Abstract
Murine leukemia virus (MLV)-derived envelope proteins containing alterations in or adjacent to the highly conserved PHQ motif present at the N terminus of the envelope surface subunit (SU) are incorporated into vector particles but are not infectious due to a postbinding block to viral entry. These mutants can be rendered infectious by the addition of soluble receptor-binding domain (RBD) proteins in the culture medium. The RBD proteins that rescue the infectivity of these defective MLV vectors can be derived from the same MLV or from other MLVs that use distinct receptors to mediate entry. We have now constructed functional immunologically reactive gibbon ape leukemia virus (GALV) envelope proteins, tagged with a feline leukemia virus (FeLV)-derived epitope tag, which are efficiently incorporated into infectious particles. Tagged GALV envelope proteins bind specifically to cells expressing the phosphate transporter protein Pit1, demonstrating for the first time that Pit1 is the binding receptor for GALV and not a coreceptor or another type of GALV entry factor. We have also determined that GALV particles bearing SU proteins with an insertion C-terminal to the PHQ motif (GALV I10) bind Pit1 but fail to infect cells. Incubation with soluble GALV RBD renders GALV I10 particles infectious, whereas incubation with soluble RBDs from MLV or FeLV-B does not. This finding is consistent with the results obtained by Lauring et al. using FeLV-T, a virus that employs Pit1 as a receptor but requires soluble FeLV RBD for entry. MLV and GALV RBDs are not able to render FeLV-T infectious (A. S. Lauring, M. M. Anderson, and J. Overbaugh, J. Virol. 75:8888-8898, 2001). Together, these results suggest that fusion-defective FeLV-T and GALV are restricted to homologous RBD rescue of infectivity.
Virus receptors are the components of cell membranes that allow viral attachment, the first step in infection. It has traditionally been thought that viruses infect and replicate in some cell types, but not others, because of the distribution of specific virus receptors on different cells. Now it is well known that although the presence of specific receptors is critical for viral entry, coreceptors and postentry factors are also important in defining the virus's host range. Some cells express the appropriate receptors but are nevertheless resistant to infection by a specific virus. In many cases, discrepancies between virus receptor distribution and virus host range can be accounted for by cell-specific expression of coreceptors or other ancillary factors required for membrane fusion and internalization of a virus after its initial attachment to its primary receptor. For example, the human CD4 molecule acts as a primary receptor for human immunodeficiency virus type 1 (HIV-1) infection when expressed in human cells but not when expressed in murine cells. When human CD4 is expressed in murine cells, it supports HIV-1 binding but not infection; infection can proceed if such cells express the appropriate coreceptor (reviewed in reference 34). Similarly, Pit1 acts as a primary receptor for feline leukemia virus type T (FeLV-T); however, infection by FeLV-T requires the presence of the FeLIX cofactor (1).
In addition to FeLV-T, the phosphate transporter Pit1 has been proposed to serve as the receptor for gibbon ape leukemia virus (GALV) (31), feline leukemia virus subgroup B (FeLV-B) (40), and murine leukemia virus (MLV) 10A1 (30, 43). GALV infects a wide variety of cell types (21) derived from diverse species (20). Because of its broad host range, GALV-based retroviral vectors have been developed for use in both in vivo and ex vivo gene transfer (29, 42). Although GALV is structurally related to MLVs, FeLVs, and other gammaretroviruses, GALV envelope proteins do not immunologically cross-react with reagents that detect these retroviral surface subunit (SU) proteins (M. V. Eiden, unpublished data). Due to the extreme lability of GALV envelope proteins, efforts to purify sufficient quantities of virus for development of the immunologically active reagents required for determining GALV particle formation and GALV receptor binding have been unsuccessful. We made a series of GALV SU proteins that have been truncated at various regions within the SU portion of the envelope downstream of the putative receptor-binding domain (RBD) and then fused to C-terminal epitope tags. Attachment of a hemagglutinin (HA) epitope tag to the C terminus of either amphotropic MLV (A-MLV) or FeLV-B SU envelope proteins generated soluble RBD proteins that efficiently bound the receptor (24). Similarly constructed truncated and tagged soluble GALV RBDs were not detectable by Western blot analyses of either transfected cell lysates or supernatants. In addition, unlike the results with MLV, for which functional chimeric envelope proteins have been created by reciprocal exchanges of portions of xenotropic, polytropic, ecotropic, and amphotropic MLV SU proteins (7, 33), none of the MLV-GALV chimeric SU proteins we have tested are incorporated into infectious particles (M. V. Eiden, unpublished data). Regions within various MLV and FeLV-B SU proteins that are specifically required for receptor binding and entry have been mapped by using a combination of SU chimeric and mutagenesis studies (2-9, 17, 18, 25-28, 32, 33, 37-39, 44-46). Similar studies have not been performed for GALV; therefore, little is known about the regions of GALV envelope protein that are involved in receptor binding, cell fusion, entry, and the pathogenicity of this virus.
Determination of the Pit1 virus-binding site has involved utilization of Pit1 mutant receptors and has been based on infectivity studies rather than binding analyses (10, 11, 13, 22, 35-39, 43; reviewed in reference 34). For example, it has been shown previously that murine MDTF (Mus dunni tail fibroblast) cells are resistant to GALV and can be rendered susceptible to GALV upon expression of the Pit1 cDNA (43). The validity of referring to Pit1 as a receptor capable of directly binding GALV has not been unequivocally determined, because heretofore GALV has not been shown to directly bind Pit1.
We report here the generation of functional tagged GALV envelope SU proteins by insertion of an 18-residue FeLV-B epitope tag, recognized by monoclonal antibody C11D8, into two separate regions of the proline rich region (PRR) of GALV; the tagged SUs are designated GALV I219 and GALV I264. Both are efficiently incorporated into infectious vector particles which bind Pit1. GALV SU proteins truncated at either residue 219 or residue 264 and bearing HA tags at their C termini retain the ability to bind Pit1. We used GALV I264 envelope as a template for mutagenesis studies in which we compared the effects of linker insertions on GALV envelope function to the effects of similar insertions in the FeLV-B envelope protein. Linker insertion mutagenesis studies of GALV SU, together with reports of previous work using MLVs as templates for linker insertion mutagenesis (17) and our present observations with FeLV-B, suggest that GALV is noticeably less tolerant of insertions in its SU region than either MLVs or FeLV-B. Finally, we introduced the C11D8 epitope into a region of GALV SU protein that has been shown in studies with mutant MLV envelopes (2-4, 12, 27, 45) to be critical for postentry virus fusion, and we assessed the effects of this mutation on GALV envelope function.
MATERIALS AND METHODS
Cell lines.
The following cell lines were used in this study: MDTF cells (23) (obtained from Olivier Danos, Institut Pasteur, Paris, France) and human embryonic kidney 293T cells, clone tsA54 (19) (obtained from Cell Genesys Inc., Foster City, Calif.). All cells were maintained in Dulbecco's modified essential medium (DMEM; Whittaker Bioproducts, Inc., Walkersville, Md.) supplemented with 5% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 4 mM glutamine.
Mutagenesis.
The sequence comprising the FeLV-B epitope, QVMTITPPQAMGPNLVLP, recognized by monoclonal antibody C11D8 (Custom Monoclonal Antibodies International, Sacramento, Calif.) was introduced into GALV envelope SU by PCR mutagenesis. GALV R213-230 was created by designing complementary overlapping synthetic oligonucleotides incorporating individual base changes necessary to convert GALV residues to FeLV-B residues. The GALV residue sequence LKITNMPAVAVGPDLVLV (amino acids 213 to 230) was replaced by the FeLV-B sequence QVMTITPPQAMGPNLVLP (amino acids 204 to 221). For the C11D8 epitope insertion mutants GALV I10, GALV I264, and GALV I219, complementary synthetic oligonucleotides encoding the C11D8 epitope (54 nucleotides) and the region of GALV SU flanking the insertion site were designed; two segments of GALV SU cDNA were amplified by using these primers plus upstream and downstream primers flanking the mutated region (oligonucleotide sequences are available upon request). These segments were annealed, after which a second round of PCR was carried out by using only the outer primers. The final PCR products were directly cloned into the TOPO TA pCR2.1 vector (Invitrogen, Carlsbad, Calif.) and sequenced by Thermo Sequenase fluorescent labeled primer cycle sequencing on an ALF Express automated sequencer (Pharmacia/Amersham, Peapack, N.J.). After confirmation of the presence of the desired mutations and the absence of unscheduled PCR mutations, the mutant envelope cDNA was subcloned into the PCIneo (Promega, Madison, Wis.) vector carrying the GALV envelope cDNA (pCIGALV) to create plasmids pCIGALV I264, pCIGALV I219, and pCIGALV I10.
A series of GALV SU constructs containing 12-mer EcoRI linkers (CCGGAATTCCGG; New England Biolabs, Beverly, Mass.) were made by using pCIGALV I264 as a template. Partial digestion of the template with EcoRV, PvuII, or SmaI was performed, followed by isolation of full-length linear DNA. This was ligated to a 100-fold molar excess of the linker by using T4 DNA ligase. The resulting clones were screened by restriction enzyme digestion with EcoRI in order to identify those containing the linker and were then sequenced as described above, in order to locate the insertions within the SU genome. FeLV-B SU envelope constructs were created in the same manner, with EcoRI linkers inserted at corresponding positions of the SU protein.
Production of retroviral vectors and viral infection.
Two days before transfection, 293T cells were seeded at a density of 106/10-cm dish. Transfection by the calcium phosphate precipitation method (Promega) was performed by using a three-plasmid system (41) including (i) a β-galactosidase genome plasmid, pkat2βgal (15); (ii) an MLV-based gag pol plasmid, pkatgagpolATG (15); and (iii) a plasmid expressing either GALV, GALV mutant, or FeLV-B 90Z envelope. Supernatant containing enveloped retroviral vectors was harvested 60 to 72 h posttransfection. For transductions, MDTF-Pit1 target cells were seeded 1 day in advance in a 24-well plate at 1.5 × 104 cells/well. Cells were exposed to a retroviral vector-containing supernatant that had been passed through a 0.45-μC Millipore (Bedford, Mass.) filter and were then adjusted to contain 10 μg of Polybrene/ml. Twenty-four hours later, the medium was changed and cells were cultured for an additional 24 to 48 h before analysis for expression of β-galactosidase by histochemical staining with X-Gal (5-bromo-4 chloro-3-indolyl-β-d-galactopyranoside), as previously described (43). Titers were determined by averaging the numbers of blue foci (BFU) obtained with vectors for each cell line tested in three or more independent experiments.
Vector particle binding assays.
Viral vector supernatants harvested from transiently transfected 293T cells were concentrated 10-fold by centrifugation at 2,000 × g at 4°C for 30 to 60 min using Ultrafree 15-100 concentrators (Millipore). MDTF or MDTF-Pit1 target cells were detached from tissue culture flasks with Cellstripper (Mediatech, Reston, Va.) cell dissociation solution; 3 × 106 cells were used for each vector to be tested. Cells were suspended in 0.3 ml of concentrated supernatant in the presence of Polybrene (10 μg/ml) and incubated at 37°C for 1 h, followed by two washes in Hanks' buffered saline solution (HBSS) containing 1% fetal bovine serum. Bound virus was detected by incubating cells in HBSS containing 1.6 μg of monoclonal antibody C11D8/μl for 1 h at 4°C; cells were washed twice in HBSS and then incubated for 1 h at 4°C in the presence of 2 μg of fluorescein-conjugated goat anti-mouse immunoglobulin G (IgG) (Pierce, Rockford, Ill.)/100 μl. Cells were fixed with 1% paraformaldehyde and then analyzed by fluorescence-activated cell sorting (FACS). Binding results from three independent experiments are expressed as the average shift in fluorescence intensity observed with vector particles bound to MDTF-Pit1 cells, minus the background value obtained on negative-control MDTF cells, normalized to the value obtained with FeLV-B bound to MDTF-Pit1 cells.
Soluble protein binding assays.
Binding assays using HA-tagged soluble FeLV-B1-144 (23), GALV1-219, or GALV1-264 SU glycoprotein were performed by harvesting supernatants from 293T cells transiently transfected, as described above, with pcDNA3.1/Zeo (Invitrogen) expressing the soluble protein. Target MDTF or MDTF-Pit1 cells were detached from tissue culture flasks with Cellstripper cell dissociation solution (Mediatech); 106 cells for each receptor cell line were suspended in 1 ml of supernatant containing soluble HA-tagged SU protein and were incubated at 37°C for 45 min. Cells were washed twice with HBSS containing 1% fetal bovine serum and then incubated in the presence of a 1:1,000 dilution of 5 mg of monoclonal antibody HA.11 (Covance/Babco, Richmond, Calif.)/ml in HBSS for 90 min at 4°C; after a wash with HBSS, cells were incubated for 1 h at 4°C in the presence of HBSS containing 2 μg of fluorescein-conjugated goat anti-mouse IgG (Pierce)/100 μl. Cells were fixed in 1% paraformaldehyde and then analyzed by FACS.
Western blot analysis of cell lysates and viral pellets.
For cell lysates, transiently transfected 293T cells were grown to confluency in a 10-cm dish and lysed in 1 ml of cold lysis buffer (150 mM NaCl, 1.0% NP-40, 50 mM Tris [pH 8.0], 10 μg of phenylmethylsulfonyl fluoride/ml, 20 μg of aprotinin/ml, 20 μl of leupeptin/ml). Cells were scraped from the 10-cm dish and centrifuged at 16,000 × g for 10 min at 4°C. The supernatant was collected and analyzed by Western blotting. Viral particles were pelleted from 8 ml of cell-free virus supernatant through a 25% sucrose gradient in TNE buffer (10 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA) in a Beckman SW41 rotor at 30,000 rpm for 2 h at 4°C. Pellets were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample loading buffer and analyzed by Western blotting. Viral pellets and cell lysates were detected by incubation with antibody C11D8 (3 μg/ml). After incubation with the second antibody, goat anti-mouse IgG conjugated to horseradish peroxidase (1:25,000) (Pierce), the signals were detected by chemiluminescence using the Renaissance Western Blot Chemiluminescence Reagent (NEN Life Science Products, Inc., Boston, Mass.) and exposure to Kodak X-Omat Blue XB-1 film.
C11D8 peptide neutralization assays.
The peptide comprising the C11D8 epitope of FeLV-B (Phoenix Pharmaceuticals, Inc., Belmont, Calif.) was added to the medium of MDTF-Pit1 target cells at different concentrations (10 to 100 μM) in order to determine if neutralization of vectors with envelopes containing the C11D8 epitope could be achieved. Before infection, target cells were incubated in the presence of a medium containing the peptide for 10 to 30 min at 35°C. The medium was removed and replaced with vectors containing the same concentration of peptide. After 3 to 15 h, the medium was removed, cells were washed with DMEM, and fresh DMEM was added; 48 h later, titers were determined by a β-galactosidase histochemical assay (43).
Infection assays using soluble SU protein.
Each of the soluble SU proteins FeLV-B1-444-HA (24), A-MLV1-448-HA (24), GALV1-264-HA RBD (residues 1 to 264, including variable region A [VRA], VRB, and the N-terminal half of the PRR), and GALV1-219-HA (residues 1 to 219, including VRA and VRB) was expressed in the supernatant of 293T cells transiently transfected with the cDNA of this protein, by using the calcium phosphate method described above. Target cells were incubated for 15 h at 35°C in the presence of GALV- or FeLV-B-enveloped viral vector particles bearing the genome for β-galactosidase, either with or without the soluble protein component. After 48 to 72 h, the number of cells infected was evaluated by the β-galactosidase histochemical assay described above.
RESULTS
Immunodetection of functional GALV SU containing the C11D8 epitope.
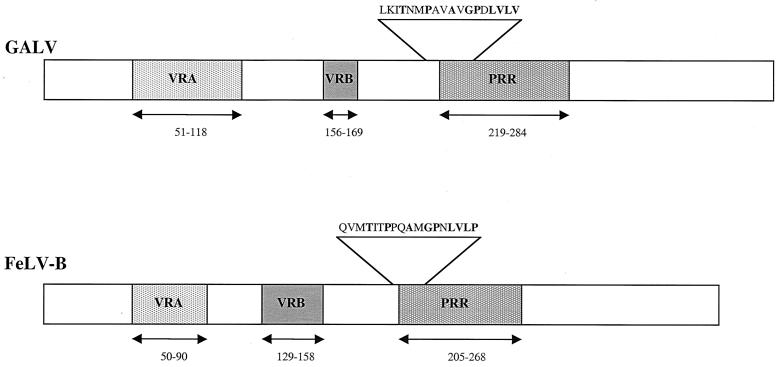
There are several well-characterized monoclonal antibodies that react with FeLVs. One of these monoclonal antibodies, C11D8, specifically interacts with residues 204 to 221 (QVMTITPPQAMGPNLVLP) within the conserved PRR of the FeLV-B envelope (14). C11D8 also reacts with a similar region in FeLV-A and FeLV-C and neutralizes infection by these viruses (16). Other gammaretroviral SUs, including those of GALV or MLVs, do not cross-react with antibody C11D8. We compared the corresponding region within GALV SU (residues 213 to 230) to the FeLV-B C11D8 epitope sequence and determined that 9 of the 18 residues are conserved between the two proteins in this region (Fig. 1). This divergence in residue sequence presumably accounts for the lack of C11D8-GALV SU immunoreactivity. Since the PRR serves similar functions in FeLV-B and GALV, we reasoned that substitution of the C11D8 epitope segment for the corresponding region of GALV SU may result in a protein that can be detected by C11D8 while retaining normal GALV envelope function.
FIG. 1.
The C11D8 epitope, QVMTITPPQAMGPNLVLP (residues 204 to 221), which derives from the amino terminus of the FeLV-B PRR, was substituted for LKITNMPAVAVGPDLVLV (residues 213 to 230) in a corresponding region of the GALV PRR. The relative positions of VRA, VRB, and the PRR within the envelope SU protein are shown.
To introduce the epitope encoding FeLV-B residues 204 to 221, we mutated the region of GALV SU between residues 213 and 230 to contain the C11D8 epitope, thereby creating GALV R213-230 (Fig. 1). GALV R213-230 SU protein can be detected with monoclonal antibody C11D8 in Western blots using cell lysates made from 293T cells transfected with the GALV R213-230 expression plasmid (Table 1). We also determined, by Western blot analysis, that vector particles efficiently incorporate GALV R213-230 envelope proteins, although the titers of these retroviral vectors are significantly lower than those of wild-type GALV vectors (Table 1).
TABLE 1.
Properties of mutant GALV SU proteins
| SU protein | Processinga | Incorporationb | Bindingc | Titer (106 BFU/ml)d |
|---|---|---|---|---|
| GALV R213-260 | + | + | ND | 0.0002 ± 0.4 |
| GALV I264 | + | + | 0.03 | 1.3 ± 0.3 |
| GALV I219 | + | + | 0.95 | 0.43 ± 0.6 |
| GALV I10 | + | + | 0.81 | 0.000001 |
| FeLV-B | + | + | 1.00 | NDe |
| GALV | ND | ND | ND | 3.8 ± 0.4 |
SU protein was detected (+) or not (−) in lysates from transiently transfected 293T cells, as described in Materials and Methods.
SU protein was present (+) or absent (−) in retroviral vector particles produced in 293T cells at 35°C.
Average shift in fluorescence intensity observed with vector particles bound to MDTF-Pit1 cells, detected by antibody C11D8 in a FACS-based binding assay, normalized to the average mean fluorescence observed with FeLV-B bound to MDTF-Pit1 cells (1.0). The average values for negative-control MDTF cells with GALV I10, GALV I219, and FeLV-B were 0.018, 0.019, and 0.016, respectively; these values were subtracted from those obtained on MDTF-Pit1 cells. Values are derived from from three independent experiments.
Titers are expressed as the average numbers of blue foci (BFU) ± standard errors of the means on MDTF-Pit1 target cells, from three experiments.
ND, not done.
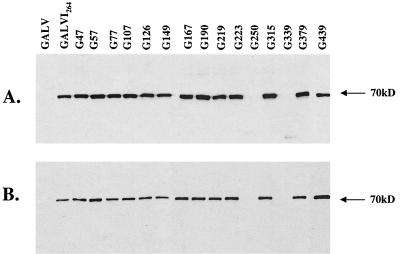
As an alternative strategy to replacing the region of the GALV PRR corresponding to the C11D8 epitope with the FeLV-derived C11D8 tag, we next directly inserted the C11D8 epitope into the GALV PRR and determined whether these mutant envelope proteins were efficiently incorporated into highly infectious GALV particles. The PRR was chosen as a site for epitope insertion on the basis of reports by others that epitopes introduced in the PRR of MLV had no adverse effects on envelope glycoprotein function (44, 45). Based on these observations, we inserted the C11D8 epitope at residue 264 in the PRR of GALV SU. The resultant GALV protein, designated GALV I264, was detected in Western blot analyses of transfected 293T cell lysates (Fig. 2A) and was efficiently incorporated into infectious particles (Table 1 and Fig. 2B). The titers obtained with GALV I264 vectors were equivalent to those obtained with wild-type GALV vectors (Table 1). Binding assays failed to detect vectors bearing GALV I264 envelopes (Table 1), suggesting that the C11D8 epitope in the PRR is occluded following GALV-receptor interaction, making it inaccessible to antibody C11D8. Occlusion of the C11D8 epitope in this region of GALV SU is not mediated by a direct association with Pit1, as appears to be the case with FeLV-B. Elder et al. have shown that incubation of FeLV-B with antibody C11D8 neutralizes infection (14). In contrast, GALV I264 infection of MDTF-Pit1 cells was not blocked by using either antibody C11D8 or C11D8 epitope peptides (data not shown). GALV I264 is a functional GALV envelope that is efficiently incorporated into infectious vector particles that react with antibody C11D8. We therefore used GALV I264 as a template for EcoRI linker insertion mutagenesis studies designed to map regions within the GALV envelope protein that are critical for envelope processing, particle incorporation, receptor binding, and entry functions.
FIG. 2.
Western blot detection, using antibody C11D8, of individual mutant GALV envelope SU proteins from transfected cell lysates (A) and in pelleted viral particles (B) harvested from the supernatant of vector-producing 293T cells. EcoRI linker insertion mutants are designated G47 through G439 (the number following “G” indicates the residue position of the insertion).
Analysis of linker insertion mutants of FeLV-B and GALV I264.
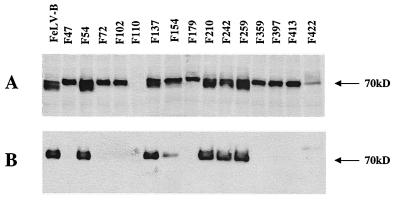
By using GALV I264 and wild-type FeLV-B as templates, 15 GALV I264 (Table 2) and FeLV-B (Table 3) SU linker insertion mutants were constructed and tested as described in Materials and Methods. Analogous regions within the FeLV-B and GALV I264 SU coding regions were chosen for the insertion of EcoRI linkers in order to more directly compare their effects on GALV and FeLV-B envelope function (Tables 2 and 3). Two of the GALV mutants, G250 and G339 (Fig. 2A), failed to be processed into mature gp70 proteins, whereas only one FeLV-B insertion mutant, F110, was not appropriately processed (Fig. 3A). All of the processed GALV mutants were incorporated into particles (Fig. 2B and Table 2). In contrast, only 7 of the 14 mutant FeLV-B glycoproteins were incorporated into particles (Fig. 3B and Table 3). Of the 13 GALV mutant SU proteins that were incorporated into particles, only particles bearing mutant G219 envelopes were infectious (Table 2). By comparison, all seven of the mutant FeLV-B envelope proteins that were incorporated into vector particles were infectious (Table 3), with titers comparable to those of wild-type FeLV-B (data not shown). We next introduced the C11D8 epitope at residue 219 within the GALV envelope protein and assessed the ability of this epitope-tagged envelope to be incorporated into infectious viral particles. As shown in Fig. 4A, GALV I 219 was efficiently incorporated into particles, and in contrast to vectors bearing GALV I264, GALV I219 particles could be detected when bound to MDTF cells expressing Pit1 (Table 1). Therefore, GALV I219 particles should prove useful reagents in GALV particle binding and entry studies.
TABLE 2.
Properties of GALV I264 linker insertion mutant SU proteins
| Mutanta | Location in SU | Processingb | Incorporationc | Infectivityd |
|---|---|---|---|---|
| G47 | N terminus | + | + | − |
| G57 | VRA | + | + | − |
| G77 | VRA | + | + | − |
| G107 | VRA | + | + | − |
| G126 | Between VRA and VRB | + | + | − |
| G149 | Between VRA and VRB | + | + | − |
| G167 | VRB | + | + | − |
| G190 | Between VRB and PRR | + | + | − |
| G219 | PRR | + | + | + |
| G223 | PRR | − | − | − |
| G250 | PRR | + | + | − |
| G315 | C terminus | + | + | − |
| G339 | C terminus | − | − | − |
| G379 | C terminus | + | + | − |
| G439 | C terminus | + | + | − |
Linkers were inserted after the residue indicated (e.g., between amino acids 47 and 48 in the first insertion mutant).
SU protein was detected (+) or not (−) in lysates from transiently transfected 293T cells, as described in Materials and Methods.
SU protein was present (+) or absent (−) in retroviral vector particles produced in 293T cells at 35°C.
Vector particles bearing mutant envelopes did (+) or did not (−) infect MDTF-Pit1 target cells.
TABLE 3.
Properties of FeLV-B linker insertion mutant SU proteins
| Mutanta | Location in SU | Processingb | Incorporationc | Infectivityd |
|---|---|---|---|---|
| F47 | N terminus | + | − | − |
| F54 | VRA | + | + | + |
| F72 | VRA | + | − | − |
| F102 | VRA | + | − | − |
| F110 | Between VRA and VRB | − | − | − |
| F137 | Between VRA and VRB | + | + | + |
| F154 | VRB | + | + | + |
| F179 | Between VRB and PRR | + | − | − |
| F210 | PRR | + | + | + |
| F242 | PRR | + | + | + |
| F259 | PRR | + | + | + |
| F359 | C terminus | + | − | − |
| F397 | C terminus | + | − | − |
| F413 | C terminus | + | − | − |
| F422 | C terminus | + | + | + |
Linkers were inserted after the residue indicated (e.g., between amino acids 47 and 48 in the first insertion mutant).
SU protein was detected (+) or not (−) in 293T cell lysates 48 to 72 h after transfection, as described in Materials and Methods.
SU protein was present (+) or absent (−) in retroviral vector particles produced in 293T cells at 35°C.
Vector particles bearing mutant envelopes did (+) or did not (−) infect MDTF-Pit1 target cells.
FIG. 3.
Western blot detection, using antibody C11D8, of individual mutant FeLV-B envelope SU proteins from transfected cell lysates (A) and in pelleted viral particles (B) harvested from the supernatant of vector-producing 293T cells. EcoRI linker insertion mutants are designated F47 through F422 (the number following “F” indicates the residue position of the insertion).
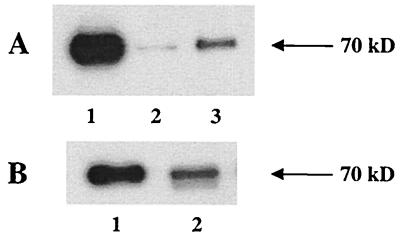
FIG. 4.
GALV envelopes containing the C11D8 epitope tag are assembled into viral particles. SU protein was detected in pelleted vector particles by Western blotting with antibody C11D8. (A) Lane 1, FeLV-B control; lane 2, GALV I10; lane 3, GALV I264. (B) Lane 1, FeLV-B control; lane 2, GALV I219.
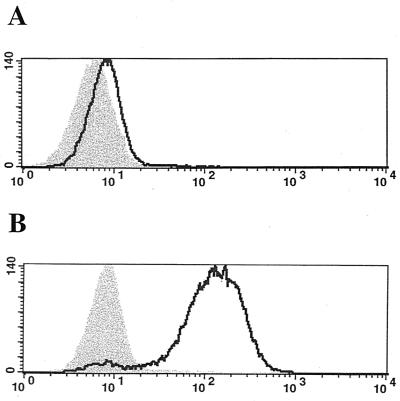
Soluble GALV SU truncated proteins containing in-frame tandem repeats of the sequence encoding the influenza virus HA epitope tag were also used as ligands in similar FACS-based binding assays. We truncated GALV SU at either residue 219, residue 264, or the carboxy terminus of SU—residue 487 and fused these envelope segments to C-terminal HA epitope tags (GALV1-219-HA, GALV1-264-HA, and GALV1-487-HA, respectively). No HA-immunoreactive GALV1-487-HA protein was detected in either 293T cell lysates or supernatants 48 to 72 h after transfections. As expected from the results for particle binding (Table 1), neither GALV1-219-HA (data not shown) nor GALV1-264-HA-tagged envelope glycoproteins bound MDTF cells (Fig. 5A); however, MDTF-Pit1 cells efficiently bound both (data not shown and Fig. 5B).
FIG. 5.
Recognition of soluble HA-tagged GALV1-264 glycoprotein by monoclonal antibody HA.11 in a FACS-based assay of binding to MDTF (A) or MDTF-Pit1 (B) cells. Shaded areas represent control MDTF cells which were not exposed to GALV; areas under solid line represent cells exposed to GALV.
GALV envelope proteins containing the C11D8 epitope at residue 10 are incorporated into particles that bind the receptor but are not infectious.
F.-L. Cosset and coworkers have demonstrated that a foreign protein segment could be introduced into the amino terminus of MLV SU at residue 5 (EMO), resulting in an envelope protein that is efficiently incorporated into particles that bind the receptor (12). However, vectors bearing EMO envelopes are not infectious, due to a postbinding block to viral entry. To determine if an analogous mutation had a similar effect on GALV envelope function, we inserted the 18-residue C11D8 epitope after residue 10 of the mature GALV SU protein and designated this mutant envelope GALV I10. The GALV I10 envelope protein was expressed in 293T cells (data not shown) and incorporated into vector particles (Fig. 4A). In a FACS-based binding assay, GALV I10-enveloped vectors did not bind MDTF cells but did efficiently bind MDTF cells expressing Pit1 (Table 1). Vectors bearing GALV I10 envelope proteins are not infectious, suggesting that entry of GALV I10 vectors into cells expressing Pit1 receptors is blocked at a postbinding stage of viral entry. These results are similar to those obtained with EMO mutants (12).
It has been shown previously that supplying homologous RBD as a soluble protein can restore infectivity to EMO (3, 4, 25, 27) and FeLV-T (24) vector particles. The infectivity of ecotropic MLV (E-MLV) fusion mutant particles harboring insertions in the N-terminal region of SU proximal to the highly conserved PHQ motif or harboring a deletion of the H residues in this motif can be restored by incubating bound particles not only with RBDs derived from E-MLV but also, alternatively, with RBDs derived from MLVs that use receptors distinct from the E-MLV receptor (3, 27). We therefore preincubated MDTF-Pit1 cells with soluble RBD proteins derived from A-MLV or FeLV-B to determine if these RBDs could render GALV I10 vectors infectious. A-MLV utilizes a related phosphate transporter, designated Pit2, as a receptor. Pit2 does not function as a GALV receptor, and Pit1 does not function as an A-MLV receptor. As mentioned above, FeLV-B can use Pit1 as a receptor. To assess the ability of RBDs to rescue GALV I10 infectivity, target cells were exposed to GALV I10 vectors that had been preincubated with either GALV, FeLV-B, or A-MLV RBDs. As expected, incubation of GALV I10 particles combined with any one of the four RBDs did not render MDTF cells susceptible to infection. Titers of approximately 105 BFU/ml were obtained when MDTF-Pit1 cells were exposed to GALV I10 in the presence of HA-tagged GALV I1-219 or GALV I1-264 (Table 4). Incubation with either A-MLV or FeLV-B soluble protein had no effect on the ability of GALV I10 particles to infect MDTF-Pit1 cells (Table 4). These data suggest that binding to Pit1 is not sufficient for RBD-mediated rescue of GALV I10 vector infectivity in trans, as only GALV RBD, but not FeLV-B RBD, can complement the postbinding entry deficiency observed with GALV I10 particles.
TABLE 4.
Effect of soluble FeLV or GALV SU on GALV I10 vector infectivity
| Vector envelopea | Target cell | Soluble SUb | Titer (105 BFU/ml)c |
|---|---|---|---|
| GALV | MDTF | None | <0.001 |
| MDTF-Pit1 | None | 50.0 ± 0.6 | |
| GALV I10 | MDTF | None | <0.001 |
| MDTF-Pit1 | None | <0.001 | |
| GALV I10 | MDTF | FeLV-B1-444 | <0.001 |
| MDTF-Pit1 | FeLV-B1-444 | <0.001 | |
| GALV I10 | MDTF | GALV1-219 | <0.001 |
| MDTF-Pit1 | GALV1-219 | 0.8 ± 0.3 | |
| GALV I10 | MDTF | GALV1-264 | <0.001 |
| MDTF-Pit1 | GALV1-264 | 1.2 ± 0.2 | |
| GALV I10 | MDTF | A-MLV1-448 | <0.001 |
| MDTF-Pit1 | A-MLV1-448 | <0.001 |
Viral vectors were produced from transiently transfected 293T cells as described in Materials and Methods. GALV I10 contains the C11D8 epitope inserted at position 10 of the SU.
Target cells were incubated with equal volumes of vector particle supernatants and medium containing soluble SU proteins.
Titers are expressed as the average numbers of blue foci (BFU) ± standard errors of the means from three experiments.
DISCUSSION
The gammaretrovirus GALV employs Pit1 as a viral entry factor (31). We have now developed epitope-tagged GALV envelope proteins that allow us to directly determine the role of Pit1 as the primary receptor for GALV. To conduct virus-receptor interaction studies, it was first necessary to develop immunologically reactive GALV envelope proteins that could be used in binding assays. We chose to use the region of FeLV-B envelope recognized by antibody C11D8 (14) as a tag for GALV SU based on several considerations. First, the C11D8 epitope recognition region is only 18 residues long, an advantage over the larger epitope tags that are commonly used, since it is less likely to disturb overall protein structure and function. Second, the C11D8 epitope is present in the PRR of FeLV-B (16), a virus that also uses Pit1 as a receptor (40). Finally, appropriate monoclonal antibodies to this epitope have been developed and used successfully in FeLV-B SU Western blotting, immunoprecipitation, and binding studies (16).
Based on the previous observation that segments within the PRR of MLVs can be replaced with the 16-residue collagen-binding domain (45), we substituted the 18-residue C11D8 epitope (residues 204 to 221 of FeLV-B) for the corresponding region of GALV SU (residues 213 to 230). GALV R213-230 envelope proteins are incorporated into vector particles, but those particles are not capable of efficiently infecting target cells (Table 1). We next inserted the C11D8 epitope into the more variable region of the GALV PRR at position 264 and determined that vectors bearing this GALV I264 envelope protein are assembled (Fig. 4A) and are highly infectious (Table 1). Surprisingly, we were not able to detect bound GALV I264-enveloped vectors by either FACS-based virus-receptor binding or immunohistochemical assays (data not shown), suggesting that this region of the envelope is not accessible to antibody once it is bound to its receptor, Pit1. We have obtained similar results with receptor-bound vectors bearing A-MLV envelope proteins with the C11D8 epitope introduced into the corresponding region of the A-MLV PRR (W. Anderson and M. V. Eiden, unpublished data). These findings suggest that SU proteins bearing C11D8 epitope tags within the PRR undergo a conformational change upon binding to the receptor, which results in the C11D8 epitope being masked and occluded from the antibody. Antibody C11D8 neutralizes FeLV-B infection; this observation suggests that the N terminus of the PRR is directly involved in FeLV-B receptor binding (16). We have now determined that, in contrast to the results obtained with FeLV-B, infection by vectors bearing GALV I264 envelope proteins is not neutralized by either the C11D8 peptide or the C11D8 antibody.
GALV I264, although not useful in monitoring GALV vector particle binding, served as a template for linker insertion mutagenesis analyses. Mutagenesis experiments were designed to compare regions within the GALV and FeLV-B SUs that tolerate insertions without loss of particle assembly, the ability to be detected when bound to a receptor, or particle infectivity. All 12 of the mutant GALV SU proteins that were appropriately processed into gp70 were also incorporated into vector particles (Table 2); however, only 1 of these mutant envelope proteins, G219, was incorporated into infectious particles (Tables 1 and 2). Similarly, 14 of the 15 mutant FeLV-B proteins were synthesized. In contrast to the results obtained with GALV mutants, 7 of the 14 mutants were incorporated into infectious particles: F54, F137, F154, F210, F242, F259, and F422 (Table 3). These findings suggest that FeLV-B SU tolerates insertions within its RBD, PRR, and C-terminal region that are not tolerated by the GALV envelope SU protein without concomitant loss of function (Tables 2 and 3). Similar studies based on mutagenesis have been undertaken using the envelope gene of Moloney murine leukemia virus (MoMLV) (17). Of the 20 linker insertions made in the SU portion of MoMLV, 11 were determined not to compromise envelope function. MoMLV VRA (79 residues) is larger than the VRAs of all other MLVs and larger than that of either FeLV-B or GALV (40 and 67 residues, respectively) (7). It has been suggested previously that not all of MoMLV VRA is required for receptor recognition and that therefore MoMLV VRA may be more tolerant of mutations than the VRAs of other gammaretroviruses (17). FeLV-B has a smaller VRA than either MoMLV or GALV, yet, as with MoMLV and in contrast to GALV, insertion mutations are tolerated in the VRAs of FeLV-B envelope glycoproteins (Tables 2 and 3). Seven MoMLV (17) and one FeLV-B envelope mutant containing insertions in the C terminus of SU were fully functional (Table 3), whereas all mutations in similar positions of GALV SU gave rise to nonfunctional envelope proteins (Table 2). With regard to mutations in the PRR, two out of four insertions in MoMLV, compared to three out of three in FeLV-B and only one out of three in GALV, resulted in fully functional SU proteins (Tables 2 and 3). Insertions in all other regions of GALV SU compromised the function of the envelope protein (Table 2). In summary, the SU proteins of MoMLV and FeLV-B, in contrast with that of GALV, tolerate insertions in the RBD, the PRR, and the extreme C terminus of SU without loss of function (17) (Tables 2 and 3).
It has been determined previously that insertions within the N-terminal region of MLV SU give rise to envelope glycoproteins that can be assembled into infectious vector particles. For example, vectors with E-MLV-based SU proteins containing either a 15-residue peptide that binds to the vitronectin receptor αγβ3 (44) or 208 residues of A-MLV envelope SU (12) introduced at the 6th residue of E-MLV SU retain their infectivity. Based on these findings, we inserted 18 residues comprising the C11D8 epitope into the analogous region of GALV SU. GALV envelopes bearing C11D8 epitope tags at the 10th residue (GALV I10) were incorporated into vectors. These vectors avidly bound the receptor but failed to infect cells, presumably due to a postbinding block to entry (Table 4). Our findings obtained with GALV I10-enveloped vectors differed from those obtained with MLV in that particles bearing GALV I10 were not infectious, whereas MLV particles bearing envelope glycoproteins with N-terminal insertions retained infectivity. MLV particles bearing envelope glycoproteins containing the 15-residue peptide insertion maintained wild-type titers when tested on NIH 3T3 cells (44), and MLV particles with the 208-residue insertion infected target cells with a diminished efficiency relative to that of wild-type MLV (12).
More recently, it has been shown that either deletion of the histidine in the PHQ motif present in the N terminus of MLV SU or insertions in regions adjacent to the PHQ motif result in fusion-defective MLV glycoproteins (2-4, 25, 27, 46). Particles bearing fusion mutant glycoproteins bind receptor, but their infectivity is blocked at a postbinding level of entry. Particle infectivity can be restored, in trans, by providing soluble RBD derived either from the wild-type form of the fusion mutant glycoprotein or from MLVs that use distinct receptors for virus binding and entry (3, 4, 25, 27). We found that particles containing GALV I10 glycoproteins, like those bearing these mutant MLV envelope proteins, were blocked at a postbinding level of entry. We therefore sought to determine if GALV I10 particle infectivity could be restored by incubation with soluble RBD derived from A-MLV, FeLV-B, or GALV. Experimental results showed that GALV I10 particle infectivity could be restored with either GALV I1-219 or GALV I1-264 RBDs but not FeLV-B1-444 or A-MLV1-448 RBDs, even though GALV and FeLV-B both utilize Pit1 for entry. Our results are similar to those reported for FeLV-T (24). FeLV-T is a naturally occurring feline retrovirus that is closely related to FeLV-B. FeLV-T, FeLV-B, and GALV use Pit1 as a primary receptor. FeLV-T SU differs from those of other gammaretroviruses in that it contains an asparagine in place of the histidine residue in its PHQ motif (1, 24). Lack of this histidine residue presumably renders FeLV-T incapable of Pit1-mediated entry in the absence of soluble FeLV RBD (1). FeLV-T infectivity is not restored by incubation with either GALV or MLV RBDs (24).
In conclusion, we have demonstrated that only MDTF cells expressing Pit1 bind GALV. We have also demonstrated that C11D8-tagged full-length GALV SU envelopes and soluble truncated GALV SUs with an HA tag can be used in Pit1 binding studies; they should prove invaluable as novel tools in the study of GALV-mediated entry. Finally, FeLV-T and GALV do not appear to use the common entry pathway shared among MLVs (3, 27). This pathway is activated by a feature conserved among MLV RBDs, but not between FeLV-B and GALV or among GALV, FeLV-T, and the MLVs. Therefore, both FeLV-T and fusion-defective GALV differ from MLVs in RBD-mediated entry.
Acknowledgments
We thank Lorraine Albritton (University of Tennessee) for helpful advice on virus binding conditions and Howard Mostowki (CBER, FDA) for FACS binding analysis. We are grateful to Peggy Faix and Steven Feldman for critical comments on the manuscript, to Ravi Murthy for helpful insights, to Adam Lauring and Julie Overbaugh for useful suggestions, and to Maria Anderson for envelope plasmids.
REFERENCES
- 1.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 2.Bae, Y., S. M. Kingsman, and A. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope glycoprotein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, A. L., R. A. Davey, and J. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett, A. L., and J. Cunningham. 2001. Receptor binding transforms the surface subunit of the mammalian C-type retrovirus envelope protein from an inhibitor to an activator of fusion. J. Virol. 75:9096-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battini, J.-L., O. Danos, and J. M. Heard. 1998. Definition of a 14-amino-acid peptide essential for the interaction between the murine leukemia virus amphotropic envelope glycoprotein and its receptor. J. Virol. 72:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battini, J.-L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battini, J.-L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battini, J.-L., P. Rodrigues, R. Muller, O. Danos., and J. M. Heard. 1996. Receptor binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J. Virol. 70:4387-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boomer, S., M. V. Eiden, C. C. Burns, and J. Overbaugh. 1997. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J. Virol 71:8116-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudry, G. J., K. B. Farrell, Y.-T. Ting, C. Schmitz, Y. S. Lie, C. J. Petropoulos, and M. V. Eiden. 1999. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J. Virol. 73:2916-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudry, G. J., and M. V. Eiden. 1997. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J. Virol. 71:8078-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosset, F.-L., F. J. Morling, Y. Takeuchi, R. A. Weiss, M. K. L. Collins, and S. J. Russell. 1995. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J. Virol. 69:6314-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eiden, M. V., K. B. Farrell, and C. A. Wilson. 1996. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to function as a gibbon ape leukemia virus receptor. J. Virol. 70:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elder, J. H., J. S. McGee, M. Munson, R. A. Houghten, W. Kloetzer, J. L. Bittle, and C. K. Grant. 1987. Localization of the neutralizing regions of the envelope gene of feline leukemia virus by using anti-synthetic peptide antibodies. J. Virol. 61:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finer, M. H., T. J. Dull, L. Qin, D. Farson, and M. R. Roberts. 1994. kat: a high-efficiency retroviral transduction system for primary human T lymphocytes. Blood 83:43-50. [PubMed] [Google Scholar]
- 16.Grant, C. K., B. J. Ernisse, O. Jarrett, and F. R. Jones. 1983. Feline leukemia virus envelope gp70 of subgroups B and C defined by monoclonal antibodies and neutralizing functions. J. Immunol. 131:3042-3049. [PubMed] [Google Scholar]
- 17.Gray, K. D., and M. J. Roth. 1993. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J. Virol. 67:3489-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han, J.-Y., P. M. Cannon, K.-M. Lai, Y. Zhao, M. V. Eiden, and W. F. Anderson. 1997. Identification of envelope residues required for the expanded host range and receptor recognition properties of 10A1 murine leukemia virus. J. Virol. 71:8103-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinzel, S., P. Krysan, M. Calos, and R. DuBridge. 1988. Use of simian virus 40 replication to amplify Epstein-Barr virus shuttle vectors in human cells. J. Virol. 62:3738-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 21.Johann, S. V., J. J. Gibbons, and B. O'Hara. 1992. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J. Virol. 66:1635-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johann, S. V., M. van Zeijl, J. Cekleniak, and B. O'Hara. 1993. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J. Virol. 67:6733-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lander, M. R., and S. K. Chattopadhyay. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J. Virol. 52:695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lauring, A. S., M. M. Anderson, and J. Overbaugh. 2001. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 75:8888-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavillette, D., B. Boson, S. J. Russell, and F. L. Cosset. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J. Virol. 75:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavillette, D., M. Maurice, C. Roche, S. J. Russell, M. Sitbon, and F. L. Cosset. 1998. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J. Virol. 72:9955-9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavillette, D., B. Boson, S. J. Russell, and F. L. Cosset. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, C.-W., and M. J. Roth. 2001. Functional characterization of the N termini of murine leukemia virus envelope proteins. J. Virol. 75:4357-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, A. D., J. V. Garcia, N. von Suhr, C. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, D. G., and A. D. Miller. 1994. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 68:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Hara, B., S. V. Johann, H. P. Klinger, D. G. Blair, H. Rubinson, K. J. Dunne, P. Sass, S. M. Vitek, and T. Robins. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1:119-127. [PubMed] [Google Scholar]
- 32.O'Reilly, L., and M. J. Roth. 2000. Second-site changes affect viability of amphotropic/ecotropic chimeric enveloped murine leukemia viruses. J. Virol. 74:899-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott, D., and A. Rein. 1992. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J. Virol 66:4632-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen, L., S. V. Johann, M. van Zeijl, F. S. Pedersen, and B. O'Hara. 1995. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J. Virol. 69:2401-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneiderman, R. D., K. B. Farrell, C. A. Wilson, and M. V. Eiden. 1996. The Japanese feral mouse PiT1 and PiT2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J. Virol. 70:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tailor, C. S., A. Nouri, and D. Kabat. 2000. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J. Virol. 74:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tailor, C. S., and D. Kabat. 1997. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J. Virol. 71:9383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tailor, C. S., Y. Takeuchi, B. O'Hara, S. V. Johann, R. A. Weiss, and M. K. Collins. 1993. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J. Virol. 67:6737-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi, Y., R. G. Vile, G. Simpson, B. O'Hara, M. K. L. Collins, and R. A. Weiss. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ting, Y.-T., C. A. Wilson, K. B. Farrell, G. J. Chaudry, and M. V. Eiden. 1998. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J. Virol. 72:9453-9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, C., M. S. Reitz, H. Okayama, and M. V. Eiden. 1989. Formation of infectious hybrid virions with gibbon ape leukemia virus and human T-cell leukemia virus retroviral envelope glycoproteins and the Gag and Pol proteins of Moloney murine leukemia virus. J. Virol. 63:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, C. A., K. B. Farrell, and M. V. Eiden. 1994. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J. Virol. 68:7697-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, B. W., P. M. Cannon, E. M. Gordon, F. L. Hall, and W. F. Anderson. 1998. Characterization of the proline-rich region of murine leukemia virus envelope protein. J. Virol. 71:5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, B. W., J. Lu, T. K. Gallaher, W. F. Anderson, and P. M. Cannon. 2000. Identification of regions in the Moloney murine leukemia virus SU protein that tolerate the insertion of an integrin-binding peptide. Virology 269:7-17. [DOI] [PubMed] [Google Scholar]
- 46.Zavorotinskaya, T., and L. Albritton. 1999. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J. Virol. 73:5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]