tatABC genes from other bacteria. Strain DADE-P was transformed with either: E. coli tatABC encoded on pUnitat2 (EcoABC), P. syringae tatABC on plasmid pUniprom-PABC (PsyABC), A. aeolicus tatA1A2BC from plasmid pFATAQ3 (AaeA1A2BC) or pBluescript (empty vector; marked as Δ in panel C). A. Growth of strains on LB medium containing 2% SDS. B. Growth of strains anaerobically on minimal glycerol TMAO medium. C. TMAO reductase activities from periplasmic fractions. *100% activity is taken as that determined from the periplasmic fraction of DADE-P carrying pUnitat2 and corresponds to 0.83μmol benzyl viologen oxidised/min/mg protein. Error bars represent the standard error of the mean (n = 3).
A summary of the results we obtained for all of the heterologous expression experiments is shown in Table 2.
Table 2.
Summary of Tat activity observed after heterologous expression of tat genes from different bacteria in E. coli tat mutant strains.
| E. coli strain | Plasmid-encoded gene(s) | Functionality of the Tat system1 |
| JARV16-P(ΔtatA/ΔtatE) | Eco tatA | ++ |
| Psy tatA | ++ | |
| Sco tatA | ++ | |
| Sco tatA(L40F) | ++ | |
| Aae tatA1 | + | |
| Aae tatA2 | + | |
| BØD-P(ΔtatB) | Eco tatB | ++ |
| Psy tatB | + | |
| Sco tatB | ++ | |
| Aae tatB | - | |
| B1LK0-P(ΔtatC) | Eco tatC | ++ |
| Psy tatC | - | |
| Sco tatC | ++ | |
| Aae tatC | - | |
| DADE-P(ΔtatABCD/ΔtatE) | Eco tatABC | ++ |
| Psy tatABC | + | |
| Aae tatA1A2tatBC | - |
1++ indicates a high level of functionality (>30% of the wild type level of periplasmic TMAO reductase activity, + indicates a lower level of functionality (<30% of the wild type level of periplasmic TMAO reductase activity, but confers the ability to grow on solid media containing SDS or TMAO), – indicates a lack of activity (no growth on SDS or TMAO-containing plates).
Heterologously expressed TatA proteins from S. coelicolor and A. aeolicus are found exclusively in the membrane fraction
One of the most striking reported differences between the Tat system of E. coli and of Gram positive (and archaeal) Tat systems is the presence of soluble forms of TatA in these latter organisms. Since we demonstrated above that the S. coelicolor and A. aeolicus TatA proteins show functionality in E. coli, it was reasonable to examine the subcellular location of these heterologously expressed TatA proteins. As shown in Fig 5A, all of the E. coli TatA protein was found in the membrane fraction. Strikingly, we also found all of the heterologously expressed S. coelicolor TatA and of the his-tagged A. aeolicus TatA1 and TatA2 proteins exclusively in the membrane fraction. Whilst we cannot rule out that any cytoplasmic forms of these heterologously-produced proteins might have been degraded, these observations indicate that at least when expressed in E. coli it is the membrane-bound protein that is functional for transport.
Figure 5.
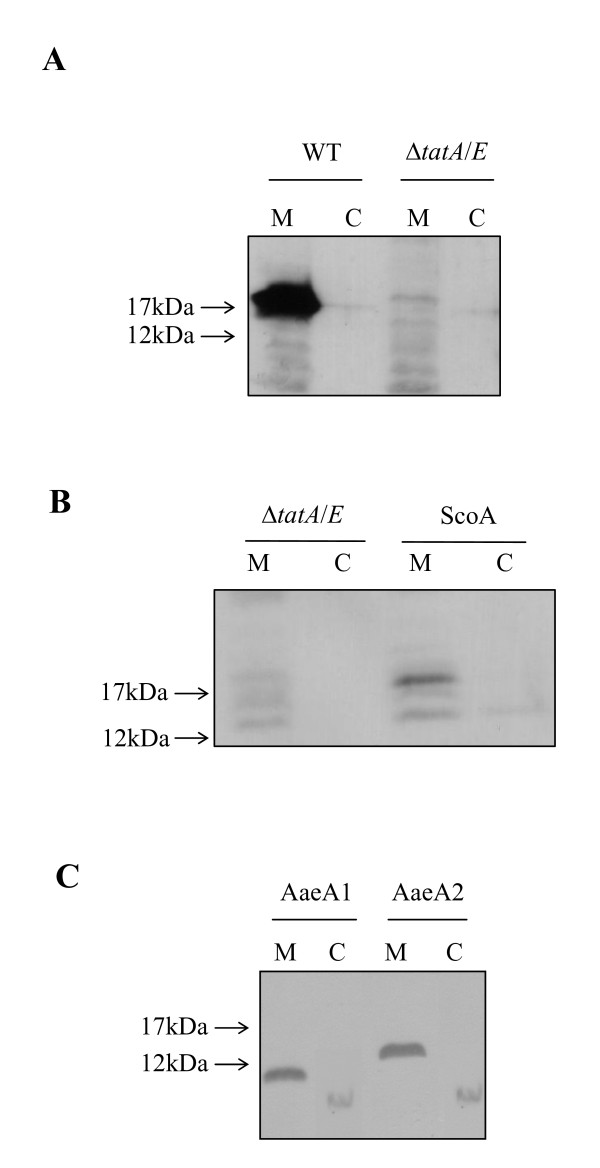
Heterologously expressed TatA proteins are located exclusively in the membrane. A. Strains MC4100 (E. coli parental strain; tat+; WT) and JARV16-P (as MC4100, ΔtatA/ΔtatE, pcnB; ΔtatA/E) were grown aerobically to late exponential phase in LB medium prior to harvesting and fractionation into membrane (M) and soluble (C) fractions. Samples were separated by SDS PAGE, electroblotted and probed with anti E. coli TatA antiserum. B. Strain JARV16-P (ΔtatA/E) or JARV16-P carrying pUniprom-SA encoding S. coelicolor TatA (ScoA) were cultured, fractionated and blotted as above, and probed with anti S. coelicolor TatA peptide antiserum. C. Strain M15 [pREP4] harboring either pQEAQ1 (AaeA1) or pQEAQ2 (AaeA2) encoding A. aeolicus TatA1 or TatA2 with C-terminal histags, respectively. Cells were cultured in LB medium until OD600 of 0.4 was reached, after which expression of the TatA protein was induced by addition of 1 mM isopropyl-β-D-galactopyranoside (IPTG) for a further 2 hours. Cells were fractionated and blotted as above, and probed with anti pentahis antiserum. For each panel, membrane and soluble material from an equivalent amount of cells was loaded.
Conclusion
In this report we have tested the ability of Tat proteins from three different eubacterial species to form complexes with E. coli Tat components. Surprisingly, we found that it was the individual Tat proteins of S. coelicolor that consistently gave the highest level of Tat activity with the cognate E. coli tat deletion strains. Indeed each of S. coelicolor TatA, TatB and TatC formed heterologous Tat systems that were able to support 40–50% of the wild type E. coli Tat activity. The P. syringae TatA protein was able to restore reasonable Tat function to the E. coli tatA/E mutant strain, but the tatB gene complemented very poorly and P. syringae TatC was apparently completely inactive.
For each of the S. coelicolor and P. syringae cross-complementation experiments, individual Tat subunits were expressed from identical constructs, with transcription being driven by the constitutive E. coli tat promoter. It is probable that the heterologously expressed S. coelicolor tat genes are translated less efficiently than the P. syringae genes because of codon bias resulting from the high G+C content of S. coelicolor DNA. Whilst we cannot completely rule out the possibility that there is a deleteriously high level of production of the individual P. syringae Tat proteins (but see below), it is none-the-less, striking that the S. coelicolor Tat proteins show a high level of cross-species activity. One possible explanation for this might be linked to the fact that the S. coelicolor has by far the largest predicted Tat secretome. Thus the S. coelicolor Tat system probably transports in excess of 150 Tat substrates, including some of the largest Tat substrates ever identified [43-45]. It therefore must recognize many different twin arginine signal peptides and as a result one might expect that the S. coelicolor Tat components are less stringent than the P. syringae system in terms of signal peptide recognition and substrate size.
It should be noted that TatC from a different pathovar of P. syringae to that tested here [but with which it shares 97% amino acid sequence identity; [46]] was reported to show at least some minimal Tat activity in the E. coli tatC strain for transport of the Tat substrates AmiA and/or AmiC [29]. However in those experiments, the expression of P. syringae tatC was from the strong ptac promoter on plasmid pTrc99A and was carried out in a pcnB+ background where plasmid copy number would be considerably higher. Thus it is likely that the levels of TatC between the two experiments would be significantly different which may account for the discrepancy in the observed activities.
It is striking to note that wherever cross-species complementation has been tested, TatA proteins always seem to retain some level of function in the heterologous host [8,47]. This suggests that the constraints on TatA function are less severe than those for TatB or TatC and is consistent with the fact that most of the interactions of heterologously expressed TatA would be self-oligomerisation to assemble into channel-forming multimers. By contrast, the constraints on cross-complementation with heterologously expressed TatB or TatC proteins are likely to be much more stringent since this would require the formation of equimolar complexes with the appropriate E. coli partner subunit, recognition of non-native signal peptides and associated conformational changes to promote assembly of the active translocase. It is notable that the A. aeolicus TatA proteins, which would normally be operating at temperatures of 80°C, retain function when expressed in E. coli cells growing at 37°C. Thus either the TatA protein does not change conformation during the transport cycle or the protein retains sufficient flexibility for function even at temperatures considerably lower than physiological.
A number of groups have reported in the literature the presence of soluble forms of the TatA and TatB proteins from Gram positive bacteria and archaea [20-22]. Fractionation and Western blotting showed quite clearly here that all of the heterologously expressed TatA proteins from S. coelicolor and A. aeolicus are found only in the membrane fraction. It is not clear why extra-membraneous forms of TatA/TatB are present in some organisms, but clearly since the S. coelicolor TatA protein [which is 100% identical to the S. lividans TatA protein; [48]] supports such a high level of Tat activity in E. coli then it is difficult to imagine how the mechanism of Tat transport at least between E. coli and Streptomycetes cannot be anything other than highly similar.
Methods
Bacterial strains and growth conditions
The E. coli strains and plasmids employed in this study are shown in Table 3. During all genetic manipulations, E. coli strains were grown aerobically in Luria-Bertani (LB) medium [49]. Concentrations of antibiotics were as described previously [6]. The growth phenotypes of mutants with TMAO as sole respiratory electron acceptor were determined on M9 minimal medium agar plates [49] supplemented with 0.5% glycerol and 0.4% TMAO and incubated in a gas jar under a hydrogen/carbon dioxide atmosphere. The SDS-resistance phenotype of mutants was tested on LB agar plates containing 2% SDS [33]. For TMAO reductase assay, cells were cultured in modified Cohen and Rickenberg medium [50], supplemented with addition of 0.2% glucose and 0.4% TMAO.
Table 3.
Strains and plasmids used in this study
| Bacterial Strains | Genotype | Source |
| MC4100 | F-ΔlacU169 araD139 rpsL150 relA1 ptsF rbs flbB5301 | [58] |
| DH5α | φ80dlacZΔM15, recA1, endA1, gyrA96, thi-1, hsdR17(rK-,mK+) supE44, relA1, deoR, Δ(lacZYA-argF) U169 | Promega |
| M15 | F-, lac, ara, gal, mtl | [59] |
| JARV16-P | As MC4100 ΔtatAΔtatE pcnB1 zad-981::Tn10d (Kmr) | [8] |
| BØD-P | As MC4100 ΔtatB pcnB1 zad-981::Tn10d (Kmr) | [8] |
| B1LK0-P | As MC4100 ΔtatC pcnB1 zad-981::Tn10d (Kmr) | This work |
| DADE-P | As MC4100 ΔtatABCD ΔtatE pcnB1 zad-981::Tn10d (Kmr) | [60] |
| Plasmids | ||
| pREP4 | KanR, lacI+ | Roche |
| pBluescript (IIKS+) | AmpR | Stratagene |
| pFAT415 | pBluescript carrying E. coli tatA | [8] |
| pFAT416 | pBluescript carrying E. coli tatB | [8] |
| pFAT417 | pBluescript carrying E. coli tatC | [8] |
| pUniprom | AmpR contains E. coli tat promoter and multiple cloning site | [51] |
| pUniprom-PA | pUniprom carrying P. syringae tatA gene | This work |
| pUniprom-PB | pUniprom carrying P. syringae tatB gene | This work |
| pUniprom-PC | pUniprom carrying P. syringae tatC gene | This work |
| pUniprom-PABC | pUniprom carrying P. syringae tatABC genes | This work |
| pUniprom-SA | pUniprom carrying S. coelicolor tatA gene | This work |
| pUniprom-SB | pUniprom carrying S. coelicolor tatB gene | This work |
| pUniprom-SC | pUniprom carrying S. coelicolor tatC gene | This work |
| pFATAQ1 | pBluescript carrying A. aeolicus tatA1and tatA2 | This work |
| pFATAQ2 | pBluescript carrying A. aeolicus tatBC | This work |
| pFATAQ3 | pBluescript carrying A. aeolicus tatA, tatA2 and tatBC | This work |
| pQE70 | C-terminal His-tag expression vector | Qiagen |
| pQEAQ1 | pQE70 carrying A. aeolicus tatA1 | This work |
| pQEAQ2 | pQE70 carrying A. aeolicus tatA2 | This work |
| pQEAQBC | pQE70 carrying A. aeolicus tatBC | This work |
| pQEAQ12 | pQE70 carrying A. aeolicus tatA1and tatA2 | This work |
Plasmid construction
Plasmids pUniprom-PA, pUniprom-PB and pUniprom-PC carry the P. syringae pv maculicola ES4326 tatA, tatB and tatC genes, respectively, under the control of the E. coli tat promoter. They were cloned following amplification with primers GCGGCCGGATCCATGGGTATTTTTGACTGG and GCGCTCTAGATTAAACC TGGTCTTTGCGG to amplify tatA, GCGCGGGGATCCATGTTCGGTATCAGC and GCGCGCTCTAGATCATGGGGCTCGCGGTGGC to amplify tatB, and GCGC GCGGATCCATGAGCGCTGATATCCCG and GCGCGCTCTAGATCACGGTGT GGTGGCGGGCGG to amplify tatC (restriction sites shown in bold) and plasmid pKS-PSMtatABC [46] as template. Each product was digested with BamHI and XbaI and cloned into pUniprom [51] that had been similarly digested. Plasmids pUniprom-SA, pUniprom-SB and pUniprom-SC carry the S. coelicolor tatA, tatB and tatC genes, respectively, under the control of the E. coli tat promoter. They were cloned following amplification with primers GCGCGCGGATCCATGTTCGGAAGGCTCG GC and GCGCGCTCTAGATCAGCGCTTGGTCGTGTC to amplify tatA, GCGCG CGGATTCGTGTTCAATGACATAGGC and GCGCGCTCTAGATCAGGTGGCG TCCATGTC to amplify tatB, and GCGCGCGGATCCATGCCGCTCGCGGAACA C and GCGCGCTCTAGATCAGGTCACGTCGTCG to amplify tatC with S. coelicolor chromosomal DNA as template. Each product was digested with BamHI and XbaI and cloned into pUniprom that had been similarly digested.
Plasmids pFATAQ1 contains the tatA1 and tatA2 genes of A. aeolicus, under control of the lac promoter. The tatA1 and tatA2 genes, whose reading frames overlap, were amplified with the following primers: GCGCGCGAATTCCCCTTAAATTATTCTC TAAGGAGGC and GCGCGCGGATCCGCTGAGTTAAGCCTCTACCTTTTCC, with A. aeolicus chromosomal DNA (a kind gift of R. Huber) as template. The product was digested with EcoRI and BamHI and cloned into pBluescript that had been digested with the same enzymes. The A. aeolicus tatB gene is not annotated on the genome [52], however, a gene encoding a protein with 23% identity (over 102 amino acid overlap) to E. coli TatB is found immediately upstream of the tatC gene (see Table 1 for identities of Tat proteins studied in this work with the paralogous E. coli protein), and we herein refer to it as tatB. Plasmid pFATAQ2 contains the tatB and tatC genes of A. aeolicus, under control of the lac promoter. The genes, whose reading frames also overlap, were amplified with primers GCGCGGATCCGGAAA GCAATCCCTATTAACGGAAG and GCGCTCTAGAGCTTATGCCTTTTGAATT TCCTTC, digested with BamHI and XbaI and cloned into pBluescript that had been pre-digested with the same enzymes. Plasmid pFATAQ3 contains an artificial operon encoding the tatA1, -A2, -B and -C genes of A. aeolicus and was generated by subcloning the tatBC coding region from pFATAQ2 by digestion with BamHI and XbaI and ligation into pFATAQ1 that had been similarly digested. For overproduction of the A. aeolicus Tat proteins with C-terminal hexahistidine affinity tags, the encoding tat genes were re-amplified and cloned into the overexpression vector pQE70. Plasmid pQEAQ1 contains the gene encoding the TatA1 protein with a C-terminal histag and was amplified using primers GCGCGCATGCACTTTCCTCTGC CGTGGC and GCGCAGATCTTTTCACCCTCCTTTTTAACTTCC, and pFATAQ1 as template. The product was digested with SphI and BglII, and cloned into similarly digested pQE70. Plasmid pQEAQ2 is analogous to pQEAQ1 but encodes a tagged version of the TatA2 protein. It was cloned in a similar manner, using primers GCGC ACATGTTTTCCCGGCGGAATATCTATG and GCGCAGATCTAGCCTCTACC TTTTCCTTCTC to amplify the encoding gene. Plasmid pQEAQ12 carries both of the A. aeolicus tatA genes, with just TatA2 supplied with the C-terminal histag. The genes were amplified using primers GCGCGCATGCACTTTCCTCTGCCGTGGC and GC GCAGATCTAGCCTCTACCTTTTCCTTCTC, the product digested with SphI and BglII, and cloned into similarly digested pQE70.
All clones obtained from PCR amplified DNA were sequenced to ensure that no mutations (other than site-specific mutations) had been introduced.
Protein methods
SDS-PAGE and immunoblotting were carried out as described [53,54] and immunoreactive bands were visualized with the ECL detection system (Amersham Biosciences). Peptide antibodies to the S. coelicolor TatA protein were raised in rabbits, using a peptide of amino acid sequence CTSSRPVTEPTDTTKR, by Davids biotechnology (Regensburg, Germany). Antibodies to E. coli TatA have been described previously [39], the anti-pentahis antibody was obtained from Qiagen. Subcellular fractions for TMAO reductase activity measurements were prepared from small (30 ml) cultures using the cold osmotic shock protocol [55]. TMAO:benzyl viologen oxidoreductase activity was measured as described previously [56]. Protein concentrations were estimated according to the method of Lowry et al. [57]. Preparation of membrane fractions was as described previously [6].
Abbreviations
Sec -general secretory pathway, Tat – Twin arginine translocation pathway, TMAO -trimethylamine-N-oxide, TorA – TMAO reductase DMSO – dimethylsulphoxide, DmsABC – membrane-bound DMSO reductase.
Authors' contributions
MGH, DG, GB, IC and DW undertook experiments, BCB and TP wrote the manuscript.
Acknowledgments
Acknowledgements
We thank Drs Frank Sargent and Govind Chandra for helpful discussion, and Prof Robert Huber of the University of Regensburg for the gift of Aquifex aeolicus chromosomal DNA. This work was supported by the CEU project LHSG-CT-2004-005257, the BBSRC through a grant-in-aid to the John Innes Centre and a BBSRC-funded PhD studentship to MGH, the Swiss National Science Foundation through the award of a fellowship to IC and the Medical Research Council via a Senior Non-Clinical Fellowship award to TP.
Contributor Information
Matthew G Hicks, Email: matthew.hicks@bbsrc.ac.uk.
David Guymer, Email: d.guymer@uea.ac.uk.
Grant Buchanan, Email: grant.buchanan@bbsrc.ac.uk.
David A Widdick, Email: david.widdick@bbsrc.ac.uk.
Isabelle Caldelari, Email: i.caldelari@uea.ac.uk.
Ben C Berks, Email: ben.berks@bioch.ox.ac.uk.
Tracy Palmer, Email: tracy.palmer@bbsrc.ac.uk.
References
- Manting EH, Driessen AJ. Escherichia coli translocase: the unravelling of a molecular machine. Mol Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- Berks BC, Palmer T, Sargent F. The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol. 2003;47:187–254. doi: 10.1016/s0065-2911(03)47004-5. [DOI] [PubMed] [Google Scholar]
- Mori H, Cline K. Post-translational protein translocation into thylakoids by the Sec and ΔpH-dependent pathways. Biochim Biophys Acta. 2001;1541:80–90. doi: 10.1016/S0167-4889(01)00150-1. [DOI] [PubMed] [Google Scholar]
- Yahr TL, Wickner WT. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 2001;20:2472–2479. doi: 10.1093/emboj/20.10.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, Thomas GH, Cole JA, Turner RJ. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/S0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- Sargent F, Bogsch E, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogsch E, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- Sargent F, Stanley NR, Berks BC, Palmer T. Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J Biol Chem. 1999;274:36073–36083. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- Jack RL, Sargent F, Berks BC, Sawers G, Palmer T. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J Bacteriol. 2001;183:1801–1804. doi: 10.1128/JB.183.5.1801-1804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J Biol Chem. 2001;276:20213–20219. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- Cline K, Mori H. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J Cell Biol. 2001;154:719–729. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami M, Luke I, Deitermann S, Eisner G, Koch HG, Brunner J, Muller M. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell. 2003;12:937–946. doi: 10.1016/S1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- Porcelli I, de Leeuw E, Wallis R, van den Brink-van der Laan E, de Kruijff B, Wallace BA, Palmer T, Berks BC. Characterisation and membrane assembly of the TatA component of the Escherichia coli twin-arginine protein transport system. Biochemistry. 2002;41:13690–13697. doi: 10.1021/bi026142i. [DOI] [PubMed] [Google Scholar]
- de Leeuw E, Granjon T, Porcelli I, Alami M, Carr SB, Müller M, Sargent F, Palmer T, Berks BC. Oligomeric properties and signal peptide binding by Escherichia coli Tat protein transport complexes. J Mol Biol. 2002;322:1135–1146. doi: 10.1016/S0022-2836(02)00820-3. [DOI] [PubMed] [Google Scholar]
- Oates J, Barrett CM, Barnett JP, Byrne KG, Bolhuis A, Robinson C. The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J Mol Biol. 2005;346:295–305. doi: 10.1016/j.jmb.2004.11.047. [DOI] [PubMed] [Google Scholar]
- Gohlke U, Pullan L, McDevitt CA, Porcelli I, de Leeuw E, Palmer T, Saibil H, Berks BC. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad Sci USA. 2005;102:10482–10486. doi: 10.1073/pnas.0503558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori J, Cline K. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J Cell Biol. 2002;157:205–210. doi: 10.1083/jcb.200202048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney-Smith C, Mori H, Cline K. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J Biol Chem. 2006;281:5476–5483. doi: 10.1074/jbc.M512453200. [DOI] [PubMed] [Google Scholar]
- Jongbloed JDH, Grieger U, Antelmann H, Hecker M, Nijland R, Bron S, van Dijl JM. Two minimal Tat translocases in Bacillus. Mol Microbiol. 2004;54:1319–1325. doi: 10.1111/j.1365-2958.2004.04341.x. [DOI] [PubMed] [Google Scholar]
- Pop O, Westermann M, Volkmer-Engert R, Schulz D, Lemke C, Schreiber S, Gerlach R, Wetzker R, Müller JP. Sequence-specific binding of prePhoD to soluble TatAd indicates protein-mediated targeting of the Tat export in Bacillus subtilis. J Biol Chem. 2003;278:38428–38436. doi: 10.1074/jbc.M306516200. [DOI] [PubMed] [Google Scholar]
- Dilks K, Giménez MI, Pohlschröder M. Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J Bacteriol. 2005;187:8104–8113. doi: 10.1128/JB.187.23.8104-8113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaeker S, Van Mellaert L, Schaerlaekens K, Van Dessel W, Vrancken K, Lammertyn E, Anné J, Geukens N. Structural organization of the twin-arginine translocation system in Streptomyces lividans. FEBS Letts. 2005;579:797–802. doi: 10.1016/j.febslet.2004.12.059. [DOI] [PubMed] [Google Scholar]
- Oates J, Mathers J, Mangels D, Kuhlbrandt W, Robinson C, Model K. Consensus structural features of purified bacterial TatABC complexes. J Mol Biol. 2003;330:277–286. doi: 10.1016/S0022-2836(03)00621-1. [DOI] [PubMed] [Google Scholar]
- Bruser T, Deutzmann R, Dahl C. Evidence against the double-arginine motif as the only determinant for protein translocation by a novel Sec-independent pathway in Escherichia coli. FEMS Microbiol Lett. 1998;164:329–336. doi: 10.1111/j.1574-6968.1998.tb13106.x. [DOI] [PubMed] [Google Scholar]
- Angelini S, Moreno R, Gouffi K, Santini C, Yamagishi A, Berenguer J, Wu L-F. Export of Thermus thermophilus alkaline phosphatase via the twin-arginine translocation pathway in Escherichia coli. FEBS Letts. 2001;506:103–107. doi: 10.1016/S0014-5793(01)02890-3. [DOI] [PubMed] [Google Scholar]
- Blaudeck N, Sprenger GA, Freudl R, Wiegert T. Specificity of signal peptide recognition in Tat-dependent bacterial protein translocation. J Bacteriol. 2001;183:604–610. doi: 10.1128/JB.183.2.604-610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop O, Martin U, Abel C, Müller JP. The twin-arginine signal peptide of PhoS and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system. J Biol Chem. 2002;277:3268–3273. doi: 10.1074/jbc.M110829200. [DOI] [PubMed] [Google Scholar]
- Snyder A, Vasil AI, Zajdowicz SL, Wilson ZR, Vasil ML. Role of the Pseudomonas aeruginosa PlcH Tat signal peptide in protein secretion, transcription, and cross-species Tat secretion system compatibility. J Bacteriol. 2006;188:1762–1774. doi: 10.1128/JB.188.5.1762-1774.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein PA, Marrichi M, Cartinhour S, Schneider DJ, DeLisa MP. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J Bacteriol. 2005;187:8450–8461. doi: 10.1128/JB.187.24.8450-8461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi FU, Feijao A, Hedges SB. A genomic timescale of prokaryotic evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol. 2004;4:44–58. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ize B, Stanley NR, Buchanan G, Palmer T. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol. 2003;48:1183–1193. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol. 2003;48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan G, de Leeuw E, Stanley NR, Wexler M, Berks BC, Sargent F, Palmer T. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol Microbiol. 2002;43:1457–1470. doi: 10.1046/j.1365-2958.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- Stanley NR, Sargent F, Buchanan G, Shi J, Stewart V, Palmer T, Berks BC. Behaviour of topological marker proteins targeted to the Tat protein transport pathway. Mol Microbiol. 2002;43:1005–1021. doi: 10.1046/j.1365-2958.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989;171:1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. PhD thesis. University of East Anglia; 2003. Characterisation of the TatA and TatB proteins of the Escherichia coli Tat translocase. [Google Scholar]
- Hicks MG, de Leeuw E, Porcelli I, Buchanan G, Berks BC, Palmer T. The Escherichia coli twin-arginine translocase: conserved residues of TatA and TatB family components involved in protein transport. FEBS Letts. 2003;539:61–67. doi: 10.1016/S0014-5793(03)00198-4. [DOI] [PubMed] [Google Scholar]
- Hicks MG, Lee PA, Georgiou G, Berks BC, Palmer T. Positive selection for loss-of-function tat mutations identifies critical residues required for TatA activity. J Bacteriol. 2005;187:2920–2925. doi: 10.1128/JB.187.8.2920-2925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F, Gohlke U, de Leeuw E, Stanley NR, Palmer T, Saibil HR, Berks BC. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur J Biochem. 2001;268:3361–3367. doi: 10.1046/j.1432-1327.2001.02263.x. [DOI] [PubMed] [Google Scholar]
- De Keersmaeker S, Van Mellaert L, Lammertyn E, Vrancken K, Anné J, Geukens N. Functional analysis of TatA and TatB in Streptomyces lividans. Biochem Biophys Res Comm. 2005;335:973–982. doi: 10.1016/j.bbrc.2005.07.165. [DOI] [PubMed] [Google Scholar]
- Jongbloed JD, Martin U, Antelmann H, Hecker M, Tjalsma H, Venema G, Bron S, van Dijl JM, Muller J. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J Biol Chem. 2000;275:41350–41357. doi: 10.1074/jbc.M004887200. [DOI] [PubMed] [Google Scholar]
- Lee PA, Buchanan G, Stanley NR, Berks BC, Palmer T. Truncation analysis of TatA and TatB defines the minimal functional units required for protein translocation. J Bacteriol. 2002;184:5871–5879. doi: 10.1128/JB.184.21.5871-5879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilks K, Rose RW, Hartmann E, Pohlschroder M. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J Bacteriol. 2003;185:1478–1483. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167–175. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdick DA, Dilks K, Chandra G, Bottrill A, Naldrett M, Pohlschroder M, Palmer T. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. 2006 doi: 10.1073/pnas.0607025103. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari I, Mann S, Crooks C, Palmer T. The Tat pathway of the plant pathogen Pseudomonas syringae is required for optimal virulence. Mol Plant-Microbe Interact. 2006;19:200–212. doi: 10.1094/MPMI-19-0200. [DOI] [PubMed] [Google Scholar]
- Bernhard M, Friedrich B, Siddiqui RA. Ralstonia eutropha TF93 is blocked in Tat-mediated protein export. J Bacteriol. 2000;182:581–588. doi: 10.1128/JB.182.3.581-588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerlaekens K, Schierová M, Lammertyn E, Geukens N, Anné J, van Mellaert L. Twin-arginine translocation pathway in Streptomyces lividans. J Bacteriol. 2001;183:6727–6732. doi: 10.1128/JB.183.23.6727-6732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Press, Cold Spring Harbor, NY; 1989. [Google Scholar]
- Cohen GN, Rickenberg HW. Concentration specifique reversible des amino acids chez Escherichia coli. Ann Inst Pasteur (Paris) 1956;91:693–720. [PubMed] [Google Scholar]
- Jack RL, Buchanan G, Dubini A, Hatzixanthis K, Palmer T, Sargent F. Coordinating assembly and export of complex bacterial proteins. EMBO J. 2004;23:3962–3972. doi: 10.1038/sj.emboj.7600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks BC, Sargent F, Palmer T. The Tat protein export pathway. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley NR, Palmer T, Berks BC. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J Biol Chem. 2000;257:11591–11596. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- Silvestro A, Pommier J, Giordano G. The inducible trimethylamine-N-oxide reductase of Escherichia coli K12: biochemical and immunological studies. Biochim Biophys Acta. 1988;954:1–13. doi: 10.1016/0167-4838(88)90049-0. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurements with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo MR, Zabin I. Beta-galactosidase from termination and deletion mutant strains. J Bacteriol. 1974;120:466–474. doi: 10.1128/jb.120.1.466-474.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PA, Greene NP, Buchanan G, Bond PJ, Punginelli C, Jack RL, Sansom MSP, Berks BC, Palmer T. Cysteine-scanning mutagenesis of the conserved domain of the twin-arginine translocase TatB component. 2006. [DOI] [PubMed]


