Abstract
Interleukin 16 (IL-16) is a chemotactic cytokine that binds to the CD4 receptor and affects the activation of T cells and replication of HIV. It is expressed as a large 67-kDa precursor protein (pro-IL-16) in lymphocytes, macrophages, and mast cells, as well as in airway epithelial cells from asthmatics after challenge with allergen. This pro-IL-16 is subsequently processed to the mature cytokine of 13 kDa. To study the expression of IL-16 at the transcriptional level, we cloned the human chromosomal IL-16 gene and analyzed its promoter. The human IL-16 gene consists of seven exons and six introns. The 5′ sequences up to nucleotide −120 of the human and murine IL-16 genes share >84% sequence homology and harbor promoter elements for constitutive and inducible transcription in T cells. Although both promoters lack any TATA box, they contain two CAAT box-like motifs and three binding sites of GA-binding protein (GABP) transcription factors. Two of these motifs are part of a highly conserved and inducible dyad symmetry element shown previously to control a remote IL-2 enhancer and the CD18 promoter. In concert with the coactivator CREB binding protein/p300, which interacts with GABPα, the binding of GABPα and -β to the dyad symmetry element controls the induction of IL-16 promoter in T cells. Supplementing the data on the processing of pro-IL-16, our results indicate the complexity of IL-16 expression, which is tightly controlled at the transcriptional and posttranslational levels in T lymphocytes.
Interleukin 16 (IL-16), originally identified in supernatants of mitogen-activated peripheral blood mononuclear cells (PBMCs) (1), is synthesized as a large 67-kDa precursor protein (2–4). This pro-IL-16 is almost exclusively expressed in lymphatic tissues (4) and contains three postsynaptic density protein, disc-large, zonulin-1 (PDZ) domains, i.e., modules involved in protein-protein interactions (5, 6). The maturation of pro-IL-16 appears to be mediated by caspase 3 leading to the bioactive IL-16, which consists of the C-terminal 121 amino acids of the precursor (4, 7). Activated CD8+ and CD4+ T cells synthesize identical levels of IL-16 transcripts, but mature IL-16 is predominantly secreted by CD8+ T lymphocytes in response to antigens, histamine, or serotonin (8, 9). In addition, human mast cells also synthesize pro-IL-16, and treatment with anaphylatoxin, C5a, or 12-O-tetradecanoylphorbol-13-acetate (TPA) increases the IL-16 mRNA level as much as 10-fold (10).
IL-16 binds to the CD4 receptor and is therefore selectively active on CD4+ T lymphocytes, monocytes, and eosinophils (11, 12). Like other cytokines, IL-16 exerts numerous and partially contradictory effects in vitro. Although it is able to render CD4+ T cells unresponsive to TCR/CD3 stimuli, it induces the expression of certain activation markers, such as IL-2 receptor subunits (13). The proliferative response of CD4+ T cells cotreated with IL-16 and IL-2 for long periods is therefore selectively enhanced (14), suggesting the possible use of IL-16 as an immune therapy to counteract conditions of CD4+ T cell deficiency such as that associated with AIDS. Moreover, IL-16 inhibits the replication of T cell and monocyte tropic HIV strains in vitro (15–17). HIV patients receiving anti-retroviral therapy display rapidly increasing IL-16 serum levels, possibly reflecting the recovery of the immune system as a consequence of the reduced viral load (18).
To learn more about the expression of IL-16, we have determined the genomic organization of the human IL-16 gene, including the structure and function of its promoter. A DNA fragment spanning the nucleotides from −408 to +88 of the IL-16 gene was found to exert constitutive promoter activity in lymphoid and nonlymphoid cells. A strong inducible promoter activity in T cells was identified within the immediate 5′ upstream DNA of approximately 90 bp. Although this promoter fragment lacks any TATA box-like sequence element, it contains three binding sites for Ets-like proteins. One of these fragments overlaps with the previously reported transcriptional start sites (4), whereas the other two sites generate a dyad symmetry element (DSE). We show here that these sites are bound by GA-binding protein (GABP) factors which, in concert with the coactivators CREB binding protein (CBP)/p300, appear to play an important role in the control of the IL-16 promoter. CBP/p300 binds to GABPα and enhances strongly the GABP mediated IL-16 promoter induction.
MATERIALS AND METHODS
Cell Culture, DNA Transfections, and Reporter Gene Assays.
Human Jurkat T cells were cultured in RPMI 1640 medium, and HeLa cells and 293 embryonic kidney cells were cultured in DMEM medium. Both media were supplemented with 10% fetal calf serum. DNA transfections were performed by using the SuperFect transfection reagent (Qiagen, Chatsworth, CA) according to the manufacturers protocol. 293 cells were transfected by using a conventional calcium phosphate coprecipitation technique. Luciferase assays were performed by using the Luciferase Assay Reagent (Promega). As transfection controls, the pSEAP2-control plasmid (CLONTECH) was cotransfected and assays were performed by using the Great EscAPe SEAP chemiluminescence detection kit (CLONTECH).
Recombinant DNA Work and Sequence Analysis.
Overlapping human IL-16 gene fragments were amplified by using human PBMC DNA, the Tth-Polymerase Mix (CLONTECH), and the following primer pairs: (5′-CTGCTGCTACCACAGGAAGACACAGCAG-3′ and 5′-GAGCTGATTCTCTGCCTGATGGA-3′; 5′-GAGCACCTAGGATCACACATC-3′ and 5′-CTGACCAGACTGAAGGGAACTGTGGTC-3′; 5′-CAGAGCTGAGAGAATATACAGAGGGTC-3′ and 5′-TTCTAGATCTGCTCCTCCTGCCAAGCT-3′; 5′-CATGTCACCATCTTACACAAGGAG-3′ and 5′-CATCACTGGCTGCAGAGGCTGAGGCTGCAG-3′; 5′-CACAGAGTGTTTCCAAATGGGCTGGCC-3′ and 5′-AGCTGTGTGTGTTATTGGCTTTGGCTTC-3′; 5′-ACCACAGCTGCTGGAGACTCCTAG-3′ and 5′-GTGGTAAGTAGTAGCAATTTTATTTGATAC-3′) The amplification products were cloned in the pGEM-T-vector (Promega) and sequenced. To minimize sequencing errors, multiple clones were sequenced for every fragment.
For the cloning of the chromosomal DNA upstream of the human IL-16 cDNA-sequence two cDNA based reverse-complementary primers, hGSP1: 5′-TCTTCTCCACCCTGGCTCGCCCTCTGT-3′ and hGSP2: 5′-ACTTGGTCTGTGGGCCGATACCAGGT-3′, were used in a nested PCR with parts of the human PromoterFinder DNA Walking kit (CLONTECH) according to the manufacturer’s protocol. Fragments of 1.7 kb (DraI library) and 1.9 kb (PvuII library) were amplified and cloned in pGEM-T. Sequencing revealed the expected 62 bp of the 5′ end of the IL-16 cDNA sequence (4) upstream of the GSP2 binding site, proving the specificity of the PCRs. In the same way, by using the murine IL-16 cDNA-specific reverse-complementary primers mGSP1: 5′-GGCACTCCTCCTTGACTCTCCTTCTGA-3′ and mGSP2: 5′-TGAAGTCTGCGTGCTGGCTGCAGGTC-3′ along with the murine PromoterFinder DNA Walking kit (CLONTECH), a 1.2-kb genomic fragment (SspI-library) containing the sequence upstream of the known cDNA (19) and 52 bp of its 5′-end was amplified.
For the construction of the promoter/reporter gene plasmids the genomic fragment from −1048 to +88 was amplified by using primers P1: 5′-CTACAGGGTACCTGGAAGGAAGCCTGCAGCCATGTC-3′, and P2: 5′-TTCAGTTATCTCGAGCTTGGTCTGTGGGGCGATACCAGGT-3′ and human DNA from PBMCs as template. The PCR product was cloned into the pGL3-Basic vector (Promega) and sequenced. Using this plasmid as template, the subfragments −408/+88, −133/+88, −90/+88, −408/+3, −408/−110 were amplified and cloned in pGL-3-Basic. All clonings were verified by sequencing.
GG-to-CC changes within the ERE (Ets-related element)-C and -D motifs and the +12 GABP-binding site were introduced into the −408/+88 promoter fragment by using the QuikChange site-directed mutagenesis system (Stratagene).
Electrophoretic Mobility-Shift Assays (EMSAs).
EMSAs were performed as described (20) by using 2 μg of nuclear protein and 0.5 μg of poly(dI-dC). In supershift EMSAs, 1 μg of antibodies (Abs) raised against GABPα, β (21) or PU.1 (20) were added to the incubations. When the DNA binding of recombinant GABP was tested, 2.5–20 ng of bacterially expressed proteins prepared as described (21) were incubated in EMSAs.
Interaction Between GABPs and p300.
Five micrograms of bacterially expressed GST/p300 fusion proteins was immobilized on glutathione agarose beads (30 μl) and incubated with 50 ng of bacterially expressed full-size GABPα, -β or -α+β (21) in 500 μl of binding buffer (20 mM Tris⋅Cl, pH 7.8/75 mM NaCl/0.5 mM EDTA/0.5 mM EGTA/5 mM NaF/0.5 mM DTT/20 μM leupeptin/0.2 mM AMSF/0.5% nonfat dry milk as blocker). After rotating for 2 h at 4°C and extensive washings with the binding buffer, bound proteins were eluted by boiling in Laemmli sample buffer, fractionated on a 12.5% polyacrylamide gel, and immunodetected by using GABPα+β-specific Abs.
RESULTS
Organization of the Human IL-16 Gene.
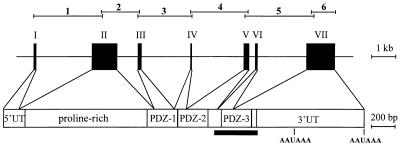
For the analysis of the chromosomal IL-16 gene, overlapping genomic DNA fragments from human PBMCs were amplified by using IL-16 cDNA-specific primer pairs and sequenced. The human IL-16 gene spans approximately 12.8 kb and consists of seven exons and six introns (Fig. 1). Exon I contains the largest part of the 5′-untranslated mRNA region. The first ATG, which corresponds to the translational start codon (4), starts on nucleotide position 51 of exon II. This exon is unusually long (1096 bp), encoding 348 aa harboring the proline-rich part and the N-terminal region of the first PDZ domain (19). Exon III and the first nucleotides of exon IV code for the remainder of the first PDZ domain. PDZ-2 is encoded by the exons IV and V. Exons VI and VII contain the sequence for PDZ-3 (found in mature IL-16). The cleavage site for the putative processing enzyme caspase 3 is encoded in exon V.
Figure 1.
Genomic organization of the human IL-16 gene. The seven exons are shown as black boxes and projected on the IL-16 mRNA. UT, untranslated 5′ and 3′ sequences; AAUAAA, poly(A) addition motifs. The fragments encoding the proline-rich region and the three PDZ domains of pro-IL-16 protein are indicated. The black bar below denotes the sequence coding for the mature IL-16 protein. The complete gene of approximately 12.8 kb was cloned and sequenced as six overlapping fragments depicted above the gene structure. The use of an alternative splice acceptor in intron 5, three nucleotides downstream of the regular site, generates a transcript with a missing alanine codon (not shown). This splice acceptor is predominantly used in mice because we were unable to find cDNAs encoding this alanine in murine PBMCs (19).
Characterization of the IL-16 Promoter.
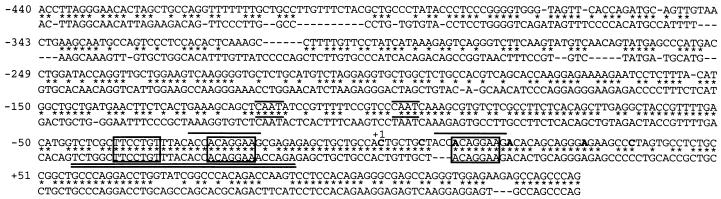
Fig. 2 shows a sequence comparison between the 5′-flanking regions of human and murine IL-16 genes spanning the nucleotides from position −440 to +130. The 5′-untranslated mRNA regions and the immediate upstream regions (up to −120) from both genes share a high sequence homology, suggesting their functional importance. Multiple transcriptional initiation sites have been identified within 40 bp 5′ of the murine and human IL-16 genes (4, 19) from which the most upstream site of human IL-16 gene was assigned to be position +1 (4). Accordingly, neither the human nor murine IL-16 gene contains a canonical TATA box. CAAT box motifs are present at the positions −98 and −118, and at +12, −25, and −39 of both genes three sites are located for the putative binding of Ets-like transcription factors. The Ets elements at +12 and −25 of the human gene form a direct repeat of 11 bp, whereas in both genes those repeats around −25 and −39 are part of a 33 bp-inverted repeat. A very similar repeat designated as DSE was recently identified as an inducible sequence element of a distal IL-2 enhancer (20).
Figure 2.
Comparison of 5′ sequences of human and murine IL-16 genes. The published start points of transcription in human cells are shown in bold (4). Nucleotide +1 corresponds to the first transcription start site of the human gene. Identical nucleotides in both sequences are indicated by ∗. Deletions indicated by dashes were introduced to obtain the highest degree of homology. The CAAT box-like sequences around positions −98 and −118 and direct repeats around +12 and −25 are overlined, the dyad symmetry element, DSE, is double-underlined, and the GABP-binding sites are boxed.
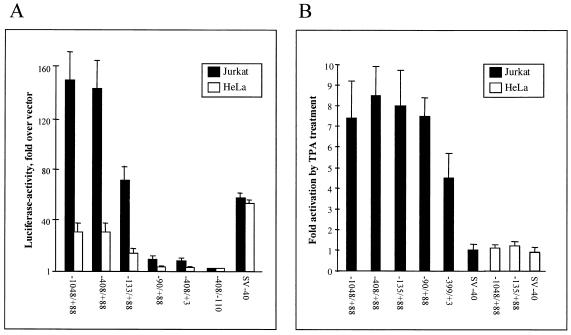
To test the IL-16 promoter activity, a DNA fragment from position −1048 to +88 and shorter subfragments were cloned in front of a luciferase reporter gene and transiently transfected into Jurkat T, cells which express considerable amounts of pro-IL-16, and HeLa cells, which express IL-16 at very low levels (unpublished results). As shown in Fig. 3A, the fragments −1048/+88 and −408/+88 exhibited a distinct promoter activity in both types of cells, being 4- to 5-fold stronger in Jurkat cells than in HeLa cells. Deletion of 5′ nucleotides up to the CAAT box-like motifs (up to −133) resulted in a 50% decrease and the removal of this motifs by deletion up to −90 in a further strong decrease of constitutive promoter activity. A similar strong decrease was observed after deleting the 3′ region between positions +3 and +88 containing the Ets element at +12, and the removal of sequences between −110 and +88 abolished any promoter activity.
Figure 3.
Promoter activity of 5′ sequences from the human IL-16 gene in Jurkat T cells and HeLa cells. The numbers below indicate the sequences of DNA fragments cloned in front of a luciferase reporter gene. The activity of a pGL3 vector (Promega) containing the SV40 early promoter is also shown. (A) Deletion analysis of the 5′ IL-16 DNA. Relative luciferase activities are given as fold activity above the activity of the promoter-less pGL3-Basic vector. The standard deviations represent the mean values from four independent experiments. (B) TPA-mediated induction of 5′ segments of IL-16 gene. Differences of luciferase activities are shown between uninduced cells and cells treated with 10 nM TPA and indicated as “Fold activation.” The standard deviations represent the mean values from three independent experiments.
To delineate the inducible promoter elements, transfected Jurkat cells and HeLa cells were induced with phorbol ester for 20 h. This resulted in a 7- to 8-fold increase of luciferase reporter gene expression in Jurkat cells, but not in HeLa cells transfected with constructs containing the IL-16 promoter region from −90 to +88 (Fig. 3B). Deletion of the sequences from +4 to +88, which include the third Ets-like protein binding site at +12 decreased the inducibility up to 50%. Because removal of the upstream region from position −1048 to −90 did not change the inducibility, the fragment from position −90 to +88 appears to harbor all sequence motifs for the induction of the IL-16 promoter in T cells.
The Binding of GABPα and -β to the Ets-Binding Sites of Proximal Promoter Region Contributes to the IL-16 Promoter Induction.
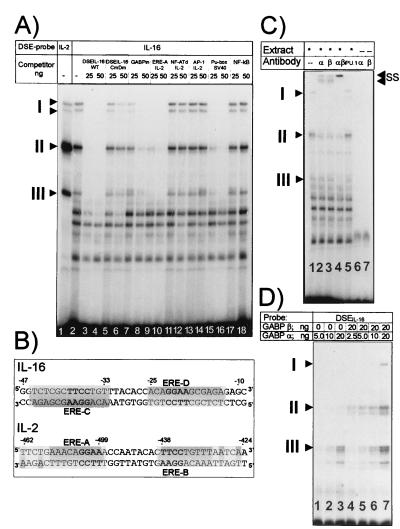
The DSE of the distal IL-2 enhancer (DSEIL-2) is bound by GABP factors (20), suggesting a binding of these factors to the IL-16 DSE (DSEIL-16) as well. To test this assumption, nuclear proteins from Jurkat cells were incubated in EMSAs by using the DSEs from the IL-16 promoter and distal IL-2 enhancer as probes. Very similar to the DSEIL-2, four specific protein–DNA complexes were generated with the DSEIL-16 which we designate as I (a and b, higher and lower, respectively), II, and III (Fig. 4A). TPA treatment of cells did not alter their appearance (data not shown). The formation of all four complexes was abolished by a 100- and 200-fold molar excess of the unlabeled probe or a similar excess of the ERE-A site from the DSEIL-2 (Fig. 4B) and the SV40 enhancer Ets site, a known GABP-binding site (20) and the +12 Ets-like site (see Fig. 4A, lanes 3+4, 7+8, 9+10, and 15+16). The competition with the +12 site indicates that this site is an additional GABP-binding site. No competition was observed when the same excess was used of a DSEIL-16 mutated in both Ets motifs (Fig. 4A, lanes 5+6) or of binding sites for NF-AT, AP-1 and NF-κB factors (Fig. 4A, lanes 11–14, 17+18).
Figure 4.
GABP factors bind to the Ets-like sequence motifs of IL-16 promoter. (A) EMSAs using nuclear proteins from Jurkat cells and the DSE from the IL-16 promoter as probe. Two micrograms of nuclear protein from noninduced Jurkat cells was incubated either with the DSEIL-2 (lane 1) or DSEIL-16 (lanes 2–18) as probes (see B) followed by electrophoresis on a native 4% polyacrylamide gel. For competition, 25 and 50 ng of the following oligonucleotides were added: lanes 3 and 4, DSEIL-16; lanes 5 and 6, DSEIL-16 mutated in both Ets motifs; lanes 7 and 8, IL-16 Ets motif at position +12; lanes 9 and 10, IL-2 ERE-A motif (see B); lanes 11 and 12, distal NF-AT site from the murine IL-2 promoter (see ref. 22); lanes 13 and 14, AP-1 binding site from the IL-2 promoter (22); lanes 15 and 16, Ets site from the SV40 enhancer (20); lanes 17 and 18, consensus NF-κB site. (B) Sequences of DSEs from the IL-16 promoter and distal IL-2 enhancer (20). The palindromic organization of Ets-related elements, EREs, is indicated in gray. (C) Supershift EMSAs with Abs raised against GABPα and -β. In lanes 1–5, 1 μg of nuclear protein from Jurkat cells was incubated with the DSEIL-16 probe, alone or with 1 μg of Ab raised against GABPα (lane 2), -β (lane 3), or with both Abs (lane 4). As a control, a Pu.1-specific Ab was added in lane 5. In lanes 6 and 7, the DSEIL-16 probe was incubated with Abs alone. SS, complexes supershifted by GABP Abs. (D) Binding of recombinant GABPα and -β to the DSEIL-16. Bacterially expressed GABPα or -β (2.5–20 ng) was incubated with a DSEIL-16 probe as indicated. I–III indicate DNA–protein complexes identical in mobility to those generated with nuclear proteins from Jurkat cells (A).
To identify the GABP proteins in the IL-16 DSE complexes, EMSA supershift assays were performed by using Abs raised against GABPα and -β. As shown in Fig. 4C (lanes 1–4), addition of these Abs resulted in the generation of several “supershift complexes” at the expense of complexes I and II. No changes in complex formation were observed when an Ab raised against Pu.1, another factor of Ets family (Fig. 4C, lane 5) was used. To confirm these results showing GABP proteins in the DSE complexes, increasing concentrations of bacterially expressed recombinant GABPα and -β proteins were incubated in EMSAs with a DSEIL-16 probe. This resulted in the generation of three retarded DNA–protein complexes (Fig. 4D) with identical mobilities to those of complexes Ia, II, and III. Whereas the rapidly running complex III was generated with GABPα alone, for the generation of complexes Ia and II, the presence of both GABPα and -β was necessary indicating the presence of both GABP proteins in complexes I and II.
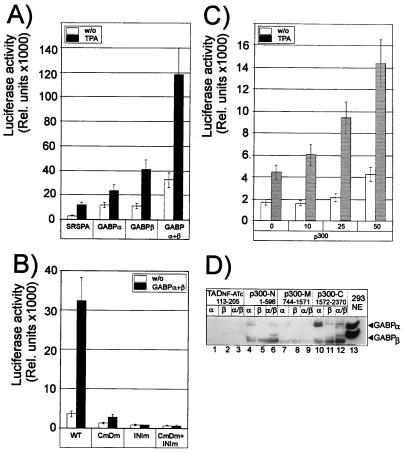
To demonstrate a functional role for GABPα and -β in the induction of IL-16 promoter, we transfected a luciferase reporter gene controlled by the IL-16 promoter (from −408 to +88) into Jurkat and 293 cells alone or with vectors expressing GABPα and -β. As shown in Fig. 5A, both GABP proteins alone exerted a moderate stimulatory effect on both the constitutive and TPA-induced promoter activity, which was significantly enhanced when both proteins were expressed together. Mutations in the GABP sites ERE-C and -D (Fig. 4B), which abolished any GABP binding (Fig. 4A, lanes 5+6) reduced significantly the stimulatory effect of GABP proteins on the constitutive and inducible activity of IL-16 promoter (Fig. 5B, CmDm). Mutation of the +12 GABP site, alone or together with the CmDm mutations, suppressed any promoter induction (Fig. 5B, INIm and CmDm+INIm), indicating a critical role for this site in the control of promoter activity.
Figure 5.
GABPα and -β stimulate, in concert with p300, the activity of IL-16 promoter. (A) GABPα and -β stimulate the IL-16 promoter. Jurkat cells were cotransfected with a luciferase reporter gene controlled by the IL-16 promoter (from −408 to +88), SRSPA-based vectors expressing GABPα or -β, or both, as indicated. The cells were left untreated or stimulated with TPA (10 ng/ml) for 24 h. (B) The GABP-binding sites within the IL-16 promoter are of crucial importance for its activity. Human 293 cells were transfected with a wild-type IL-16 promoter/luciferase gene construct or mutated IL-16 promoter/luciferase constructs alone or together with vectors expressing GABPα and -β. CmDm, mutations in the ERE-C and ERE-D motifs (see Fig. 4B); INIm, mutations within the +12 GABP site which overlaps the transcriptional initiation site. (C) CBP/p300 enhance the activity of the IL-16 promoter in a dose-dependent manner. 293 cells were cotransfected with a luciferase reporter gene controlled by the IL-16 promoter (from −408 to +88) and 0–50 ng of a p300 expression vector as indicated. (D) Interaction between GABPα and p300. Equal amounts of bacterially expressed glutathione S-transferase/p300 fusion proteins spanning N-terminal, middle, or C-terminal portions of p300 immobilized on glutathione agrose beads were incubated with 500 ng of bacterially expressed GABPα, -β or -α+β. The bound proteins were eluted and immunodetected by using a mix of GABPα+β-specific Abs. As a control for p300/GABP interaction, a glutathione S-transferase fusion protein containing the N-terminal transacting domain of NF-ATc (TADNF-ATc, 113–205) was used. In lane 13, 50 μg of nuclear proteins from 293 cells were fractionated. The appearance of a weak GABPα band in lane 11 is the result of an accidental contamination of the GABPβ protein preparation with GABPα. The appearance of a second GABPα band in many lanes is probably a GABPα cleavage product. The lowest strong band is most likely the result of the crossreactivity of GABP Abs with a bacterial protein binding unspecifically to p300.
The Cofactors CBP/p300 Bind to GABP Proteins and Enhance the Induction of IL-16 Promoter.
Ets-1, the prototype of Ets factors, was shown to cooperate with the nuclear coactivators CBP/p300 (23, 24). To investigate whether CBP/p300 plays a similar role in the activation of IL-16 promoter by GABP factors, increasing amounts of vectors expressing p300 or CBP were cotransfected into 293 cells with the luciferase reporter gene controlled by the IL-16 promoter. Overexpressing p300 (Fig. 5C) or CBP (data not shown) resulted in a dose-dependent increase in the constitutive and inducible IL-16 promoter activity, suggesting that this cofactor participates in the activation of the IL-16 promoter. To demonstrate that the GABP factors are the targets of p300-mediated activation of the IL-16 promoter, binding of p300 to GABP proteins was tested. Bacterially expressed, full-length GABPα and -β were incubated with GST/p300 proteins spanning large portions of the N-terminal, central, and C-terminal regions of p300. As shown in Fig. 5D, GABPα bound specifically to the C-terminal part of p300 and much more weakly to its N-terminal part. No binding to the N-terminal transactivation domain of NF-ATc (TAD-NFATc) used as negative control was observed. GABPβ was only found in the GST/p300-bound fraction when incubated together with GABPα (Fig. 5D, lane 12), indicating an indirect, GABPα-mediated recruitment of GABPβ. These results demonstrate a direct interaction between GABPα and the C-terminal part of p300.
DISCUSSION
In this study we have described the structure of the human IL-16 gene, including its promoter. The IL-16 gene consists of seven exons. Exons II–VII encode the pro-IL-16 precursor protein, whereas the mature IL-16 is encoded by sequences of the last three exons. The IL-16 gene is almost exclusively expressed in lymphocytes and epidermis cells of the respiratory tract after allergic stimulation (4, 25, 26). It has been shown that the activation of lymphocytes by antigens, PHA, and other mitogens strongly increases the transcription of IL-16 (1, 10). We have identified the immediate upstream region of the IL-16 gene (reaching up to nucleotide position −90), along with the first 88 bp of the untranslated mRNA region, as the most important DNA fragment for the control of inducible expression of IL-16 gene in T lymphocytes. This region contains three GGAA core motifs for the binding of Ets-like factors around the positions −39, −25, and +12. The two upstream sites at −39 and −25 form a 33-bp DSE that resembles the DSE motifs within a distal IL-2 enhancer (see Fig. 4B) and the CD18 promoter. As shown previously for GABP binding to the DSE of the distal IL-2 enhancer (20) the GABP proteins α and β are the most prominent factors binding to the DSE of IL-16 promoter in nuclear protein extracts from T cells. Because the introduction of mutations into these GABP binding sites (Fig. 4B) reduced significantly the induction of promoter in Jurkat cells (Fig. 5A), one has to conclude that the binding of GABP factors to both sites is of crucial importance for the induction of IL-16 promoter in T cells.
GABPα and -β form heterodimeric (α–β) and heterotetrameric (α2–β2) complexes when they bind to the DSE of the distal IL-2 enhancer (20, 21). The same heteromeric complexes were generated with the DSEIL-16 as probe (Fig. 4 A and D). However, approximately 3-fold more tetrameric than dimeric complexes were formed on the DSEIL-16 than DSEIL-2 (determined by dividing the amount of tetrameric complexes by that of dimeric complexes according to phosphoimager data, see also lanes 1 and 2 in Fig. 4A). This may be caused by the specific structure of DSEIL-16, in which the GGAA core sequences, although forming a palindrome (as in the DSEIL-2) are not directed to each other (as in the DSEIL-2) but are directed in opposite directions. This configuration appears to facilitate the formation of heterotetramers, probably through the leucine zipper motifs in GABPβ (see refs. 27 and 28 for a discussion) and appears to enhance transcription. We have shown previously for the activity of multiples of DSEIL-2 that the generation of heterotetramers gives rise to a strong activation of transcription compared with the relatively weak activity of multiple repeats of identical EREs to which GABPα and -β bind as heterodimers (20).
The transcriptional induction of IL-16 promoter by GABP factors is distinctly enhanced by the cofactors CBP/p300, which interact with GABPα (Fig. 5D). CBP and p300 are nuclear proteins that were shown to bind to and to increase the activity of numerous transcription factors by exerting adaptor functions and histone acetylase activity (29). Moreover, they provide a molecular platform for a large number of potentially important associated proteins (30). Interestingly, several inducible transcription factors, such as AP-1/c-Jun, NF-kB/p65, and the NF-AT factors NF-ATp and c, that are activated after lymphocyte activation, were shown to cooperate with CBP/p300 (31, 32). For NF-ATc, it has been demonstrated that CBP/p300 is able to synergize with multiple protein kinase cascades in the transcriptional activation of this factor. This may also be true for the activation of GABPs that are a nuclear target of Ras-Raf-Erk and JNK/SAP kinase cascades (21, 33). Although a direct phosphorylation of GABPα by Erk2 has been demonstrated in vitro and in vivo (33), the phosphorylation of CBP/p300 or of associated proteins by these mitogen-activated protein kinases provides a further level of regulation through which the induction of IL-16 promoter by GABP factors may be controlled in T cells.
The third Ets-like binding site at position +12 appears to be a further GABP-binding site. Deletion of the region containing this site decreased both the constitutive and inducible activation of IL-16 promoter (see Fig. 3A). Many TATA-less RNA polymerase II promoters contain typical initiator elements, i.e., short conserved sequences that are bound by proteins of the transcriptional machinery. Some of these promoters, including the CD3ɛ, CD4, CD11c, CD18, CD72, IL-2Rβ, TGFβ RII, Ets-2, murine laminin B2, type IV collagenase, thymidylate synthetase, and cytochrome c oxidase subunit IV promoters possess Ets-like binding motifs near their transcriptional start sites, suggesting a new class of initiator elements (34–45). Recently, two PEA3 motifs that are bound by GABP factors were shown to function as a minimal transcriptional initiator element when placed in front of a luciferase gene (46). Thus, the +12 GABP binding site of IL-16 promoter might act as a similar initiator element starting transcription from the TATA-less IL-16 gene.
The CAAT box-like sequences located between −133 and −90 are needed for the full IL-16 promoter activity in both Jurkat T cells and HeLa cells. Additional positively acting sequence element(s) is/are located in the region between −408 and −133, because deletion of this segment leads to a 50% decrease of promoter activity. However, the sequence elements and the transcription factors binding to and controlling the activity of these elements remain to be identified.
Taken together, the structure of the inducible IL-16 promoter differs markedly from that of typical interleukin promoters (such as the IL-2 and IL-4 promoters), which are predominantly activated on antigen-mediated stimulation in peripheral CD4+ T cells. It will be interesting to determine the physiological differences reflected in the architecture of these two types of lymphokine promoters.
Acknowledgments
We thank Olga Reimers and Martin Selbert for excellent technical assistance. For gifts of reagents we are very much indebted to Drs. Angelika Hoffmeyer, Egbert Flory, and Richard Eckner. We are grateful to Dr. Steve Norley for help in the preparation of the manuscript. Parts of this work were supported by Grant 1506/TG04 from the Federal Ministry of Health and the Heinz Kuthe de Mouson legacy (to R.K.) and the SFB 465 (Würzburg) (to E.S.).
ABBREVIATIONS
- Ab
antibody
- CBP
CREB binding protein
- EMSA
electrophoretic mobility-shift assay
- PBMC
peripheral blood mononuclear cells
- PDZ
postsynaptic density protein, disc-large, zonulin-1
- TPA
12-O-tetradecanoylphorbol-13-acetate
- DSE
dyad symmetry element
- ERE
Ets-related element
- IL
interleukin
Footnotes
References
- 1.Center D M, Cruikshank W. J Immunol. 1982;128:2563–2568. [PubMed] [Google Scholar]
- 2.Bazan J F, Schall T J. Nature (London) 1996;381:29–30. doi: 10.1038/381029a0. [DOI] [PubMed] [Google Scholar]
- 3.Bannert N, Baier M, Werner A, Kurth R. Nature (London) 1996;381:30. doi: 10.1038/381030a0. [DOI] [PubMed] [Google Scholar]
- 4.Baier M, Bannert N, Werner A, Lang K, Kurth R. Proc Natl Acad Sci USA. 1997;94:5273–5277. doi: 10.1073/pnas.94.10.5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyle D A, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 6.Cho K O, Hunt C A, Kennedy M B. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Center D, Wu M H, Cruikshank W W, Yuan J, Andrews D W, Kornfeld H. J Biol Chem. 1998;273:1144–1149. doi: 10.1074/jbc.273.2.1144. [DOI] [PubMed] [Google Scholar]
- 8.Laberge S, Cruikshank W W, Kornfeld H, Center D M. J Immunol. 1995;155:2902–2910. [PubMed] [Google Scholar]
- 9.Laberge S, Cruikshank W W, Beer D J, Center D M. J Immunol. 1996;156:310–315. [PubMed] [Google Scholar]
- 10.Rumsaeng V, Cruikshank W W, Foster B, Prussin C, Kirshenbaum A S, Davis T A, Kornfeld H, Center D M, Metcalfe D D. J Immunol. 1997;159:2904–2910. [PubMed] [Google Scholar]
- 11.Cruikshank W W, Center D M, Nisar N, Wu M, Natke B, Theodore A C, Kornfeld H. Proc Natl Acad Sci USA. 1994;91:5109–5113. doi: 10.1073/pnas.91.11.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Center D M, Kornfeld H, Cruikshank W W. Immunol Today. 1996;17:476–481. doi: 10.1016/0167-5699(96)10052-i. [DOI] [PubMed] [Google Scholar]
- 13.Cruikshank W W, Lim K, Theodore A C, Cook J, Fine G, Weller P F, Center D M. J Immunol. 1996;157:5240–5248. [PubMed] [Google Scholar]
- 14.Parada N A, Center D M, Kornfeld H, Rodriguez W L, Cook J, Vallen M, Cruikshank W W. J Immunol. 1998;160:2115–2120. [PubMed] [Google Scholar]
- 15.Baier M, Werner A, Bannert N, Metzner K, Kurth R. Nature (London) 1995;378:563. doi: 10.1038/378563a0. [DOI] [PubMed] [Google Scholar]
- 16.Zhou P, Goldstein S, Devadas K, Tewari D, Notkins A L. Nat Med. 1997;3:659–664. doi: 10.1038/nm0697-659. [DOI] [PubMed] [Google Scholar]
- 17.Idziorek T, Khalife O, Billaut-Mulot E, Hermann M, Aumercier M, Mouton Y. Clin Exp Immunol. 1989;112:84–91. doi: 10.1046/j.1365-2249.1998.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bisset L R, Rothen M, Joller-Jemelka H I, Dubs R W, Grob P J, Opravil M. AIDS. 1997;11:485–491. doi: 10.1097/00002030-199704000-00012. [DOI] [PubMed] [Google Scholar]
- 19.Bannert N, Adler H S, Werner A, Baier M, Kurth R. Immunogenetics. 1998;47:390–397. doi: 10.1007/s002510050374. [DOI] [PubMed] [Google Scholar]
- 20.Avots A, Hoffmeyer A, Flory E, Cimanis A, Rapp U R, Serfling E. Mol Cell Biol. 1997;17:4381–4389. doi: 10.1128/mcb.17.8.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmeyer A, Avots A, Flory E, Weber C K, Serfling E, Rapp U. J Biol Chem. 1998;273:10112–10119. doi: 10.1074/jbc.273.17.10112. [DOI] [PubMed] [Google Scholar]
- 22.Serfling E, Avots A, Neumann M. Biochim Biophys Acta. 1995;1263:181–200. doi: 10.1016/0167-4781(95)00112-t. [DOI] [PubMed] [Google Scholar]
- 23.Janknecht R, Hunter T. Nature (London) 1996;383:22–23. doi: 10.1038/383022a0. [DOI] [PubMed] [Google Scholar]
- 24.Yang C, Shapiro L H, Rivera M, Kumar A, Brindle P K. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laberge S, Ernst P, Ghaffar O, Cruikshank W W, Kornfeld H, Center D M, Hamid Q. Am J Respir Cell Mol Biol. 1997;17:193–202. doi: 10.1165/ajrcmb.17.2.2750. [DOI] [PubMed] [Google Scholar]
- 26.Hessel E M, Cruikshank W W, Van Ark I, De Bie J J, Van Esch B, Hofman G, Nijkamp F P, Center D M, Van Oosterhout A J. J Immunol. 1998;160:2998–3005. [PubMed] [Google Scholar]
- 27.Batchelor A H, Piper D E, de la Brousse F C, McKnight S L, Wolberger C. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 28.Graves B J. Science. 1998;279:1000–1001. doi: 10.1126/science.279.5353.1000. [DOI] [PubMed] [Google Scholar]
- 29.Eckner R. Biol Chem. 1996;377:685–688. [PubMed] [Google Scholar]
- 30.Torchia J, Glass C, Rosenfeld M G. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 31.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J-S, See R H, Deng T, Shi Y. Mol Cell Biol. 1996;16:4312–4326. doi: 10.1128/mcb.16.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flory E, Hoffmeyer A, Smola U, Rapp U R, Bruder J T. J Virol. 1996;70:2260–2268. doi: 10.1128/jvi.70.4.2260-2268.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clevers H C, Dunlap S, Wileman T E, Terhorst C. Proc Natl Acad Sci USA. 1988;85:8156–8160. doi: 10.1073/pnas.85.21.8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmon P, Giovane A, Wasylyk B, Klatzmann D. Proc Natl Acad Sci USA. 1993;90:7739–7743. doi: 10.1073/pnas.90.16.7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Böttinger E P, Shelley C S, Farokhzad O C, Arnaout M A. Mol Cell Biol. 1994;14:2604–2615. doi: 10.1128/mcb.14.4.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosmarin A G, Caprio D G, Kirsch D G, Handa H, Simkevich C P. J Biol Chem. 1995;270:23627–23633. doi: 10.1074/jbc.270.40.23627. [DOI] [PubMed] [Google Scholar]
- 38.Ying H, Chang J F, Parnes J R. J Immunol. 1998;160:2287–2296. [PubMed] [Google Scholar]
- 39.Lin J X, Bhat N K, John S, Queale W S, Leonard W J. Mol Cell Biol. 1993;13:6201–6210. doi: 10.1128/mcb.13.10.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markiewicz S, Bosselut R, Le Deist F, de Villartay J P, Hivroz C, Ghysdael J, Fischer A, de Saint Basile G. J Biol Chem. 1996;271:14849–14855. doi: 10.1074/jbc.271.25.14849. [DOI] [PubMed] [Google Scholar]
- 41.Mavrothalassitis G J, Papas T S. Cell Growth Differ. 1991;2:215–224. [PubMed] [Google Scholar]
- 42.Ogawa K, Burbelo P D, Sasaki M, Yamada Y. J Biol Chem. 1988;263:8384–8389. [PubMed] [Google Scholar]
- 43.Huhtala P, Chow L T, Tryggvason K. J Biol Chem. 1990;265:11077–11082. [PubMed] [Google Scholar]
- 44.Jolliff K, Li Y, Johnson L F. Nucleic Acids Res. 1991;19:2267–2274. doi: 10.1093/nar/19.9.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virbasius J V, Scarpulla R C. Mol Cell Biol. 1991;11:5631–5638. doi: 10.1128/mcb.11.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu M, Yang X Y, Schmidt T, Chinenov Y, Wang R, Martin M E. J Biol Chem. 1997;272:29060–29067. doi: 10.1074/jbc.272.46.29060. [DOI] [PubMed] [Google Scholar]