Abstract
Bovine leukemia virus (BLV) and human T-cell lymphotropic virus type 1 (HTLV-1) belong to the genus of deltaretroviruses. Their entry into the host cell is supposed to be mediated by interactions of the extracellular (SU) envelope glycoproteins with cellular receptors. To gain insight into the mechanisms governing this process, we investigated the ability of SU proteins to interact with specific ligands. In particular, by affinity chromatography, we have shown that BLV SU protein specifically interacted with zinc ions. To identify the protein domains involved in binding, 16 peptides distributed along the sequence were tested. Two of them appeared to be able to interact with zinc. To unravel the role of these SU regions in the biology of the virus, mutations were introduced into the env gene of a BLV molecular clone in order to modify residues potentially interacting with zinc. The fusogenic capacity of envelope mutated within the first zinc-binding region (104 to 123) was completely abolished. Furthermore, the integrity of this domain was also required for in vivo infectivity. In contrast, mutations within the second zinc-binding region (218 to 237) did not hamper the fusogenic capacity; indeed, the syncytia were even larger. In sheep, mutations in region 218 to 237 did not alter infectivity or viral spread. Finally, we demonstrated that the envelope of the related HTLV-1 was also able to bind zinc. Interestingly, zinc ions were found to be associated with the receptor-binding domain (RBD) of Friend murine leukemia virus (Fr-MLV) SU glycoprotein, further supporting their relevance in SU structure. Based on the sequence similarities shared with the Fr-MLV RBD, whose three-dimensional structure has been experimentally determined, we located the BLV zinc-binding peptide 104-123 on the opposite side of the potential receptor-binding surface. This observation supports the hypothesis that zinc ions could mediate interactions of the SU RBD either with the C-terminal part of SU, thereby contributing to the SU structural integrity, or with a partner(s) different from the receptor.
Bovine leukemia virus (BLV) and human T-cell lymphotropic virus type 1 (HTLV-1) belong to the genus of deltaretroviruses (70). HTLV-1 is the etiologic agent of adult T-cell leukemia (40, 58, 76, 83) and HTLV-1-associated myelopathy or tropical spastic paraparesis (36, 43, 56). BLV causes enzootic bovine leukemia, a chronic and contagious disease that evolves over an extended period of time, with tumors developing in a small number of infected cattle but in the majority of infected sheep (12, 46, 52, 54). One of the main interests of the BLV system is that it allows direct experimentation in cattle and sheep as a model for HTLV-1-induced pathogenesis (77, 78).
Like other retroviruses, HTLV-1 and BLV enter the host cell through interactions of their envelope glycoproteins with specific cellular receptors, a process leading to fusion of the virus and cell membranes and followed by delivery of the viral genome into the cytoplasm. To date, these receptors remain unknown despite several extensive attempts (1, 34, 35, 41, 48, 64). Nevertheless, the HTLV-1 receptor is a widely expressed protein (44, 55, 68, 71). Integrins and adhesion molecules are involved in HTLV-1 biology, but these proteins are not the primary receptor (22, 38, 39). For BLV, a candidate receptor has been isolated (3), but its murine homolog was later shown to encode an adaptor protein associated with intracellular vesicles (69).
The HTLV-1 and BLV envelopes are multimeric complexes composed of a surface receptor-binding subunit (SU; gp46 for HTLV-1 and gp51 for BLV) associated with a transmembrane protein (TM; gp21 for HTLV-1 and gp30 for BLV). Similar to other retroviruses, binding of SU protein to the receptor(s) triggers conformational changes resulting in projection of the TM amino-terminal fusion peptide into the target cell membrane. HTLV-1 gp21 contains in its cytoplasmic domain a YSLI motif that is essential for cell-to-cell transmission and involved in a postfusion step (25). BLV gp30 harbors, in its cytoplasmic domain, similar YXXL motifs sharing similarities with the immunoreceptor tyrosine-based inhibition and activation motifs (ITIM and ITAM, respectively) (17, 23, 62). These gp30 YXXL motifs are implicated in vitro in signal transduction pathways (6), are required for in vivo infection and maintenance of viral loads (79), and play critical roles in viral entry and incorporation of envelope into the virions (42). Furthermore, BLV gp30 has been shown to interact physically with phosphatase SHP-1 (18).
Conservation of TM proteins from several viruses, as supported by sequence alignments and crystallographic studies, underlines common features in the structure and the mode of action of retroviral fusion proteins (15, 32, 33, 47). In particular, the HTLV-1 gp46 and BLV gp51 heads possibly interacting with the cellular receptor have been predicted to adopt a structure corresponding to the overall topology of constant immunoglobulin domains (15).
In 1997, Fass and coworkers published the structure of the receptor-binding domain (RBD) of a mammalian C-type retrovirus glycoprotein (31). This RBD encompasses the amino-terminal half of the Friend murine leukemia virus (Fr-MLV) SU glycoprotein and reveals a conserved immunoglobulin-like antiparallel β-sheet framework, in which is embedded a variable subdomain proposed to serve as the receptor-binding surface. Interestingly, zinc ions were found to be associated with the crystals. Nevertheless, the presence of these ions could be due to the crystallization conditions, and the relevance of the zinc ion-binding sites was unclear.
In the present report, we have shown that the BLV SU glycoprotein specifically interacts with zinc ions. To identify envelope SU regions involved in this binding, peptides distributed along the gp51 sequence were tested, and two of them were able to interact with zinc. In order to position one of these peptides, we aligned the BLV gp51 RBD domain with the corresponding Fr-MLV RBD. To gain insight into the role of these regions in the biological properties of the virus, mutations were introduced to modify residues potentially interacting with zinc, and the effect of these mutations on the fusogenic capacity and in vivo infectivity was tested. Finally, we have shown that the HTLV-1 envelope SU protein also interacted with zinc ions.
MATERIALS AND METHODS
Zinc affinity chromatography and ELISA.
Affinity chromatography was performed with 400 μl of Chelating Sepharose Fast Flow (Amersham Biosciences). Columns were used either uncharged (as a negative control) or after loading with 5 mg of ZnCl2/ml in aqueous solution. Chromatography was performed according to the manufacturer's instructions. The load and wash solution was 20 mM sodium phosphate-500 mM NaCl-0.5% Tween 20, pH 7.5. The elution solution had the same composition except that EDTA was added at a concentration of 250 mM.
Fetal lamb kidney (FLK) cells constitutively expressing BLV were cultivated in minimal essential medium (MEM) (Life Technologies) supplemented with 10% inactivated fetal calf serum and antibiotics. The supernatant was harvested after 3 days, and 300 μl was mixed with 750 μl of load and wash buffer. An aliquot of 200 μl was taken as a control. The mix was loaded on the top of the metal affinity column. An 800-μl flowthrough fraction was collected, and after extensive washes with 5 volumes of column, elution buffer was used to collect two fractions of eluate (400 μl). All fractions were precipitated by addition of 0.5 volume of a 30% polyethylene glycol (PEG) 8000-0.4 M NaCl solution. The samples were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12.5% polyacrylamide), transferred to Immobilon-P membranes (Millipore Corporation), and analyzed by Western blotting with antibodies directed against the gp51 protein (A, B, B′, C, D, D′, and E [10]). Secondary antibodies coupled with alkaline phosphatase were incubated and subsequently revealed after incubation with CDP-Star Western blot chemiluminescence reagent (NEN Life Science). Alternatively, the titers of gp51 SU glycoproteins present in the fractions were determined by an enzyme-linked immunosorbent assay (ELISA) procedure as previously described (60).
MT2 HTLV-1-producing cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (Life Technologies) and antibiotics. The protocol used for chromatography is described above. The antibody used for Western blot was the gp46-specific MF2 antibody (51).
Synthetic gp51 peptides (5 to 25 μg) were diluted in 250 μl of load and wash solution, 30-μl aliquots were taken, and the samples were loaded onto 200-μl chelating columns. The 200-μl flowthrough, wash, and eluate fractions were harvested, and their titers for gp51 peptides were determined by ELISA with specific rabbit antisera (59). The aliquots and the fractions recovered from the affinity columns were diluted to 820 μl with phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM NaH2PO4, 1.47 mM KH2PO4, pH 7.4). Ninety-six-well immunoplates (Maxisorb; Nunc) were coated at room temperature with the mixtures (eight 100-μl wells for each fraction). Four hours later, the wells were washed twice with PBS-0.2% Tween 80, and serial threefold dilutions of the peptide-specific rabbit antisera were added to the wells in the presence of bovine serum albumin (0.67%) and Tween 80 (1.33%). After an overnight incubation at 4°C and three washes, the presence of bound antibodies was revealed by protein A-linked peroxidase (100 ng per well for 30 min). After four washes with water, peroxidase activity was determined by adding 100 μl of 3,3′,5,5-tetramethyl-benzidine microwell peroxidase substrate (KPN). The reactions were arrested after 15 min of incubation in the dark by adding 100 μl of 2 N H2SO4, and the optical densities were measured at 450 and 690 nm.
Comparative sequence analysis, and relationships with three-dimensional structures.
Sequences of SU proteins from BLV-HTLV-1 and Fr-MLV are difficult to align with current alignment procedures because they have diverged too much. For this reason, we used hydrophobic cluster analysis (14), which adds information about secondary structures. Indeed, the use of a two-dimensional helical representation of protein sequences (see the inset in Fig. 3A) allows us to contour hydrophobic amino acids (V, I, L, M, F, Y, and W) into hydrophobic clusters whose positions statistically match those of regular secondary structures (82). These clusters and their successive correspondences into globular domains are much more conserved than sequences, constitute robust signatures of folds, and allow us to recognize distant but significant relationships.
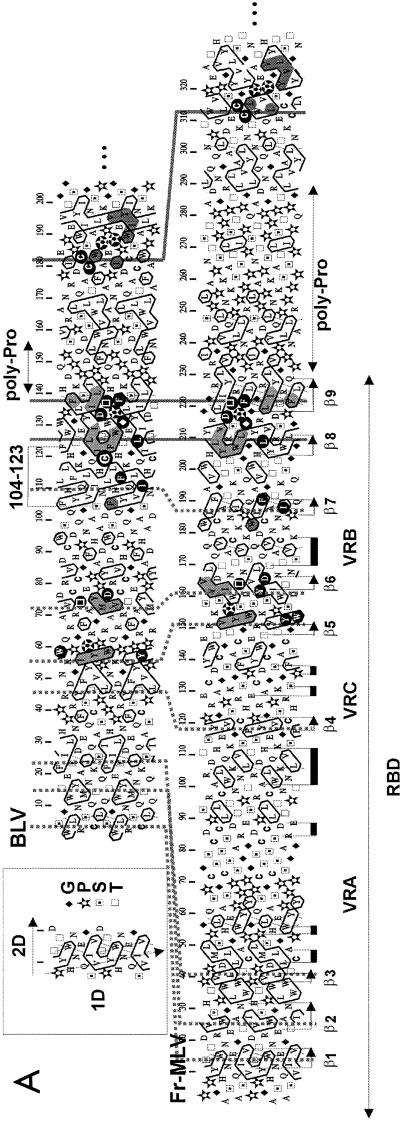
FIG. 3.
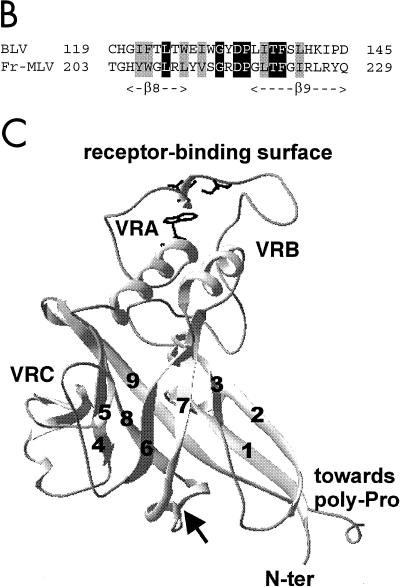
Sequence-structure relationships of the BLV gp51 protein. (A) Hydrophobic cluster analysis of the SU N-terminal ends. Sequences are shown on a duplicated α-helical net, where hydrophobic amino acids (V, I, L, F, M, Y, and W) are contoured (14). These residues form clusters that statistically correspond to the internal faces of regular secondary structures (α-helices and β-strands) (82). The way to read sequences (1D) and secondary structures (2D) and the symbols used for particular amino acids are indicated in the upper left inset. The Fr-MLV receptor-binding domain (RBD) and its secondary structures (arrows for the β-sheets and black boxes for the α-helices) are indicated below the Fr-MLV sequence. The position of the BLV 104-123 peptide is shown, and the C119 residue is circled. Both Fr-MLV and BLV RBD sequences end with a region rich in proline residues (poly-Pro), indicating the presence of a hinge separating distinct domains. A region located just before the hinge is particularly well conserved between Fr-MLV and BLV and encompasses strands β8 and β9 of the Fr-MLV RBD. Identities are indicated in white on a black background, and cluster similarities are shaded in gray. Sequences upstream of β8 and β9 are less conserved, and the variable regions (VRA to VRC), poor in hydrophobic residues, are reduced in BLV gp51. Proposed correspondences between the different strands of the immunoglobulin-like domain are indicated with stippled lines. (B) Alignment of the BLV and Fr-MLV SU sequences encompassing strands β8 and β9 of the Fr-MLV RBD structure. Cysteine 119 is located just upstream of strand β8. Identities and cluster similarities are indicated as in panel A. (C) Ribbon representation of the three-dimensional structure of the Fr-MLV RBD, corresponding to the N-terminal part of the SU protein (PDB 1AOL [31]). Strands are labeled 1 to 9, and variable regions VRA to VRC are indicated. Following the alignment presented in A, the cysteine present in peptide 104-123 (circled in panel A) should be located between strands β7 and β8 (see arrow). The position of the receptor-binding surface is shown, as well as three amino acids (S84, D86, and W102) which were shown to be critical for receptor binding and infection (24).
Crystallographic data were taken from the Protein Data Bank (8). Visualization of three-dimensional structures was performed with a Swiss-Pdb viewer (37).
Plasmid constructions.
Plasmid pBLVIX, which contains the wild-type provirus BLV344, was described elsewhere (80). To construct other recombinant proviruses, mutations were first introduced into the wild-type envelope gene (AF503581) cloned into plasmid pCMVenv (79, 80). These mutations were performed by site-directed mutagenesis with a two-step PCR procedure essentially as described previously (79). The following sense (S) and complementary (C) oligonucleotides that contain the selected mutations were used: C119A-S, 5′-TCTCAAACAAGCTCATGGAATTTT-3′; C119A-C, 5′-AAAATTCCATGAGCTTGTTTGAGA-3′; C179A+C182A-S, 5′-CCCAGACGCTGCTATAGCTTGGGAACC-3′; C179A+C182A-C, 5′-GGTTCCCAAGCTATAGCAGCGTCTGGG-3′; H229A+H230A-S, 5′-CCCAAGGATGGGCCGCCCCTTCCCAGA-3′; and H229A+H230A-C, 5′-TCTGGGAAGGGGCGGCCCATCCTTGGG-3′.
A first round of PCRs allowed the amplification of two fragments encompassing the env gene, a 5′-end insert (with the T7 primer and one of the complementary C oligonucleotides) and a 3′-end fragment (with the SP6 primer and the corresponding sense oligonucleotide). After PCR, the two inserts were migrated onto an agarose gel, purified with the Sephaglas bandprep kit (Amersham Biosciences), and amplified in a second round of PCR with the SP6 and T7 oligonucleotides. The resulting DNAs, which contain the entire env gene sequence, were then digested with the HindIII and XbaI restriction endonucleases and introduced into the corresponding sites of the pGem7 plasmid (Promega). In order to verify the presence of the desired substitutions and the absence of Taq DNA polymerase errors, the mutated fragments were sequenced by the dideoxy chain termination procedure with a set of primers located along the env gene (T7 sequencing kit; Amersham Biosciences).
The mutated env sequences were then cloned in the NcoI and XbaI sites of plasmid pBLVIX to generate pBLVC119A, pBLVC179A+C182A, and pBLVH229A+H230A. Restriction analysis and nucleotide sequencing were performed to verify the integrity of the resulting constructs. To obtain the proviruses mutated within both regions (pBLVC119A+H229A+H230A and pBLVC179A+C182A+H229A+H230A), site-directed mutagenesis was performed at residues H229 and H230 on plasmids pGem7envC119A and pGem7envC179A+C182A, respectively. The resulting fragments were reintroduced into the proviruses as described above. These plasmids were then purified by centrifugation on a cesium chloride gradient as described elsewhere (65).
Plasmids pSFVenvIX and pSFVenvHind, which encode the wild-type BLV env gene, are isogenic to the pSFVenvWT (33). To construct the plasmids that express the mutated envelopes (pSFVenvC119A, pSFVenvC179A+C182A, pSFVenvH229A+H230A, pSFVenvC119A+H229A+H230A, and pSFVenvC179A+C182A+H229A+H230A), the NcoI-BamHI env fragments (at positions 4925 to 6997, following the numeration of Rice et al. [63]) were isolated from the recombinant proviruses and inserted in the corresponding sites of pSFVenvWT.
Expression of recombinant BLV envelopes into BHK cells and syncytium formation assay.
General procedures for the Semliki Forest virus (SFV) expression system were described previously (50). pSFV plasmids containing the wild-type or recombinant envelope gene were linearized with the SpeI restriction endonuclease and used as templates for in vitro transcription (33). The purified RNAs were then electroporated into 4 × 106 baby hamster kidney (BHK) cells by two consecutive pulses (830 V, 40 μF, maximum resistance) with a Cellject electroporation system (Eurogentec). After transfection, the cells were immediately diluted in 24 ml of G-MEM medium (Life Technologies) complemented with 10% heat-inactivated serum. Three and a half million cells were cultivated at 37°C for 60 h for the Western blot experiments.
For the syncytium formation assay, the remaining cells (5 × 105) were plated into 5-cm dishes. After 4 h of cultivation to allow their attachment to the dishes, these cells were cocultivated for 20 h with 2 × 106 CC81 indicator cells in order to evaluate their fusion capacity. After culture, the cells were washed with PBS and fixed for 10 min at room temperature in a solution containing 25% acetic acid and 75% methanol. After three washes with water, the multinucleated cells were colored with Giemsa's azur eosin methylene blue for microscopy (Merck) in 50% glycerol-50% methanol solution. Syncytia containing more than 4 nuclei were counted under the light microscope in 10 different fields at ×100 magnification, and the numbers of nuclei were counted for each mutant envelope protein within 20 syncytia at ×200 magnification.
In vivo infection, proviral loads, and semiquantitative PCR.
The recombinant pBLV plasmids were mixed with a cationic lipid and injected into sheep as described previously (81). The sheep were maintained under controlled conditions at the Veterinary and Agrochemical Research Center (Machelen, Belgium). Sera were collected weekly and analyzed for BLV seroposivity by immunodiffusion and ELISA (60).
To evaluate the proviral loads, blood samples (500 μl) were mixed with an equal volume of freshly prepared lysis buffer [0.32 M sucrose, 10 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 1% Triton X-100]. The samples were centrifuged for 2 min, and the pellets were resuspended in 500 μl of lysis buffer by vortexing. This step was repeated until the pellets were clear. The samples were then resuspended in 500 μl of PCR buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl, pH 8.3) and incubated with 2 μl of proteinase K (15 mg/ml) for 1 h at 50°C. Five-microliter aliquots were amplified by PCR in the presence of 200 μM each of the four deoxynucleotide triphosphates, 200 ng of primers, and 1 U of Taq DNA polymerase (Roche). The two oligonucleotides used were PCREA (5′-TCCTGGCTACTAACCCCCCCGT-3′ at position 4560 according to the numeration of Rice et al. [63]) and DREX4 (5′-CCCCAACCAACAACACTTGCTT-3′ at position 7060). The reaction mixtures were overlaid with 75 μl of mineral oil, denatured for 5 min at 94°C, and amplified by 29 cycles of PCR (30 s at 94°C, 45 s at 57°C, and 2 min at 72°C). After PCR, the samples were analyzed by Southern blot hybridization with a BLV probe (SacI insert from plasmid pBLV344).
To ensure that the viruses which propagated in vivo were not revertants, the envelope sequences were amplified by 36 PCR cycles with the oligonucleotides PCREA and DREX4. The env genes were then directly sequenced with the primer 5360 (5′-TGGGTCAACACGTCCTTGT-3′, position 5360 according to the numeration of Rice et al. [63]) with the double-stranded DNA cycle sequencing system (Life Technologies).
RESULTS
BLV envelope SU glycoprotein binds zinc.
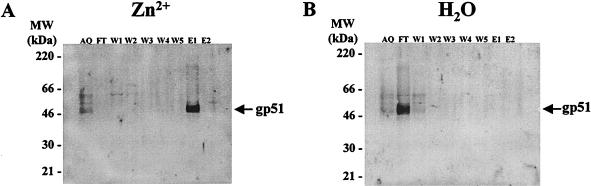
In order to evaluate the capacity of BLV SU protein to interact with zinc, cell culture supernatants from BLV-producing FLK cells were chromatographed on a zinc affinity column, and the resulting fractions (aliquot, flowthrough, wash, and eluate) were concentrated by PEG precipitation. The concentrated fractions were analyzed by gel electrophoresis, followed by immunoblotting with antibodies specific for gp51. The BLV SU envelope glycoprotein was able to specifically interact with the zinc column as it was only recovered in the eluate fraction (E1, Fig. 1A). To eliminate the possibility of a nonspecific gp51 binding to the Sepharose fast flow gel, FLK cell supernatant was chromatographed on an uncharged column matrix. It appeared that no binding occurred on the chelating matrix devoid of zinc (Fig. 1B).
FIG. 1.
BLV envelope SU glycoprotein interacts with zinc. (A) Chelating Sepharose beads charged with Zn2+ ions were incubated with cell culture supernatant from BLV-producing FLK cells. An aliquot (AQ) was taken before loading the column. The flowthrough fraction (FT) was harvested, and the column was washed five times with load and wash buffer (20 mM sodium phosphate, 500 mM NaCl, 0.5% Tween 20, pH 7.5). Wash fractions W1 to W5 were harvested. Finally, elution fractions (E1 and E2) were collected after addition of elution buffer (the same composition as load and wash buffer except that EDTA was added at a concentration of 250 mM). The material was concentrated by PEG precipitation and migrated onto an SDS-polyacrylamide gel. After electroblotting of the proteins, the nitrocellulose membrane was first incubated with monoclonal antibodies directed against BLV gp51 and subsequently with a secondary antibody conjugated to alkaline phosphatase. The presence of the BLV envelope SU protein (gp51) was then visualized by chemiluminescence. (B) As a control, the chelating Sepharose fast flow gel was incubated with water and the procedure described for panel A was followed.
Metals such as Cu2+, Co2+ and Mn2+ can potentially replace Zn2+ in protein interactions (66). In order to find ions that could replace zinc for the gp51 binding, we performed similar experiments with a matrix charged with other divalent ions. It appeared that gp51 specifically binds to Sepharose fast flow gel charged with Co2+, Cu2+, and Ni2+ but not with Mn2+, Fe2+, Pb2+, Cd2+, Mg2+, or Ca2+ ions (data not shown).
Identification of peptides within the gp51 sequence exhibiting zinc ion binding activities.
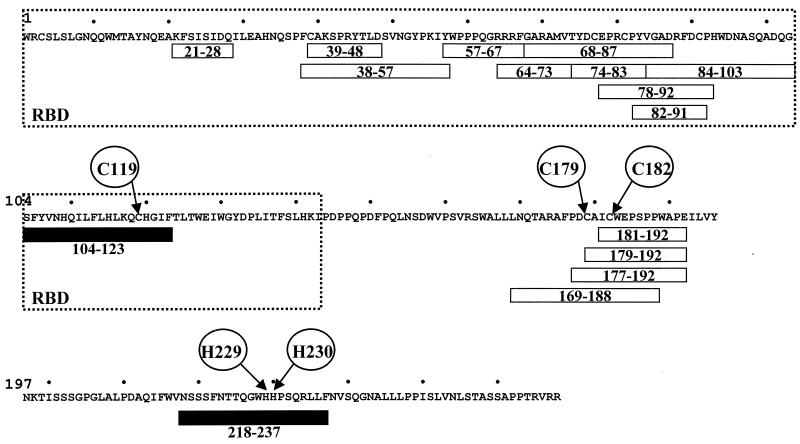
In order to identify SU envelope regions involved in Zn2+ binding, peptides distributed along the gp51 sequence were tested for their ability to bind chelating Sepharose fast flow gel charged with Zn2+ ions. A series of peptides (21-28, 38-57, 39-48, 57-67, 64-73, 68-87, 74-83, 78-92, 82-91, 84-103, 104-123, 169-188, 177-192, 179-192, 181-192 and 218-237; see Fig. 2) were tested for their ability to bind zinc ions. The concentrations of the peptides in the column fractions were determined by an ELISA procedure with specific antisera (see Materials and Methods). Among the 16 peptides tested, 2 of them, peptides 104-123 and 218-237, were able to bind zinc (Fig. 2 and data not shown). Peptide 104-123 contains three histidine residues and one cysteine (C119) likely to bind zinc. Peptide 218-237 does not include any cysteines but harbors two histidine residues (H229 and H230). Like the entire gp51 glycoprotein, these two peptides were also able to bind to Ni2+, Co2+, and Cu2+ ions (data not shown).
FIG. 2.
Two peptides within BLV SU glycoprotein specifically bind zinc. The sequence of BLV SU protein is presented without considering the signal peptide (residue 1 corresponding to the precursor cleavage site) (15). A series of 16 peptides encompassing the SU protein (peptides 21-28, 38-57, 39-48, 57-67, 64-73, 68-87, 74-83, 78-92, 82-91, 84-103, 104-123, 169-188, 177-192, 179-192, 181-192, and 218-237) (16, 59) were tested for their ability to bind chelating Sepharose charged with Zn2+ ions. The concentration of the peptides in the different fractions was determined by an ELISA procedure with specific antisera (13, 16, 59). Peptides 104-123 and 218-237 were able to specifically interact with zinc ions (black boxes), while other peptides exhibited no zinc-binding activity (open boxes). Within peptides 104-123 and 218-237, residues C119, H229, and H230, potentially interacting with zinc, are indicated. Conserved amino acids C179 and C182 are also circled. Localization of the BLV receptor-binding domain (RBD), deduced from the sequence alignment with the Fr-MLV RBD, is surrounded.
Sequence analysis of the BLV gp51 protein.
To date, the only oncoviral SU protein for which experimental information about atomic structure is available is that of Fr-MLV (31). The information is, however, limited to its N-terminal receptor-binding domain (RBD), leaving the C-terminal part as yet unexplored at the structural level. Therefore, we compared the BLV gp51 sequence to that of Fr-MLV in order to localize the position of the BLV zinc-binding peptides on a three-dimensional model predicted by inference relative to Fr-MLV. However, as the two proteins are highly divergent and could not be aligned by current alignment procedures, we used bidimensional hydrophobic cluster analysis to assess the putative structural similarities between the BLV gp51 and the Fr-MLV gp70.
The procedure of alignment was helped by the consideration of other retroviral SU sequences, particularly the HTLV-1 gp46. The Fr-MLV gp70 and BLV gp51 sequences both possess a central region rich in proline residues (poly-Pro, Fig. 3A), which ends the Fr-MLV RBD sequence and indicates the presence of a hinge separating distinct domains. Some striking correspondences of hydrophobic clusters between the BLV gp51 and the Fr-MLV gp70 sequences (shaded in Fig. 3A), accompanied by sequence identities (white letters on a black background), were observed just before and after the poly-Pro hinge. Upstream of the poly-Pro hinge, striking similarities were indeed observed in the gp70 region, which includes strands β8 and β9 (Fig. 3A and B). In addition, a putative correspondence was also noticed between the BLV gp51 sequence and Fr-MLV strands β5, β6, and β7 (Fig. 3A), which together form, with strands β8 and β9, the core of the Fr-MLV RBD antiparallel β-sandwich (Fig. 3C). This observation strongly supported a shared three-dimensional fold for the two RBD.
The amino-terminal region of the RBD domains, however, appeared quite different in these retroviral SU proteins. Indeed, variable regions present in Fr-MLV (VRA to VRC) were obviously absent or very limited in length in BLV (Fig. 3A). A small pocket at one end of the Fr-MLV RBD, containing amino acids of the VRA region, was shown to serve as a specific receptor-binding surface (top of Fig. 3C) (24, 31). The regions located downstream of the poly-Pro hinge also presented striking similarities. In particular, gp51 cysteines C179 and C182 were strictly conserved within the sequence of the Fr-MLV gp70 (Fig. 2 and 3A) and of other oncoviral SUs. Unfortunately, the corresponding gp70 cysteines are not included in the Fr-MLV RBD crystal (Fig. 2 and 3A) (31).
On the basis of the observed sequence similarities between Fr-MLV and BLV SUs, we then visualized the structure of the Fr-MLV RBD to tentatively position the BLV zinc-binding peptide 104-123 and the residue C119 within. The well-conserved strands β8 and β9 constitute the central part of the β-sheet (Fig. 3C). Peptide 104-123 encompassed strands β7 and β8 (Fig. 3A), and cysteine 119 was predicted to be located just two amino acids upstream of strand β8 (Fig. 3A and B). Therefore, residue C119 can be located within the loop between strands β7 and β8 (see the arrow on Fig. 3C), on the opposite side from the variable regions VRA and VRB and from the hydrophobic receptor-binding surface (top of Fig. 3C).
Mutation of the cysteine residue within BLV domain 104-123 destroys cell fusion.
In order to gain insight into the role of the zinc-binding regions (104 to 123 and 218 to 237) in the biological properties of the BLV envelope, a series of mutations were introduced in the env gene of an infectious BLV provirus (81). Mutations were designed to replace amino acids potentially involved in zinc interaction with alanine residues. The cysteine at position 119 and the histidines at positions 229 and 230 were therefore replaced by alanines, generating mutants C119A and H229A+H230A. In parallel, the role of conserved cysteine residues 179 and 182 was also investigated (C179A+C182A mutant), although 4 peptides overlapping this region did not interact with zinc ions (Fig. 2). Simultaneous mutations in the cysteines and in peptide 218-237 were also performed, leading to mutants C119A+H229A+H230A and C179A+C182A+H229A+H230A.
To analyze the functional activities of the mutated viruses, the recombinant envelopes were cloned in the SFV expression vector and expressed in BHK cells. The influence of the mutations within the zinc-interacting regions on the fusogenic capacities of the BLV envelope complex was analyzed. The experimental protocol is based on the cocultivation of BHK cells, which express the envelope proteins, with CC81 indicator cells, which are highly susceptible to fusion. When the two cell types make contact, the envelope molecules expressed at the surface of BHK cells bind to the CC81 lipid bilayer, and large multinucleated cells or syncytia are formed.
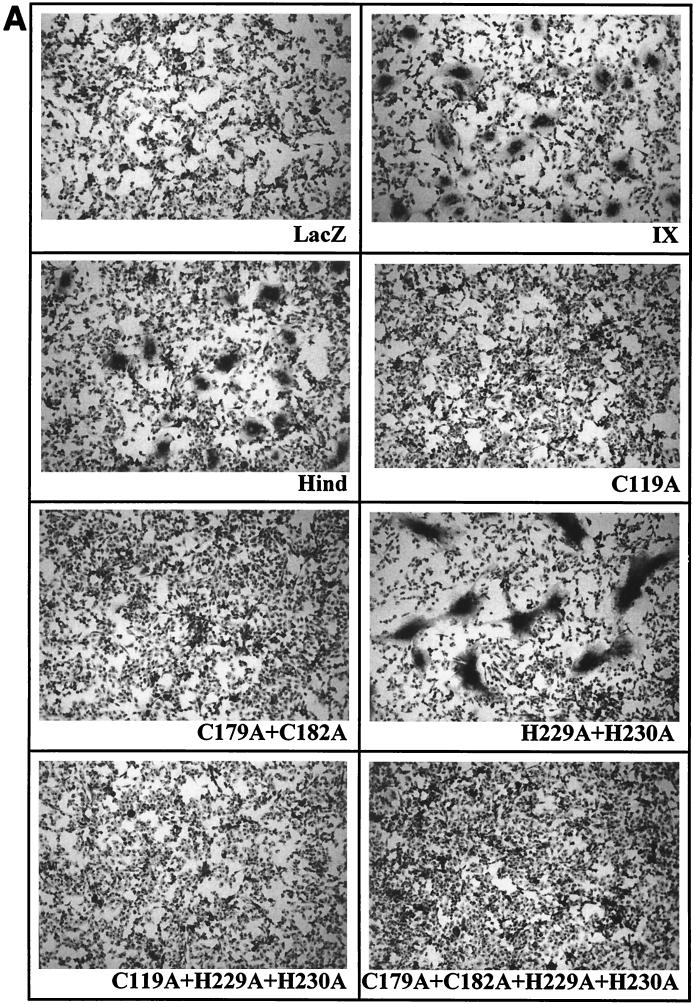
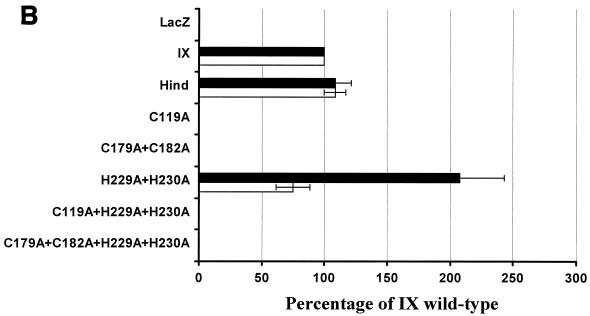
To facilitate counting, the syncytia were fixed, colored with Giemsa solution, and visualized microscopically (Fig. 4A). As positive controls, two wild-type envelope proteins (IX and Hind) were used. The numbers of syncytia were determined in 10 different microscope fields, and the numbers of nuclei were counted in 20 syncytia. Results were normalized to the levels of the wild-type IX envelope, arbitrarily set at 100% (Fig. 4B). As a negative control, no syncytia were observed when BHK cells transfected with the pSFVLacZ vector and expressing the lacZ gene were mixed with CC81 cells (illustrated in Fig. 4A and quantified in Fig. 4B).
FIG. 4.
Fusogenic capacity of recombinant BLV envelope proteins. (A) The fusogenic capacity of the different mutants was tested by cocultivation of CC81 indicator cells with BHK cells expressing the BLV envelope proteins. The wild-type (IX and Hind) and mutated (C119A, C179A+C182A, H229A+H230A, C119A+H229A+H230A, and C179A+C182A+H229A+H230A) envelope genes were cloned into the pSFV expression vector (pSFVenv plasmids). BHK cells were transfected with RNAs transcribed from the different pSFVenv vectors and cocultivated for 20 h with CC81 indicator cells. As a negative control, cells were also transfected with a pSFV vector expressing the lacZ gene. After fixation, the nuclei were colored with Giemsa, and the syncytia were visualized microscopically at ×100 magnification. The diameter of the syncytia induced by the wild-type envelope proteins was in the range of 100 to 200 μm. (B) The numbers of syncytia counted in 10 different microscope fields (open bars) were arbitrarily normalized to the wild-type levels (IX = 100%). In addition, the mean number of nuclei within 20 syncytia was evaluated for each mutant envelope protein (black bars). The data represent mean values of three independent experiments.
The C119A mutation within peptide 104-123 completely impaired the capacity of the envelope to form syncytia (Fig. 4A and B, C119A). Similarly, the mutations of the conserved cysteines C179 and C182 completely destroyed the BLV envelope's fusogenic capacity. In contrast, mutant H229A+H230A retained the ability to carry out envelope-mediated fusion. For this particular mutant, the number of syncytia was similar to the wild-type levels. Nevertheless, the size of the syncytia, i.e., the mean number of nuclei per syncytium, was drastically increased compared to the wild-type levels (Fig. 4A and B). Finally, the H229A+H230A mutation had no effect on the C119A or the C179A+C182A phenotype: indeed, mutants C119A+H229A+H230A and C179A+C182A+H229A+H230A also lost their fusogenic capacity.
These data led us to conclude that the integrity of cysteine C119 was required for gp51-induced cell fusion, in contrast to residues H229 and H230. Furthermore, the region that includes cysteines 179 and 182 was also essential for fusogenic capacity.
Expression of envelope SU mutants.
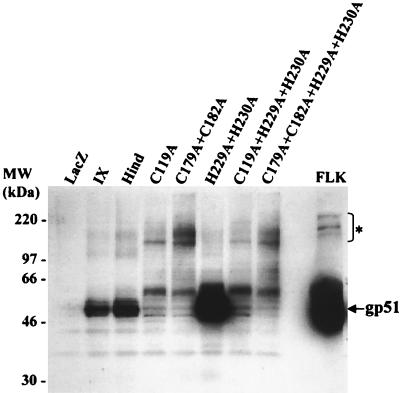
To determine whether the lack of fusogenic capacity of some mutants could be attributed to impaired expression or processing of the envelopes, we compared the expression levels of the mutated SU proteins. BHK cells expressing the different mutated envelopes were harvested, and their lysates were analyzed by Western blotting with a mix of monoclonal antibodies directed against the gp51 molecule. As a negative control, a lysate from BHK cells expressing LacZ did not yield a specific signal (Fig. 5, LacZ). In contrast, mutant H229A+H230A expressed a BLV SU glycoprotein migrating at 51 kDa, the wild-type molecular mass defined by an FLK lysate (Fig. 5, FLK) and BHK cells expressing the recombinant wild-type BLV envelope (Fig. 5, IX and Hind). Furthermore, for mutant H229A+H230A, the amount of gp51 protein was increased, as illustrated in Fig. 5 and as titrated by indirect enzyme-linked immunosorbent assay (data not shown). The other mutants expressed small amounts of fully processed gp51 molecules, indicating that the substitutions inhibited the cleavage of the precursor or the stability of the envelope. Of note, unprocessed precursors were also observed in the FLK positive control.
FIG. 5.
Expression of wild-type and mutant gp51 proteins with the SFV system. After in vitro transcription of the different pSFVenv vectors (wild-type and mutants), the corresponding purified RNAs were transfected into BHK cells by electroporation. The cells were cultivated for 60 h and harvested, and cell lysates corresponding to 1.5 million cells were added per well of an SDS-polyacrylamide gel. The proteins were transferred onto a nitrocellulose membrane and incubated in the presence of a mixture of monoclonal antibodies directed against the BLV gp51 protein. After incubation with an anti-mouse immunoglobulin antibody coupled to peroxidase, the presence of the gp51 proteins was visualized by chemiluminescence. Supernatant from BLV-producing FLK cells was used as a positive control. The unprocessed precursors are indicated (*).
From these Western blot experiments, we concluded that mutations C119A and C179A+C182A within the gp51 protein drastically impaired envelope processing.
Effect of mutations in the context of an infectious molecular clone.
The BLV system provides a unique opportunity to analyze the behavior of recombinant viruses in vivo, supporting the use of BLV as model for the related virus HTLV-1 (78). To assess the potential role of the residues located within the zinc-binding regions, the mutated SU genes were introduced into an infectious and pathogenic molecular clone (pBLV344), yielding proviruses pBLV-C119A, pBLV-C179A+C182A, pBLV-H229A+H230A, pBLV-C119A+H229A+H230A, and pBLV-C179A+C182A+H229A+H230A.
To evaluate the ability of the recombinant viruses to express viral genes in cell culture, the wild-type and mutated proviruses were first introduced by transient transfection into D17 canine osteosarcoma cells. Four micrograms of proviral DNA was transfected into 5 × 105 cells together with the pLTRLuc reporter, which contains the BLV long terminal repeat cloned upstream of the luciferase gene (53). At 2 days posttransfection, half of the cells were harvested, and the luciferase activity in the lysates was determined. The remaining cells were used to determine the expression of the major p24gag antigen by an ELISA method. From these experiments, it appeared that all the recombinant proviruses were able to synthesize the Tax transactivator and the p24 major capsid protein at wild-type levels (data not shown).
To assess viral infectivity, the wild-type and mutated proviruses were then injected intradermally into sheep. Sera were collected weekly and analyzed for the presence of BLV antibodies by both immunodiffusion and ELISA (60). As expected, the wild-type provirus efficiently induced seroconversion within two of two injected animals (Fig. 6A). In contrast, mutants C119A and C179A+C182A were not infectious after injection into sheep (Fig. 6A). Similarly, proviruses harboring multiple mutations (C119A+H229A+H230A and C179A+C182A+H229A+H230A) were not infectious. Finally, mutation H229A+H230A still allowed infection in vivo, since both sheep injected with this mutant seroconverted (Fig. 6A).
FIG. 6.
In contrast to histidines 229 and 230, cysteine 119 is essential for infectivity in vivo. (A) To assess infectivity, recombinant proviruses carrying mutated gp51 genes were constructed, mixed with cationic liposomes, and injected intradermally into sheep. The infectious potential was evaluated by two criteria: the presence of antibodies directed towards the gp51 protein (as measured by ELISA and immunodiffusion) and PCR amplification of viral sequences. (B) To evaluate the proviral loads in sheep 2674 and 2675 infected with the H229A+H230A mutant, DNA was extracted from 500-μl aliquots of peripheral blood at 6 months postseroconversion and amplified by 29 cycles of PCR. Of note, the lymphocyte counts for the different animals were all in the normal range (3,000 to 5,000 per mm3). The samples were then analyzed by Southern blot hybridization with a BLV probe. DNAs from sheep 114 and 117 (uninfected [N.I.] animals) and 292 and 293 (infected with the wild-type virus) were used as negative and positive controls, respectively. As a standard for quantification, serial dilutions of the lysates (1:10 and 1:100) were amplified in parallel.
To evaluate the efficiency of viral propagation of this mutant, the proviral loads were analyzed by semiquantitative PCRs. To this end, blood samples were collected by jugular venipuncture at 6 months postseroconversion, and the corresponding BLV env sequences were amplified from their lysates by 29 cycles of PCR. The amplicons were migrated onto an agarose gel and analyzed by Southern blot hybridization with a BLV probe (Fig. 6B). As a control for PCR contamination, no viral sequence could be detected with blood samples from uninfected sheep (sheep 114 and 117). As expected, viral sequences (a 2.5-kb fragment) were successfully amplified from blood samples corresponding to sheep 292 and 293, infected with the wild-type BLV provirus. Serial dilutions of these positive controls supported the semiquantitative character of the amplification. When the same protocol was applied to lysates from sheep 2674 and 2675 infected with the H229A+H230A provirus, it appeared that the 2.5-kb proviral fragment was also amplified, its level of amplification being close to the wild-type level (Fig. 6B). Similar results were obtained with blood samples collected 3.5 months after seroconversion (data not shown). Therefore, the H229A+H230A mutation in the BLV envelope did not drastically affect proviral loads in sheep.
In order to confirm these PCR amplification data, viral expression was estimated ex vivo by titration of the major capsid p24 protein. Purified peripheral blood mononuclear cells from sheep were cultured for 48 h, and the titers of the p24 antigen were determined from the cells or in the supernatants by the ELISA procedure. In agreement with our PCR amplification data, similar amounts of p24 protein were measured in peripheral blood mononuclear cell cultures from sheep infected with either the wild-type virus or the H229A+H230A mutant virus (data not shown).
Finally, to ensure that the viruses propagating within animals 2674 and 2675 were not revertants, the corresponding envelope genes were amplified from the blood lysates by 36 cycles of PCR, and the amplicons were directly sequenced. The viruses propagating in these animals indeed harbored the H229A+H230A mutation (data not shown).
We concluded that the mutation within the gp51 zinc-binding region 104-123 (mutation C119A) hampered infection in vivo while mutations within the zinc-binding domain 218-237 (mutant H229A+H230A) still allowed viral infectivity and spread in vivo.
HTLV-1 envelope SU glycoprotein also has zinc ion-binding capacity.
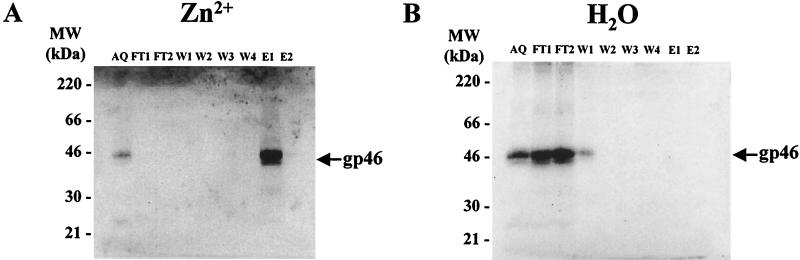
To our knowledge, direct binding of any retroviral SU envelope to zinc ions has never been reported. Therefore, we asked if this observation is unique to BLV SU protein or can be extended to other retroviral envelopes. In order to test binding of the HTLV-1 SU protein to zinc, MT2 cells were cultured and the corresponding supernatant was mixed with load and wash buffer and chromatographed on an affinity column charged with Zn2+. The resulting fractions were concentrated by PEG precipitation and analyzed by gel electrophoresis and immunoblotting with an antibody specific for the HTLV-1 gp46 protein. No binding was detected on the chelating matrix that had not been charged with zinc (Fig. 7B), but the HTLV-1 SU envelope glycoprotein was efficiently retained in the presence of zinc ions (Fig. 7A).
FIG. 7.
Binding of HTLV-1 envelope SU glycoprotein to zinc. Chelating Sepharose fast flow gel was charged with Zn2+ ions (A) or water (B) and packed into a column. The columns were loaded with MT2 cell culture supernatant mixed with load and wash buffer, and the procedure described for Fig. 1 was followed. The HTLV-1 gp46 glycoprotein was detected by incubation with the MF2 monoclonal antibody before incubation with a secondary antibody conjugated to alkaline phosphatase. AQ, aliquot; FT, flowthrough; W, wash; E, eluate.
We concluded that, like BLV SU, HTLV-1 SU protein also interacted with zinc, underlining a common property of these two deltaretroviruses.
DISCUSSION
The presence of three zinc ions per crystal of Fr-MLV RBD was puzzling; crystals did not grow readily in zinc acetate instead of calcium acetate and were clustered instead of rodlike or needlelike, but nevertheless, there were no significant differences between the protein structures refined against data from zinc- or calcium-containing crystals (31). In this paper, we demonstrated direct binding of two retroviral SU proteins to zinc ions. The identification of two peptides having zinc-binding abilities within the BLV gp51 sequence further supported this observation. However, we cannot exclude that other regions of the gp51 protein may be able to bind zinc, and conversely, peptides that bind zinc on their own may not be able to bind it within some conformation(s) of the entire gp51.
On the basis of the alignment of part of the gp51 sequence with that of Fr-MLV gp70, we predicted that the C119 amino acid (located within the zinc ion-binding peptide 104-123) should be situated in a loop linking strands β7 and β8, the strands constituting the core of the immunoglobulin-like sandwich. This loop is on the opposite site of variable regions VRA and VRB, which should be very limited in length in the corresponding BLV sequence and could determine the receptor specificity. The localization of the C119 amino acid on the opposite site of the RBD receptor-binding surface contrasts with the localization of the amino acids bound by the zinc atoms in the Fr-MLV RBD structure. Indeed, two of the zinc atoms bound by Fr-MLV SU are associated with Asp86 and one is associated with His55, both of these residues being located on the RBD receptor-binding surface (31).
The model for the BLV gp51 head established by Callebaut et al. in 1994 predicted the position of cysteine 119 in a loop that does not belong to the receptor-binding domain (15). Whatever the spatial position of this amino acid, its mutation is sufficient to impair envelope processing and, consequently, fusogenic capacity and in vivo infectivity. The region encompassing amino acids C179 and C182 is also required for these biological functions. This CXXC motif is highly conserved across a broad range of distantly related retroviruses. It resembles the motif present at the active site of thiol-disulfide exchange enzymes and has been proposed to be implicated in a labile disulfide bond between the SU and TM subunits of the Fr-MLV envelope protein complex (57). For BLV, it is possible that these cysteine mutations affect the SU/TM disulfide bonds observed by Johnston and Radke (45).
For the H229A+H230A mutant, the envelope exhibited an increased fusion activity in terms of size of syncytia, probably because of increased amounts of gp51 protein (compared to wild-type levels). In vivo, a better fusogenic capacity can be an advantage, but an increased amount of gp51 antigen would also trigger a better host immune response. The spread of the virus in its host seems to be a balance between these two phenomena: the H229A+H230A mutant virus was infectious, and its proviral loads were close to that of the wild-type virus. Furthermore, sheep 2674 injected with this virus died with lymph node tumors 1 year after injection, as commonly observed with the wild-type virus (78). Residues 229 and 230 are located in the carboxy-terminal half of the protein, and the corresponding crystallographic data are not available for Fr-MLV. Considering the position of strand β9 and the probable position of the following poly-Pro hinge on the Fr-MLV RBD structure (Fig. 3C), the rest of the SU protein and, consequently, the zinc-ion binding region 218 to 237 may be located close to the C119 region. The zinc-binding site could therefore constitute the interface between the two parts of the BLV SU protein separated by the poly-Pro hinge (whereas the zinc-binding sites were not integral to the structure for the Fr-MLV RBD [31]). This hypothesis is reinforced by the observation that, in the amino-terminal portion of the Fr-MLV RBD (and thus in a position proximal to the predicted one of the BLV C119; Fig. 3C), a conserved histidine residue has been identified as mediating a critical postbinding step in infection by interacting between the RBD and the remainder of the SU and/or TM protein (2, 4, 5, 49).
These experiments and our observation, i.e., the probable localization of amino acids critical for zinc binding at the opposite side of the receptor-binding site, are coherent with the hypothesis that zinc is essential to promote a functional interaction between the RBD and the rest of the SU and/or TM protein. Nevertheless, it is still possible that zinc mediates the primary interaction of the SU envelope with the receptor. Zinc has indeed been shown in the literature to mediate other receptor-ligand interactions: the binding of human growth hormone to the human prolactin receptor (19) and of the p58 killer cell immunoglobulin-related receptor (KIR) to HLA (61). Like the BLV TM protein, KIRs have cytoplasmic tails containing copies of immunoreceptor tyrosine-based motifs (ITAM or ITIM) (62, 73, 75). Like the BLV TM envelope protein, the p58 KIR cytoplasmic tail binds the protein phosphatase SHP-1 (11). Moreover, the overall fold of growth hormone and KIR domains resembles immunoglobulin-like domains, similar to that of the Fr-MLV RBD (9, 26, 29, 30, 67).
Nevertheless, Fan et al. (27) have shown that zinc is not required either for the proper folding of p58 KIR or for the specific binding of HLA by the KIR, suggesting that the requirement for zinc is subsequent to binding. In fact, zinc ions are known to undergo relatively rapidly ligand exchange reactions (7). So, it is not surprising that the interaction between KIRs and their ligands shows unusually fast association and dissociation rates (74). Similarly, the interaction between human growth hormone and its extracellular receptor does not absolutely require zinc ions but is accommodated by the presence of a zinc-binding site that potentiates 8,000-fold Zn2+ dependence toward binding (19). Therefore, zinc may be required for multimerization of the envelope proteins and be critical for the formation of clusters of envelope and receptor molecules.
Vales-Gomez et al. have recently established that the formation of clusters between KIR and HLA-C molecules is subsequent to a Zn2+-induced multimerization of the KIR molecules (72). Furthermore, a covalently linked KIR dimer binds more tightly to HLA than the wild-type monomer, indicating that the KIR C-terminal stem could stabilize oligomerization (27, 28). Similarly, we speculate that the H229A+H230A mutation within the BLV envelope stem consolidates envelope oligomerization and confers a kinetic advantage for the fusion process.
Finally, zinc may mediate the interaction, occurring post-receptor-envelope binding, of the BLV or HTLV-1 envelope with one or more unknown factors. HTLV-1-induced fusion is indeed a multistep process that is susceptible to inhibition at different stages of the fusion pathway after receptor binding (20). The interaction of HTLV-1 envelope with the target cell membrane is either weak or unstable, and one or more cellular factors appear to be required (21). Zinc may be involved in the binding of such factors with the envelope SU proteins. Our structural model locates the BLV zinc-binding region 104-123 on the opposite side from the hydrophobic Fr-MLV receptor-binding surface and therefore supports the hypothesis that zinc ions are probably not implicated in the early SU/receptor stage of interaction.
The precise role of the zinc ions in the fusion process remains to be determined. In our hands, attempts to test the influence of zinc-chelating agents such as TPEN (Molecular Probes) on syncytium formation capacity were unsuccessful because of the high cell toxicity of these compounds. Further experiments (i.e., the use of recombinant purified proteins) will be necessary to identify proteins interacting with zinc ions and SU glycoproteins.
In conclusion, we have shown here a direct binding of the BLV and HTLV-1 envelope SU proteins to zinc ions and we have identified two peptides distributed along the BLV gp51 sequence that are able to interact with zinc. Based on the Fr-MLV RBD structure, we have predicted that the first zinc-binding peptide should be located on the opposite side of the potential receptor-binding surface, suggesting that zinc ions mediate interactions either with the rest of the SU and/or TM protein or with partners different from the receptor.
Acknowledgments
This work was supported by the Belgian Fonds de la Recherche Scientifique Médicale, the FB Assurances, the Service de Programmation pour la Politique Scientifique SSTC P4/30, the Fédération Belge Contre le Cancer, the Action de Recherche Concertée du Ministère de la Communauté Française 98/03-224 and 99/03-247, the Fondation Internationale Brachet, and the Programme FEDER Région Wallonne-Commission Européenne. J.-S.G., C.V.L., L.W., and R.K. are members of the Belgian Fonds National de la Recherche Scientifique (FNRS). D.D. is a postdoctoral fellow of the Belgian Région Wallonne (grant 991/4202).
We thank M. Nuttinck, T. Peremans, J. M. Londes, and G. Vandendaele for technical help. We are grateful to D. Londos-Gagliardi for providing the gp46-specific MF2 antibody, to C. Bangham and S. Daenke for providing the MT2 cell line, and to J. M. Cunningham for helpful discussion.
REFERENCES
- 1.Agadjanyan, M. G., K. E. Ugen, B. Wang, W. V. Williams, and D. B. Weiner. 1994. Identification of an 80-kilodalton membrane glycoprotein important for human T-cell leukemia virus type I and type II syncytium formation and infection. J. Virol. 68:485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban, J., D. Portetelle, C. Altaner, B. Horion, D. Milan, V. Krchnak, A. Burny, and R. Kettmann. 1993. Isolation and characterization of a 2.3-kilobase-pair cDNA fragment encoding the binding domain of the bovine leukemia virus cell receptor. J. Virol. 67:1050-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett, A. L., and J. M. Cunningham. 2001. Receptor binding transforms the surface subunit of the mammalian C-type retrovirus envelope protein from an inhibitor to an activator of fusion. J. Virol. 75:9096-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett, A. L., R. A. Davey, and J. M. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaufils, P., D. Choquet, R. Z. Mamoun, and B. Malissen. 1993. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 12:5105-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg, J. M., and Y. Shi. 1996. The galvanization of biology: a growing appreciation for the roles of zinc. Science 271:1081-1085. [DOI] [PubMed] [Google Scholar]
- 8.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyington, J. C., S. A. Motyka, P. Schuck, A. G. Brooks, and P. D. Sun. 2000. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature 405:537-543. [DOI] [PubMed] [Google Scholar]
- 10.Bruck, C., S. Mathot, D. Portetelle, C. Berte, J. D. Franssen, P. Herion, and A. Burny. 1982. Monoclonal antibodies define eight independent antigenic regions on the bovine leukemia virus (BLV) envelope glycoprotein gp51. Virology 122:342-352. [DOI] [PubMed] [Google Scholar]
- 11.Burshtyn, D. N., A. M. Scharenberg, N. Wagtmann, S. Rajagopalan, K. Berrada, T. Yi, J. P. Kinet, and E. O. Long. 1996. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity 4:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callahan, R., M. M. Lieber, G. J. Todaro, D. C. Graves, and J. F. Ferrer. 1976. Bovine leukemia virus genes in the DNA of leukemic cattle. Science 192:1005-1007. [DOI] [PubMed] [Google Scholar]
- 13.Callebaut, I., A. Burny, V. Krchnak, H. Gras-Masse, B. Wathelet, and D. Portetelle. 1991. Use of synthetic peptides to map sequential epitopes recognized by monoclonal antibodies on the bovine leukemia virus external glycoprotein. Virology 185:48-55. [DOI] [PubMed] [Google Scholar]
- 14.Callebaut, I., G. Labesse, P. Durand, A. Poupon, L. Canard, J. Chomilier, B. Henrissat, and J. P. Mornon. 1997. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell. Mol. Life Sci. 53:621-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callebaut, I., D. Portetelle, A. Burny, and J. P. Mornon. 1994. Identification of functional sites on bovine leukemia virus envelope glycoproteins using structural and immunological data. Eur. J. Biochem. 222:405-414. [DOI] [PubMed] [Google Scholar]
- 16.Callebaut, I., V. Voneche, A. Mager, O. Fumiere, V. Krchnak, M. Merza, J. Zavada, M. Mammerickx, A. Burny, and D. Portetelle. 1993. Mapping of B-neutralizing and T-helper cell epitopes on the bovine leukemia virus external glycoprotein gp51. J. Virol. 67:5321-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantor, G. H. 1996. A potential proline-rich motif upstream of the immunoreceptor tyrosine-based activation motif in bovine leukemia virus gp30, Epstein-Barr virus LMP2A, herpesvirus papio LMP2A, and African horsesickness virus VP7. Virology 220:265-266. [DOI] [PubMed] [Google Scholar]
- 18.Cantor, G. H., S. M. Pritchard, O. Orlik, G. A. Splitter, W. C. Davis, and R. Reeves. 1999. Bovine leukemia virus transmembrane protein gp30 physically associates with the down-regulatory phosphatase SHP-1. Cell. Immunol. 193:117-124. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham, B. C., S. Bass, G. Fuh, and J. A. Wells. 1990. Zinc mediation of the binding of human growth hormone to the human prolactin receptor. Science 250:1709-1712. [DOI] [PubMed] [Google Scholar]
- 20.Daenke, S., and S. Booth. 2000. HTLV-1-induced cell fusion is limited at two distinct steps in the fusion pathway after receptor binding. J. Cell Sci. 113:37-44. [DOI] [PubMed] [Google Scholar]
- 21.Daenke, S., and S. Booth. 2000. Molecular mechanisms affecting HTLV type 1-dependent fusion at the cell membrane: implications for inhibiting viral transmission. AIDS Res. Hum. Retrovir. 16:1731-1736. [DOI] [PubMed] [Google Scholar]
- 22.Daenke, S., S. A. McCracken, and S. Booth. 1999. Human T-cell leukaemia/lymphoma virus type 1 syncytium formation is regulated in a cell-specific manner by ICAM-1, ICAM-3 and VCAM-1 and can be inhibited by antibodies to integrin beta2 or beta7. J. Gen. Virol. 80:1429-1436. [DOI] [PubMed] [Google Scholar]
- 23.Daeron, M. 1996. Building up the family of ITIM-bearing negative coreceptors. Immunol. Lett. 54:73-76. [DOI] [PubMed] [Google Scholar]
- 24.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J. Virol. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delamarre, L., C. Pique, A. R. Rosenberg, V. Blot, M. P. Grange, I. Le Blanc, and M. C. Dokhelar. 1999. The Y-S-L-I tyrosine-based motif in the cytoplasmic domain of the human T-cell leukemia virus type 1 envelope is essential for cell-to-cell transmission. J. Virol. 73:9659-9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Vos, A. M., M. Ultsch, and A. A. Kossiakoff. 1992. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 255:306-312. [DOI] [PubMed] [Google Scholar]
- 27.Fan, Q. R., D. N. Garboczi, C. C. Winter, N. Wagtmann, E. O. Long, and D. C. Wiley. 1996. Direct binding of a soluble natural killer cell inhibitory receptor to a soluble human leukocyte antigen-Cw4 class I major histocompatibility complex molecule. Proc. Natl. Acad. Sci. USA 93:7178-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan, Q. R., E. O. Long, and D. C. Wiley. 2000. A disulfide-linked natural killer cell receptor dimer has higher affinity for HLA-C than wild-type monomer. Eur. J. Immunol. 30:2692-2697. [DOI] [PubMed] [Google Scholar]
- 29.Fan, Q. R., E. O. Long, and D. C. Wiley. 2001. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat. Immunol. 2:452-460. [DOI] [PubMed] [Google Scholar]
- 30.Fan, Q. R., L. Mosyak, C. C. Winter, N. Wagtmann, E. O. Long, and D. C. Wiley. 1997. Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors. Nature 389:96-100. [DOI] [PubMed] [Google Scholar]
- 31.Fass, D., R. A. Davey, C. A. Hamson, P. S. Kim, J. M. Cunningham, and J. M. Berger. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277:1662-1666. [DOI] [PubMed] [Google Scholar]
- 32.Gallaher, W. R., J. M. Ball, R. F. Garry, M. C. Griffin, and R. C. Montelaro. 1989. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res. Hum. Retrovir. 5:431-440. [DOI] [PubMed] [Google Scholar]
- 33.Gatot, J. S., I. Callebaut, J. P. Mornon, D. Portetelle, A. Burny, P. Kerkhofs, R. Kettmann, and L. Willems. 1998. Conservative mutations in the immunosuppressive region of the bovine leukemia virus transmembrane protein affect fusion but not infectivity in vivo. J. Biol. Chem. 273:12870-12880. [DOI] [PubMed] [Google Scholar]
- 34.Gavalchin, J., N. Fan, M. J. Lane, L. Papsidero, and B. J. Poiesz. 1993. Identification of a putative cellular receptor for HTLV-I by a monoclonal antibody, Mab 34-23. Virology 194:1-9. [DOI] [PubMed] [Google Scholar]
- 35.Gavalchin, J., N. Fan, P. G. Waterbury, E. Corbett, B. D. Faldasz, S. M. Peshick, B. J. Poiesz, L. Papsidero, and M. J. Lane. 1995. Regional localization of the putative cell surface receptor for HTLV-I to human chromosome 17q23.2-17q25.3. Virology 212:196-203. [DOI] [PubMed] [Google Scholar]
- 36.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407-410. [DOI] [PubMed]
- 37.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 38.Hildreth, J. E. 1998. Syncytium-inhibiting monoclonal antibodies produced against human T-cell lymphotropic virus type 1-infected cells recognize class II major histocompatibility complex molecules and block by protein crowding. J. Virol. 72:9544-9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hildreth, J. E., A. Subramanium, and R. A. Hampton. 1997. Human T-cell lymphotropic virus type 1 (HTLV-1)-induced syncytium formation mediated by vascular cell adhesion molecule-1: evidence for involvement of cell adhesion molecules in HTLV-1 biology. J. Virol. 71:1173-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinuma, Y., K. Nagata, M. Hanaoka, M. Nakai, T. Matsumoto, K. I. Kinoshita, S. Shirakawa, and I. Miyoshi. 1981. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. USA 78:6476-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imai, T., K. Fukudome, S. Takagi, M. Nagira, M. Furuse, N. Fukuhara, M. Nishimura, Y. Hinuma, and O. Yoshie. 1992. C33 antigen recognized by monoclonal antibodies inhibitory to human T cell leukemia virus type 1-induced syncytium formation is a member of a new family of transmembrane proteins including CD9, CD37, CD53, and CD63. J. Immunol. 149:2879-2886. [PubMed] [Google Scholar]
- 42.Inabe, K., M. Nishizawa, S. Tajima, K. Ikuta, and Y. Aida. 1999. The YXXL sequences of a transmembrane protein of bovine leukemia virus are required for viral entry and incorporation of viral envelope protein into virions. J. Virol. 73:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobson, S., C. S. Raine, E. S. Mingioli, and D. E. McFarlin. 1988. Isolation of an HTLV-1-like retrovirus from patients with tropical spastic paraparesis. Nature 331:540-543. [DOI] [PubMed] [Google Scholar]
- 44.Jassal, S. R., R. G. Pohler, and D. W. Brighty. 2001. Human T-cell leukemia virus type 1 receptor expression among syncytium-resistant cell lines revealed by a novel surface glycoprotein-immunoadhesin. J. Virol. 75:8317-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnston, E. R., and K. Radke. 2000. The SU and TM envelope protein subunits of bovine leukemia virus are linked by disulfide bonds, both in cells and in virions. J. Virol. 74:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kettmann, R., D. Portetelle, M. Mammerickx, Y. Cleuter, D. Dekegel, M. Galoux, J. Ghysdael, A. Burny, and H. Chantrenne. 1976. Bovine leukemia virus: an exogenous RNA oncogenic virus. Proc. Natl. Acad. Sci. USA 73:1014-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lando, Z., P. Sarin, M. Megson, W. C. Greene, T. A. Waldman, R. C. Gallo, and S. Broder. 1983. Association of human T-cell leukaemia/lymphoma virus with the Tac antigen marker for the human T-cell growth factor receptor. Nature 305:733-736. [DOI] [PubMed] [Google Scholar]
- 49.Lavillette, D., A. Ruggieri, S. J. Russell, and F. L. Cosset. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 51.Londos-Gagliardi, D., M. H. Armengaud, F. Freund, R. Dalibart, E. Moze, S. Huet, E. Legrand, and B. J. Guillemain. 1997. Antibodies directed against a variable and neutralizable region of the HTLV-I envelope surface glycoprotein. Leukemia 11(Suppl. 3):38-41. [PubMed] [Google Scholar]
- 52.Mammerickx, M. 1970. Sur l'utilisation du mouton pour les expériences sur la leucose bovine. Exp. Anim. 3:285-293. [Google Scholar]
- 53.Merezak, C., C. Pierreux, E. Adam, F. Lemaigre, G. G. Rousseau, C. Calomme, C. Van Lint, D. Christophe, P. Kerkhofs, A. Burny, R. Kettmann, and L. Willems. 2001. Suboptimal enhancer sequences are required for efficient bovine leukemia virus propagation in vivo: implications for viral latency. J. Virol. 75:6977-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller, J. M., L. D. Miller, C. Olson, and K. G. Gillette. 1969. Virus-like particles in phytohemagglutinin-stimulated lymphocyte cultures with reference to bovine lymphosarcoma. J. Natl. Cancer Inst. 43:1297-1305. [PubMed] [Google Scholar]
- 55.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 56.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i:1031-1032. [DOI] [PubMed]
- 57.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Portetelle, D., C. Dandoy, A. Burny, J. Zavada, H. Siakkou, H. Gras-Masse, H. Drobecq, and A. Tartar. 1989. Synthetic peptides approach to identification of epitopes on bovine leukemia virus envelope glycoprotein gp51. Virology 169:34-41. [DOI] [PubMed] [Google Scholar]
- 60.Portetelle, D., M. Mammerickx, and A. Burny. 1989. Use of two monoclonal antibodies in an ELISA test for the detection of antibodies to bovine leukaemia virus envelope protein gp51. J. Virol. Methods 23:211-222. [DOI] [PubMed] [Google Scholar]
- 61.Rajagopalan, S., C. C. Winter, N. Wagtmann, and E. O. Long. 1995. The Ig-related killer cell inhibitory receptor binds zinc and requires zinc for recognition of HLA-C on target cells. J. Immunol. 155:4143-4146. [PubMed] [Google Scholar]
- 62.Reth, M. 1989. Antigen receptor tail clue. Nature 338:383-384. [PubMed] [Google Scholar]
- 63.Rice, N., R. Stephens, and R. Gilden. 1987. Sequence analysis of the bovine leukemia virus genome, p. 115-144. In A. Burny and M. Mammerickx (ed.), Enzootic bovine leukosis and bovine leukemia virus. Nijhoff, Boston, Mass.
- 64.Sagara, Y., C. Ishida, Y. Inoue, H. Shiraki, and Y. Maeda. 1998. 71-kilodalton heat shock cognate protein acts as a cellular receptor for syncytium formation induced by human T-cell lymphotropic virus type 1. J. Virol. 72:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 66.Sarkar, B. 1995. Metal replacement in DNA-binding zinc finger proteins and its relevance to mutagenicity and carcinogenicity through free radical generation. Nutrition 11:646-649. [PubMed] [Google Scholar]
- 67.Somers, W., M. Ultsch, A. M. De Vos, and A. A. Kossiakoff. 1994. The X-ray structure of a growth hormone-prolactin receptor complex. Nature 372:478-481. [DOI] [PubMed] [Google Scholar]
- 68.Sutton, R. E., and D. R. Littman. 1996. Broad host range of human T-cell leukemia virus type 1 demonstrated with an improved pseudotyping system. J. Virol. 70:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suzuki, T., and H. Ikeda. 1998. The mouse homolog of the bovine leukemia virus receptor is closely related to the delta subunit of adaptor-related protein complex AP-3, not associated with the cell surface. J. Virol. 72:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tidona, C. A., and G. Darai. 2002. The Springer index of viruses. Springer-Verlag, Heidelberg, Germany.
- 71.Trejo, S. R., and L. Ratner. 2000. The HTLV receptor is a widely expressed protein. Virology 65:6468-6477. [DOI] [PubMed] [Google Scholar]
- 72.Vales-Gomez, M., R. A. Erskine, M. P. Deacon, J. L. Strominger, and H. T. Reyburn. 2001. The role of zinc in the binding of killer cell Ig-like receptors to class I MHC proteins. Proc. Natl. Acad. Sci. USA 98:1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vales-Gomez, M., H. Reyburn, and J. Strominger. 2000. Interaction between the human NK receptors and their ligands. Crit. Rev. Immunol. 20:223-244. [PubMed] [Google Scholar]
- 74.Vales-Gomez, M., H. T. Reyburn, M. Mandelboim, and J. L. Strominger. 1998. Kinetics of interaction of HLA-C ligands with natural killer cell inhibitory receptors. Immunity 9:337-344. [DOI] [PubMed] [Google Scholar]
- 75.Vivier, E., and M. Daeron. 1997. Immunoreceptor tyrosine-based inhibition motifs. Immunol. Today 18:286-291. [DOI] [PubMed] [Google Scholar]
- 76.Weiss, R. A. 1992. Retroviruses and human cancer. Semin. Cancer Biol. 3:321-328. [PubMed] [Google Scholar]
- 77.Willems, L., A. Burny, D. Collete, O. Dangoisse, F. Dequiedt, J. S. Gatot, P. Kerkhofs, L. Lefebvre, C. Merezak, T. Peremans, D. Portetelle, J. C. Twizere, and R. Kettmann. 2000. Genetic determinants of bovine leukemia virus pathogenesis. AIDS Res. Hum. Retrovir. 16:1787-1795. [DOI] [PubMed] [Google Scholar]
- 78.Willems, L., A. Burny, D. Collete, O. Dangoisse, J. S. Gatot, P. Kerkhofs, L. Lefebvre, C. Merezak, D. Portetelle, J. C. Twizere, and R. Kettmann. 1999. Bovine leukemia virus as a model for human T-cell leukemia virus. Curr. Top. Virol. 1:139-167. [DOI] [PubMed] [Google Scholar]
- 79.Willems, L., J. S. Gatot, M. Mammerickx, D. Portetelle, A. Burny, P. Kerkhofs, and R. Kettmann. 1995. The YXXL signaling motifs of the bovine leukemia virus transmembrane protein are required for in vivo infection and maintenance of high viral loads. J. Virol. 69:4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Willems, L., R. Kettmann, F. Dequiedt, D. Portetelle, V. Voneche, I. Cornil, P. Kerkhofs, A. Burny, and M. Mammerickx. 1993. In vivo infection of sheep by bovine leukemia virus mutants. J. Virol. 67:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Willems, L., D. Portetelle, P. Kerkhofs, G. Chen, A. Burny, M. Mammerickx, and R. Kettmann. 1992. In vivo transfection of bovine leukemia provirus into sheep. Virology 189:775-777. [DOI] [PubMed] [Google Scholar]
- 82.Woodcock, S., J. P. Mornon, and B. Henrissat. 1992. Detection of secondary structure elements in proteins by hydrophobic cluster analysis. Protein Eng. 5:629-635. [DOI] [PubMed] [Google Scholar]
- 83.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 79:2031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]