Abstract
Herpesvirus entry into cells and herpesvirus-induced cell fusion are related processes in that virus penetration proceeds by fusion of the viral envelope and cell membrane. To characterize the human herpesvirus 8 (HHV-8) glycoproteins that can mediate cell fusion, a luciferase reporter gene activation assay was used. Chinese hamster ovary (CHO) cells expressing the HHV-8 glycoproteins of interest along with a luciferase reporter gene under the control of the T7 promoter were cocultivated with human cells transfected with T7 RNA polymerase. Because HHV-8 glycoprotein B (gB) expressed in CHO cells localizes to the perinuclear region, a truncated form of gB (designated gBMUT) that lacks putative endocytosis signals was constructed by deletion of the distal 58 amino acids of the cytoplasmic tail. HHV-8 gBMUT was expressed efficiently on the surface of CHO cells. HHV-8 gB, gH, and gL could mediate the fusion of CHO cells with two different human cell types, embryonic kidney cells and B lymphocytes. Substituting gBMUT for gB significantly enhanced the fusion of CHO cells with human embryonic kidney cells but not B lymphocytes. Thus, two human cell types known to be susceptible to HHV-8 entry were also suitable targets for cell fusion induced by HHV-8 gB, gH, and gL. For human embryonic kidney cells and B cells at least, optimal fusion was noted with the expression of all three HHV-8 glycoproteins.
Herpesvirus glycoproteins expressed in the virion envelope mediate the initial attachment of the virus to the cell surface and the subsequent penetration of the nucleocapsid into the cell cytoplasm (14, 62). For herpesviruses, entry and virus-induced cell fusion are related processes in that penetration proceeds by fusion of the viral envelope with the cell membrane and requires many of the same viral glycoproteins as virus-induced cell fusion (62, 63).
By use of a variety of techniques to detect viral nucleic acids or proteins, human herpesvirus 8 (HHV-8) has been detected in B cells (3, 16), endothelial cells (10, 16, 55, 64), monocytes (8), and epithelial cells (16) in vivo. Cell types that HHV-8 can infect in vitro include human primary B cells (7, 57), macrophages (7), epithelial cells (19, 21, 57, 70, 71), endothelial cells (7, 18, 44, 57), fibroblasts (70), and a variety of human carcinoma cells (57, 70), along with owl monkey kidney cells (43, 57) and hamster kidney cells (57). Serial infection of a human embryonic kidney cell line (293 cells) with HHV-8 derived from primary Kaposi's sarcoma explants and the ability of human cytomegalovirus (HCMV) to activate HHV-8 lytic replication in latently infected human fibroblasts have been reported (19, 70). However, inoculating cells with HHV-8 in vitro typically results in abortive or latent infections rather than productive infections (7, 21, 43, 44, 57). The difficulty in sustaining a productive HHV-8 infection in vitro limits the ease and extent to which the functional domains of HHV-8 glycoproteins can be studied.
The herpesviruses studied to date, including HHV-8, encode glycoprotein B (gB), gH, and gL homologues (43, 60, 62). Both gB and gH are highly conserved (22, 23, 51, 62), and all three appear to be essential for herpesvirus infectivity (20, 26, 31, 42, 56, 59, 62). Only alphaherpesviruses, such as herpes simplex virus (HSV) type 1 (HSV-1), HSV-2, and pseudorabies virus (PrV), express gD (62). HHV-8, a type 2 gammaherpesvirus, does not appear to encode a gD homologue.
HHV-8 gB is a type I membrane protein 845 amino acids in length (5, 53). The first 23 amino acids are most likely a cleavable signal sequence. Predicted near the C terminus are two hydrophobic regions, with the last domain most likely the transmembrane domain. The hydrophilic ectodomain contains numerous N-glycosylation sites and a putative proteolytic cleavage site. HHV-8 gB possesses a putative heparan-binding domain, and the glycoprotein can interact with heparan sulfate-like moieties (1), consistent with the observation that the binding of HHV-8 to cells involves an interaction with heparan sulfate (2, 6).
The cellular distribution and processing of HHV-8 gB depend on the cell type in which the glycoprotein is expressed. In Chinese hamster ovary (CHO) cells, HHV-8 gB tagged with a hemagglutinin (HA) epitope localizes to the perinuclear region without any apparent movement to the periphery (53). In Vero cells, HHV-8 gB is expressed throughout the cytoplasm (5). Although a proteolytic cleavage site is predicted, HHV-8 gB is not cleaved when expressed transiently in CHO cells, simian virus 40 (SV40)-transformed African green monkey kidney cells (COS-1 cells), or Epstein-Barr virus (EBV)-infected lymphoblastoid cells (5, 53). In the HHV-8-infected body cavity-based lymphoma cell line BCBL-1, however, gB is expressed on the cell surface and is cleaved (1, 5). The envelope of virions purified from BCBL-1 cells expresses cleaved gB (1).
In most herpesviruses, the correct transport of gH from the endoplasmic reticulum to the cell membrane and incorporation into virions require the coexpression of gL (17, 27, 29, 37, 61, 73). One exception is PrV gL, which is required for viral infectivity but not for the incorporation of gH into virion particles (31). However, the absence of gL results in a different form of processing of the N-glycans on PrV gH. A third glycoprotein, gp42, associates with the EBV gH-gL complex and is required for viral entry into B cells but not epithelial cells (36). In addition, the correct glycosylation but not the cell surface expression of EBV gH requires the coexpression of gp42 (35). Preliminary work with HHV-8 has shown that the cell surface expression of gH in B cells also requires the coexpression of gL (S. I. Gerber, R. M. Longnecker, P. G. Spear, and P. E. Pertel, Abstr. Infect. Dis. Soc. Am. 36th Annu. Meet., abstr. 59, 1998, Clin. Infect. Dis. 27:931). HHV-8 does not appear to encode a gp42 homologue.
The expression of HSV-1 or HSV-2 gB, gD, and gH-gL in cells susceptible to infection by these viruses is sufficient to induce cell-cell fusion (45, 52, 69). For the alphaherpesvirus PrV, the fusion of at least some susceptible cell types can occur with only three glycoproteins, gB, gH, and gL (32). This finding is consistent with previous studies showing that, although PrV gD is essential for viral penetration into cells, gD-negative PrV mutants can still form plaques (50, 56). Similarly, transient expression of EBV gB, gH-gL, and gp42 in CHO cells is necessary and sufficient to induce fusion with the human B-cell line Daudi (24).
In addition to gB, another HHV-8 envelope glycoprotein, K8.1A, can interact with cellular heparan sulfate (6, 72). K8.1A and K8.1B are generated by splicing of the HHV-8 open reading frame K8.1 (12, 13, 54). This open reading frame is a positional homologue of EBV gp350/220 (12, 54), the glycoprotein that mediates the initial attachment of EBV to B cells by interaction with cellular CD21 (46, 66). The expression of EBV gp350 is not required for the fusion of CHO and Daudi cells (24).
Described below is a quantitative and efficient cell fusion assay used to identify the HHV-8 glycoproteins that are necessary and sufficient to mediate cell-cell fusion. Two human cell types known to be susceptible to HHV-8 entry, embryonic kidney cells and B lymphocytes, were also suitable targets for cell fusion induced by HHV-8 gB, gH, and gL. The fusion assay described here should allow further investigation into the functional domains of HHV-8 glycoproteins and possibly the cellular factors that are important in mediating cell fusion.
MATERIALS AND METHODS
Cells.
CHO cells were obtained from the American Type Culture Collection (Rockville, Md.) and were passaged in Ham's F12 medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% fetal bovine serum (FBS) and antibiotics. 293T cells are human embryonic kidney cells expressing the SV40 large T antigen (15) and were passaged in Dulbecco's modified Eagle medium (MEM) supplemented with 10% FBS and antibiotics. Vero cells are an adult African green monkey cell line (American Type Culture Collection) and were passaged in medium 199 supplemented with 5% FBS and antibiotics. BCBL-1 is an HHV-8-infected body cavity-based lymphoma cell line (58) obtained from the AIDS Research and Reference Reagent Program (Rockville, Md.). BJAB is an EBV-negative lymphoma B-cell line (41). Daudi is an EBV-infected Burkitt's lymphoma cell line (30). The lymphoblastoid cell lines LCL-1, LCL-2, and LCL-10 were transformed by wild-type EBV, while 6.16, KC.1, and M.2 were transformed by gB-negative EBV mutants (26, 34, 38). All B-cell lines were passaged in RPMI 1640 medium containing 10% FBS and antibiotics, except for BCBL-1 cells, which were passaged in RPMI 1640 medium containing 10% FBS, 5 × 10−5 M 2-mercaptoethanol, and antibiotics.
Construction of expression vectors.
Y. Kawaoka (University of Wisconsin, Madison) provided expression plasmid pCAGGS/MCS (33). Plasmid pCAGT7 expresses T7 RNA polymerase under the control of the chicken β-actin promoter and the HCMV immediate-early enhancer (49), while plasmid pT7EMCLuc expresses the firefly luciferase gene under the control of the T7 promoter. Y. Matsuura (National Institute of Infectious Diseases, Tokyo, Japan) provided both plasmids.
Amplification of the glycoprotein open reading frames of interest from the HHV-8-infected cell line BCBL-1 was done by PCR with Vent polymerase (New England Biolabs, Beverly, Mass.) as described previously (53). HHV-8 DNA was isolated from BCBL-1 cells by using a Trizol reagent kit (Gibco BRL). Amplification of the open reading frames was done in duplicate independent reactions to exclude the presence of PCR-introduced mutations. PCR products were desalted and concentrated with a QIAEX II gel extraction kit (Qiagen, Valencia, Calif.). The HHV-8 gL open reading frame was amplified by using a BamHI restriction endonuclease site-tagged sense primer (5′-TTGGATCCGTCTGAGCAGCGAGAGCAG-3′) and an EcoRI-tagged antisense primer (5′-TGAATTCATGGGGATCTTTGCGCTATTTG-3′); introduced restriction endonuclease sites are shown in bold type. The amplified fragment was digested with BamHI and EcoRI and then inserted between the BglII and EcoRI sites of pCAGGS/MCS, generating pPEP109. Nucleotide sequencing of the gL open reading frame revealed no polymorphisms compared to the sequence derived from HHV-8-infected cell line BC-1 (60).
The HHV-8 gH open reading frame was amplified by using a BamHI restriction endonuclease site-tagged sense primer (5′-TTGGATCCATGCAGGGTCTAGCCTTCTTGGC-3′) and a BamHI-tagged antisense primer (5′-TTGGATCCGGTCGAACTGATATGTGACGG-3′). After digestion with BamHI, the PCR fragment was inserted into the BamHI site of pSG5 (Stratagene, La Jolla, Calif.) and screened for orientation, generating pPEP39. Nucleotide sequencing of the gH open reading frame revealed one silent polymorphism at base position 867 (T to C) and two polymorphisms at base positions 524 (T to A) and 982 (A to G) that result in isoleucine-to-asparagine and threonine-to-alanine substitutions, respectively. The gH open reading frame was subsequently excised from pPEP39 with BamHI, inserted into the BglII site of pCAGGS/MCS, and screened for orientation, generating pPEP103.
Construction of the plasmid containing HHV-8 gB (pPEP29) was described previously (53). The gB gene was excised by digestion with EcoRI and BglII and inserted between these sites of pCAGGS/MCS, generating pPEP102. To truncate the HHV-8 gB open reading frame, part of the cytoplasmic tail was PCR amplified by using a sense primer (5′-GCTCATTGGTTACCGGATTC-3′) and a BglII restriction endonuclease site- and stop codon-tagged antisense primer (5′-GGTAGATCTAGATTTCCTCCCGTGTTG-3′); the introduced restriction endonuclease site is shown in bold type, while the stop codon is underlined. The PCR fragment was digested with BglII and SphI and inserted between these sites of pPEP102, generating pPEP120. Nucleotide sequence analysis of the PCR-amplified region revealed no mutations.
The construction of plasmids containing HSV-1 strain KOS [HSV-1(KOS)] gB (pPEP98), HSV-1(KOS) gD (pPEP99), HSV-1(KOS) gH (pPEP100), and HSV-1(KOS) gL (pPEP101) was previously described (52).
The Northwestern University Biotechnology Facility and the Great Lakes Regional Center for AIDS Research constructed the primers and performed the nucleotide sequencing.
Cell enzyme-linked immunosorbent assay (CELISA).
Subconfluent CHO cells were transfected with plasmids expressing HHV-8 glycoproteins by using Lipofectamine reagent in Opti-MEM (Gibco BRL). Individual wells were transfected with 0.5 to 2.0 μg of each expression vector, keeping the total amount of DNA constant by the addition of empty vector DNA. After incubation at 37°C in 5% CO2 for 8 h, the transfection mixture was removed and Ham's F12 medium containing 10% FBS was added. The cells were incubated for an additional 4 h before they were detached with 0.25% trypsin-1 mM tetrasodium EDTA (Gibco BRL) and replated on 96-well plates. After incubation for 18 h, cells were washed with phosphate-buffered saline (PBS). For detection of intracellular and cell surface glycoprotein expression, cells were fixed with PBS containing 2% formaldehyde and 0.2% gluteraldehyde, permeabilized with buffer containing 2 mM magnesium chloride, 0.02% (octylphenoxy)polyethoxyethanol, and 0.01% deoxycholic acid, and then incubated with primary antibody diluted in PBS containing 3% bovine serum albumin; for detection of cell surface glycoprotein expression, cells were incubated with primary antibody first and then fixed. Subsequently, cells were sequentially incubated with biotinylated goat anti-mouse immunoglobulin G (IgG) conjugate (Sigma, St. Louis, Mo.) and streptavidin-horseradish peroxidase conjugate (Amersham Pharmacia Biotech, Piscataway, N.J.). After the addition of 50 mM phosphate-citrate buffer containing 0.03% sodium perborate and 3,3′,5,5′-tetramethylbenzidine (Sigma) dissolved in dimethyl sulfoxide, optical density (OD) readings at 370 nm were obtained with a plate spectrophotometer (SpectraMax 250; Molecular Devices, Sunnyvale, Calif.).
Antibody production.
Antibodies specific for HHV-8 glycoproteins were produced by immunizing BALB/c mice intradermally with HHV-8 gB or HHV-8 gH and gL expression vectors as previously described for HSV-1 gB (39, 65). For immunization, 50 μg of each expression vector was injected. The Animal Care and Use Committee of Northwestern University approved the protocol.
Fusion assays.
Cell fusion was quantified and visualized by using a luciferase reporter gene activation assay (49). Subconfluent effector cells in six-well plates were transfected with vectors expressing the glycoproteins of interest and a vector expressing luciferase under the control of the T7 promoter. Target cells in six-well plates were transfected with a vector expressing T7 RNA polymerase. After incubation at 37°C in 5% CO2 for 10 h, the transfection mixture was removed. Target cells were subsequently detached with trypsin-EDTA and then overlaid on the effector cells at a 1:1 ratio. B-cell lines were transfected by electroporation. Briefly, 107 cells were resuspended in 400 μl of RPMI 1640 medium containing 50 μg of the T7 RNA polymerase expression vector. Cells were electroporated by using a Gene Pulser electroporator (Bio-Rad Laboratories), transferred to 25-cm2 flasks containing RPMI 1640 medium, and then incubated for 10 h at 37°C in 5% CO2. Approximately 106 viable B cells were overlaid on the effector cells.
Luciferase activity was quantified by using a luciferase reporter assay system (Promega, Madison, Wis.) 6 to 48 h after cocultivation. Cells were washed with PBS and then lysed with passive lysis buffer. After the supernatant was collected, beetle luciferin substrate was added, and luminosity readings were obtained by using a TD-20/20 luminometer (Turner Designs; Promega). The supernatant was diluted 1:10 in water for all readings. To visualize cell fusion, an antiluciferase immunoassay was used. Cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.1% Triton X-100 (Sigma) in PBS, and then incubated with antiluciferase antibody (Promega) diluted to 20 μg/ml in PBS containing 3% bovine serum albumin. Subsequently, cells were sequentially incubated with biotinylated goat anti-mouse IgG, streptavidin-β-galactosidase conjugate (Sigma), and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) substrate (Sigma) dissolved in and diluted to 1.0 mg/ml in ferricyanide buffer [3 mM K3Fe(CN)6, 3.125 mM K4Fe(CN)6]. Cells were counterstained with 0.1% nuclear fast red (Sigma). Photographs of the cells were taken with an Olympus digital camera.
Immunoprecipitation.
Subconfluent CHO cells were transfected with plasmids expressing HHV-8 glycoproteins as described above. The cells were subsequently washed, incubated with MEM depleted of both methionine and cysteine for 20 min at 37°C in 5% CO2, and then incubated for 2 h with depleted MEM containing [35S]Met and [35S]Cys (Amersham Pharmacia Biotech). The cells were washed with ice-cold PBS and then lysed with RIBA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing protease inhibitors (2 μg of aprotinin/ml, 2 μg of leupeptin/ml, and 1 μg of pepstatin A/ml) for 30 min on ice. After centrifugation, the cleared lysates were preabsorbed with mouse serum and a mixture of protein G- and protein A-Sepharose (Amersham Pharmacia Biotech). The lysates were then incubated at 4°C with either mouse anti-HHV-8 gB polyclonal sera or anti-HHV-8 gH-gL sera before collection of the immune complexes by the addition of protein G- and protein-A Sepharose. After the addition of SDS sample buffer, the samples were boiled for 5 min and then loaded onto an SDS-polyacrylamide gel (N,N′-methylene-bisacrylamide) (Bio-Rad Laboratories). Following electrophoresis, the gel was fixed with a methanol-acetic acid solution, soaked in Amplify (Amersham Pharmacia Biotech), and then dried. Radiolabeled proteins were visualized after exposure to autoradiography film (Amersham Pharmacia Biotech).
Statistical analysis.
Comparison of means was done by two-tailed t tests. Correction of all P values for multiple comparisons was done by the Bonferroni procedure.
Nucleotide sequence accession number.
The gH nucleotide sequence was deposited in GenBank under accession number AF448055.
RESULTS
Cell surface expression of HHV-8 glycoproteins.
To investigate if HHV-8 glycoproteins could induce cell-cell fusion, the open reading frames for several HHV-8 glycoproteins, including gB, gH, and gL, were inserted into plasmid pCAGGS/MCS. This plasmid permits the expression of inserts under the control of the chicken β-actin promoter and HCMV immediate-early enhancer (47). The reasons for testing these three glycoproteins were as follows. (i) HSV-induced cell fusion requires the expression of only gB, gD, and gH-gL (45, 52, 69). (ii) Although PrV gD is required for viral entry (50, 56), PrV-induced cell fusion requires the expression of only gB, gH, and gL (32). (iii) EBV-induced cell fusion requires the expression of gB, gH, gL, and gp42 (24). (iv) HHV-8 encodes homologues of gB, gH, and gL but not gD and gp42 (43, 60).
Antibodies specific for HHV-8 glycoproteins were produced by immunizing mice with HHV-8 gB or HHV-8 gH and gL expression vectors as previously described by others for HSV-1 gB (39, 65). The specificities of the generated antibodies were confirmed by CELISA. When fixed cells were used to detect total glycoprotein expression, mouse anti-HHV-8 gB polyclonal sera (diluted 1:1,000) could detect gB expressed in CHO cells, while anti-HHV-8 gH-gL sera (diluted 1:1,000) could detect gH and gH-gL. Mouse anti-HHV-8 gB sera could not detect expressed gH or gH-gL, nor could mouse anti-HHV-8 gH-gL sera detect expressed gB (data not shown).
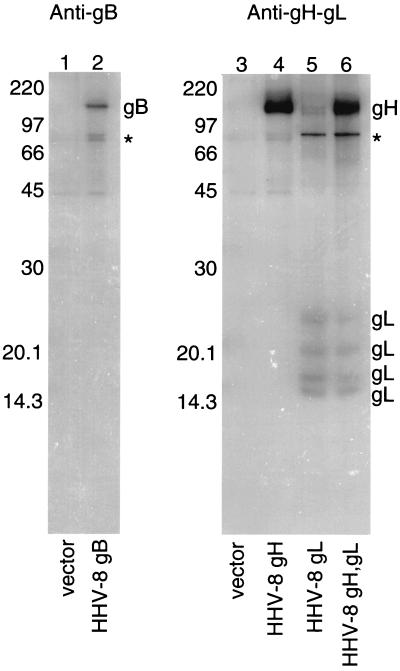
To further confirm the specificity of the antibodies, mouse polyclonal sera were used to immunoprecipitate radiolabeled proteins from CHO cells transfected with HHV-8 glycoproteins. As shown in Fig. 1, anti-gB sera could immunoprecipitate an approximately 120-kDa protein from CHO cells expressing HHV-8 gB (lane 2) but not from cells transfected with a vector (lane 1). The apparent size of this protein is similar to that previously described for HHV-8 gB and HA-tagged gB expressed in CHO cells (53). Anti-gH-gL sera could immunoprecipitate an approximately 120-kDa protein from CHO cells transfected with HHV-8 gH (Fig. 1, lane 4) or with gH and gL (lane 6) but not from cells transfected with gL (lane 5) or with a vector (lane 3). Similarly, anti-gH-gL sera could immunoprecipitate 15-, 18-, 21-, and 24-kDa proteins from cells transfected with gL (Fig. 1, lane 5) or with gH and gL (lane 6) but not from cells transfected with gH (lane 4) or with a vector (lane 3). The apparent sizes of these proteins are similar to those obtained after expression of HA-tagged gH (120 kDa) or HA-tagged gL (18 to 28 kDa) in B cells (Gerber et al., Abstr. Infect. Dis. Soc. Am. 36th Annu. Meet.).
FIG. 1.
Immunoprecipitation of HHV-8 gB, gH, and gL. CHO cells were transfected with plasmids expressing the indicated HHV-8 glycoproteins. The cells were radiolabeled 24 h after transfection and then lysed before immunoprecipitation with the indicated mouse polyclonal sera. In the left panel (lanes 1 and 2), radiolabeled proteins were immunoprecipitated with anti-HHV-8 gB sera. In the right panel (lanes 3 to 6), proteins were immunoprecipitated with anti-HHV-8 gH-gL sera. Two bands (designated with an asterisk) were consistently visualized in all six lanes on the original autoradiographs at between approximately 70 and 80 kDa, although the intensities varied.
Wild-type HHV-8 gB expressed in CHO cells localizes to the perinuclear region (53). Contained within the cytoplasmic tail of gB are two endocytic sorting signals, a tyrosine-based signal of the form Yxxφ (where φ represents a hydrophobic amino acid and x is any amino acid) and a dileucine-containing signal (Fig. 2) (40). To potentially enhance the cell surface expression of gB, the last 58 amino acids of the gB cytoplasmic tail were deleted. The predicted transmembrane domain was not deleted. The truncated form of gB was designated gBMUT. Detection of HHV-8 glycoprotein expression was done by CELISA. Total expression was detected by fixing and permeabilizing cells transfected with the glycoproteins of interest before incubation with the primary antibody. Cell surface expression was detected by incubating the transfected cells with the primary antibody before fixing. Transfected CHO cells expressed similar total amounts of gB and gBMUT (Fig. 3B), but only gBMUT was detected in appreciable amounts on the cell surface (Fig. 3A).
FIG. 2.
Amino acid sequence of the final 120 amino acids of HHV-8 gB. The amino acid sequence of HHV-8 gB was derived from the nucleotide sequence obtained from published reports (53, 60). The predicted transmembrane domain is in bold letters. The Yxxφ (YKPL) and dileucine endocytosis motifs are boxed. The 58 amino acids deleted in the truncated form of gB (designated gBMUT) are underlined.
FIG. 3.
Enhanced cell surface expression of HHV-8 gBMUT. CHO cells were transfected with plasmids expressing HHV-8 gB or gBMUT, replated on 96-well dishes, and incubated for 18 h. (A) For detection of cell surface glycoprotein expression, cells were first incubated with primary antibody and then fixed. (B) For detection of intracellular and cell surface glycoprotein expression, cells were fixed, permeabilized, and then incubated with primary antibody. Subsequently, cells were incubated with biotinylated goat anti-mouse IgG and streptavidin-horseradish peroxidase conjugate. After the addition of substrate, OD readings at 370 nm were obtained. Shown are the means and standard deviations for four replicate samples. In panel A, the difference between the OD readings obtained for cells transfected with a vector alone and cells transfected with gBMUT was statistically significant (P < 0.001; two-tailed t test), while the difference between the readings obtained with the vector alone and gB was not (P = 0.155). In panel B, the differences between the readings obtained with the vector alone and gB and gBMUT were both statistically significant (P < 0.001 and P = 0.013, respectively).
Preliminary work with HHV-8 has shown that the cell surface expression of gH in B cells also requires the coexpression of gL (Gerber et al., Abstr. Infect. Dis. Soc. Am. 36th Annu. Meet.). In these studies, HA-tagged HHV-8 gH and gL were transfected either alone or in combination into BJAB cells (an EBV-negative lymphoma B-cell line), and cell surface proteins were subsequently labeled with biotin. After collection of the biotin-labeled proteins, immunoblotting with an anti-HA monoclonal antibody demonstrated that the cell surface expression of HA-tagged HHV-8 gH required the coexpression of HA-tagged gL. Figure 4 shows the results of a CELISA to detect the total and cell surface expression of HHV-8 gH and gL transfected alone or in combination into CHO cells. Detection of gH and gL on the cell surface by anti-gH-gL sera was optimal when both glycoproteins were coexpressed (Fig. 4A). Significantly less gH was detected on the cell surface in the absence of gL, and no cell surface gL was detected in the absence of gH. The anti-gH-gL sera detected the total expression of gH and gL when the glycoproteins were transfected into CHO cells alone or in combination (Fig. 4B).
FIG. 4.
Detection of gH and gL on the cell surface. The detection of gH and gL on the cell surface was optimal when both glycoproteins were coexpressed. CHO cells were transfected with plasmids expressing the indicated HHV-8 glycoproteins. Detection of cell surface (A) and total (B) glycoprotein expression was done by using anti-gH-gL sera as described in the legend to Fig. 3. Shown are the means and standard deviations for three to eight replicate samples. In panel A, the differences between the OD readings obtained for cells transfected with a vector alone and cells transfected with gH or with gH plus gL were both statistically significant (P < 0.001; two-tailed t test), as was the difference between the readings obtained with gH and with gH plus gL (P < 0.001). The difference between the readings obtained with the vector and with gL was not statistically significant (P = 0.774). In panel B, the differences between the readings obtained with the vector and those obtained with gH, gL, or gH plus gL were all statistically significant (P = 0.015, P = 0.007, and P = 0.002, respectively). The difference between the readings obtained with gH and with gH plus gL was not statistically significant (P = 0.632).
The cell surface expression of gBMUT (Fig. 5, left panel) and gH-gL (Fig. 5, right panel) was demonstrated when all three glycoproteins were transfected into CHO cells together. Again, the specificities of the mouse polyclonal sera were confirmed when mouse anti-HHV-8 gB polyclonal sera detected the surface expression of gBMUT but not gH-gL (Fig. 5, left panel), while anti-HHV-8 gH-gL sera detected the surface expression of gH-gL but not gBMUT (Fig. 5, right panel). Less cell surface expression of gBMUT and gH-gL was noted when all three glycoproteins were expressed together rather than individually. These results may represent differences in expression, exposure on the cell surface, or ability to detect exposure.
FIG. 5.
Cell surface expression of HHV-8 glycoproteins. CHO cells were transfected with plasmids expressing the indicated HHV-8 glycoproteins. After cells were replated on 96-well dishes and incubated for 18 h, they were incubated with primary antibody. Subsequently, cells were fixed and then incubated with biotinylated goat anti-mouse IgG and streptavidin-horseradish peroxidase conjugate. After the addition of substrate, OD readings at 370 nm were obtained. Shown are the means and standard deviations for four replicate samples. In the left panel, the differences between the OD readings obtained for cells transfected with a vector alone and those obtained for cells transfected with gBMUT alone or with gBMUT, gH, and gL were both statistically significant (P < 0.001 or P = 0.005, respectively; two-tailed t test), while the differences between readings obtained with the vector and those obtained with gH plus gL or with a combination of gB, gH, and gL were not (P = 1.0). In the right panel, the differences between the readings obtained with the vector and those obtained with gH plus gL, a combination of gB, gH, and gL, or a combination of gBMUT, gH, and gL were all statistically significant (P < 0.001). The difference between the readings obtained with the vector and with gBMUT was not statistically significant (P = 0.986).
HHV-8 glycoprotein-mediated fusion of CHO and 293T cells.
The cells that HHV-8 can infect in vitro include the human embryonic kidney cell line 293 and primary B cells (7, 19, 21, 57, 70, 71). The techniques used to confirm the ability of HHV-8 to infect these cells have included detection of viral DNA by PCR (19, 71), viral RNA by reverse transcriptase PCR (7, 57, 71), viral proteins by immunofluorescence (70), and green fluorescent protein expressed by a recombinant HHV-8 mutant (70). Because the efficient fusion of cells by HSV-1 glycoproteins requires the expression of entry receptors (52), it was reasoned that cells susceptible to HHV-8 entry may also be able to fuse with CHO cells expressing the correct set of HHV-8 glycoproteins. The following experiments were done with 293 cells expressing the SV40 large T antigen (293T cells) instead of 293 cells. The results obtained with 293 cells were similar but less reproducible (data not shown).
To detect cell fusion, a luciferase reporter gene activation assay was used (49). Effector cells transfected with the luciferase reporter gene under the control of the T7 promoter and the HHV-8 glycoproteins of interest were cocultivated with target cells transfected with T7 RNA polymerase. In these and subsequent experiments, the effector cells were CHO cells expressing HHV-8 glycoproteins. Because the contents of the effector and target cells must mix in order for the T7 RNA polymerase to transcribe the luciferase gene, the level of luciferase activity represents the extent of fusion between the effector and target cells.
To estimate the numbers of cells that expressed all three glycoproteins, an immunoassay with a pool of mouse anti-gB and anti-gH-gL polyclonal sera was used. CHO cells were transfected with each glycoprotein alone or in combination under the same conditions as those used for the fusion assay. Twenty-four hours after transfection, the cells were fixed and permeabilized before sequential incubation with mouse sera, mouse-specific biotinylated anti-IgG conjugate, streptavidin-β-galactosidase conjugate, and X-Gal substrate. Cells were counterstained with nuclear fast red. The total number of cells and the number of blue cells were counted in three representative high-power fields. The percentage of blue cells after transfection with a vector alone was 0.2% (range, 0.0 to 0.6%). In contrast, the percentages of blue cells were 6.2% (4.3 to 9.2%), 8.0% (6.8 to 9.1%), 6.7% (2.8 to 9.9%), and 7.0% (3.7 to 9.0%) after transfection of gB, gH, or gL or of all three glycoproteins together, respectively. The percentages of blue cells detected among cells transfected with all three glycoproteins were 7.3% (6.6 to 7.8%) and 6.1% (4.7 to 7.2%) when anti-gB sera and anti-gH-gL sera were used, respectively. Based on these results, the transfection efficiency was between 6 and 8%, and successfully transfected cells expressed all three glycoproteins.
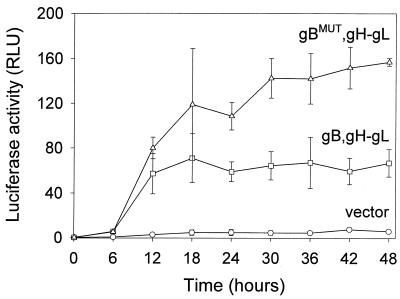
Figure 6 shows the results obtained when effector cells transfected with (i) gB, gH, and gL, (ii) gBMUT, gH, and gL, or (iii) a vector were cocultivated with target 293T cells and the expression of luciferase activity was monitored over time. Significant luciferase activity occurred after cocultivation of 293T target cells with effector cells expressing gB and gH-gL but not the vector. Substituting gBMUT for gB significantly enhanced the level of luciferase activity. Maximal luciferase activity occurred approximately 18 to 24 h after cocultivation and persisted at 48 h. Because luciferase mRNA and protein degrade over time (67), the persistence of luciferase activity implies that cell-cell fusion was continuing over the 48 h.
FIG. 6.
Kinetics of cell fusion as measured by luciferase production. Luciferase activity was measured at the indicated times after cocultivation of effector and target cells. CHO effector cells were transfected with plasmids expressing the indicated glycoproteins or a control vector and the luciferase reporter plasmid. 293T target cells were transfected with T7 RNA polymerase. Shown are mean relative luminosity units (RLU) and standard deviations for six replicate samples obtained on two separate occasions.
Efficient fusion of CHO and 293T cells requires gB and gH-gL.
As shown in Fig. 7, efficient luciferase activity required the expression of HHV-8 gB and gH-gL. Consistent with the results shown in Fig. 6, substituting gBMUT for gB resulted in significantly enhanced luciferase activity. Of note, cocultivating 293T cells with effector cells expressing only gH-gL resulted in an approximately 30-fold increase in mean luciferase activity compared to that in cells transfected with a vector. Similarly, cocultivating 293T target cells with CHO effector cells expressing only gB or gBMUT resulted in about five- or sevenfold increases in mean luciferase activity, respectively. Although these increases were all reproducible and statistically significant, efficient luciferase activity required the expression of all three HHV-8 glycoproteins. Luciferase activity increased over 130- and 200-fold when effector cells were transfected with gB, gH, and gL or with gBMUT, gH, and gL, respectively. Cocultivating target cells with effector cells expressing only gH or gL resulted in an approximately fivefold increase in mean luciferase activity (data not shown). When preincubated with effector cells, neither anti-gH-gL nor anti-gB polyclonal sera (diluted 1:50) could inhibit cell fusion (data not shown). Because HHV-8 gB, gH, and gL could induce the fusion of CHO and 293T cells, it is possible that the antibodies in the mouse sera did not recognize epitopes involved in cell fusion.
FIG. 7.
Efficient cell fusion requires HHV-8 gB and gH-gL. CHO effector cells were transfected with plasmids expressing the indicated glycoproteins or a control vector and the luciferase reporter plasmid. 293T target cells were transfected with T7 RNA polymerase. After cocultivation for 24 h, luciferase activity was measured. Shown are the mean relative luminosity units (RLU) and standard deviations for six replicate samples obtained on two separate occasions. Mean differences between values for the vector and those for gBMUT, gH plus gL, a combination of gB, gH, and gL, or a combination of gBMUT, gH, and gL were all statistically significant (P < 0.001; two-tailed t test). The difference between the vector and gB values was also statistically significant (P = 0.012). Additional experiments done independently resulted in similar findings (data not shown). For comparison, cocultivation of 293T cells with CHO effector cells expressing HSV-1 glycoproteins gB, gD, and gH-gL resulted in a mean RLU of 226.2.
Visualization of the cells expressing luciferase was performed by immunostaining with an antiluciferase antibody. Scattered luciferase-expressing cells were noted after cocultivation of 293T cells with effector cells transfected with a vector. Significantly more luciferase-expressing cells were noted after cocultivation of effector cells expressing HHV-8 gB and gH-gL or HHV-8 gBMUT and gH-gL. The number of blue cells correlated with luciferase activity (Table 1 and Fig. 7). At least 80% of the blue cells observed after cocultivation of 293T cells with effector cells transfected with the HHV-8 glycoproteins had two to four nuclei, demonstrating that the expression of luciferase resulted from cell fusion. Cell fusion mediated by HSV-1 gB, gD, and gH-gL was more efficient. The number of luciferase-expressing cells observed was larger with effector cells expressing HSV-1 glycoproteins. In addition, the polykaryocytes tended to be larger and to contain more nuclei. Figure 8 shows representative examples of the visualized cells.
TABLE 1.
Viral glycoproteins required for cell fusion, as quantified by an antiluciferase immunoassaya
| Glycoprotein(s) expressed in CHO effector cells | No. of blue cellsb |
|---|---|
| Vector | 3.7 ± 2.9 |
| HHV-8 gB | 6.3 ± 5.8 |
| HHV-8 gBMUT | 10.3 ± 3.2 |
| HHV-8 gH-gL | 28.7 ± 11.6 |
| HHV-8 gB and gH-gL | 81.3 ± 21.9 |
| HHV-8 gBMUT and gH-gL | 194.3 ± 41.1 |
| HSV-1 gB, gD, and gH-gL | 547.3 ± 189.7 |
CHO effector cells were transfected with plasmids expressing the indicated viral glycoproteins and the luciferase reporter plasmid. 293T target cells were transfected with T7 RNA polymerase. Visualization of cells expressing luciferase was done with an antiluciferase immunoassay. Shown are the mean number of blue cells per 500,000 cells and standard deviations from three replicate samples obtained on two separate occasions.
Mean differences between values for the vector and those for a combination of gB, gH, and gL, for a combination of gBMUT, gH, and gL, and for a combination of HSV-1 gB, gD, gH, and gL were all statistically significant (P = 0.022, P = 0.008, and P = 0.046, respectively; two-tailed t test). Mean differences between values for the vector and those for gB, gBMUT, or gH plus gL were not statistically significant (P = 1.0, P = 0.334, and P = 0.134, respectively). Cocultivation of target cells with effector cells expressing only gH or gL resulted in no significant increase in the number of luciferase-expressing cells (data not shown).
FIG. 8.
Visualization of cells that express luciferase. CHO effector cells transfected with plasmids expressing the indicated glycoproteins along with the luciferase reporter plasmid were cocultivated for 24 h with 293T target cells transfected with T7 RNA polymerase. After cells were fixed and permeabilized, detection of luciferase expression was performed by immunostaining with an antiluciferase antibody. For comparison, results obtained after cocultivation of 293T cells with effector cells expressing HSV-1 glycoproteins gB, gD, and gH-gL are shown.
HHV-8 gB and gH-gL can induce the fusion of CHO and B cells.
Experiments similar to those described above were performed to test the ability of HHV-8 gB, gH, and gL to mediate the fusion of CHO cells with other cell lines. BJAB is an EBV-negative immortalized lymphoma B-cell line, while LCL-10 is a lymphoblastoid cell line transformed with wild-type EBV (34, 41). As shown in Fig. 9, HHV-8 gB, gH, and gL could mediate the fusion of both BJAB and LCL-10 cells with CHO cells. In contrast to the results obtained with 293T cells, however, substituting gBMUT for gB did not increase luciferase activity. Testing of additional immortalized B-cell lines, including an EBV-positive Burkitt's lymphoma cell line (Daudi) and several lymphoblastoid cell lines transformed with either wild-type EBV (LCL-1 and LCL-2) or gB-negative EBV (6.16, KC.1, and M.2) (26, 30, 34, 38), resulted in similar findings (data not shown). As before, luciferase activity was optimal when all three glycoproteins were coexpressed (data not shown).
FIG. 9.
HHV-8 gB and gH-gL can mediate fusion of CHO and B cells. CHO effector cells were transfected with plasmids expressing the indicated glycoproteins or a control vector and the luciferase reporter plasmid. Target B cells were electroporated with T7 RNA polymerase. Luciferase activity was measured 24 h after cocultivation of effector and target cells. Shown are the mean relative luminosity units (RLU) and standard deviations for three replicate samples. In the left panel, differences between values for the vector and those for a combination of gB, gH, and gL or a combination of gBMUT, gH, and gL were both statistically significant (P < 0.001; two-tailed t test), while the difference between values for gB, gH, and gL and for gBMUT, gH, and gL was not (P = 0.219). In the right panel, differences between values for the vector and those for a combination of gB, gH, and gL or for a combination of gBMUT, gH, and gL were both statistically significant (P = 0.002; two-tailed t test), while the difference between the values for gB, gH, and gL and for gBMUT, gH, and gL was not (P = 0.920). Similar results were obtained in additional experiments done independently (data not shown).
When effector CHO cells expressing HHV-8 gB and gH-gL were cocultivated with Vero or CHO cells, increases in luciferase activity were also noted. However, the relative increases were considerably smaller than those noted with 293T or B cells. Figure 10 shows that approximately 6- and 15-fold increases were noted after cocultivation of effector CHO cells expressing gB and gH-gL with target Vero and CHO cells, respectively. Substituting gBMUT for gB did not enhance luciferase activity for either cell line. For comparison, the relative increases noted when 293T or BJAB cells were used as the target cell lines were calculated from the data presented in Fig. 7 or 9, respectively. Cocultivating 293T cells with effector CHO cells expressing gB and gH-gL or gBMUT and gH-gL resulted in approximately 130- or 200-fold increases in luciferase activity, respectively. For BJAB cells, the increases were approximately 50-fold for both gB and gH-gL and gBMUT and gH-gL. The observation that Vero and CHO cells functioned poorly compared to 293T and B cells as targets for HHV-8 glycoprotein-mediated cell fusion provides support for the hypothesis that this fusion is a receptor-mediated process. It is possible that either the receptor used differs between the cell types or that the same receptor is expressed but at different levels.
FIG. 10.
Fusion of various cell lines mediated by HHV-8 glycoproteins. CHO effector cells were transfected with plasmids expressing the indicated glycoproteins or a control vector and the luciferase reporter plasmid. CHO or Vero target cells were transfected with T7 RNA polymerase. After cocultivation of the effector and target cells, luciferase activity was measured. Shown are the means and standard deviations for calculated fold increases for three replicate samples. The fold increases (or relative luciferase activities) were calculated as the ratio of values obtained with effector cells expressing the indicated glycoproteins to values obtained with effector cells transfected with the vector. For comparison, relative luciferase activities were calculated from the results presented in Fig. 7 and 9 when 293T and BJAB cells were used as target cells. Using the unadjusted mean luciferase values, the differences obtained with CHO target cells between values for the vector and those for a combination of gB, gH, and gL or a combination of gBMUT, gH, and gL were both statistically significant (P < 0.001; two-tailed t test). The differences obtained with Vero target cells between values for the vector and those for a combination of gB, gH, and gL or a combination of gBMUT, gH, and gL were also both statistically significant (P = 0.002 and P < 0.001, respectively). The differences between values for gB, gH, and gL and those for gBMUT, gH, and gL with both CHO and Vero target cells were not statistically significant (P = 0.871 and P = 1.0, respectively).
DISCUSSION
The fusion of CHO cells with two human cell types (293T and B cells) was induced by three HHV-8 glycoproteins, gB, gH, and gL. For 293T cells and B cells, optimal fusion was noted with the expression of all three HHV-8 glycoproteins. The fusion noted after the expression of these glycoproteins occurred even though efficient cell surface expression of gB by CHO cells was not detected. It is possible that the antibodies or the assay used in these studies was not sufficiently sensitive to detect low-level expression of gB on the cell surface. It is more likely, however, that efficient cell surface expression of gB is not required for mediating cell fusion.
Consistent with these results, efficient cell fusion induced by the alphaherpesvirus PrV requires the expression of gB, gH, and gL but not gD (32). In contrast, cell fusion induced by two other alphaherpesviruses, HSV-1 and HSV-2, requires the expression of all four glycoproteins, gB, gD, and gH-gL (45, 52, 69), and cell fusion induced by the type 1 gammaherpesvirus EBV requires gB, gH-gL, and gp42 (24). HHV-8, a type 2 gammaherpesvirus, does not appear to encode gD or gp42 homologues.
Significant fusion was also noted when effector CHO cells expressing HHV-8 gH-gL were cocultivated with 293T target cells. This result is consistent with the ability of varicella-zoster virus gH-gL to mediate the fusion of transfected HeLa and human melanoma cells (17). In addition, minimal but reproducible fusion was induced when HHV-8 gB or the truncated form of gB, gBMUT, was expressed alone. These results imply that both gH-gL and gB possibly interact with cellular factors expressed by 293T cells. Although speculative, it is possible that gH-gL or gB mediates cell fusion by interacting with one or more cellular factors that also can function as receptors for viral entry. This scenario would be similar to the observation that efficient fusion of CHO cells induced by HSV-1 glycoproteins requires the expression of an entry receptor (52).
Enhanced cell fusion was noted when HHV-8 gBMUT was expressed with gH-gL. Previously, it was shown that gB localizes to the perinuclear region of CHO cells (53). In contrast, a truncated form of gB lacking the distal 58 amino acids of the cytoplasmic tail (gBMUT) was expressed efficiently on the surface of CHO cells. Located within the sequences of the deleted amino acids are two potential endocytic sorting signals (dileucine and Yxx℘ motifs) (40). Deletion of one or both of the sequences may be required for efficient cell surface expression of gB in CHO cells. Interestingly, HHV-8 gB is expressed throughout the cytoplasm of Vero cells (5), implying that intracellular processing of gB is cell type dependent. Wild-type PrV gB, which encodes both motifs, aggregates in the cell cytoplasm of rabbit kidney cells (RK13 cells), while a truncated form that has these sequences deleted (PrV gB-008) localizes to the plasma membrane (48). Similar motifs are located in the cytoplasmic tails of HCMV gB, varicella-zoster virus gB, and HSV-1 gB, yet all three glycoproteins are expressed on the plasma membrane (9, 25, 28, 48, 68).
In addition to facilitating cell surface expression in CHO cells, removal of the last 58 amino acids of HHV-8 gB enhanced its ability to mediate the fusion of CHO and 293T cells. Removing part of the C-terminal tail of both PrV gB and HSV-1 gB significantly alters the fusogenicity of these glycoproteins (4, 11, 32, 48). The expression of the truncated form of PrV gB (gB-008) and gH is sufficient for cell fusion (32). It is not clear, however, if the effect of the deletion is to enhance cell surface expression, to remove a putative block to cell fusion, or to enhance the fusogenicity of gB.
The studies presented here used a quantitative and efficient cell fusion assay to identify the HHV-8 glycoproteins that could mediate the fusion of CHO cells with 293T and B cells. Although only HHV-8 gB, gH, and gL were tested, these three glycoproteins were found to be sufficient and necessary to induce cell fusion under the described conditions. However, it is possible that additional viral glycoproteins may well influence the ability of HHV-8 glycoproteins to induce cell fusion. Further work to characterize any additional viral factors that can influence cell fusion is needed.
For herpesviruses, entry and virus-induced cell fusion are related processes. Penetration proceeds by fusion of the viral envelope with the cell membrane. Entry and cell-cell fusion require many of the same viral glycoproteins. The unavailability of high titers of virus and the limited susceptibility of cultured cells limit the study of HHV-8 glycoproteins. The fusion assay described here has been used to study the viral glycoproteins and cellular factors required for cell fusion induced by other herpesviruses, including HSV-1 and EBV (24, 52). This assay should facilitate investigation into the functional domains of HHV-8 glycoproteins and the cellular factors that are important in mediating cell fusion and possibly viral entry. Ultimately, further work with additional assays is necessary to prove that these HHV-8 glycoproteins are sufficient and necessary for fusion and viral entry in vivo.
Acknowledgments
I thank N. Susmarski for help with tissue culture, L. Kwan for help with CELISA, Y. Kawaoka for pCAGGS/MCS, and Y. Matsuura for pCAGT7 and pT7EMCLuc. I also thank A. Aiyar, N. Clipstone, R. Longnecker, P. Spear, and C. Waltenbaugh for technical advice and support.
A clinical scientist award from the Doris Duke Charitable Foundation helped to support this work.
REFERENCES
- 1.Akula, S. M., N. P. Pramod, F.-Z. Wang, and B. Chandran. 2001. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology 284:235-249. [DOI] [PubMed] [Google Scholar]
- 2.Akula, S. M., F.-Z. Wang, J. Vieira, and B. Chandran. 2001. Human herpesvirus 8 interaction with target cells involves heparan sulfate. Virology 282:245-255. [DOI] [PubMed] [Google Scholar]
- 3.Ambroziak, J. A., D. J. Blackbourn, B. G. Herndier, R. G. Glogau, J. H. Gullett, A. R. McDonald, E. T. Lennette, and J. A. Levy. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582-583. [DOI] [PubMed] [Google Scholar]
- 4.Baghian, A., L. Huang, S. Newman, S. Jayachandra, and K. G. Kousoulas. 1993. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J. Virol. 67:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baghian, A., M. Luftig, J. B. Black, Y.-X. Meng, C.-P. Pau, T. Voss, P. E. Pellett, and K. G. Kousoulas. 2000. Glycoprotein B of human herpesvirus 8 is a component of the virion in a cleaved form composed of amino- and carboxyl-terminal fragments. Virology 269:18-25. [DOI] [PubMed] [Google Scholar]
- 6.Birkmann, A., K. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackbourn, D. J., E. Lennette, B. Klencke, A. Moses, B. Chandran, M. Weinstein, R. G. Glogan, M. H. Witte, D. L. Way, T. Kutzkey, B. Herndier, and J. A. Levy. 2000. The restricted cellular host range of human herpesvirus 8. AIDS 14:1123-1133. [DOI] [PubMed] [Google Scholar]
- 8.Blasig, C., C. Zietz, B. Haar, F. Neipel, S. Esser, N. H. Brockmeyer, E. Tschachler, S. Colombini, B. Ensoli, and M. Sturzl. 1997. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J. Virol. 71:7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bold, S., M. Ohlin, W. Garten, and K. Radsak. 1996. Structural domains involved in human cytomegalovirus glycoprotein B-mediated cell-cell fusion. J. Gen. Virol. 77:2297-2302. [DOI] [PubMed] [Google Scholar]
- 10.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. D. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 11.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 12.Chandran, B., C. Bloomer, S. R. Chan, L. Zhu, E. Goldstein, and R. Horvat. 1998. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology 249:140-149. [DOI] [PubMed] [Google Scholar]
- 13.Chandran, B., M. S. Smith, D. M. Koelle, L. Corey, R. Horvat, and E. Goldstein. 1998. Reactivities of human sera with human herpesvirus-8-infected BCBL-1 cells and identification of HHV-8-specific proteins and glycoproteins and the encoding cDNAs. Virology 243:208-217. [DOI] [PubMed] [Google Scholar]
- 14.Cooper, N. R. 1994. Early events in human herpesvirus infection of cells, p. 365-388. In E. Wimmer (ed.), Cellular receptors for animal viruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 15.DuBridge, R. B., P. Tang, H. C. Hsia, P.-M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupin, N., C. Fisher, P. Kellam, S. Ariad, M. Tulliez, N. Franck, E. Van Marck, D. Salmon, I. Gorin, J.-P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duus, K. M., C. Hatfield, and C. Grose. 1995. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology 210:429-440. [DOI] [PubMed] [Google Scholar]
- 18.Flore, O., S. Rafii, S. Ely, J. J. O'Leary, E. M. Hyjek, and E. Cesarman. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma associated herpesvirus. Nature 394:588-592. [DOI] [PubMed] [Google Scholar]
- 19.Foreman, K. E., J. Friborg, W.-P. Kong, C. Woffendin, P. J. Polverini, B. J. Nickoloff, and G. J. Nabel. 1997. Propagation of a human herpesvirus from AIDS-associated Kaposi's sarcoma. N. Engl. J. Med. 336:163-171. [DOI] [PubMed] [Google Scholar]
- 20.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friborg, J., Jr., W.-P. Kong, C. C. Flowers, S. L. Flowers, Y. Sun, K. E. Foreman, B. J. Nickoloff, and G. J. Nabel. 1998. Distinct biology of Kaposi's sarcoma-associated herpesvirus from primary lesions and body cavity lymphomas. J. Virol. 72:10073-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goltz, M., H. Broll, A. Mankertz, W. Weigelt, H. Ludwig, H.-J. Buhk, and K. Borchers. 1994. Glycoprotein B of bovine herpesvirus 4: its phylogenetic relationship to gB equivalents of the herpesviruses. Virus Genes 9:53-59. [DOI] [PubMed] [Google Scholar]
- 23.Gompels, U. A., M. A. Craxton, and R. W. Honess. 1988. Conservation of glycoprotein H (gH) in herpesviruses: nucleotide sequence of the gH gene from herpesvirus saimiri. J. Gen. Virol. 69:2819-2829. [DOI] [PubMed] [Google Scholar]
- 24.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 25.Heineman, T. C., N. Krudwig, and S. L. Hall. 2000. Cytoplasmic domain signal sequences that mediate transport of varicella-zoster virus gB from the endoplasmic reticulum to the Golgi. J. Virol. 74:9421-9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrold, R. E., A. Marchini, S. Fruehling, and R. Longnecker. 1996. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J. Virol. 70:2049-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, D. C., and P. G. Spear. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1102-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khattar, S. K., S. van Drunen Littel-van den Harke, S. K. Attah-Poku, L. A. Babiuk, and S. K. Tikoo. 1996. Identification and characterization of a bovine herpesvirus-1 (BHV-1) glycoprotein gL which is required for proper antigenicity, processing, and transport of BHV-1 glycoprotein gH. Virology 219:66-76. [DOI] [PubMed] [Google Scholar]
- 30.Klein, E., G. Klein, J. S. Nadkarni, J. J. Nadkarni, H. Wigzell, and P. Clifford. 1968. Surface IgM-kappa specificity on a Burkitt lymphoma cell in vivo and in derived culture lines. Cancer Res. 28:1300-1310. [PubMed] [Google Scholar]
- 31.Klupp, B. G., W. Fuchs, E. Weiland, and T. C. Mettenleiter. 1997. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J. Virol. 71:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobasa, D., M. E. Rodgers, K. Wells, and Y. Kawaoka. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J. Virol. 71:6706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, S. K., and R. Longnecker. 1997. The Epstein-Barr virus glycoprotein 110 carboxy-terminal tail domain is essential for lytic virus replication. J. Virol. 71:4092-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Q., C. Buranathai, C. Grose, and L. M. Hutt-Fletcher. 1997. Chaperone functions common to nonhomologous Epstein-Barr virus gL and varicella-zoster virus gL proteins. J. Virol. 71:1667-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, D. X., U. A. Gompels, J. Nicholas, and C. Lelliott. 1993. Identification and expression of the human herpesvirus 6 glycoprotein H and interaction with an accessory 40K glycoprotein. J. Gen. Virol. 74:1847-1857. [DOI] [PubMed] [Google Scholar]
- 38.Longnecker, R., C. L. Miller, X.-Q. Miao, A. Marchini, and E. Kieff. 1992. The only domain which distinguishes Epstein-Barr virus latent membrane protein 2A (LMP2A) from LMP2B is dispensable for lymphocyte infection and growth transformation in vitro; LMP2A is therefore nonessential. J. Virol. 66:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manickan, E., R. J. Rouse, Z. Yu, W. S. Wire, and B. T. Rouse. 1995. Genetic immunization against herpes simplex virus. Protection is mediated by CD4+ T lymphocytes. J. Immunol. 155:259-265. [PubMed] [Google Scholar]
- 40.Marsh, M., and H. T. McMahon. 1999. The structural era of endocytosis. Science 285:215-220. [DOI] [PubMed] [Google Scholar]
- 41.Menezes, J., W. Leibold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22:276-284. [PubMed] [Google Scholar]
- 42.Miethke, A., G. M. Keil, F. Weiland, and T. C. Mettenleiter. 1995. Unidirectional complementation between glycoprotein B homologues of pseudorabies virus and bovine herpesvirus 1 is determined by the carboxy-terminal part of the molecule. J. Gen. Virol. 76:1623-1635. [DOI] [PubMed] [Google Scholar]
- 43.Moore, P. S., S. J. Gao, G. Dominguez, E. Cesarman, O. Lungu, D. M. Knowles, R. Garber, P. E. Pellett, D. J. McGeoch, and Y. Chang. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J. Virol. 70:549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moses, A. V., K. N. Fish, R. Ruhl, P. P. Smith, J. G. Strussenberg, L. Zhu, B. Chandran, and J. A. Nelson. 1999. Long-term infection and transformation of dermal microvascular endothelial cells by human herpesvirus 8. J. Virol. 73:6892-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muggeridge, M. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH, and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 46.Nemerow, G. R., C. Mold, V. K. Schwend, V. Tollefson, and N. R. Cooper. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-200. [DOI] [PubMed] [Google Scholar]
- 48.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 50.Peeters, B., N. De Wind, M. Hooisma, F. Wagenaar, A. Gielkens, and R. Moormann. 1992. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J. Virol. 66:894-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pereira, L. 1994. Function of glycoprotein B homologues of the family Herpesviridae. Infect. Agents Dis. 3:9-28. [PubMed] [Google Scholar]
- 52.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 53.Pertel, P. E., P. G. Spear, and R. Longnecker. 1998. Human herpesvirus-8 glycoprotein B interacts with Epstein-Barr virus (EBV) glycoprotein 110 but fails to complement the infectivity of EBV mutants. Virology 251:402-413. [DOI] [PubMed] [Google Scholar]
- 54.Raab, M.-S., J.-C. Albrecht, A. Birkman, S. Yaguboglu, D. Lang, B. Fleckenstein, and F. Neipel. 1998. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J. Virol. 72:6725-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rauh, I., and T. C. Mettenleiter. 1991. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J. Virol. 65:5348-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renne, R., D. Blackbourn, D. Whitby, J. Levy, and D. Ganem. 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Redes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 59.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spaete, R. R., K. Perot, P. I. Scott, J. A. Nelson, M. F. Stinski, and C. Pachl. 1993. Coexpression of truncated human cytomegalovirus gH with the UL115 gene product or the truncated human fibroblast growth factor receptor results in transport of gH to the cell surface. Virology 193:853-861. [DOI] [PubMed] [Google Scholar]
- 62.Spear, P. G. 1993. Entry of alphaherpesviruses into cells. Semin. Virol. 4:167-180. [Google Scholar]
- 63.Spear, P. G. 1993. Membrane fusion induced by herpes simplex virus, p. 201-232. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press, Inc., Boca Raton, Fla.
- 64.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang, D.-C., M. DeVit, and S. A. Johnston. 1992. Genetic immunization is a simple method for eliciting an immune response. Nature 356:152-154. [DOI] [PubMed] [Google Scholar]
- 66.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping and endocytosis. Cell 50:203-213. [DOI] [PubMed] [Google Scholar]
- 67.Thompson, J. F., L. S. Hayes, and D. B. Lloyd. 1991. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene 103:171-177. [DOI] [PubMed] [Google Scholar]
- 68.Tugizov, S., E. Maidji, J. Xiao, Z. Zheng, and L. Pereira. 1998. Human cytomegalovirus glycoprotein B contains autonomous determinants for vectorial targeting to apical membranes of polarized epithelial cells. J. Virol. 72:7374-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vieira, J., O. Hearn, L. E. Kimball, B. Chandran, and L. Corey. 2001. Activation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J. Virol. 75:1378-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vieira, J., M. L. Huang, D. M. Koelle, and L. Corey. 1997. Transmissible Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi's sarcoma. J. Virol. 71:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, F.-Z., S. M. Akula, N. P. Pramod, L. Zeng, and B. Chandran. 2001. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J. Virol. 75:7517-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yaswen, L. R., E. B. Stephens, L. C. Davenport, and L. M. Hutt-Fletcher. 1993. Epstein-Barr virus glycoprotein gp85 associates with the BKRF2 gene product and is incompletely processed as a recombinant protein. Virology 195:387-396. [DOI] [PubMed] [Google Scholar]