Abstract
Entry by retroviruses is mediated through interactions between the viral envelope glycoprotein and the host cell receptor(s). We recently identified two host cell proteins, FeLIX and Pit1, that are necessary for infection by cytopathic, T-cell-tropic feline leukemia viruses (FeLV-T). Pit1 is a classic multiple transmembrane protein used as a receptor by several other simple retroviruses, including subgroup B FeLV (FeLV-B), and FeLIX is a secreted cellular protein expressed from endogenous FeLV-related sequences (enFeLV). FeLIX is nearly identical to FeLV-B envelope sequences that encode the N-terminal half of the viral surface unit (SU), because these FeLV-B sequences are acquired by recombination with enFeLV. FeLV-B SUs can functionally substitute for FeLIX in mediating FeLV-T infection. Both of these enFeLV-derived cofactors can efficiently facilitate FeLV-T infection only of cells expressing Pit1, not of cells expressing the related transport protein Pit2. We therefore have used chimeric Pit1/Pit2 receptors to map the determinants for cofactor binding and FeLV-T infection. Three distinct determinants appear to be required for cofactor-dependent infection by FeLV-T. We also found that Pit1 sequences within these same domains were required for binding by FeLIX to the Pit receptor. In contrast, these determinants were not all required for receptor binding by the FeLV-B SU cofactors used in this study. These data indicate that cofactor binding is not sufficient for FeLV-T infection and suggest that there may be a direct interaction between FeLV-T and the Pit1 receptor.
Retroviral entry requires a specific interaction between the viral envelope glycoprotein and a cell surface receptor. The envelope protein is synthesized as a precursor protein that is cleaved into surface (SU) and transmembrane (TM) subunits by a cellular protease. The TM anchors the SU to the viral membrane and plays an important role in fusion between the viral and host cell membranes. The SU contains the receptor binding domain (RBD) and is therefore the major viral determinant for cell tropism. Binding of the SU to the receptor triggers structural rearrangements within the envelope glycoprotein that activate the fusion peptide within the TM subunit. Host-range and receptor binding studies have mapped the murine leukemia virus (MLV) RBD to the N terminus of the SU (8-11, 15, 16, 35, 39, 42), and structural studies suggest that the variable regions (VRA and VRB) within the RBD are organized into three disulfide bonded loops (21). The major receptor binding determinants of the feline leukemia virus (FeLV) envelope have also been localized to the N terminus of SU (4, 12, 29, 50-52). For viruses such as MLV and most FeLVs, it is thought that only one receptor is required for both binding and activation of the fusion machinery.
While the role of the SU in receptor binding has long been recognized, recent data indicate that it may also participate in postbinding events in viral entry (5, 30, 31, 59). For example, mutation of an N-terminal histidine in MLV results in an SU that can bind a receptor but not mediate infection. Infection by these defective envelopes can be restored when soluble SU fragments encompassing the RBD are supplied in trans. In the MLV system, rescue can be mediated by several distinct retroviral SU fragments as long as receptors for both the soluble SU and the viral envelope are present on target cells (31). It has been suggested that interactions between the N-terminal RBD of the soluble protein and a C-terminal domain of the viral SU may be important for activation of the fusion machinery by defective MLVs (6, 7, 30). Relatively little is known about receptor-mediated events subsequent to binding and fusion, although several studies have suggested roles for receptor clustering, cytoskeletal rearrangements, and receptor signaling in the entry process (11, 22, 27, 28, 47, 49, 56).
Although known retroviral receptors are diverse in structure and function, the receptors for the simple mammalian retroviruses, such as MLV and FeLV, share several common features. Most of the identified receptors code for multiple transmembrane proteins, and many are involved in cellular transport of amino acids or other small molecules (44). The phosphate transport proteins, Pit1 and Pit2, serve as receptors for five different type C mammalian retroviruses (3, 37, 38, 41, 54, 55, 58). These proteins are 62% identical to each other at the amino acid level and have been predicted to contain 10 transmembrane domains with five extracellular loops (37, 41, 55). Gibbon ape leukemia virus (GALV), subgroup B FeLV (FeLV-B), and 10A1 murine leukemia virus all use Pit1 as a receptor, and the construction of chimeric Pit1/Pit2 proteins has facilitated the identification of receptor domains necessary for infection by GALV and FeLV-B (13, 17, 32, 34, 38, 45, 51, 52). Key receptor determinants have also been inferred by testing closely related Pit proteins from different species for receptor function (14, 18, 19, 25, 33, 46, 48, 53, 57, 58). Many of these studies have identified region A, a stretch of nine amino acids in Pit1 and Pit2 that is highly polymorphic among species, as an important receptor determinant for usage by GALV and FeLV-B (25, 53). However, chimeric analyses have identified domains outside of region A that are also necessary for infection by these viruses (13, 17, 25, 32, 33, 38, 45, 52).
The Pit receptor determinants are thought to interact specifically with different SU subdomains. Genetic studies of FeLV-B receptor tropism suggest that VRA binds to C-terminal domains of Pit1 that were originally defined as loops 4 and 5, while VRB may interact with sequences in the N-terminal half of Pit1 (51, 52). While these data indicate a role for particular Pit1 residues in viral infection, work from our laboratories indicates that FeLV-Bs can also utilize Pit2 proteins as receptors and that differential Pit tropism is determined by subtle differences in the length or composition of the enFeLV-derived sequences within the SU. For example, two closely related FeLV-B envelopes, 90Z and Gardner-Arnstein (GA), use human Pit1 (HuPit1) as a receptor but differ in their ability to infect cells using the Pit2 receptor (12, 50, 54). The 90Z envelope can also efficiently utilize the feline Pit2 protein (FePit2), and a chimeric envelope containing the presumed 90Z RBD in an FeLV-A backbone (90ZRBD) can recognize both HuPit2 and FePit2 (4, 12). In contrast, the Gardner-Arnstein envelope does not infect cells expressing HuPit2 and utilizes the FePit2 receptor with reduced efficiency (4, 50). However, substitution of an arginine for a glutamine in the receptor binding domain of the GA SU (GARBD,73Q→R) restores HuPit2 usage and leads to more efficient infection of cells expressing FePit2 (4, 50).
Like FeLV-B, FeLV-T utilizes Pit1 as a receptor, but members of this FeLV subgroup also require a second protein, FeLIX, for infection (3). FeLIX is a truncated, secreted protein expressed from an enFeLV. Consistent with the recombinatorial origin of FeLV-B envelopes, FeLIX is nearly identical to FeLV-B within the presumed RBD, and like FeLV-B, FeLIX binds Pit1 (4, 29). Indeed, FeLV-B SUs can functionally substitute for FeLIX and mediate FeLV-T infection of cells expressing Pit1 (3). In our studies to date, we have found that only those cofactors with RBDs derived from enFeLV can facilitate infection by FeLV-T. Soluble SUs from amphotropic MLV (A-MLV), GALV, and FeLV-A are not able to facilitate infection in the same way, despite the fact that A-MLV can bind Pit2 and GALV can bind Pit1 (20, 29). Unlike FeLV-B, which can utilize both Pit1 and Pit2, FeLV-T is specific in its requirement for Pit1 even when the dual-tropic FeLV-B SUs are supplied as cofactors (29).
The observation that cofactor binding is not sufficient for infection indicates that these receptors may be required for other aspects of FeLV-T entry. To better understand the mechanism of entry, we have used chimeric and mutant Pit receptors to identify regions of Pit1 that are required for FeLV-T infection. Our data indicate that Pit1 residues in at least two distinct domains are required for FeLV-T infection. When FeLIX was used as a cofactor, the Pit1 determinants for FeLIX binding and FeLV-T infection were identical. However, in testing the cofactor activity of two distinct FeLV-B SUs, we were able to identify domains of Pit1 that are dispensable for cofactor binding but are necessary for FeLV-T infection. These data are discussed in terms of models for FeLV-T entry.
MATERIALS AND METHODS
Cell culture.
293T human embryonic kidney fibroblasts were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 0.25 μg of amphotericin B per ml (complete DMEM). Stable mus dunni tail fibroblast (MDTF) cell lines expressing wild-type, chimeric, and mutant Pit receptor proteins were generated by transducing cells with vesicular stomatitis virus G protein-enveloped vectors containing the appropriate cDNA. HuPit1, HuPit2, and hamster Pit2 (HaPit2) have been described previously (19, 58). The K1, K2, K7, C1, C2, C3 (formerly EEEGG, GGGEE, GGGAA, GAGGG, GGAGG, and GAAGG, respectively), and HuPit2 K522E constructs have also been described previously (19, 32). HuPit1 D550K and HuPit1 D550T were created by changing the aspartate residue at position 550 to a lysine or threonine, respectively, using PCR mutagenesis; the PCR product was subcloned into a TA cloning vector (Invitrogen, San Diego, Calif.), sequenced, and then subcloned into the pLNSX plasmid (36). All MDTF-Pit cell lines were maintained in complete DMEM containing 0.6 mg of G418/ml.
Constructs for expression of HA-tagged FeLIX and FeLV-B SUs.
CS2-FeLV-B-GARBD-SU-HA, CS2-FeLV-B-GARBD,73Q→R-SU-HA, CS2-FeLV-A-61E-SU-HA, and CS2-FeLIX-HA have been described previously (29, 50). These constructs encode the open reading frame for the envelope protein, including the signal peptide, with a C-terminal deletion that removes the last 10 amino acids of the SU and the entire TM domain such that the SU is shed from the cell. Two copies of the hemagglutinin (HA) epitope are in frame at the C terminus of the SU. CS2-FeLV-T-61C-SU-HA was made in a similar manner. Briefly, a fragment encoding envelope amino acids 1 to 435 of FeLV-T-61C was amplified by PCR from the proviral clone EECC (43) using primers containing SacI sites at their 5′ termini. SacI-digested PCR product was ligated into the SacI site of CS2-HA, and clones were verified by DNA sequence analysis.
Preparation of viral supernatants and SU conditioned media.
Viral pseudotypes containing MLV genomes with the lacZ reporter gene were generated by transient transfection of 293T cells using a calcium phosphate protocol as described previously (29). For FeLV-B pseudotypes, cells were transfected with equal amounts of an FeLV Gag-Pol expression construct (61E-LTR-Δpsi-gag-pol), a retroviral genome encoding lacZ (pRT43.2Tnlsβgal1), and an FeLV-B envelope expression construct (pcDNA3.1-GARBDenv and pcDNA3.1-GARBD,73Q→Renv) (50). For FeLV-T pseudotypes, cells were transfected with equal amounts of EECC-Δpsi (40) and pRT43.2Tnlsβgal1. Conditioned media containing HA-tagged soluble retroviral surface units and FeLIX-HA were also generated by transient transfection of 293T cells. In this case, cells were plated at a density of 2 × 106 cells per 10-cm dish 24 h prior to transfection. Ten micrograms of CS2-FeLV-B-GARBD-SU-HA, CS2-FeLV-B-GARBD,73Q→R-SU-HA, CS2-FeLV-A-61E-SU-HA, CS2-FeLV-T-61C-SU-HA, or CS2-FeLIX-HA plasmid were transfected per 10-cm dish, and cell supernatants were harvested 48 h posttransfection.
Immunoprecipitation and Western blotting.
Detection of HA-tagged FeLIX and FeLV-B SU in conditioned media was performed as described previously (29). Briefly, cell supernatants were precleared and then immunoprecipitated with an ascites concentrate of monoclonal antibody HA.11 (Covance, Berkeley, Calif.) and protein A-Sepharose. One half of each immunoprecipitate was resolved on a sodium dodecyl sulfate-10% polyacrylamide gel, and the gel was transferred to a blotting membrane. Western blot analysis was performed using a rabbit polyclonal HA.11 primary antibody (Covance) and horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Bio-Rad, Hercules, Calif.).
Infection assays.
Infections were performed as described previously (29). Briefly, target cells were plated approximately 24 h prior to infection. On the day of infection, the culture medium was replaced with new medium containing 4 μg of Polybrene/ml. In cases in which FeLIX or SU conditioned medium was used, these supernatants were diluted 1:10 in new medium (i.e., 100 μl of conditioned medium in an 1,100-μl total volume). Cells were infected with a range of dilutions of viral pseudotypes that packaged the murine retroviral genome pRT43.2Tnlsβgal1 and stained for β-galactosidase expression 48 h postinfection, as described previously (26).
Flow cytometric binding assay.
Analysis of receptor binding by FeLIX-HA, FeLV-B-GARBD-SU-HA, FeLV-B-GARBD,73Q→R-SU-HA, FeLV-A-61E-SU-HA, and FeLV-T-61C-SU-HA to cells expressing Pit receptors was performed as described previously (29). MDTF-Pit cells were detached from the dish and incubated with supernatant from cells expressing secreted SU proteins. Bound SU was detected using an ascites concentrate of monoclonal antibody HA.11 (Covance) followed by R-phycoerythrin-conjugated goat antimouse antibody (DAKO, Carpinteria, Calif.). Cells were analyzed using a fluorescence-activated cell sorter (FACS) (Becton Dickinson, San Diego, Calif.).
RESULTS
Sequences in the N terminus of Pit1 regulate FeLV-T infectivity.
We have previously shown that both FeLIX and the FeLV-B SU can efficiently mediate FeLV-T infection of cells expressing Pit1, but not Pit2 (29). For the present study, we used as cofactors both FeLIX and the GARBDSU, which contains the receptor binding domain of the Gardner-Arnstein clone of FeLV-B in an FeLV-A envelope backbone (Fig. 1A), because they can bind to Pit1 but not Pit2. For comparison, we also included the GARBD,73Q→R SU in our analysis of FeLV-T cofactors, because this SU binds to both Pit1 and Pit2 (50). We generated conditioned media containing soluble HA-tagged versions of these three proteins by transient transfection of 293T cells. As shown in Fig. 1B, we obtained fairly equivalent expression of FeLIX, GARBD SU, and GARBD73Q→R SU in these supernatants, similar to what we observed in previous experiments.
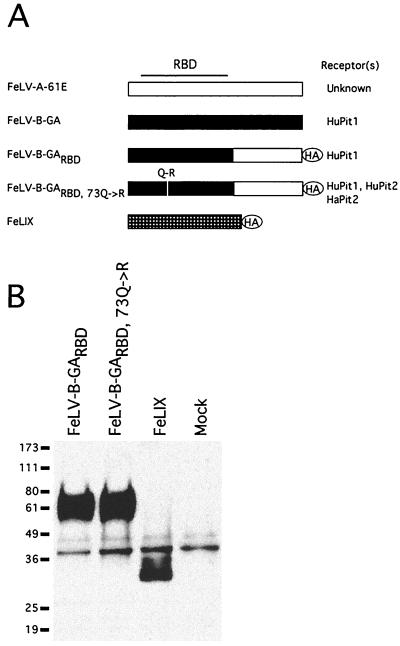
FIG. 1.
Structure and expression of FeLV-T cofactors. (A) Schematic of enFeLV-derived SU fragments used in this study. FeLV-B-GA is the SU from the Gardner-Arnstein molecular clone. FeLV-B-GARBD is a chimeric SU with the N-terminal half, including the RBD, derived from the Gardner-Arnstein molecular clone (shown in black) and the C-terminal half derived from FeLV-A-61E (shown in white) (50). FeLV-B-GARBD,73Q→R encodes a glutamine-to-arginine substitution at position 73 of SU within the RBD (50). The FeLIX open reading frame is also shown schematically (hatched box) to illustrate its relation to the FeLV-B SU. The codons for FeLIX are 97% identical to those for GARBD within the portion of the envelope gene coding for the mature SU (3). The presumed FeLV RBD is indicated at the top of the figure, and the receptor specificity of each envelope is indicated to the right (50). The location of the C-terminal HA epitope tags are indicated. (B) Detection of HA-tagged retrovirus SUs in conditioned media. Human embryonic kidney 293T cells were transfected with constructs expressing the indicated HA-tagged SU proteins, and cell supernatants were harvested 48 h posttransfection. SUs were immunoprecipitated from 1 ml of each supernatant using a monoclonal antibody directed against the HA epitope (29). One-half of each immunoprecipitate was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting using a polyclonal antibody directed against the same HA epitope. “Mock” indicates cell supernatant from mock-transfected cells. Molecular mass markers (in kilodaltons) are indicated to the left. The supernatants shown here were used for all experiments shown in Fig. 3 and 4.
In order to map the receptor determinants for FeLV-T infection, we utilized a panel of MDTF cell lines that express receptor chimeras derived from HuPit1 and either HuPit2 or HaPit2 sequences (Fig. 2). These chimeras were designed to assay determinants predicted to lie within extracellular loops based on the original published data (25). However, an alternative topology for the Pit proteins has recently been proposed (19a). For this reason and because these chimeras were not exchanged at precise boundaries, we refer here to domains of the Pit proteins rather than extracellular or intracellular loops. These domains are shown in Fig. 2. We measured receptor tropism using an assay that detects a single cycle of FeLV-T infection with the cofactors (FeLIX, GARBD SU, GARBD,73Q→R SU) supplied as conditioned medium at the time of infection (29). Consistent with our previous reports (3, 29), both FeLIX and the GARBD SU could mediate FeLV-T infection of cells expressing HuPit1 but not those expressing human or hamster Pit2 (Fig. 3). Despite the fact that viruses pseudotyped with the GARBD,73Q→R envelope can infect both HuPit1 and HuPit2 cells (50), the GARBD,73Q→R SU, when acting as a cofactor, could only mediate FeLV-T infection of cells expressing HuPit1.
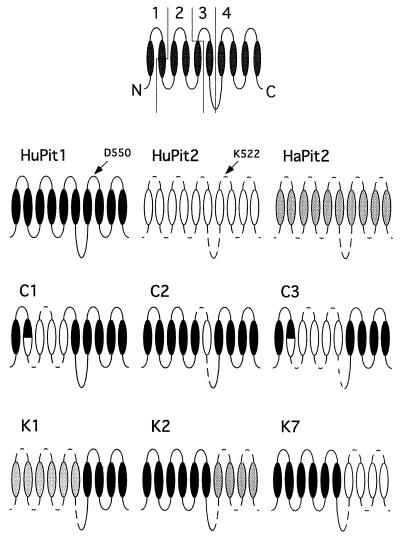
FIG. 2.
Schematic of chimeric Pit receptors used in this study. Orientation of the Pit receptor structure is based on the original predicted topology indicated at the top of the figure. This schematic shows the domains (1-4) as they are referred to in the text. The putative transmembrane domains (ovals) and extracellular and intracellular loops (lines) are shown. For HuPit1, the domains are as follows: domain 1 (amino acids 1 to 65), domain 2 (amino acids 66 to 215), domain 3 (amino acids 216 to 390 in the case of the “C” chimeras and 216 to 427 in the case of the “K” chimeras), and domain 4 (amino acids 391 to 680 in the case of the “C” chimeras and 428 to 680 in the case of the “K” chimeras). For chimeric receptors, HuPit1 sequences are shown as black ovals and solid lines. HuPit2 sequences are shown as white ovals and dashed lines. HaPit2 sequences are shown as gray ovals and dashed lines. The names are indicated above each receptor schematic. The first amino acid in region A is indicated for HuPit1 (aspartic acid, D550) and HuPit2 (lysine, K522) (25, 53). Receptors used in this study but not shown include HuPit1 D550K, HuPit1 D550T, and HuPit2 K522E.
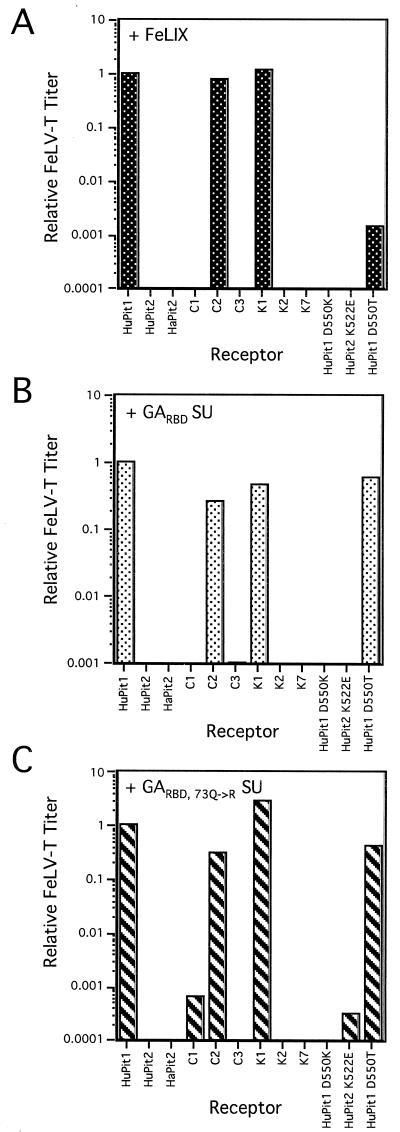
FIG. 3.
FeLV-T infection of cells expressing chimeric Pit receptors. MDTF cells expressing the indicated receptors (x axis in each panel) were challenged with viral particles, pseudotyped with the FeLV-T SU, that packaged the gene for β-galactosidase. Conditioned media containing FeLIX (A), GARBD SU (B), or GARBD,73Q→R SU (C) were diluted 1:10 in fresh media and added at the time of infection. Titers were calculated as β-galactosidase focus-forming units per milliliter. For each receptor and cofactor pair, the titers were normalized to those obtained with the same cofactor on cells expressing HuPit1 (y axis in each panel, log scale). A negative result (no bar shown) indicates that no β-galactosidase-positive foci were seen with 100 μl of viral supernatant. Data are representative of at least two independent experiments.
Four chimeras were tested that allowed us to determine whether there are domains in the N-terminal half of Pit1 that affect FeLV-T receptor specificity—C1, C2, C3, and K1. FeLV-T was able to infect cells expressing a HuPit1 receptor containing HaPit2 sequences in domains 1, 2, and 3 when either FeLIX or the GA-derived SUs were used as cofactors (see Fig. 3, K1). Therefore, HaPit2 sequences are tolerated in these regions. However, cells expressing a HuPit1 receptor containing N-terminal HuPit2 sequences, in domains 2 and 3, and HuPit1 sequences elsewhere were resistant to FeLV-T infection when either FeLIX or the GA-derived SUs were used as cofactors (see Fig. 3, C3). Because the first domain (amino acids 1 to 65 in HuPit1 and 1 to 50 in HuPit2) of HaPit2 and HuPit2 are identical at the amino acid level (58), these data indicate that there is an N-terminal Pit determinant (domain 2 and/or 3) for FeLV-T infection and that HaPit2 sequences also encode this determinant while HuPit2 sequences are not tolerated. We therefore focused on chimeras containing domains 2 and 3 individually to further localize this determinant. FeLV-T was able to utilize a chimeric HuPit1 receptor containing HuPit2 sequences in domain 3 with all three cofactors (see Fig. 3, C2). In contrast, cells expressing a chimeric HuPit1 receptor with HuPit2 sequences in domain 2 remained largely resistant to infection, although a very low level of infection was observed with the GARBD,73Q→R SU (see Fig. 3, C1). These data suggest that HuPit1 sequences in domain 2 (amino acids 66 to 215 in HuPit1) encode a key determinant for FeLV-T infection. Furthermore, both HuPit1 and HaPit2 contain this determinant, whereas HuPit2 sequences are not tolerated.
Region A is a determinant of FeLV-T infectivity.
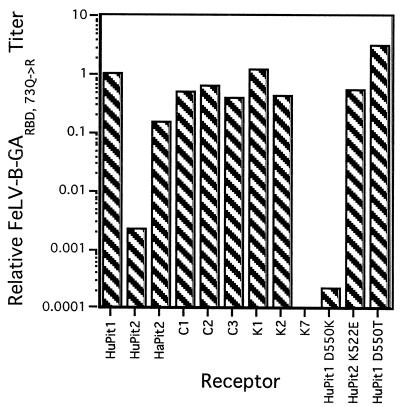
We next tested chimeric HuPit1 receptors with Pit2 substitutions at the C terminus of the protein. Cells expressing a HuPit1 receptor with HuPit2 sequences in domain 4 were not infectible by FeLV-T with any of the cofactors tested (see K7, Fig. 3). Similarly, none of the cofactors could mediate FeLV-T infection using HuPit1 receptors containing HaPit2 sequences in this region of the receptor (see K2, Fig. 3). However, evidence from infection studies with FeLV-B (shown below) and A-MLV verifies that these chimeric receptors are expressed at the cell surface and are functional for entry. Therefore, these data indicate that there is also at least one C-terminal HuPit1 determinant for FeLV-T infection.
Domain 4 of Pit1 and Pit2 contains region A, a stretch of nine amino acids that is highly polymorphic among species and contains determinants for FeLV-B and GALV infection (25, 53). In contrast, the remainder of domain 4 is much more conserved among Pit1 and Pit2 proteins. We therefore hypothesized that at least one of the C-terminal determinants identified above is a single amino acid difference within region A. HuPit1 encodes an aspartic acid at position 550, while HuPit2 has a lysine at the analogous position (amino acid 522, Fig. 2). When we tested cells expressing a HuPit1 receptor containing an aspartic acid-to-lysine substitution at position 550, we found that they were resistant to FeLV-T infection (see HuPit1 D550K, Fig. 3). In contrast, all three cofactors could mediate infection of cells expressing a HuPit1 receptor with a threonine substitution at position 550, although FeLV-T titers were considerably lower with FeLIX as cofactor (see HuPit1 D550T, Fig. 3). While these results suggest that an acidic or neutral residue at HuPit1 position 550 is an important FeLV-T receptor determinant, other residues in region A and/or sequences at the N terminus may also be required (see above). FeLV-T was not able to efficiently infect cells that express a HuPit2 receptor encoding a glutamic acid residue at the analogous position (see HuPit2 K522E), further supporting a model in which there are multiple domains of the Pit1 receptor that are required for FeLV-T infection. Together, these studies of the HuPit1 C terminus demonstrate that, as in the case of GALV and FeLV-B (25, 53), the first position of region A is a key determinant for FeLV-T infection.
There are Pit1 determinants for FeLV-T entry in addition to those required for cofactor binding.
Our previous results suggested that cofactor binding to the Pit1 receptor was necessary but not sufficient for FeLV-T infection (29). Therefore, the Pit1 receptor determinants identified above could play a role in either cofactor binding or subsequent events in FeLV-T infection. To define binding determinants and distinguish between these two possibilities, we analyzed cofactor binding to the chimeric and mutant receptors using a flow-cytometric assay (4, 29, 50). In this assay, cells are incubated in conditioned medium containing the cofactor in question (see Fig. 1B), and bound cofactor is detected with fluorescently labeled antibodies. We could detect binding of cofactor to receptor for all cofactor-Pit receptor combinations that were able to mediate FeLV-T infection (Fig. 4 and data not shown). This is consistent with a requirement for cofactor-receptor interactions in FeLV-T entry.
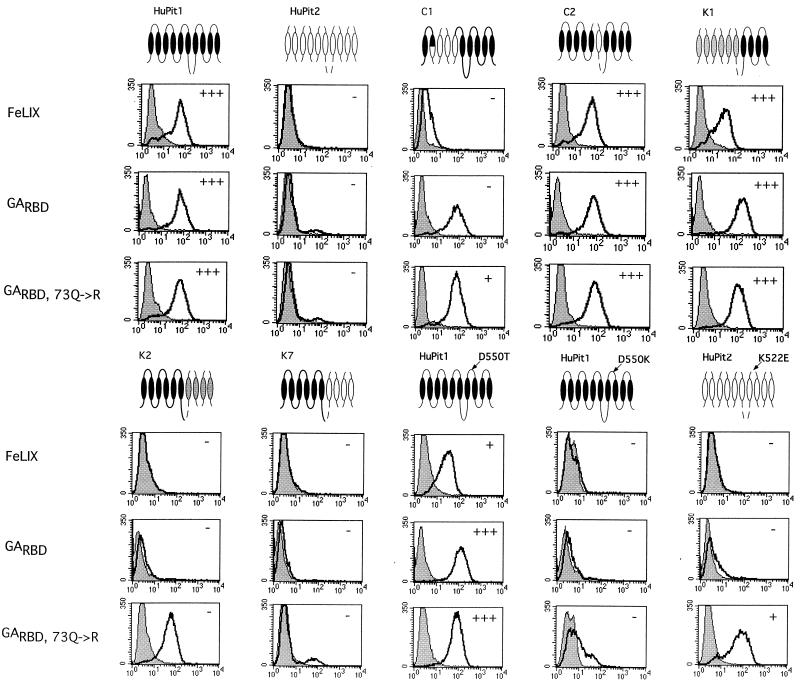
FIG. 4.
Receptor binding properties of HA-tagged SU cofactors. MDTF cells expressing the indicated receptors (top of figure) were incubated with 500 μl of conditioned medium containing the SUs indicated (left of figure) as described in reference 29. Bound SUs were detected by staining with a monoclonal antibody directed against the HA epitope. In all cases the x axis is fluorescence intensity (log scale) and the y axis is cell number. Mock samples (shaded profiles) are cells incubated in standard medium and stained with the same antibody. Data are representative of at least three independent experiments. The ability of each receptor to support infection by FeLV-T with the corresponding receptor and soluble cofactor, as shown in Fig. 3, is represented for convenience here as + or − in the upper right corner of each panel. +++ indicates a relative FeLV-T titer of 0.1 to 1.0 for the indicated cofactor and receptor compared to titers with the same cofactor on HuPit1 cells. ++ indicates a relative titer of 0.01 to 0.1, + indicates a relative titer of 0.0001 to 0.01, and − indicates that no β-galactosidase-positive foci were seen with 100 μl of viral supernatant.
In the case of FeLIX, there was a complete concordance between receptor determinants for cofactor binding and FeLV-T infection. For example, we could not detect efficient binding by FeLIX to cells expressing the C1 receptor (HuPit2 domain 2), suggesting that HuPit1 amino acids 66 to 215 are required for binding by FeLIX (Fig. 4). We were also unable to detect binding to receptors containing HuPit2 or HaPit2 sequences in domain 4 (K7 and K2), implicating residues in the C-terminal half of Pit1 in FeLIX binding. FeLIX was able to bind to the HuPit1 D550T receptor, yet other region A substitutions were not tolerated (see HuPit1 D550K and HuPit2 K522E). Therefore, our data suggest that HuPit1 sequences in both domain 2 and region A are necessary for FeLIX binding.
We obtained somewhat different results with the GARBD SU cofactor. Like FeLIX, the GARBD SU was not able to efficiently bind receptors with Pit2 sequences in domain 4 (K7 and K2) and, among the region A mutants tested, was only able to bind to HuPit1 D550T (Fig. 4). These results indicate that the C-terminal receptor determinants are similar for FeLIX and the GARBD SU. Unlike FeLIX, we did observe significant binding of the GARBD SU to the C1 receptor, which contains HuPit2 sequences in domain 2. However, cells expressing the C1 receptor were not infectible by FeLV-T when the GARBD SU was used as a cofactor (see Fig. 3, above). Because the GARBD SU can bind to C1 but does not mediate FeLV-T infection through the same receptor, HuPit1 sequences in domain 2 are likely required for aspects of FeLV-T infection other than cofactor binding. Therefore, while this region of Pit1 may be required for both FeLIX binding and subsequent events in FeLIX-mediated FeLV-T entry, it appears to be necessary only for aspects of infection other than cofactor binding when the GARBD is used as the FeLV-T cofactor.
We previously have shown that the GARBD,73Q→R SU can utilize HuPit2, HaPit2, and FePit2 as receptors in addition to Pit1 (4, 50). Consistent with these observations, we found that the GARBD,73→R SU was able to bind to many of the chimeric receptors containing Pit2 sequences. We found no specific requirement for HuPit1 sequences at the N terminus of the receptor since the GARBD,73Q→R SU could bind to the K1 receptor, which encodes HaPit2 sequence in domains 1, 2, and 3 (data not shown). Similarly, we also observed binding to the C1 and C2 receptors, which have HuPit2 sequences in domains 2 and 3, respectively (Fig. 4 and data not shown). Unlike the related GARBD SU, the GARBD,73Q→R SU could bind to a receptor with HaPit2 sequences in domain 4 (K2), reflecting its broader receptor tropism. However, the GARBD,73Q→R SU could neither bind to nor mediate FeLV-T infection through the K7 receptor, which has HuPit2 sequences in the same domains (Fig. 4). These data suggest that there is a binding determinant for the GARBD,73Q→R SU in the C-terminal half of HuPit1 and that HaPit2 sequences encode this same determinant whereas HuPit2 sequences do not. Neither of these C-terminal chimeras could support FeLV-T infection when the GARBD,73Q→R cofactor was used. The ability of the K2 chimera, encoding HaPit2 sequences in domain 4, to support GARBD,73Q→R binding but not FeLV-T infection suggests that, like domain 2, these C-terminal determinant(s) are required for events in entry other than cofactor binding.
Of the receptors bearing mutations in region A, FeLV-T was able to utilize only HuPit1 D550T, independent of the cofactor used. Consistent with this observation, we were able to detect binding of all three cofactors to this receptor (Fig. 4). Although the mean fluorescence intensity of the GARBD,73Q→R SU on HuPit1 D550T appears greater than on HuPit1 in the profiles shown, this small difference was not reproducible from experiment to experiment. In contrast, we could not detect FeLIX binding to either HuPit1 D550K or HuPit2 K522E. Similarly, we detected little to no binding by the GARBD SU to these receptors. We did observe binding of the GARBD,73Q→R SU to HuPit1 D550K, although the shift in fluorescence intensity was weaker than with HuPit1 or HuPit1 D550T. The GARBD,73Q→R SU could efficiently bind to HuPit2 K522E, indicating that a single region A substitution in HuPit2 can enhance binding of this cofactor. While we observed GARBD,73Q→R SU cofactor binding to all region A mutants, the GARBD,73Q→R SU was not able to mediate FeLV-T infection of cells expressing HuPit1 D550K or HuPit2 K522E. These data suggest that the residue at position 550 is a key receptor determinant for events in FeLV-T entry other than cofactor binding. It appears that FeLV-T requires either an acidic residue or a neutral residue in this first position of region A. An acidic residue in region A is not in itself sufficient, however, since the HuPit2 K522E receptor could support cofactor binding but not infection by FeLV-T. Therefore, other HuPit1 domains, such as domain 2, are required for FeLV-T entry.
Chimeric Pit proteins that do not support FeLV-T infection are functional FeLV-B receptors.
By testing the ability of the chimeric receptors both to bind cofactor and to mediate FeLV-T infection, we have been able to identify regions in Pit1 that are dispensable for cofactor binding but necessary for infection. The binding studies also show that these receptors are expressed on the cell surface. However, these experiments do not rule out the possibility that the mutant or chimeric receptors are no longer able to carry out events such as envelope and receptor activation as a result of the engineered changes. To examine this further, we measured FeLV-B infection on cells expressing these mutant and chimeric receptors. Using a single-cycle-of-infection assay, we found that all of the receptors to which the GARBD,73Q→R SU could bind were able to support infection by FeLV-B-GARBD,73Q→R (Fig. 5). FeLV-B could infect cells expressing receptors containing Pit2 substitutions within either the N-terminal (K1, C1, C2, and C3) or C-terminal determinants (K2) identified above. While FeLV-B-GARBD,73Q→R was not able to utilize either the HuPit1 D550K or K7 receptors, these receptors were able to support infection by other retroviruses. Specifically, MDTF-HuPit1 D550K cells could support infection by GALV, and Chinese hamster ovary cells expressing K7 could be infected by A-MLV (data not shown). Therefore, all of the receptors used in this study are functional for retroviral entry, and chimeric receptors with Pit2 sequences in domains 2 and 4 are able induce the necessary SU- and receptor-mediated events in entry by subgroup B FeLV. The inability of these same receptors to support FeLV-T infection in the presence of FeLV-B SU cofactors suggests that these determinants are required for interactions specific to FeLV-T.
FIG. 5.
Infection of cells with viral particles pseudotyped with the FeLV-B SU. MDTF cells expressing the indicated receptors (x axis in each panel) were challenged with FeLV-B-GARBD,73Q→R pseudotypes that packaged the gene for β-galactosidase. Titers were calculated as β-galactosidase focus-forming units per milliliter. For each receptor, the titers were normalized to those on cells expressing HuPit1 (y axis in each panel, log scale). A negative result (no bar shown) indicates that no β-galactosidase-positive foci were seen with 100 μl of viral supernatant. Data are representative of at least three independent experiments.
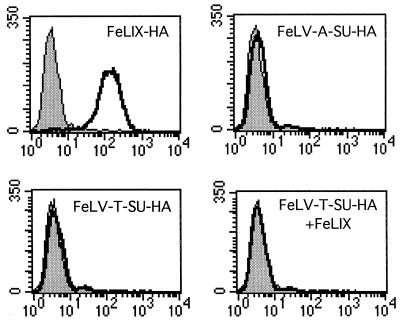
FeLV-T SU binding to HuPit1 cannot detected by FACS analyses.
Our identification of domains specifically required for FeLV-T infection could indicate a role for HuPit1 in FeLV-T envelope binding. We therefore assayed receptor binding by the FeLV-T SU using our flow-cytometric assay. Conditioned media containing HA-tagged FeLV-T and FeLV-A SUs were generated as described above and expression verified by immunoprecipitation (data not shown). As before, FeLIX was able to bind cells expressing the HuPit1 receptor but not the parental MDTFs (Fig. 6). Consistent with the cellular tropism of FeLV-A, the FeLV-A SU did not bind to either MDTF or MDTF-HuPit1 cells (40). Surprisingly, we were unable to detect significant binding of the FeLV-T SU to cells expressing HuPit1 even when FeLIX was added to the binding reaction. These studies do not rule out the possibility that FeLV-T binds weakly to Pit1, because our previous studies suggest that there are cases where FeLVs can infect at a reduced level via a specific receptor, but this low-level binding cannot be detected by FACS (4). These studies do indicate that receptor binding by FeLV-T is much weaker than binding by either FeLV-B or FeLIX. However, we are mindful that we have analyzed receptor binding under only one experimental condition, where free virus and cofactor are added simultaneously to permissive cells.
FIG. 6.
MDTF-HuPit1 cells were incubated with 500 μl of conditioned medium containing FeLIX, FeLV-A SUs, or FeLV-T SUs in a 1-ml total volume as described in reference 29. Bound SUs were detected by staining with a monoclonal antibody directed against the HA epitope. For the FeLV-T SU plus FeLIX binding reactions, cells were incubated with 500 μl of FeLV-T SU conditioned media and 500 μl of conditioned media containing an untagged version of FeLIX. In all cases the x axis is fluorescence intensity (log scale) and the y axis is cell number. Mock samples (shaded profiles) are cells incubated in standard media and stained with the same antibody.
DISCUSSION
Cytopathic, T-cell-tropic FeLV-T isolates are the only known examples of naturally arising, simple retroviruses that require two host cell proteins for entry into target cells (3). In our previous studies, we showed that FeLV-T is specific in its requirement for the Pit1 transmembrane receptor and a cofactor containing an RBD derived from enFeLV sequence (29). These studies showed that cofactor binding to the transmembrane receptor is necessary but not sufficient for infection and that the binding affinity of these two proteins does not appear to be a primary determinant of FeLV-T tropism. Together, these data suggested that entry by FeLV-T is dependent on a series of interactions among Pit1, the cofactor (e.g., FeLIX), and the FeLV-T envelope. In an attempt to better understand these requisite intermolecular interactions, we utilized a panel of chimeric and mutant Pit proteins to map receptor determinants for FeLV-T infection. We also used three closely related enFeLV-derived cofactors that would be expected to be found within infected animals (4, 29). By assaying both cofactor binding and the ability of FeLV-B virus to infect using each receptor, we were able to define Pit1 determinants for FeLV-T infection that are not required for cofactor binding or receptor activation by FeLV per se but are specifically required for FeLV-T infection. These data imply that there is a direct role for Pit1 residues in binding or postbinding events mediated specifically by the FeLV-T envelope protein.
A strength of our approach was that we were able to independently measure the binding properties and cofactor activity of closely related enFeLV-derived envelope fragments that differ in their ability to bind to Pit2. Using these assays, we were able to identify three distinct classes of Pit1 determinants. The first lies within the C-terminal half of the protein and contains a key determinant for cofactor binding; none of the cofactors could bind to receptors with HuPit2 sequences in domain 4, and only one, the GARBD,73Q→R SU, could bind when HaPit2 sequences were substituted. Variation within region A is likely to play some role in the differential effect of HuPit2 versus HaPit2 substitutions, since sequence differences between these two orthologs cluster within this subdomain. The second set of receptor mutants revealed a Pit1 determinant in domain 2 that is important for both cofactor binding and FeLV-T infection. Substitutions within this region impaired FeLIX binding but not binding by the GA-derived SU cofactors, suggesting that the binding determinants of these enFeLV-derived cofactors differ. We cannot distinguish whether these differences result from amino acid differences in the RBD of these cofactors or whether they result from the fact that FeLIX lacks the C-terminal domain. Recent studies by Lavillette and colleagues suggest that a C-terminal domain in the MLV SU is important in postbinding events in infection (30). However, because there are differences in binding between GARBD and GARBD,73Q→R, which share identical C-terminal sequences, at least some of the distinctions in binding reflect the amino acid change in the RBD. Our data also suggest that domain 2 of Pit1 is necessary for FeLV-T infection, because binding by the GA-derived cofactors was not sufficient for viral entry.
Finally, a third class of receptor mutants was identified that affects only FeLV-T infection. This conclusion is based on the observation that all three cofactors could bind to HuPit1 D550T, which contains a substitution within region A, while cells expressing this receptor were almost completely resistant to FeLV-T infection in the presence of FeLIX. These HuPit1 D550T cells were, however, susceptible to infection when the GA-derived cofactors were tested, implying that this infection determinant is cofactor dependent. We interpret these results with caution, however, since the observed differences in FeLV-T infectivity with the three cofactors could also be due to subtle differences in cofactor binding affinity that are beneath the resolution of our flow cytometric assay. Our previous studies suggest that the amount of cofactor used is at or near saturation (29). The charge of the residue at position 550 also appears to be key, since none of the cofactors could mediate infection of cells expressing the HuPit1 D550K receptor mutant. In this case, the mutation affects cofactor binding as well. Importantly, all of the receptors used in this study were functional for retroviral entry, suggesting that the determinants we have identified are specific for FeLV-T infection.
Our results suggest that Pit1 plays a role in FeLV-T entry, in addition to cofactor binding and receptor activation. However, we do not detect binding between the FeLV-T SU and Pit1 under conditions where we can detect binding between the cofactor and Pit1. This suggests either that FeLV-T does not bind to Pit1 directly or that binding is weak. Indeed, our previous studies suggest that this binding assay cannot detect lower-affinity interactions between FeLV envelopes and receptors that nonetheless permit entry (4), and similar observations have been made for MLV (1). Thus, these experiments do not allow us to conclude whether FeLV-T binds directly to Pit1, but they do indicate that binding to Pit1 by FeLV-T, if it occurs, is a much lower-affinity interaction than binding by FeLV-B SU or FeLIX. Interestingly, this is in contrast to defective MLVs, which efficiently bind receptor but require soluble SUs to rescue infection.
On the basis of our analyses of chimeric receptors that bind cofactor but do not permit FeLV-T entry, we hypothesize that Pit1 is required for FeLV-T envelope binding, either directly or as part of a ternary complex with the soluble cofactor. A role for Pit1 in viral recruitment would, in part, explain the observed receptor specificity of FeLV-T. This specificity differs from what has been reported in the MLV system, where viruses pseudotyped with defective envelopes could be rescued by several different soluble SU proteins as long as receptors for both the viral envelope and the SU cofactor were expressed in target cells (31). These data suggest that receptor binding by a soluble SU can activate fusion by the defective MLV envelope through its receptor in trans. In the case of the naturally arising FeLV-T variant, we have shown that several other SU-receptor pairs are unable to mediate FeLV-T infection through Pit1 even when Pit1 and the receptor used by the SU cofactor are coexpressed in the target cell membrane (29). Importantly, we found that an FeLV-B SU that binds equally well to both Pit1 and Pit2 can only mediate FeLV-T infection through Pit1, indicating that FeLV-T is not simply being recruited into the complex by the cofactor. One model to explain these data is that Pit1 plays a role in both cofactor and viral envelope binding. In this model, binding by the cofactor to Pit1 would activate fusion by the FeLV-T envelope, also bound to Pit1. These interactions could potentially occur either on separate Pit1 proteins or as part of a ternary complex with each Pit1 protein occupied by both cofactor and viral envelope. Indeed, some of our data imply that interactions between the FeLV-T envelope and the cofactor are necessary for Pit1 binding. We have found that only enFeLV-derived cofactors, and not other envelopes that bind to Pit receptors, such as A-MLV or GALV, are able to mediate FeLV-T infection (29). Our identification here of a region A determinant for infection that is cofactor dependent could suggest that cofactor-envelope interactions compensate for weaker interactions between the viral envelope protein and the Pit1 receptor. It is therefore interesting that data from the MLV system indicate that soluble SUs may activate fusion by interacting with a C-terminal domain in the viral envelope protein (6, 7, 30).
The human and simian immunodeficiency viruses (HIV and SIV) are the only other known examples of retroviruses that require two host cell proteins for infection, and as such may inform our understanding of entry by FeLV-T. In most cases, entry by these primate lentiviruses requires successive interactions with the type I transmembrane protein CD4 and a multiple transmembrane chemokine receptor, such as CCR5 or CXCR4 (44). After binding to CD4, the viral envelope undergoes a conformational change, exposing a binding site for the chemokine coreceptor, and binding to this coreceptor activates the viral envelope to a fusogenic state (24). Consistent with the catalytic role of CD4 in HIV and SIV entry, soluble forms of CD4 can facilitate infection of CD4-negative cells (2). Like CD4, both membrane bound (FeLV-B envelope proteins) and soluble (FeLIX) cofactors can mediate infection by FeLV-T even though FeLIX and CD4 both probably act at the target cell membrane in a natural infection. Our data would be consistent with a model in which the FeLV-T cofactor performs a similar catalytic function in entry. In one such model, receptor binding by FeLIX induces a conformational change in Pit1 that exposes a binding site for the FeLV-T envelope. This ordered binding model is in contrast to the trans-activation or ternary complex models discussed above, which appear to explain entry by defective MLVs. Alternatively, the cofactor may trigger structural rearrangements in the FeLV-T envelope protein, exposing a Pit1 binding site; to date, the data are consistent with either of these models of cofactor-induced conformational changes. As discussed above, we have been unable to detect stable binding by the FeLV-T SU to cells expressing Pit1, even in the presence of FeLIX, using FACS analyses. Interestingly, it has not been possible to detect binding by the HIV-1 SU to the CXCR4 coreceptor under conditions where binding to CCR5 can be detected, and other approaches have been required to detect this interaction (23). Thus, it will be important to develop sensitive, quantitative methods to directly examine protein-protein interactions between the FeLV-T envelope and both FeLIX and Pit1.
In this study, we have differentiated between cofactor binding to Pit1 and productive FeLV-T infection. Importantly, we have also controlled for the possibility that the mutations in the Pit1 receptors affect cell surface expression or their ability to undergo receptor activation. We have found a role for Pit1 in both cofactor binding and infection, and our results further suggest that Pit1 could potentially serve as a receptor for the FeLV-T envelope protein. FeLV-T is just one of five simple retroviruses that are known to use either Pit1 or Pit2 for entry (3, 37, 38, 41, 54, 55, 58). It is interesting that domain 2 and/or region A have been identified as important Pit receptor determinants for infection by GALV, A-MLV, and FeLV-B (25, 51-53) and that region A may play a role in postbinding events in FeLV-B entry (18a). The fact that these domains are also important for infection by FeLV-T suggests that there may be common receptor-mediated events in entry by these viruses. Therefore, while certain aspects of FeLV-T entry appear to be specific to this subgroup, further dissection of the intermolecular interactions and receptor determinants for FeLV-T may serve to elucidate fundamental principles of retroviral infection.
Acknowledgments
We thank A. Dusty Miller for helpful discussion and Maria Anderson, Claire Hankenson, and Jennifer Riddell for technical assistance.
This work was supported by NIH grant CA 51080. A.S.L. was supported by NIH training grant 2 T32 CA09229, an ARCS Fellowship, and the Paul Allen Foundation. H.H.C. was supported by the Paul Allen Foundation.
REFERENCES
- 1.Albritton, L. M., J. W. Kim, L. Tseng, and J. M. Cunningham. 1993. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J. Virol. 67:2091-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan, J. S., J. Strauss, and D. W. Buck. 1990. Enhancement of SIV infection with soluble receptor molecules. Science 247:1084-1088. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, M. M., A. S. Lauring, S. Roberston, C. Dirks, and J. Overbaugh. 2001. Feline Pit2 functions as a receptor for subgroup B feline leukemia viruses. J. Virol. 75:10563-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, A. L., and J. M. Cunningham. 2001. Receptor binding transforms the surface unit of the mammalian C-type retrovirus envelope protein from an inhibitor to an activator of fusion. J. Virol. 75:9096-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnett, A. L., R. A. Davey, and J. M. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Battini, J. L., O. Danos, and J. M. Heard. 1998. Definition of a 14-amino-acid peptide essential for the interaction between the murine leukemia virus amphotropic envelope glycoprotein and its receptor. J. Virol. 72:428-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battini, J. L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battini, J. L., J. M. Heard, and O. Danos. 1992. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 66:1468-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battini, J. L., P. Rodrigues, R. Muller, O. Danos, and J. M. Heard. 1996. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J. Virol. 70:4387-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boomer, S., M. Eiden, C. C. Burns, and J. Overbaugh. 1997. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J. Virol. 71:8116-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudry, G. J., and M. V. Eiden. 1997. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J. Virol. 71:8078-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudry, G. J., K. B. Farrell, Y. T. Ting, C. Schmitz, S. Y. Lie, C. J. Petropoulos, and M. V. Eiden. 1999. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J. Virol. 73:2916-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davey, R. A., C. A. Hamson, J. J. Healey, and J. M. Cunningham. 1997. In vitro binding of purified murine ecotropic retrovirus envelope surface protein to its receptor, MCAT-1. J. Virol 71:8096-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davey, R. A., Y. Zuo, and J. M. Cunningham. 1999. Identification of a receptor-binding pocket on the envelope protein of friend murine leukemia virus. J. Virol. 73:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyer, K., F. S. Pedersen, and L. Pedersen. 2000. A 13-amino-acid Pit1-specific loop 4 sequence confers feline leukemia virus subgroup B receptor function upon Pit2. J. Virol. 74:2926-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eglitis, M. A., M. V. Eiden, and C. A. Wilson. 1993. Gibbon ape leukemia virus and the amphotropic murine leukemia virus 4070A exhibit an unusual interference pattern on E36 Chinese hamster cells. J. Virol. 67:5472-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eiden, M. V., K. B. Farrell, and C. A. Wilson. 1996. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J. Virol. 70:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Farrell, K. B., J. L. Russ, R. K. Murthy, and M. V. Eiden. 2002. Reassessing the role of region A in Pit1-mediated viral entry. J. Virol. 76:7683-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrell, K. B., Y. T. Ting, and M. V. Eiden. 2002. Fusion-defective gibbon ape leukemia virus vectors can be rescued by homologous but not heterologous soluble envelope proteins. J. Virol. 76:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fass, D., R. A. Davey, C. A. Hamson, P. S. Kim, J. M. Cunningham, and J. M. Berger. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 Å resolution. Science 277:1662-1666. [DOI] [PubMed] [Google Scholar]
- 22.Haywood, A. M. 1994. Virus receptors: binding, adhesion strengthening, and changes in viral structure. J. Virol. 68:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman, T. L., G. Canziani, L. Jia, J. Rucker, and R. W. Doms. 2000. A biosensor assay for studying ligand-membrane receptor interactions: binding of antibodies and HIV-1 Env to chemokine receptors. Proc. Natl. Acad. Sci. USA 97:11215-11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman, T. L., and R. W. Doms. 1998. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS 12:S17-S26. [PubMed] [Google Scholar]
- 25.Johann, S. V., M. van-Zeijl, J. Cekleniak, and B. O'Hara. 1993. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J. Virol. 67:6733-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kizhatil, K., and L. M. Albritton. 1997. Requirements for different components of the host cell cytoskeleton distinguish ecotropic murine leukemia virus entry via endocytosis from entry via surface fusion. J. Virol. 71:7145-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klasse, P. J., R. Bron, and M. Marsh. 1998. Mechanisms of enveloped virus entry into animal cells. Adv. Drug Delivery Rev. 34:65-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauring, A. S., M. M. Anderson, and J. Overbaugh. 2001. Specificity in receptor usage by FeLV-T: implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 75:8888-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavillette, D., B. Boson, S. J. Russell, and F. L. Cosset. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J. Virol 75:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavillette, D., A. Ruggieri, S. J. Russell, and F. L. Cosset. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leverett, B. D., K. B. Farrell, M. V. Eiden, and C. A. Wilson. 1998. Entry of amphotropic murine leukemia virus is influenced by residues in the putative second extracellular domain of its receptor, Pit2. J. Virol. 72:4956-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundorf, M. D., F. S. Pedersen, B. O'Hara, and L. Pedersen. 1999. Amphotropic murine leukemia virus entry is determined by specific combinations of residues from receptor loops 2 and 4. J. Virol. 73:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundorf, M. D., F. S. Pedersen, B. O'Hara, and L. Pedersen. 1998. Single amino acid insertion in loop 4 confers amphotropic murine leukemia virus receptor function upon murine Pit1. J. Virol. 72:4524-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, A. D., D. G. Miller, J. V. Garcia, and C. M. Lynch. 1993. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 217:581-599. [DOI] [PubMed] [Google Scholar]
- 37.Miller, D. G., R. H. Edwards, and A. D. Miller. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 91:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, D. G., and A. D. Miller. 1994. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 68:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan, R. A., O. Nussbaum, D. D. Muenchau, L. Shu, L. Couture, and W. F. Anderson. 1993. Analysis of the functional and host range-determining regions of the murine ecotropic and amphotropic retrovirus envelope proteins. J. Virol. 67:4712-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moser, M., C. Burns, S. Boomer, and J. Overbaugh. 1998. The host range and interference properties of two closely related feline leukemia virus variants suggest that they use distinct receptors. Virology 242:366-377. [DOI] [PubMed] [Google Scholar]
- 41.O'Hara, B., S. V. Johann, H. P. Klinger, D. G. Blair, H. Rubinson, K. J. Dunn, P. Saas, S. M. Vitek, and T. Robins. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1:119-127. [PubMed] [Google Scholar]
- 42.Ott, D., and A. Rein. 1992. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J. Virol. 66:4632-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Overbaugh, J., P. R. Donahue, S. L. Quackenbush, E. A. Hoover, and J. I. Mullins. 1988. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science 239:906-910. [DOI] [PubMed] [Google Scholar]
- 44.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedersen, L., S. V. Johann, M. van-Zeijl, F. S. Pedersen, and B. O'Hara. 1995. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J. Virol. 69:2401-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen, L., M. van Zeijl, S. V. Johann, and B. O'Hara. 1997. Fungal phosphate transporter serves as a receptor backbone for gibbon ape leukemia virus. J. Virol. 71:7619-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodrigues, P., and J. M. Heard. 1999. Modulation of phosphate uptake and amphotropic murine leukemia virus entry by posttranslational modifications of PIT-2. J. Virol. 73:3789-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneiderman, R. D., K. B. Farrell, C. A. Wilson, and M. V. Eiden. 1996. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J. Virol. 70:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siess, D. C., S. L. Kozak, and D. Kabat. 1996. Exceptional fusogenicity of Chinese hamster ovary cells with murine retroviruses suggests roles for cellular factor(s) and receptor clusters in the membrane fusion process. J. Virol. 70:3432-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugai, J., M. Eiden, M. M. Anderson, N. Van Hoeven, C. D. Meiering, and J. Overbaugh. 2001. Identification of envelope determinants for feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or Pit2. J. Virol. 75:6841-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tailor, C. S., and D. Kabat. 1997. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J. Virol. 71:9383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tailor, C. S., A. Nouri, and D. Kabat. 2000. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J. Virol. 74:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tailor, C. S., Y. Takeuchi, B. O'Hara, S. V. Johann, R. A. Weiss, and M. K. Collins. 1993. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J. Virol. 67:6737-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeuchi, Y., R. G. Vile, G. Simpson, B. O'Hara, M. K. Collins, and R. A. Weiss. 1992. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J. Virol. 66:1219-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Zeijl, M., S. V. Johann, E. Closs, J. Cunningham, R. Eddy, T. B. Shows, and B. O'Hara. 1994. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA 91:1168-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, H., R. Paul, R. E. Burgeson, D. R. Keene, and D. Kabat. 1991. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J. Virol. 65:6468-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson, C. A., K. B. Farrell, and M. V. Eiden. 1994. Comparison of cDNAs encoding the gibbon ape leukaemia virus receptor from susceptible and non-susceptible murine cells. J. Gen. Virol. 75:1901-1908. [DOI] [PubMed] [Google Scholar]
- 58.Wilson, C. A., K. B. Farrell, and M. V. Eiden. 1994. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J. Virol. 68:7697-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zavorotinskaya, T., and L. M. Albritton. 1999. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J. Virol. 73:5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]