Abstract
The Dsi1 locus was identified as a common integration site for Moloney murine leukemia virus (MLV) in rat thymic lymphomas, but previous efforts to identify a gene affected by these insertions were unsuccessful. We considered the Runx3 gene a potential candidate on the basis of genetic mapping which showed that Dsi1 and Runx3 are closely linked on mouse chromosome 4 and the precedent of the related Runx2 gene, which emerged recently as a Myc-collaborating gene activated by retroviral insertion in thymic lymphomas of CD2-MYC mice. We now report the physical mapping of the Dsi1 locus to a site 30 kb upstream of the distal (P1) promoter of the murine Runx3 gene. Comparison with the syntenic region of human chromosome 1 shows that the next gene is over 250 kb 5′ to Runx3, suggesting that Runx3 may be the primary target of retroviral insertions at Dsi1. Screening of CD2-MYC lymphomas for rearrangements at Dsi1 revealed a tumor cell line harboring an MLV provirus at this locus, in the orientation opposite that of Runx3. Proviral insertion was associated with very high levels of expression of Runx3, with a preponderance of transcripts arising at the P1 promoter. These results confirm that Runx3 is a target of retroviral insertions at Dsi1 and indicate that Runx3 can act as an alternative to Runx2 as a Myc-collaborating gene in thymic lymphoma.
Murine leukemia virus (MLV)-induced tumors have provided many examples of cellular proto-oncogene disruption by proviral integration. Moreover, as integration by these agents appears to be effectively random, the identification of preferred sites of clonal integration in tumor DNA has often served as an important first clue to the proximity of a gene whose deregulated expression confers a selective advantage on the tumor cell (reviewed in reference 16). The Dsi1 locus was identified as a novel common retroviral integration site in Moloney MLV-induced rat thymomas. Chromosomal localization studies placed Dsi1 on mouse chromosome 4 and suggested that it was distinct from any of the previously identified proto-oncogenes or common integration sites (9, 43). However, attempts to identify a gene affected by insertions at Dsi1 were inconclusive, despite the presence of prominent DNase I-hypersensitive sites and regions of evolutionary sequence conservation around this site (43).
Since these early studies were performed, knowledge of the mouse genome has grown considerably, and the number of genes identified as targets for activation (or inactivation) by retroviral insertional mutagenesis has also grown. Our recent studies identified Runx2 as a frequent target for proviral insertion in T-cell lymphomas following MLV infection of CD2-MYC mice (36). The Runx2 gene is itself oncogenic when overexpressed and shows particularly strong synergy with Myc (41); retroviral infection of CD2-Runx2 mice leads to acceleration of lymphoma onset, with a very high hit rate for integration at either c-Myc or N-Myc genes (7). MLV insertions at Runx2 were found to activate the distal (P1) promoter (36), which is normally active only in a restricted range of nonlymphoid tissues, including bone and testis (34, 36).
The Runx family (Runx1 to Runx3) of mammalian transcription factors control key events in developmental regulation and cell fate specification (reviewed in reference 35). The three Runx products (also known as core-binding factor α chains) have a common DNA-binding domain, runt, defined by homology with the Drosophila pair-rule gene Run (17), and interact with a common heterodimeric binding partner (Cbfβ) which both increases binding affinity and protects its α chain partners from proteolytic degradation (15, 28, 46). The Runx genes have a broadly similar exon-intron structure and a complex transcriptional pattern, resulting from alternative initiation, splicing, and polyadenylation (1, 2, 12, 23, 24, 26, 33). Two alternative transcription start sites have been described, a 5′-distal (P1) and a 3′-proximal (P2) site (13, 33, 36), which generate the two major N-terminal isoforms (P1/MASN and P2/MRIPV). Localization of the two alternative promoters reveals that they are separated by a large distance, 160, 90, and 30 kb for RUNX1, RUNX2, and RUNX3, respectively .
The Runx3 gene has been mapped to murine chromosome 4 (8) within the same chromosomal domain as Dsi1. However, the presence of a number of recombinants between these loci in interspecific backcross mice indicated that their physical linkage might not be close. To explore this relationship further, we examined the physical linkage of Dsi1 and Runx3 in murine P1-derived bacteriophage artificial chromosome (PAC) recombinant libraries. Our study shows that these loci are in fact very closely linked and implicate Runx3 as the target for insertions at Dsi1.
MATERIALS AND METHODS
Transgenic mice and cell lines.
The CD2-MYC transgenic mice were generated as described previously (37). The lymphoma cell lines T1i, T14i, T29i, T48i, T82i and T85i were established from thymic lymphomas induced following neonatal MLV infection of CD2-MYC mice. The murine T-cell line EL4 was obtained from the European Collection of Animal Cell Cultures (Salisbury, England).
PACs.
A mouse genomic PAC library (RPC121) (30) consisting of approximately 128,899 clones and gridded in 4 by 4 arrays on a nylon membrane was obtained from the United Kingdom Human Genome Mapping Project Resource Centre. The seven filters were hybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C overnight, with [α-32P]dCTP-labeled ds1-SR and Runx3 probes. Following washing at high stringency (3 times for 20 min each, 60°C in 0.5× SSC with 0.5% SDS) and exposure to X-ray film, positive clones were identified and glycerol stabs were obtained from the United Kingdom Human Genome Mapping Project Resource Centre. PAC DNA was isolated using the Qiagen plasmid purification maxi kit with modifications described by the manufacturers.
Southern and Northern analysis.
Preparation of high-molecular-weight DNA and total RNA from tumor-derived cell lines and subsequent DNA and RNA hybridization analysis were as previously described (41).
RESULTS AND DISCUSSION
Dsi1 locus maps ≈30 kb upstream of the P1 promoter region of murine Runx3 gene.
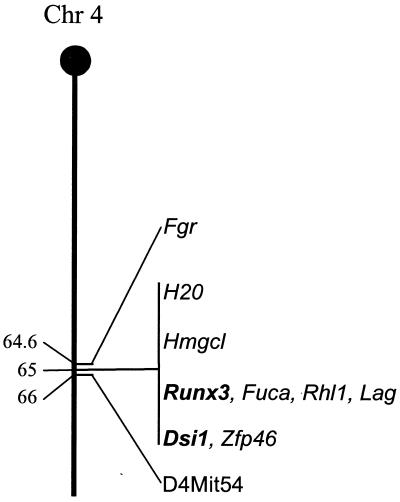
Previous genetic linkage studies with recombinant inbred mice located both Dsi1 and Runx3 within a 3.3-centimorgan (cM) region on mouse chromosome 4 (8), while a recent search of the genetic linkage maps of mouse chromosome 4 (http://www.informatics.jax.org) placed these two loci much closer, within 0.2 cM of each other (Fig. 1). To explore the possibility that the loci are in very close proximity, we screened a murine PAC library (RPC121; United Kingdom Human Genome Mapping Project Resource Centre) with probes specific for Dsi1 (ds1-SR [43]) and Runx3 (exon 1, specific for the 5′ untranslated region [UTR] of the P1 promoter region, and exon 5, immediately downstream of the runt domain). A number of clones were isolated, of which two, PAC 448-L9 and PAC 568-D12, were positive for the three probes used.
FIG. 1.
Genetic linkage map of murine chromosome 4, illustrating close proximity of the Dsi1 locus and Runx3 gene. Also indicated are additional genes and anchor markers mapped in the same region. Distances are in centimorgans from the centromere.
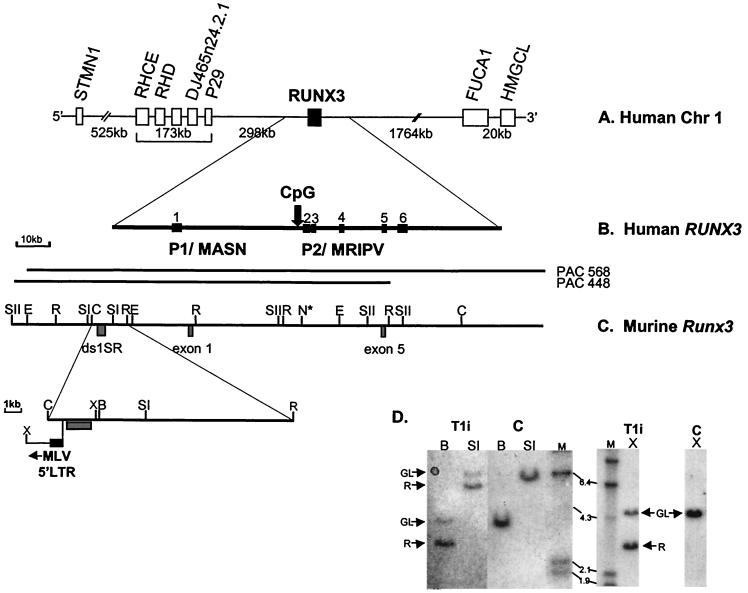
The PAC clones were characterized by pulsed-field gel electrophoresis (PFGE) of restriction digests (data not shown), and preliminary restriction mapping of the clones identified a single 105-kb ClaI fragment positive for ds1-SR and Runx3 exon 1 and exon 5. Further analysis demonstrated that the ds1-SR and exon 1 probes lie on adjacent genomic EcoRI fragments (Fig. 2C). This analysis revealed that the Dsi1 locus is only 30 kb upstream of the P1 region of the Runx3 gene.
FIG. 2.
Comparative physical map of human RUNX3 and murine Runx3 genes. (A) Physical map of human chromosome 1 spanning 2.7 Mb around the RUNX3 gene, based on public database information (http://genome.ucsc.edu). All of the genes indicated are transcribed in the same orientation as RUNX3. (B) Genomic structure of the human RUNX3 gene. The locations of the distal P1 and proximal P2 promoter regions are indicated. Vertical arrow, P2-associated CpG island. (C) Physical map of murine Runx3 gene. PAC 448-L9 and PAC568-D12 were isolated from the murine RPC121 PAC library. ds1-SR and the Runx3 exon 1 and exon 5 probes are indicated. Only restriction sites mapped with the probes are shown. N* indicates the presence of multiple NotI sites and the inferred position of P2-associated CpG island from draft murine genome sequence (accession number AF169246). Expanded region shows the position of the integrated MLV in T1i. Restriction enzyme abbreviations: SI, SstI; SII, SstII; E, EagI; N, NotI; C, ClaI; R, EcoRI; X, XhoI; B, BamHI. (D) Southern analysis of MLV-infected CD2-MYC lymphomas. Genomic DNA (20 μg; T1i cell line and control [C] nontransgenic thymus) was digested with BamHI (B), SstI (SI), and XhoI (X) and hybridized with the ds1-SR probe. GL, germ line; R, rearranging fragment. HindIII-digested λ DNA was used as molecular size markers (sizes shown in kilobases).
The PAC clones were mapped further with several rare-cutting endonucleases (NotI, EagI, BssHI, and SstII) that specifically cleave CpG sequences. Clusters of these endonuclease cleavage sites identify the presence of CpG islands, which are associated with the 5′ ends of mammalian genes (6). PFGE and conventional agarose gel electrophoresis of the resulting digests generated only large restriction fragments following hybridization with the ds1-SR and Runx3 exon 1 and exon 5 probes, revealing no evidence of CpG clusters in the regions covered by the probes. However, analysis of the draft murine genome sequence around the P2-proximal transcriptional start site sequence (accession number AF169246) reveals a cluster of sites ≈15 kb upstream of exon 5 (Fig. 2C). This CpG island is conserved across species and is found in the equivalent region of the human RUNX3 gene (Fig. 2B) (24). In contrast, no CpG island was observed in the vicinity of the upstream P1 promoters of either the murine Runx3 gene or its human homologue.
Comparison of the overall physical structure of the region around the human and murine Runx3 genes reveals a high degree of organizational conservation (Fig. 1 and 2A). A number of genes in both species map to within similar regions around the Runx3 gene (FUCA1 and Fuca, RHCE and Rhl1, and STMN1 and Lag). Inspection of the draft human genome sequence 5′ to the RUNX3 gene reveals that the closest neighbor lies 298 kb upstream. Assuming that the overall genomic architecture is conserved between mammalian species, Runx3 will be by far the closest gene to the Dsi1 locus.
Rearrangement of the murine Dsi1 locus due to provirus insertion: mapping of the inserted element and its orientation with respect to Runx3.
Previous attempts to analyze the consequences of insertions at the rat Dsi1 locus were confounded by the fact that only limited amounts of primary tumor material were available for RNA analyses (43). We therefore decided to look for further examples of proviral insertion at the Dsi1 locus, with emphasis on tumor-derived cell lines. Analysis of a series of cell lines derived from rat thymic lymphomas provided no further examples (S. Bear and P. Tsichlis, personal communication).
In view of the frequent activation of the related Runx2 gene by MLV insertion in CD2-MYC mouse lymphomas (7), we decided to screen a panel of these tumors and derived cell lines for Dsi1 rearrangements by Southern analysis. Screening with the ds1-SR probe revealed rearranging genomic bands in tumor T1i (Fig. 2D). It is interesting that the intensity of the germ line bands observed in the T1i cell line DNA are submolar with respect to control DNA. This was not the case in the original tumor, where germ line and rearranging bands were equimolar (data not shown), suggesting that in vitro propagation of cells derived from the primary tumor has allowed the expansion of a subpopulation of tumor cells which have amplified the rearranged allele, possibly due to chromosomal duplication.
Analysis of the restriction digests mapped the MLV integration immediately upstream of the ds1-SR region. The transcriptional orientation of the inserted element was determined by digestion with XhoI, which cuts asymmetrically within the Moloney MLV genome. Analysis of the T1i XhoI digestion pattern (Fig. 2D) reveals a rearranging 3-kb fragment, consistent with proviral integration in the opposite transcriptional orientation to the Runx3 gene.
Vijaya et al. (43) reported regions of evolutionary sequence conservation detected by the ds1-SR probe. Our analysis of the human genome draft sequence reveals a fragment of chromosome 1 which displays 88.7% homology over 293 bp (with a number of gaps in the alignment) to the rat ds1-SR sequence. The region of chromosome 1 identified lies 35.5 kb upstream of the P1 region of the RUNX3 gene. This analysis reveals that the proviral insertion mapped in the murine T-cell lymphoma T1i and in the rat thymomas RT13-1, RT2-2, and RT10-2 studied by Vijaya et al. lie in the same orientation with respect to the ds1-SR sequence. In T1i, restriction analysis locates the viral 5′ long terminal repeat (LTR) ≈30 kb upstream of the Runx3 P1 promoter. Additional screening of the tumor series with the Runx3 exon 1 probe revealed no further genomic rearrangements. However, proviral insertion at Dsi1/Runx3 appears to be a rare event, unlike insertion at Runx2, where analysis of the same tumor series revealed viral insertion in 16 of 58 tumors examined (5).
T1i cell line expresses very high levels of Runx3 transcripts originating from the P1 promoter.
The integration of MLV upstream and in the opposite orientation to the Runx3 P1 promoter suggested that this might result in cis activation mediated by viral LTR enhancers (19). In order to address this possibility, Northern blot analysis was performed on RNA from MLV-infected CD2-MYC tumors and derived cell lines. The probes used included a Runx3 exon 5 probe, which detects both P1- and P2-derived transcripts, and a Runx3 exon 1 probe, which detects P1-derived transcripts only (Fig. 3A). The exon 5 probe detected a 4-kb transcript across the range of cell lines examined. This observation is in agreement with previous studies, which demonstrated the widespread expression of Runx3 in a variety of hematopoietic cell lines (21, 25) and tissues (33). Two cell lines derived from tumors T1i and T14i showed markedly increased levels of this transcript (Fig. 3B). Even more strikingly, the exon 1 probe revealed abundant transcripts only in two cell lines, T1i and T14i. Densitometry analysis confirmed that the levels of P1-derived transcript were indeed increased in both T1i and T14i when normalized with respect to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. 3C). Because of the unique specificity of the two probes, this analysis clearly demonstrates that P1-derived transcripts are generally far less abundant than P2-derived transcripts. Of interest is the observation that the P1 promoter is active in normal mouse thymus, albeit at lower levels than P2. The sizes of the transcripts detected by the Runx3 probes revealed no abnormality, suggesting that the overexpressed mRNA may be otherwise intact.
FIG. 3.
Expression of murine Runx3 transcripts. (A) Schematic representation of Runx3 exon structure and derived RNA transcripts. Solid boxes represent exons 1 to 6, and open boxes indicate 5′ and 3′ untranslated sequences. Exon 1- and exon 5-specific probes are indicated by hatched and grey boxes, respectively. (B) Northern blot analysis of 20 μg of total RNA from T1i, T14i T29i, T48i, T82i, T85i, and EL4 cells and from control nontransgenic thymus (Thy) was performed using Runx3 probes, exon 1 (upper) and exon 5 (lower), and GAPDH to control for RNA loading. The blot was hybridized sequentially with the exon 1, exon 5, and GAPDH probes, with stripping between hybridizations. The exon 1 probe detects only those transcripts derived from the P1 promoter, whereas the exon 5 probe detects both P1- and P2-derived transcripts. (C) Graphic representation of expression levels of P1-specific transcripts normalized with respect to GAPDH expression.
The unusual pattern of expression in T1i is consistent with the interpretation that MLV insertion at Dsi1 has resulted in activation of the Runx3 P1 promoter. This mode of activation appears similar to that at Runx2, where MLV insertions upstream of and in the opposite orientation to the P1 promoter have been observed (36). The reason for high expression of Runx3 from P1 in tumor T14i is not yet clear. Screening of ≈40 kb of genomic sequence around the Dsi1 and Runx3 distal promoter regions has so far failed to reveal evidence of rearrangement. It is conceivable, however, that longer-range activation can occur, as such effects have been noted at the Mlvi1 and Cb1/Fim3 loci, which appear to activate the c-Myc and Evi1 genes from a greater distance (4, 20, 40).
Runx genes: a family of interchangeable Myc-collaborating genes?
Our previous studies have shown that Runx2 overexpression in the T-cell compartment predisposes to lymphoma development and is strongly synergistic with MYC (41). Moreover, both c- and N-myc are the preferred targets for provirus insertion following analysis of insertion sites in MLV-induced lymphomas in CD2-Runx2 transgenic mice (7). Together, these data strongly suggest that the Runx2-Myc combination favors tumor development in the T-cell compartment. The results presented in this study confirm Runx3 as a target for retroviral insertion at Dsi1 and indicate that Runx3 can act as an alternative to Runx2 as a Myc-collaborating gene in thymic lymphoma.
In mammals, three Runx genes have been identified, compared to one gene (runt) in Drosophila, for which studies have illuminated the essential roles of these factors in development (39). The three mammalian genes have distinct lineage-specific functions, as revealed by knockout phenotypes and analysis of normal expression patterns. Mice null for Runx1 or Cbfb display embryonic lethality and a profound block in definitive hematopoiesis (29, 44, 45). This observation is complemented by studies which show expression of Runx1 in a wide range of hematopoietic cells (34). Although low levels of Runx2 expression have been detected in T cells and in nonlymphoid cells, such as 3T3 and Buffalo rat liver cells, the major site of expression is in cells of the bone-forming lineages (11, 36). Mice null for Runx2 show a complete lack of bone formation (18, 31). Runx3 is more widely expressed (21, 25) and, like Runx1 and Runx2, also appears to play an important role in tissue development. Homozygous deletion of Runx3 leads to a profound defect in gastrointestinal tract differentiation (Y. Ito, personal communication).
However, all three genes are expressed in normal thymus, albeit at different levels, and both Runx2 and Runx3 appear to act as targets for MLV activation by a similar mechanism. This situation appears to be analogous to c-Myc and N-Myc, essential genes which are expressed in a tissue-specific manner (for a review, see reference 22) but can be activated by MLV in lymphomas in an apparently interchangeable fashion (10, 27, 42).
A common feature of MLV insertion at Runx2 and Runx3 appears to be the specific overexpression of P1 isoforms. This could be the result of selection for efficient activation, as the P1 and P2 promoters differ in structure and temporal regulation. However, there is also evidence to suggest that both the Runx1 and Runx2 P1 isoforms differ from their P2 counterparts in biological function (3, 14, 38) and that Runx1 N-terminal isoforms differ in both translational efficiency (32) and DNA-binding affinity (38). Future studies will focus on the significance of Runx3 promoter shift and P1 isoform overexpression for thymic development and lymphomagenesis.
Acknowledgments
The Cancer Research Campaign and Leukemia Research Fund provided support for this work.
We thank the United Kingdom Human Genome Resource Centre for murine PAC clones and David Steffen for providing the ds1-SR probe. We thank Alma Jenkins for excellent technical assistance and Karen Blyth for critical reading of the manuscript.
REFERENCES
- 1.Ahn, M.-Y., S.-C. Bae, M. Maruyama, and Y. Ito. 1996. Comparison of the human genomic structure of the runt domain-encoding PEBP2/CBFα family. Gene 168:279-280. [DOI] [PubMed] [Google Scholar]
- 2.Bae, S. C., E. Takahashi, Y. W. Zhang, E. Ogawa, K. Shigesada, Y. Namba, M. Satake, and Y. Ito. 1995. Cloning, mapping and expression of PEBP2 alpha C, a third gene encoding the mammalian Runt domain. Gene 159:245-248. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, C., A. Javed, J. Y. Choi, J. Green, V. Rosen, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2001. Differential regulation of the two principal Runx2/Cbfa1 N-terminal isoforms in response to bone morphogenetic protein 2 during development of the osteoblast phenotype. Endocrinology 142:4026-4039. [DOI] [PubMed] [Google Scholar]
- 4.Bartholomew, C., and J. N. Ihle. 1991. Retroviral insertions 90 kilobases proximal to the evi-1 myeloid transforming gene activate transcription from the normal promoter. Mol. Cell. Biol. 11:1820-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxter, E., K. Blyth, E. R. Cameron, and J. C. Neil. 2001. Selection for loss of p53 function in T-cell lymphomagenesis is alleviated by Moloney murine leukemia virus infection in myc transgenic mice. J. Virol. 75:9790-9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bird, A. 1987. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 3:342-347. [Google Scholar]
- 7.Blyth, K., A. Terry, N. Mackay, F. Vaillant, M. Bell, E. R. Cameron, J. Neil, and M. Stewart. 2001. Runx2: a novel oncogenic effector revealed by in vitro complementation and retroviral tagging. Oncogene 20:295-302. [DOI] [PubMed] [Google Scholar]
- 8.Calabi, F., M. Rhodes, P. Williamson, and Y. Boyd. 1995. Identification and chromosomal mapping of a third mouse runt-like locus. Genomics 26:607-610. [DOI] [PubMed] [Google Scholar]
- 9.Ceci, J. D., L. D. Siracusa, N. A. Jenkins, and N. G. Copeland. 1989. A molecular linkage map of mouse chromosome 4 including the localization of several proto-oncogenes. Genomics 5:699-709. [DOI] [PubMed] [Google Scholar]
- 10.Dolcetti, R., S. Rizzo, V. de Re, G. Feriotto, and M. Biocchi. 1989. N-myc activation by proviral insertion in MCF247-induced murine T-cell lymphomas. Oncogene 8:1009-1014. [PubMed] [Google Scholar]
- 11.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 12.Geoffroy, V., D. A. Corral, L. Zhou, B. Lee, and G. Karsenty. 1998. Genomic organization, expression of the human CBFA1 gene, and evidence for an alternative splicing event affecting protein function. Mamm. Genome 9:54-57. [DOI] [PubMed] [Google Scholar]
- 13.Ghozi, M., Y. Bernstein, V. Negreanu, D. Levanon, and Y. Groner. 1996. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc. Natl. Acad. Sci. USA 93:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harada, H., S. Tagashira, M. Fujiwara, S. Ogawa, T. Katsumata, A. Yamaguchi, T. Komori, and M. Nakatsuka. 1999. Cbfa1 isoforms exert functional differences in osteoblast differentiation. J. Biol. Chem. 274:6972-6978. [DOI] [PubMed] [Google Scholar]
- 15.Huang, G., K. Shigesada, K. Ito, H. Wee, T. Yokomizo, and Y. Ito. 2001. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 20:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonkers, J., and A. Berns. 1996. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim. Biophys. Acta 1287:29-57. [DOI] [PubMed] [Google Scholar]
- 17.Kania, M. A., A. S. Bonner, J. B. Duffy, and J. P. Gergen. 1990. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 4:1701-1713. [DOI] [PubMed] [Google Scholar]
- 18.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y.-H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 19.Kung, H.-J., C. Boerkel, and T. H. Carter. 1991. Retroviral mutagenesis of cellular oncogenes: a review with insights into the mechanisms of insertional activation. Curr. Top. Microbiol. Immunol. 171:1-25. [DOI] [PubMed] [Google Scholar]
- 20.Lazo, P. A., J. S. Lee, and P. N. Tsichlis. 1990. Long-distance activation of the Myc protooncogene by provirus insertion in Mlvi-1 or Mlvi-4 in rat T-cell lymphomas. Proc. Natl. Acad. Sci. USA 87:170-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le, X., Y. Groner, S. M. Kornblau, Y. Gu, W. N. Hittleman, D. Levanon, K. Mehta, R. B. Arlinghaus, and K. S. Chang. 1999. Regulation of AML2/CBFA3 in hematopoietic cells through the retinoic acid receptor alpha-dependent signaling pathway. J. Biol. Chem. 274:21651-21658. [DOI] [PubMed] [Google Scholar]
- 22.Lemaitre, J., R. S. Buckle, and M. Mechali. 1996. c-Myc in the control of cell proliferation and embryonic development. Adv. Cancer Res. 70:95-144. [DOI] [PubMed] [Google Scholar]
- 23.Levanon, D., Y. Bernstein, V. Negreanu, M. Ghozi, I. Baram, R. Aloya, D. Goldenberg, D. Lotem, and Y. Groner. 1996. A large variety of alternatively spliced and differentially expressed messenger RNAs are encoded by the human acute myeloid leukemia gene AML1. DNA Cell Biol. 15:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Levanon, D., G. Glusman, T. Bangsow, E. Ben-Asher, D. A. Male, N. Avidan, C. Bangsow, M. Hattori, T. D. Taylor, S. Taudien, K. Blechschmidt, N. Shimizu, A. Rosenthal, Y. Sakaki, D. Lancet, and Y. Groner. 2001. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene 262:23-33. [DOI] [PubMed] [Google Scholar]
- 25.Levanon, D., V. Negreanu, Y. Bernstein, I. Bar-Am, L. Avivi, and Y. Groner. 1994. AML1, AML2, and AML3, the human members of the runt domain gene family: cDNA structure, expression, and chromosomal localization. Genomics 23:425-432. [DOI] [PubMed] [Google Scholar]
- 26.Miyoshi, H., M. Ohira, K. Shimizu, K. Mitani, H. Hirai, T. Imai, K. Yokoyama, E. Soeda, and M. Ohki. 1995. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 23:2762-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroy, T., S. Verbeek, A. Ma, P. Achacoso, A. Berns, and F. Alt. 1991. E mu N- and E mu L-myc cooperate with E mu pim-1 to generate lymphoid tumors at high frequency in double-transgenic mice. Oncogene 6:1941-1948. [PubMed] [Google Scholar]
- 28.Ogawa, E., M. Inuzuka, M. Maruyama, M. Satake, M. Naito-Fujimoto, Y. Ito, and K. Shigesada. 1993. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology 194:314-331. [DOI] [PubMed] [Google Scholar]
- 29.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 30.Osoegawa, K., M. Tateno, P. Woon, E. Frengen, A. Mammoser, J. Catanese, Y. Hayashizaki, and P. de Jong. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10:116-128. [PMC free article] [PubMed] [Google Scholar]
- 31.Otto, F., A. P. Thornell, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. H. Stamp, R. S. P. Beddington, S. Mundlos, B. R. Olsen, P. B. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765-771. [DOI] [PubMed] [Google Scholar]
- 32.Pozner, A., D. Goldenberg, V. Negreanu, S.-Y. Le, O. Elroy-Stein, D. Levanon, and Y. Groner. 2000. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell. Biol. 20:2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rini, D., and F. Calabi. 2001. Identification and comparative analysis of a second runx3 promoter. Gene 273:13-22. [DOI] [PubMed] [Google Scholar]
- 34.Satake, M., S. Nomura, Y. Yamagachi-Iwai, Y. Takahama, Y. Hashimoto, M. Niki, Y. Kitamura, and Y. Ito. 1995. Expression of the runt domain-encoding PEBP2α genes in T cells during thymic development. Mol. Cell. Biol. 15:1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speck, N. A., T. Stacy, Q. Wang, T. North, T. L. Gu, J. D. Miller, M. Binder, and M. Marin-Padilla. 1999. Core-binding factor: a central player in hematopoeisis and leukemia. Cancer Res. 59:S1789-S1793. [PubMed] [Google Scholar]
- 36.Stewart, M., A. Terry, M. O'Hara, K. Blyth, E. Baxter, E. Cameron, D. Onions, and J. C. Neil. 1997. Proviral insertions induce the expression of bone-specific isoforms of PEBP2αA (CBFA1): evidence for a new myc collaborating gene. Proc. Natl. Acad. Sci. USA 94:8646-8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart, M. A., E. Cameron, S. Toth, M. Campbell, R. McFarlane, D. Onions, and J. C. Neil. 1993. Conditional expression and oncogenicity of a human c-myc transgene linked to a human CD2 dominant control region. Int. J. Cancer 53:1023-1030. [DOI] [PubMed] [Google Scholar]
- 38.Telfer, J. C., and E. V. Rothenberg. 2001. Expression and function of a stem cell promoter for the murine CBFA2 gene: distinct roles and regulation in natural killer and T cell development. Dev. Biol. 229:363-382. [DOI] [PubMed] [Google Scholar]
- 39.Tracey, W. D., and N. A. Speck. 2000. Potential role for RUNX1 and its orthologs in determining hematopoietic cell fate. Cell Dev. Biol. 11:337-342. [DOI] [PubMed] [Google Scholar]
- 40.Tsichlis, P. N., D. Shepherd, and S. E. Bear. 1989. Activation of the mlvi-1/mis-1/pvt-1 locus in Moloney murine leukemia virus-induced T-cell lymphomas. Proc. Natl. Acad. Sci. USA 86:5487-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaillant, F., K. Blyth, A. Terry, M. Bell, E. R. Cameron, J. Neil, and M. Stewart. 1999. A full-length Cbfa1 gene product perturbs T-cell development and promotes lymphomagenesis in synergy with MYC. Oncogene 18:7124-7134. [DOI] [PubMed] [Google Scholar]
- 42.van Lohuizen, M., S. Verbeek, P. Krimpenfort, J. Domen, C. Saris, T. Radaszkiewicz, and A. Berns. 1989. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell 56:673-682. [DOI] [PubMed] [Google Scholar]
- 43.Vijaya, S., D. L. Steffen, C. Kozak, and H. L. Robinson. 1987. Dsi-1, a region with frequent proviral insertions in Moloney murine leukemia virus-induced rat thymomas. J. Virol. 61:1164-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Q., T. Stacy, J. D. Miller, A. F. Lewis, T. L. Gu, X. Huang, J. H. Bushweller, J. C. Bories, F. W. Alt, G. Ryan, P. P. Liu, A. Wynshaw-Boris, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell 87:697-708. [DOI] [PubMed] [Google Scholar]
- 46.Wang, S., Q. Wang, B. E. Crute, I. N. Melnikova, S. R. Keller, and N. A. Speck. 1993. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 13:3324-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]