Abstract
Hepadnaviral reverse transcription requires template switches for the genesis of relaxed circular (RC) DNA, the major genomic form in virions. Two template switches, primer translocation and circularization, are required during the synthesis of the second, or plus, strand of DNA. Studies of duck hepatitis B virus (DHBV) indicate that in addition to the requirement for repeated sequences at the donor and acceptor sites, template switching requires at least three other cis-acting sequences, 5E, M, and 3E. In this study we analyzed a series of variant heron hepatitis B viruses (HHBV) in which the regions of the genome that would be expected to contain 5E, M, and 3E were replaced with DHBV sequence. We found that all single and double chimeras were partially defective in the synthesis of RC DNA. In contrast, the triple chimera was able to synthesize RC DNA at a level comparable to that of unchanged HHBV. These results indicate that the three cis-acting sequences, 5E, M, and 3E, need to be compatible to contribute to RC DNA synthesis, suggesting that these sequences interact during plus-strand synthesis. Second, we found that the defect in RC DNA synthesis for several of the single and double chimeric viruses resulted from a partial defect in primer translocation/utilization and a partial defect in circularization. These findings indicate that the processes of primer translocation and circularization share a mechanism during which 5E, M, and 3E interact.
Hepatitis B viruses infect the livers of a number of mammalian and avian species, resulting in acute and chronic liver diseases, such as hepatitis, cirrhosis, and hepatocellular carcinoma (for reviews see references 3, 8, and 24). Hepadnaviruses are small (∼3 kb), enveloped, circular double-stranded DNA viruses that replicate through an RNA intermediate via reverse transcription (27). Reverse transcription takes place within the viral nucleocapsid in the cytoplasm of the initially infected cell (27). The coencapsidation of pregenomic RNA (pgRNA) and the viral reverse transcriptase (P protein) into a nascent nucleocapsid (1, 2, 10) and the initiation of DNA synthesis (28, 29) are the first two steps in DNA replication and are likely to occur simultaneously. Both steps use the same trans factor, P protein, and cis element, epsilon (21, 29). After polymerization of 4 nucleotides (nt), the nascent minus-strand DNA switches templates to a position near the 3′ end of the pgRNA (20, 28, 29). Minus-strand DNA synthesis resumes with RNase H activity of the P protein (4, 22) degrading the pgRNA that has been copied into DNA (27). Minus-strand synthesis proceeds to the 5′ end of pgRNA template, resulting in a full-length minus strand (Fig. 1A) (13, 23, 30). The final RNase H cleavage leaves a short segment of RNA (18 or 19 nt) that will be the primer for the initiation of plus-strand synthesis (Fig. 1A) (12, 14). The 3′ end of the primer contains a 12-nt sequence called direct repeat 1, or DR1. A second copy of these 12 nt, called DR2, is found near the 5′ end of the minus strand. After completion of the minus strand, the replication pathway divides into two pathways that lead toward two different double-stranded DNA species (26). The dominant pathway leads to the synthesis of relaxed circular (RC) DNA (Fig. 1A through E). In this pathway the plus-strand primer translocates to DR2, where it will prime plus-strand DNA synthesis (Fig. 1B) (12). DR2 is within 50 nt of the 5′ end of the minus strand, and synthesis proceeds to the 5′ end of the template (Fig. 1C). To allow elongation past the 5′ end of the minus strand, the final template switch, called circularization, occurs (Fig. 1D) (12). The minus strand is terminally redundant for 7 or 8 nt (13). This redundancy, called r, is the donor and acceptor site for circularization. Once the 3′ end of the nascent plus strand anneals to 3′r, plus-strand synthesis resumes. Elongation will ultimately yield RC DNA (Fig. 1E). The second form of double-stranded DNA arises when the plus-strand synthesis initiates from DR1 (Fig. 1F). This type of synthesis, called in situ priming, yields a duplex linear (DL) form of the genome (26).
FIG. 1.
Synthesis of plus-strand DNA for DHBV. HHBV is likely to employ an identical strategy, except that the sequence of r is 5′-GTAATCT-3′. (A) Completion of synthesis of minus-strand DNA and generation of the RNA primer for the synthesis of plus-strand DNA. Black lines represent minus-strand DNA. White ovals labeled P represent the P protein, which is covalently attached to the 5′ terminus of the minus strand. The rectangles that contain nucleotide sequences represent the direct repeats, DR1 and DR2. The final RNase H cleavage during the synthesis of minus-strand DNA generates the RNA primer used for the initiation of plus-strand DNA synthesis. The primer is 18 or 19 nt and contains the DR1 at its 3′ end. (B) Primer translocation. For a majority of minus-strand DNA templates the plus-strand primer is translocated from DR1 to DR2. (C) Initiation of plus-strand DNA synthesis from DR2. DNA synthesis initiates from DR2 and proceeds to the 5′ end of template copying 5′r. Thin parallel lines represent plus-strand DNA. (D) Circularization. To permit elongation past the 5′ end of minus-strand DNA, the 3′ end of the nascent plus strand anneals to 3′r at the 3′ end of minus-strand DNA. (E) Resumption and elongation of plus-strand DNA synthesis yields RC DNA. (F) A small subset of minus-strand DNA templates have plus-strand DNA synthesis initiated from DR1. This is called in situ priming and generates a DL form of the genome.
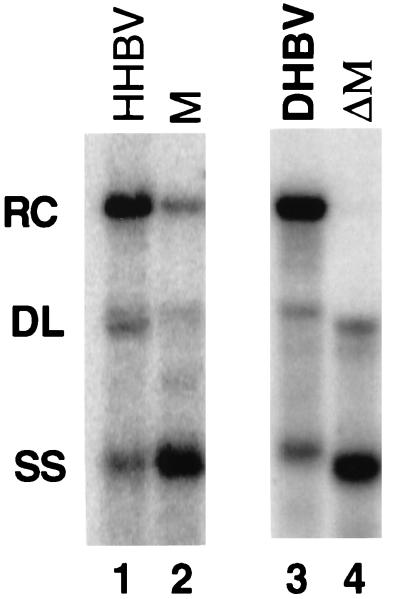
DNA replicative intermediates isolated from capsids from cells infected or transfected with duck hepatitis B virus (DHBV) or heron hepatitis B virus (HHBV) reveals, when analyzed by Southern blotting, a characteristic pattern of three major bands: RC DNA, DL DNA, and a 3-kb minus-strand species (SS) of DNA (Fig. 2, lanes 1 and 3). These three main bands are found at characteristic proportions for wild-type HHBV and DHBV, with RC DNA being the most abundant (Fig. 2, lanes 1 and 3). When one of the steps in plus-strand synthesis is inhibited, the proportions of the three forms is altered. For example, variants of DHBV have been described in which the identity of DR1 and DR2 has been altered (14). This variant synthesizes DL DNA instead of RC DNA. A variant in which the identity of r was changed accumulated an SS species at the expense of RC DNA (15). A region M variant of DHBV accumulated SS DNA at the expense of RC DNA (9). Thus, Southern blotting can reveal whether a viral variant is defective in template switching during plus-strand DNA synthesis.
FIG. 2.
When HHBV contains an M region from DHBV, less RC DNA is synthesized. Southern blotting was performed on viral DNA extracted from cytoplasmic capsids from transfected LMH cells. Lane 1, wild-type HHBV; lane 2, M chimera; lane 3, wild-type DHBV; lane 4, a variant of DHBV that is deficient in M function because nt 724 to 832 have been deleted. The viral DNA in lanes 1 and 2 was detected with a probe derived from nt 1165 to 2057 of HHBV that was specific for minus-strand DNA. Lanes 3 and 4 were hybridized with a genome-length DHBV probe that was specific for minus-strand DNA. Positions of RC, DL, and SS DNA are indicated.
In addition to DR1, DR2, 5′r, and 3′r, the viral genome contains other cis-acting sequences that are important for plus-strand DNA replication. The analysis of DHBV reverse transcription has identified three additional cis-acting sequences that participate in plus-strand DNA synthesis (9). One of these sequences, called M, lies near the middle of the minus strand. The other two regions, called 3E and 5E, are near the 3′ and 5′ ends of the minus-strand template, respectively. We have used chimeras of HHBV and DHBV to study the mechanism by which the three cis-acting sequences, 3E, M, and 5E, contribute to the synthesis of plus-strand DNA. Our analyses demonstrate that 3E, M, and 5E contribute to both plus-strand template switches, indicating that the mechanisms of primer translocation and circularization share a common component. In addition, our analyses demonstrate that 3E, M, and 5E need to be derived from the same virus to function properly, indicating that 3E, M, and 5E interact during plus-strand DNA synthesis. This work provides evidence for a template structure in which regions M, 3E, and 5E interact to juxtapose the ends of the minus-strand template to facilitate plus-strand primer utilization and circularization.
MATERIALS AND METHODS
Viruses and molecular clones.
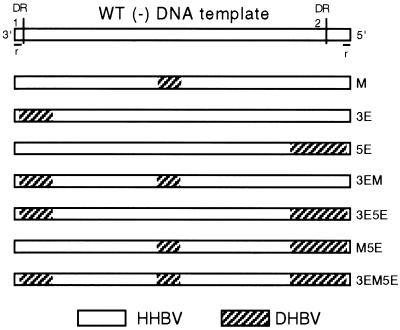
Molecular clones were derived from HHBV4 (25; accession number M22056) and DHBV16 (17; accession number K01834). The pgRNA of HHBV4 and its chimeric derivatives were expressed from a plasmid that contained 1.4 tandem copies of HHBV4 DNA (19). The pgRNA expressed from these plasmids was null for C and P protein production. A 4-nt insertion at the unique HindIII site (nt 38) prevented expression of C protein, and a 4-nt deletion at the unique BstXI site (nt 1439) ablated expression of P protein (19). Expression plasmids for both HHBV and DHBV replication proteins have been described previously (9, 19). In each of the chimeric clones, HHBV sequence was replaced with an analogous segment of DHBV16 sequence (Fig. 3). The M chimera was replaced between nt 718 to 823. To make its molecular clone, oligonucleotide-directed site-specific mutagenesis (11) was used to introduce an EcoRV site at nt 718 and an AatII site at nt 823 into HHBV4. Subsequently, an EcoRV to AatII restriction fragment from DHBV16 was introduced into HHBV4. DNA sequencing was performed to verify the absence of unwanted mutations. The individual 3E and 5E variants were constructed with restriction enzyme sites common to DHBV and HHBV. Standard procedures were used to make these molecular clones, and details describing their construction will be provided upon request. The molecular clone of the 3E variant expressed a pgRNA whose first 253 nt (nt 2535 to 2787 [BspLU11I site]) were DHBV, and the remainder were HHBV. Therefore, the 3E chimera expressed a minus-strand DNA that was replaced with 253 nt of DHBV sequence at its 3′ end. The molecular clone of the 5E variant expressed a pgRNA whose first 2,699 nt was HHBV followed by an insertion of DHBV sequence beginning at the SgrAI site (nt 2206) to its 3′ end. Therefore, the 5E chimera expressed a HHBV minus-strand DNA that was replaced with 338 nt of DHBV sequence at its 5′ end. The plasmids that expressed each of the double and triple chimeras were generated by replacing the appropriate restriction fragments with analogous fragments from individual single chimeras. Lastly, chimeras that contained either DHBV 3E or 5E substitutions had the DHBV r sequence change to the HHBV version. DHBV and HHBV r sequences differ: 5′-GTAATTCT-3′ versus 5′-GTAATCT-3′, respectively.
FIG. 3.
Structure and names of single, double, and triple chimeric viruses. Each rectangle is a linear representation of minus-strand DNA. The top rectangle represents wild-type (WT) HHBV with its 5′ and 3′ ends, with DR1, DR2, and r indicated. In each chimera the region replaced by DHBV sequence is indicated by diagonal shading. The name of each chimera is indicated on the right side.
Cell cultures, transfections, and isolation of replicative intermediates.
The chicken hepatoma cell line LMH was used in all cell culture experiments (7). Transfections were performed by the calcium phosphate method of Chen and Okayama (5). Typically, 6 μg of plasmid DNA was transfected into LMH cells that were at 50 to 70% of confluency on 60-mm-diameter plates. In all transfections, the ratio of protein donor plasmid to pgRNA expression plasmid was 1:1. Viral DNA was isolated from cytoplasmic core particles 3 days after the transfection. Transfected cells were lysed in 0.5 ml of a solution containing 10 mM Tris, 1 mM EDTA, and 0.2% NP-40 (pH 8.0). Nuclei and cellular debris were removed by pelleting in a microcentrifuge at 4°C. Cytoplasmic lysates were treated with a solution containing 22.5 μg of DNase I, 10 μg of RNase A, and 6 mM magnesium acetate for 1 h at 37°C, followed by a 1-h treatment at 37°C with 200 μg of pronase in a solution containing 5 mM EDTA, 100 mM NaCl, and 0.4% sodium dodecyl sulfate. Final purification of viral DNA was done by phenol-chloroform extraction and ethanol precipitation.
Southern blotting.
DNA electrophoresis (1.25% agarose in Tris-borate-EDTA buffer) and transfer to Hybond-N (Amersham) were carried out by using standard methods (18). Typically, 1/10 to 1/5 of a single transfection was analyzed. Hybridizations were carried out by the methods of Church and Gilbert (6). The radiolabeled probe was minus-strand specific and was derived from HHBV nt 1165 to 2057. All chimeras contained HHBV sequence over these nucleotides. Autoradiography was performed with a Molecular Dynamics PhosphorImager 445SI. The data in Tables 1 and 3 were derived by measuring the levels of RC, DL, and SS DNA for a virus and expressing each of the three DNA forms as a percentage of the total. A deficiency in primer translocation or circularization will lead to altered proportions of the three DNA forms.
TABLE 1.
Proportions of replicative intermediates measured by Southern blotting
| DNA form | % DNA form (mean ± SD) ina:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| HHBV | 3E | 5E | M | 3EM | 3E5E | M5E | 3EM5E | |
| RC | 59 ± 5 | 52 ± 11 | 30 ± 4 | 15 ± 5 | 41 ± 9 | 9 ± 4 | 31 ± 4 | 57 ± 6 |
| DL | 15 ± 2 | 12 ± 3 | 14 ± 1 | 8 ± 2 | 11 ± 4 | 8 ± 2 | 13 ± 1 | 13 ± 2 |
| SS | 26 ± 4 | 36 ± 9 | 56 ± 4 | 77 ± 6 | 48 ± 7 | 83 ± 5 | 56 ± 4 | 30 ± 5 |
Values are based on measurements made on intracellular viral DNA isolated from independent transfections; 24 for HHBV, 17 for 3E, 9 for 5E, 16 for M, 16 for 3EM, 15 for 3E5E, 9 for M5E, and 22 for 3EM5E. Two-sample separate variance t test was used to detect differences in means between HHBV and mutant values (mean percentage) for RC DNA. Statistical tests were carried out with Systat software. P values are as follows: 3E, 0.01; 5E, <0.001; M, <0.001; 3EM,<0.001; 3E5E, <0.001; M5E, <0.001; 3EM5E, 0.109.
TABLE 3.
Proportions of replicative intermediates measured by Southern blotting of chimeras replicated with DHBV P and C
| DNA form | % DNA form (mean ± SD) ina:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| DHBV | HHBV | 3E | 5E | M | 3EM | 3E5E | M5E | 3EM5E | |
| RC | 72 ± 8 | 33 ± 1 | 23 ± 1 | 11 ± 5 | 3 ± 1 | 15 ± 5 | 4 ± 2 | 14 ± 6 | 38 ± 7 |
| DL | 11 ± 2 | 14 ± 1 | 6 ± 2 | 16 ± 3 | 11 ± 3 | 10 ± 1 | 10 ± 2 | 12 ± 2 | 15 ± 1 |
| SS | 17 ± 7 | 53 ± 1 | 71 ± 1 | 73 ± 7 | 86 ± 4 | 76 ± 6 | 86 ± 4 | 74 ± 6 | 47 ± 7 |
Values are based on measurements made on intracellular viral DNA isolated from three independent transfections.
Primer extension analysis.
A detailed description of the primer extension analysis, its underlying rationale, and calculation of values derived from this analysis can be found in Loeb and Tian (16). Briefly, the primer extension reactions used Vent exo− thermostable DNA polymerase and 10 reaction cycles. Each sample was analyzed in three different primer extension reactions. An internal standard was added to each sample of viral DNA to permit the comparison of the level of viral DNA from the different reactions. The internal standard was the 320-nt HinfI fragment (nt 2318 to 2636) of HHBV DNA, which was from a plasmid. Each measurement of viral DNA was normalized to the level of the internal standard. The first primer extension reaction measured the level of minus-strand DNA in the sample, which defines the value C. The primer used in this reaction, called primer C, was derived from HHBV nt 2423 to 2439. From the second primer extension reaction the value B was derived, which was the level of plus-strand DNA that had initiated from DR2 and had extended at least to the 5′ end of the minus-strand template. The primer used in this reaction, called primer B, was derived from the sequence that is represented by the first 18 nt of minus-strand DNA. The third reaction was used to derive the value A, which represented the level of plus-strand DNA initiating from DR2 that had successfully circularized and elongated by at least an additional 85 nt. This primer, called primer A, was derived from HHBV nt 2605 to 2628. From these three primary values, ratios (expressed as percentages) were derived from three pair-wise comparisons of these three measurements (Table 2). The percentages in Table 2 reflect the efficiencies of priming from DR2 (B/C), priming from DR2, and circularization (A/C). The value A/B represents the efficiency of circularization.
TABLE 2.
Efficiency of plus-strand DNA synthesis as determined by primer extension of chimeric viruses replicated with HHBV P and C proteinsa
| Plus-strand eventb | Steps shown in Fig. 1 | % Efficiency (mean ± SD [P value]) for:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| HHBV | 3E | 5E | M | 3EM | 3E5E | M5E | 3EM5E | ||
| Priming from DR2c | A to C | 67 ± 15 (1.0) | 62 ± 20 (0.6) | 47 ± 14 (0.02) | 40 ± 16 (0.01) | 63 ± 12 (0.6) | 37 ± 12 (0.001) | 73 ± 22 (0.6) | 71 ± 14 (0.7) |
| Priming from DR2 and circularizationd | A to E | 30 ± 4 (1.0) | 25 ± 2 (0.02) | 17 ± 3 (<0.001) | 6 ± 1 (<0.001) | 18 ± 5 (0.001) | 5 ± 2 (<0.001) | 14 ± 5 (0.003) | 34 ± 5 (0.1) |
| Circularizatione | C to E | 45 | 40 | 31 | 15 | 29 | 14 | 19 | 48 |
Values are based on measurements made with intracellular viral DNA isolated from the following numbers of independent transfections: for HHBV, 7; for 3E, 5; for 5E, M, 3EM, 3E5E, M5E, and 3EM5E, 6. Two-sample separate variance t test was used to detect differences in means between HHBV and mutant values for priming from DR2 and for priming from DR2 and circularization. Statistical tests were carried out with Systat software.
See Materials and Methods for an explanation.
B/C value.
A/C value.
A/B value.
RESULTS
Aim, experimental design, and rationale.
Our aim was to learn how the three cis-acting elements, 3E, 5E, and M, contribute to the synthesis of plus-strand DNA. Our strategy was to use chimeras of HHBV and DHBV as tools to study the function of 3E, 5E, and M. A previous study indicated that an HHBV variant that contained an approximately 1,000-nt substitution of DHBV sequence within the middle of the minus strand had a cis-acting defect during the synthesis of plus-strand DNA (19). These findings indicated that the study of HHBV/DHBV chimeras could be informative. We asked whether 3E, 5E, and M need to be derived from the same virus for the synthesis of RC DNA synthesis. The necessity for 3E, 5E, and M to be compatible would suggest that these three cis-acting sequences interact, either directly or indirectly, to contribute to the synthesis of RC DNA synthesis. Our first goal was to determine whether we could use chimeras as tools to study the function of 3E, M, and 5E. Throughout our studies a common experimental strategy was used to examine the synthesis of viral DNA. LMH cells were transfected with two plasmids to initiate viral DNA synthesis. One plasmid expressed pgRNA that served as the initial template for DNA synthesis. Molecular clones that expressed pgRNA were modified to not express the viral C and P proteins, which are necessary for viral DNA synthesis. Instead, the replication proteins were translated from a variant pgRNA transcribed from the second plasmid which, due to a cis-defect in encapsidation, did not serve as a template for DNA synthesis (18). Several days after transfection of LMH cultures, viral DNA was isolated from cytoplasmic extracts and plus-strand DNA synthesis was evaluated by Southern blotting and primer extension.
HHBV containing DHBV sequence from nt 718 to 823 is defective for the synthesis of RC DNA.
A previous analysis indicated that a derivative of HHBV containing DHBV sequence from nt 403 to 1364 had a cis-acting defect for the synthesis of RC DNA (19). This analysis indicated that cis-acting sequences for RC DNA synthesis were within the altered region and that the DHBV version of these sequences could not function in the context of the remainder of HHBV template when acted on by HHBV C and P proteins. An independent study of DHBV indicated that removal of nt 724 to 832 resulted in a cis-acting defect for the synthesis of RC DNA (9). No other cis-acting sequences for RC DNA synthesis were found between nt 403 to 1364 in the analysis of DHBV, although the region between nt 455 to 723 could not be analyzed due to the presence of the second encapsidation signal. On the basis of both of these analyses the existence of cis-acting element M was proposed. We wanted to know if HHBV that only contained a DHBV M region was defective for RC DNA synthesis. To this end we constructed and analyzed an HHBV variant that contained DHBV sequence from nt 718 to 823, named M chimera (Fig. 3). Southern blotting of viral DNA isolated from cytoplasmic capsids indicated that M chimera supported the synthesis of substantially less RC DNA than did HHBV (Fig. 2, lanes 1 and 2, and Table 1). As a comparison, Southern blotting of DHBV and the variant of DHBV with a deletion of nt 724 to 832 can be seen in lanes 3 and 4 of Fig. 2. This analysis indicated that HHBV and DHBV have the same cis-acting sequence, called M, located at similar positions within their genomes. The M chimera pregenome did not support the synthesis of normal levels of RC DNA when it was replicated with DHBV C and P protein (Table 3). With this result in hand we asked whether region M needed to be compatible with region 3E and/or 5E for the synthesis of normal levels of RC DNA. To answer this question we analyzed a series of variants of HHBV that were substituted by DHBV either singly, doubly, or triply with M, 3E, and 5E (Fig. 3).
Single and double chimeras are partially defective for RC DNA accumulation.
Although previous studies of DHBV indicated the presence of the 3E and 5E cis-acting sequences, the extent and boundaries of these elements were not precisely determined (9). Because of this uncertainty we substituted relatively large sections of sequence when constructing the 3E and the 5E chimeras (Fig. 3). The minus-strand DNA expressed by the 3E chimera was replaced with 253 nt of DHBV sequence, from nt 2787 to the 3′ end (nt 2535). Minus-strand DNA has a terminal redundancy, named r, that participates in the template switch to circularize the genome (Fig. 1D and E). DHBV and HHBV r sequences differ: 5′-GTAATTCT-3′ versus 5′-GTAATCT-3′, respectively. Therefore, a chimera such as 3E would have a DHBV 3′r and a HHBV 5′r. To eliminate the potential that nonidentical r sequences would negatively affect circularization and therefore the synthesis of RC DNA, the DHBV r sequence in the 3E chimera was changed to match that of its HHBV counterpart by deleting a single nucleotide. The 5E chimera expressed a HHBV minus-strand DNA that was replaced by 338 nt of DHBV sequence from nt 2206 to the 5′ end (nt 2542). The DHBV little r sequence was changed to the HHBV sequence. Southern blotting of DNA extracted from cytoplasmic capsids of the 5E chimera indicated a reduction in the proportion of RC DNA (Fig. 4 and Table 1) and an increase in the proportion of SS DNA (56%, versus 26% for wild type). The 3E chimera synthesized slightly less RC DNA than unchanged HHBV (Fig. 4 and Table 1). Although the magnitude of the defect for both the 3E and 5E chimeras was not as great as that observed for the M chimera, these results indicated that analysis of the double and triple chimeras could be informative. Southern blot analysis of DNA extracted from cytoplasmic capsids of the doubly substituted viruses, 3E5E, 3EM, and M5E (Fig. 3), indicated that each made less RC DNA than the unchanged virus (Fig. 4 and Table 1). The proportion of RC DNA synthesized by 3EM and M5E was 41 and 31%, respectively (versus 59% for wild type). More strikingly, the proportion of RC DNA made by the 3E5E virus was only 9%. The magnitude of the defect for the 3E5E double chimera was substantially greater than that of each of the two constituent single substitutions, 3E and 5E. In general in the Southern blot analyses, when a chimera synthesized less RC DNA a concomitant increase in the proportion of SS DNA was seen (Fig. 4 and Table 1). Overall, the two viruses with the greatest defect in the synthesis of RC DNA were the M and 3E5E chimeras. This observation suggested that both ends of the minus-strand template need to be compatible with its middle for normal RC DNA synthesis.
FIG. 4.
Southern blotting of chimeras. The triple chimera, 3EM5E, is restored in its ability to synthesize RC DNA. Southern blotting was performed on viral DNA extracted from cytoplasmic capsids from transfected LMH cells. The blot was hybridized with a probe derived from nt 1165 to 2057 of HHBV that was specific for minus-strand DNA. WT, wild type.
The triple chimeric virus synthesizes RC DNA efficiently.
Southern blotting of intracellular DNA synthesized by the 3EM5E virus indicated RC DNA levels had been impressively restored. The proportion of RC DNA in the 3EM5E chimera was 57%, versus 59% for wild type (Fig. 4 and Table 1). These results are evidence that 3E, M, and 5E need to be compatible with each other to synthesize normal levels of RC DNA and suggest that the 3E, M, and 5E cis-acting sequences interact to contribute to the mechanism of RC DNA synthesis. In this experiment the pgRNA of 3EM5E was replicated with HHBV C and P proteins. Therefore, the ability of 3EM5E to synthesize normal levels of RC DNA indicated that an incompatibility between the HHBV replication proteins and the DHBV 3E, M, and 5E sequences did not exist, suggesting that the defect seen with single and double chimeras was not due to an incompatibility with the HHBV C and/or P protein.
M region contributes to both template switches during plus-strand DNA synthesis.
Although the M chimera virus was defective for RC DNA formation, Southern blot analyses did not indicate which step(s) in plus-strand DNA synthesis was inhibited. The accumulation of SS DNA in the Southern blot analysis suggested two likely possibilities. Plus-strand primers normally destined for DR2 were not utilized, resulting in the accumulation of full-length minus-strand DNA. Another possible reason for the accumulation of SS DNA would be failure to circularize plus strands that had primed from DR2. An analysis based on primer extension has been used to measure the extent to which a variant virus has primed plus-strand DNA from DR2 and then the extent to which those plus strands have circularized (9, 15, 16). This analysis relies on making three measurements with three different primer extension reactions and is illustrated in Fig. 5. The primer extension analysis was performed on unchanged HHBV and the M chimera variant and is summarized in Table 2. The level of plus-strand DNA initiating from DR2 normalized to the level of minus strand for the region M mutant was about 60% of that of the unchanged HHBV (Table 2, priming from DR2 value, 40% versus 67%). In addition, the M chimera had a defect in circularization (Table 2, circularization value). Only 15% of the plus strands that had initiated from DR2 and elongated to the 5′ end of minus-strand DNA had circularized. This result is in contrast to that for unchanged HHBV, which had circularized 45% of its plus-strand DNA primed from DR2. The cumulative effect of the two deficiencies was an 80% reduction, relative to values for wild-type HHBV, in the fraction of minus-strand DNA templates that were primed from DR2 and subsequently circularized. The M chimera virus did not support a measurable increase in plus-strand priming from DR1 as judged by Southern blotting (Table 1). Thus, the primer extension analysis of M chimera virus indicated that the reduction in the level of RC DNA was the result of two defects during plus-strand DNA synthesis. First, there was a reduction in the level of plus-strand DNA initiating from DR2, suggesting an inhibition of primer translocation/utilization without increased priming in situ. Second, for the plus strands that had translocated and initiated from DR2 a partial reduction in the level of circularization was measured. These results indicated that region M contributes to the processes of primer utilization/translocation and circularization, suggesting that the mechanisms of these two template switches share a common feature. Primer extension analysis of the 3E5E chimera revealed similar trends. 3E5E supported initiation of plus-strand DNA synthesis at 55% of the level of unchanged HHBV (Table 2). In addition, for 3E5E only 14% of the plus strands of DNA that initiated from DR2 circularized, in contrast to 45% for the wild type (Table 2). The net effect was an 83% reduction relative to values for unchanged HHBV in the percentage of minus-strand DNA templates that had plus-strand priming from DR2 and subsequently circularizing. In summary, the primer extension analysis indicated that for the M and 3E5E chimera viruses, both template switches during the synthesis of plus-strand DNA were affected.
FIG. 5.
The strategy to measure the extent to which a virus carries out primer translocation/utilization and circularization by primer extension. Three primers, named C, B, and A, are used. Primer C measures the level of minus-strand DNA. Primer B measures the level of plus-strand DNA initiated from DR2 and elongated to at least the 5′ end of the minus strand. Primer A measures the level of plus-strand DNA initiating from DR2 that has successfully circularized. (A) RC DNA will be detected with all three primers. (B) A replicative intermediate inhibited for circularization. It will be detected with primers C and B but not with primer A. (C) A replicative intermediate inhibited for primer translocation/utilization. It will be detected with primer C but not with primers A and B.
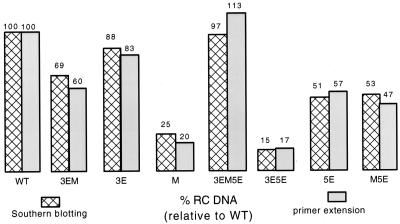
Primer extension analysis of the remaining single and double chimeras indicated that each of the variants were defective, to various degrees, for primer translocation/utilization and/or circularization, although the magnitude of the sum of both defects for the remaining single and double chimeras was less than that seen for the M or 3E5E viruses. In addition, in general a good agreement was seen between the results of the primer extension and those of the Southern blotting analyses. A decrease in the level of plus-strand DNA primed from DR2 and circularized as measured by primer extension was paralleled with a decrease in the proportion of RC DNA seen by Southern blotting (Fig. 6). The rank order of the ability of variants to prime plus-strand DNA synthesis from DR2 and subsequently circularize their genomes judged by primer extension was similar to the rank order of RC DNA synthesis as judged by Southern blotting. Lastly, the 3EM and M5E viruses primarily had defects in circularization.
FIG. 6.
Results from Southern blotting and primer extension analysis corroborate. Comparison of the proportion of RC DNA as determined by Southern blotting or the level of priming from DR2 and circularization as determined by primer extension between HHBV and the chimeras. In this comparison, all values are relative to that of the wild type (WT), which has been normalized to 100.
Primer extension analysis of the 3EM5E virus indicated that it carried out primer translocation/utilization and circularization at least as well as our unchanged HHBV. The findings from this analysis were consistent with those of the Southern blot studies and reinforce the interpretation that 3E, M, and 5E need to be compatible with each other for their proper function.
Replication of the chimeric viruses with DHBV P and C proteins results in more severe defects in plus-strand synthesis.
In the above analyses the chimeras were replicated with HHBV C and P proteins. We asked whether the magnitude of the defect in RC DNA synthesis for the chimeric viruses could be lessened if they were replicated with DHBV C and P protein. To answer this question we cotransfected expression plasmids for either HHBV, DHBV, or the various chimeric pgRNAs that were deficient for C and P protein production with a plasmid that expressed DHBV C and P protein. Replicative intermediates were isolated after 3 days, and Southern blotting was performed. A summary of this analysis is presented in Table 3. All of the chimeric viruses synthesized less RC DNA when replicated with DHBV replication proteins than when replicated with the HHBV counterparts. Consistent with this trend, when the HHBV pgRNA was reverse transcribed by the DHBV proteins a 50% decrease in the proportion of RC DNA was seen compared to that for replication with its endogenous proteins.
DISCUSSION
Our analysis shows that 3E, M, and 5E need to be compatible with each other for proper function. The requirement for compatibility strongly suggests that these cis-acting sequences interact with each other to carry out their function. Our analysis does not tell us the nature of the interaction or whether it is direct or indirect. The most pronounced mutant phenotypes were seen when the sequence at ends of the minus-strand template was from a different virus than the sequence of the M region (the 3E5E and M chimeras). Primer extension analysis of the 3E5E and M variants indicated that 3E, M, and 5E contribute to both primer translocation/utilization and circularization, indicating that the mechanisms of these template switches have a common component. Based on these conclusions we propose a model. The role of M is to interact simultaneously with 3E and 5E to position the ends of the minus-strand template to facilitate primer translocation and circularization.
The use of chimeric HHBV/DHBV viruses as tools to study plus-strand DNA synthesis has been informative. In particular, the M, 3E5E, and 3EM5E viruses all display striking phenotypes that lend themselves to straightforward interpretations. But the chimeric approach is not without limitations. In general, a mutant phenotype indicates that an important mechanism has been affected and suggests that the substituted sequence contains an important cis-acting sequence. But lack of a mutant phenotype cannot be interpreted to indicate the absence of the cis-acting element. It could mean that a cis-acting sequence is present in the substituted region but no functional incompatibility exists. For example, the small defect measured for the 3E virus cannot be interpreted to indicate that HHBV does not have a 3E element. In light of all of our results a more likely interpretation is that DHBV 3E is functional within the context of an HHBV M and 5E.
The ability of the 3EM5E triple chimeric virus to synthesize RC DNA at wild-type levels when replicated with HHBV P and C proteins means that either the replication proteins, C and P, do not interact with 3E, M, or 5E in a sequence-specific manner or that they do interact in a sequence-specific manner and an incompatibility between the HHBV replication proteins and the DHBV cis-elements does not exist. In a different analysis, we found that when the HHBV pregenome was replicated with DHBV proteins a 50% decrease in the proportion of RC DNA was seen (Table 3). This was not the case with the reciprocal complementation. A DHBV pgRNA replicated with HHBV replication proteins displayed normal levels of RC DNA (data not shown). The reason underlying the incompatibility between the DHBV replication proteins and the HHBV pgRNA is not clear, but it does not appear to be operating through 3E, M, and 5E.
Acknowledgments
We thank Bill Sugden for critical review of the manuscript.
This work was supported by National Institutes of Health grants R29 GM50263, P01 CA22443, P30 CA07175, and T32 CA09135 and American Cancer Society grant JFRA-651.
REFERENCES
- 1.Bartenschlager, R., M. Junker-Neipmann, and H. Schaller. 1990. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 64:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and H. Schaller. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 11:3413-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chisari, F. V., and C. Ferrari. 1995. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 13:29-60. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y., and P. L. Marion. 1996. Amino acids essential for RNase H activity of hepadnaviruses are also required for efficient elongation of minus-strand viral DNA. J. Virol. 70:6151-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condreay, L. D., C. E. Aldrich, L. Coates, W. S. Mason, and T. T. Wu. 1990. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J. Virol. 64:3249-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 9.Havert, M. B., and D. D. Loeb. 1997. cis-acting sequences in addition to donor and acceptor sites are required for template switching during synthesis of plus-strand DNA for duck hepatitis B virus. J. Virol. 71:5336-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch, R. C., J. E. Lavine, L. J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viurses are required for genomic RNA packaging as well as for reverse transcription. Nature 344:552-555. [DOI] [PubMed] [Google Scholar]
- 11.Kunkel, T. A., J. D. Roberts, and R. A. Zabour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 12.Lien, J.-M., Aldrich, C. E., and W. S. Mason. 1986. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J. Virol. 57:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lien, J. M., D. J. Petcu, C. E. Aldrich, and W. S. Mason. 1987. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J. Virol. 61:3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeb, D. D., R. C. Hirsch, and D. Ganem. 1991. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses; implication for other reverse transcribing elements. EMBO J. 10:3533-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeb, D. D., K. J. Gulya, and R. Tian. 1997. Sequence identity of the terminal redundancies on the minus-strand DNA template are necessary but not sufficient for the template switch during hepadnaviral plus-strand DNA synthesis. J. Virol. 71:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeb, D. D., and R. Tian. 2001. Mutations that increase in situ priming also decrease circularization for duck hepatitis B virus. J. Virol. 75:6492-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandart, E., A. Kay, and F. Galibert. 1984. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J. Virol. 49:782-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Mueller-Hill, K., and D. D. Loeb. 1996. Previously unsuspected cis-acting sequences for DNA replication revealed by characterization of a chimeric heron/duck hepatitis B virus. J. Virol. 70:8310-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nassal, M., and A. Rieger. 1996. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 70:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollack, J., and D. Ganem. 1994. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 68:5579-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radziwill, G., W. Tucker, and H. Schaller. 1990. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J. Virol. 64:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seeger, C., D. Ganem, and H. E. Varmus. 1986. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science 232:477-484. [DOI] [PubMed] [Google Scholar]
- 24.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprengel, R., E. Kaleta, and H. Will. 1988. Isolation and characterization of a hepatitis B virus endemic in herons. J. Virol. 62:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staprans, S., D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Summers, J., and W. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 28.Tavis, J. T., S. Perri, and D. Ganem. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, G., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Will, H., W. Reiser, T. Weimer, E. Pfaff, M. Büscher, R. Sprengel, R., Cattaneo, and H. Schaller. 1987. Replication strategy of human hepatitis B virus. J. Virol. 61:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]