Abstract
Virus infections induce changes in the expression of host cell genes. A global knowledge of these modifications should help to better understand the virus/host cell interactions. To obtain a more comprehensive view of the rainbow trout response to a viral infection, we used the subtractive suppressive hybridization methodology in the viral hemorrhagic septicemia model of infection. We infected rainbow trout leukocytes with viral hemorrhagic septicemia virus (VHSV), and total RNA from infected and mock-infected cells was compared at 40 h postinfection. Twenty-four virus-induced genes were ultimately retrieved from the subtracted cDNA library, and their differential expression was further confirmed by semiquantitative reverse transcription-PCR and Northern blot analysis. Among these sequences, three were already described as VHSV-induced genes. Eight sequences with known homologs were extended to full-length cDNA using 5′ and 3′ rapid amplification of cDNA ends, and they were subsequently divided into three functional subsets. Four genes were homologous to mammalian interferon responsive genes, three were similar to chemo-attractant molecules (CXC chemokine, galectin), and two had nucleic acid binding domains. All of the virus-induced genes were also induced by rainbow trout interferon, indicating that the interferon pathway is the predominant component of the anti-VHSV response. They were also expressed in vivo in experimentally infected fish, indicating their biological relevance in natural infection.
The host response to viral infection represents a complex coordination of gene products, which are precisely tuned to activate or inactivate specific pathways and finally counteract the effects of viral gene products. A view of the components participating in this first line of defense will certainly help in understanding the pathological issues of typical viral infections. With the use of large-scale screening of mRNA changes, it is possible to define changes in gene expression that underlie the host response to viral pathogens and to gain specific insights into the molecular nature of the host pathways that govern viral pathogenesis. In this regard, viral hemorrhagic septicemia constitutes an interesting model to study the host response to viral infection. Indeed, this disease is caused by a virus with a small set of genes and develops in a lower vertebrate, and these characteristics could assist in the elucidation of the generic biochemical pathways involved in either immune or pathological outcomes.
Viral hemorrhagic septicemia is an important viral disease occurring in rainbow trout Oncorhynchus mykiss in Europe. Clinical signs of the disease caused by this virus consist of severe hemorrhages on the skin, muscles, and internal organs and an overall systemic infection leading to high mortality in juvenile fish. The causative agent is a single-stranded RNA virus, which belongs to the family Rhabdoviridae, genus Novirhabdovirus. The nonsegmented negative-strand RNA genome of the viral hemorrhagic septicemia virus (VHSV) is now completely resolved (41). It encodes six proteins: the nucleocapsid N (4), the polymerase-associated protein P, the matrix protein M (3), the transmembrane glycoprotein G (43), a nonstructural protein (NV) (2), and the RNA-dependent RNA polymerase L.
The rainbow trout is able to mount a strong specific immune response against the virus. Protection is specifically afforded by neutralizing antibodies, which are directed against the viral glycoprotein (6, 31). The induction of T cells specifically reactive to the virus was also recently demonstrated (7). Mx and major histocompatibility complex (MHC) class II gene expression was up-regulated at the site of injection of a DNA vaccine expressing the VHSV glycoprotein (6, 26), indicating that cells of the immune system and components of the nonspecific response were activated. However, little is still known about the antiviral nonspecific response in teleosts. The induction of an interferon-like activity by viruses was described in rainbow trout several years ago, but neither the protein nor the gene have been identified in any fish species. In mammals, type I interferon plays a key role in the host response to viral infection. More than 100 interferon-responsive genes have been described to date (15, 46), and this highlights the diversity of the cell response mechanisms selected by viruses. Many of these interferon-induced genes remain poorly characterized, but some have a strong antiviral activity. The best-characterized interferon-induced antiviral pathways involve the Mx, the double-stranded RNA (dsRNA)-dependent protein kinase, and the 2′,5′ oligo(A) synthetase system. However, the complex nature of the interferon-mediated resistance to viruses (50) remains to be further investigated. In teleosts, several homologs of mammal interferon-induced genes have been cloned (8, 44, 47), and interferon-stimulating responsive elements have been identified in the promoter region of new virus-induced genes in rainbow trout (9, 14).
In order to take a broad view of the changes following VHSV infection, we have used large-scale screening of changes in mRNA from rainbow trout leukocytes that have been subjected to VHSV infection. One such method, differential display reverse transcription (RT)-PCR, led previously to the identification of VHSV-induced genes, vig-1 (8) and vig-2 (9). Both vig-1 and vig-2 were also responsive to rainbow trout interferon, but only vig-1 showed direct induction by the viral glycoprotein. To expand our screening to a larger set of genes, we used the methodology of suppressive subtractive hybridization (SSH) (16) in this study, which resulted in a cDNA library enriched with gene transcripts differentially expressed. Screening this library with complex forward and reverse subtracted probes led to the identification of 24 differentially expressed sequences, which included vig-1, vig-2, and Mx3. This set of transcripts represents the first cluster of rainbow trout genes descriptive of the leukocyte response to VHSV infection. From this cluster, eight new virus-induced genes were further characterized, and all were shown to respond in vitro to rainbow trout interferon and in vivo to VHSV experimental infection.
MATERIALS AND METHODS
Fish and leukocyte preparation.
Rainbow trout were raised in the Jouy-en-Josas experimental fish facility. The fish were sacrificed by overexposure to 2-phenoxyethanol diluted 1:1,000. The entire kidneys were removed aseptically and dissected. Cells from the head kidney (pronephros) of a single fish were deposited on a Ficoll solution (Lymphocyte separation medium [d = 1.077]; Eurobio, Les Ullis, France) and centrifuged 10 min at 900 × g. The leukocyte fraction at the Ficoll-medium interface was collected and divided into two aliquots of 4.5 × 107 cells. Cells from one aliquot were incubated for 40 h with strain 07-71 of VHSV (1 PFU per cell). The second aliquot was used as a control and incubated with cell culture medium.
RNA isolation, cDNA synthesis, and subtraction.
Infected and control cells were scraped from the culture flask and centrifuged (1,500 × g, 5 min, 4°C), and total RNA was extracted with the Trizol reagent (Life Technologies) according to the manufacturer's instructions. cDNA was synthesized from total RNA using the SMART PCR cDNA Synthesis Kit (Clontech). In short, the process included the use of a modified oligo(dT) primer (CDS), which primed the first-strand synthesis reaction using reverse transcriptase. When the mRNA cap was detected, the enzyme added extra nucleotides that served as a template for a SMART oligonucleotide. The product of the reverse transcription was full-length, single-stranded cDNA, including the 5′ end of the mRNA along with sequences complementary to the SMART oligonucleotide, which allowed for complete cDNA amplification with universal primers. To compare the two populations of resulting cDNA, the method of SSH was then performed using the PCR-Select cDNA Subtraction Kit (Clontech) (Fig. 1). Briefly, the cDNAs from the tester (with virus) and driver (without virus) were digested with RsaI, and the tester cDNA was then ligated to either of two different cDNA adaptors. During a first hybridization, excess driver was added to tester cDNA samples, which were then denatured and allowed to anneal. This step allowed for an equalization of high- and low-abundance sequences. Concurrently, differentially expressed sequences were significantly enriched. In the second hybridization, the two primary hybridization samples were mixed without denaturation. As a result, the remaining subtracted, equalized single-stranded tester cDNAs reassociated to form hybrids with a different adaptor on each end. To further select for differentially expressed sequences, denatured driver cDNA was again added to these hybrid samples. These forward-subtracted samples were then used in PCR to amplify the differentially expressed sequences. Reverse subtraction with the tester sample representing the cDNA without virus and the driver as the cDNA with virus was also performed for subsequent screening analyses.
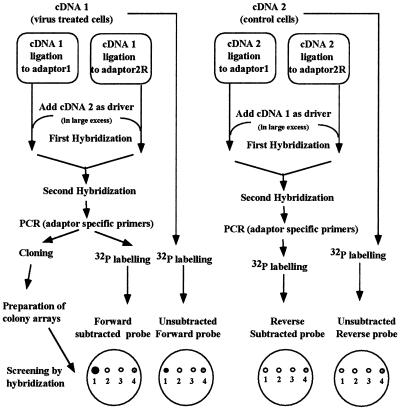
FIG. 1.
Subtraction and screening protocols. cDNA subtracted library was constructed by cloning of the forward-subtracted PCR product: cDNA1(from virus-treated cells) was ligated to adaptors and saturated using cDNA2 (from control cells). The probes used for screening were obtained from original cDNA1 (Unsubtracted Forward probe) and cDNA2 (Unsubtracted Reverse probe), from forward-subtracted PCR product (Forward subtracted probe), and for reverse-subtracted PCR product (Reverse subtracted probe). Colonies hybridizing with Forward subtracted and Unsubtracted forward probes, but not with Reverse subtracted or with unsubtracted reverse probes, were selected (colony 1 in the figure).
Construction of the subtracted cDNA library.
The PCR products from the subtraction procedures were purified with Sephacryl S-400 columns (Pharmacia) and then cloned using the TOPO T/A Cloning Kit (Invitrogen) to create the subtracted-cDNA library. Briefly, 1, 3, and 4 μl of the forward-subtracted-cDNA were each ligated to the TOPO vector and then transformed into competent Escherichia coli TOP 10F′ cells. The transformed E. coli cells were plated onto Luria-Bertani (LB) agar containing ampicillin (100 mg/ml) and 5-bromo-4-chloro-3-indolyl-d-galactoside (X-Gal) and incubated at 37°C overnight. Individual white colonies were picked and incubated in a 96-well microtiter plate (100 μl of LB-ampicillin per well) overnight at 37°C.
Screening the subtracted cDNA library.
The subtracted library was analyzed for differential expression by utilizing the PCR-Select Differential Screening Kit (Clontech) according to the manufacturer's instructions. In this step, the subtracted cDNA library was hybridized with forward- and reverse-subtracted cDNA and with unsubtracted cDNA probes (Fig. 1). First, four separate nylon membranes were placed onto LB-ampicillin agar plates. The overnight broth cultures of white colonies from the subtracted cDNA library were then transferred from the 96-well plate onto the four nylon membranes using a multichannel pipette. Thus, the clones were similarly disposed onto each of the four membranes. The agar plates containing the nylon membranes were incubated overnight at 37°C. The membranes were each transferred onto a piece of Whatman paper presaturated with a denaturation solution (0.5 M NaOH, 1.5 M NaCl) for a 4-min incubation and then onto Whatman paper presaturated with a neutralization solution (0.5 M Tris-HCl [pH 7.4], 1.5 M NaCl) for 4 min. Each membrane was allowed to dry, and the DNA was fixed by baking for 2 h at 80°C. Membranes were prehybridized with a 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) solution containing calf thymus solution (1:1,000) as a blocking agent diluted in the ExpressHyb hybridization solution (Clontech). From the four similar membranes, one was hybridized with a forward-subtracted probe, one was hybridized with a reverse-subtracted probe, one was hybridized with an unsubtracted tester probe, and the last one was hybridized with an unsubtracted driver probe. The corresponding 32P-labeled probes were made from cDNAs or PCR products with the Ready prime II labeling system (Amersham Pharmacia, Little Chalfont, United Kingdom). Membranes were incubated with the probes overnight at 72°C. The samples were then washed in low-stringency (2× SSC-0.5% sodium dodecyl sulfate) and high-stringency (0.2× SSC-0.6% sodium dodecyl sulfate) solutions and exposed to X-ray film with an intensifying screen overnight at −80°C.
Sequence determination.
Colonies that showed a differential response by reacting with the forward-subtracted probe only or the forward-subtracted and unsubtracted tester probes only were selected for analysis. These colonies were grown overnight in LB-ampicillin broth, and the plasmid was purified with a plasmid miniprep spin kit (Nucleospin; Macherey-Nagel, Düren, France). Purified plasmids were subjected to automated sequencing with direct and reverse universal primers.
Semiquantitative RT-PCR assay.
Unique sequences identified by the differential screening procedure and grouped by cross-comparison analysis were analyzed for differential expression by semiquantitative RT-PCR (18) using primers designed specifically for each differential sequence. PCR conditions were as follows: 94°C for 5 min, 94°C for 45 s and 50 to 65°C for 45 s, and 72°C for 1.5 min for 25 to 33 cycles, followed by 72°C for 5 min. Primers used are indicated in Table 1.
TABLE 1.
PCR primers for amplification of virus induced candidate genes
| Primer | Sequence (5′→3′) | Annealing temp (°C) | PCR product size (bp) |
|---|---|---|---|
| A29D | CATCAGTCTCAGCGATGATTCAAA | 55 | 225 |
| A29R | TGCTTGCCCTCGTGAATCAG | ||
| B44D | CAGTTTCTCGGAGTGCATCACA | 55 | 174 |
| B44R | CAGAGACACTTCAGCACCTTC | ||
| B6D | TGTCTCCTGATGTGCCTCAC | 55 | 613 |
| B6R | CATTCCTTTGCCTAGGGCATCT | ||
| A83D | CGATGTCACTGCAGGGTTGGAG | 50 | 91 |
| A83R | ACCGGCTGTCGTTCTCCAGAGT | ||
| A14D | GAACAGAGAGCACCATGAGAGAG | 50 | 187 |
| A14R | TGGAAGATTTTATGAATGTCTCAA | ||
| B32D | AGGACATGAGGGCTTTCTGCTT | 55 | 328 |
| B32R | GGACCAGGACCAGTTCTGTTGT | ||
| B143D | TCCTGGGATCTGTTGACAGAAA | 55 | 428 |
| B143R | CACAAGATGAAACAACCCAGGA | ||
| B305D | GTACCCCTCAACCCTGGTCTC | 55 | 192 |
| B305R | TTTACGAAGAGCTGCAGACTGG | ||
| B126D | GGTGGCCGATTAGCCAAATTAT | 55 | 414 |
| B126R | GCCTTCTCATTGAGCTTCTCCA | ||
| B153D | TTTGGGTCCACTATCGGAGA | 50 | 193 |
| B153R | CCCAACCCAGGAGTTATGAA | ||
| B68D | CCTGAGGAAAAACCAAGTGTCTTCA | 50 | 150 |
| B68R | CGTGCATGCATACTTACATGCTT | ||
| B17D | ATCAGTGGGGCATTTGCAAGGTTCATG | 55 | 352 |
| B17R | CCACCTGTCTGCAGATGCCATGTTT | ||
| B225D | ACATTTCAGGGTAGCTGCTTGC | 55 | 238 |
| B225R | CAGCACAAGTAAACATGGTTCG | ||
| B12D | GGGACCAGCATTAAGAGGGAAC | 55 | 265 |
| B12R | TGGCAGTACAAAGACCACAGT | ||
| B203D | TGCAAGAGCGCATTTGTGAGTTTCC | 55 | 255 |
| B203R | GCCACCCCAGCACGCTTGA | ||
| B88D | TCTTGCAGACTGTTGTGGTTGA | 55 | 591 |
| B88R | CTCTGACCTCAGTATGCGCTGT | ||
| B51D | CCAGGAGGCACTAGACAAGG | 65 | 200 |
| B51R | GGAGCATACTGATCGTCAG | ||
| B319D | TGGATTGTCGGCTATGTTGA | 65 | 203 |
| B319R | TTGAGGACCACAAACCCTTC | ||
| B21D | TGATCATCACCCCATCCTTT | 55 | 191 |
| B21R | TGTCTTGTCTGCCCTGTTCA | ||
| B160D | AGTTTCCCGAGTTGAGCAGA | 65 | 199 |
| B160R | AATCGATCCTTGACCGACTC |
Northern blot analysis.
Total RNA (10 μg) from rainbow trout leukocytes exposed to virus and from the mock exposure were fractionated in formaldehyde-agarose gel (1.2%) in phosphate buffer and blotted onto nylon membrane (Zetaprobe; Bio-Rad). Blots were probed with 32P-labeled cDNA probes corresponding to each candidate genes. To generate the probes, relevant PCR products were separated in agarose gel, and purified using the Nucleospin Extract kit (Macherey-Nagel). Random priming was performed from 60 ng DNA using the Ready prime II labeling system (Amersham Pharmacia). A PhosphorImager (Molecular Dynamics) was used to quantify the signal. Membranes were then stripped and rehybridized with an actin probe to control the amount of mRNA loaded in the gel.
Isolation of full-length cDNA.
5′-rapid amplification of cDNA ends (5′-RACE) and 3′-RACE were performed using the SMART RACE cDNA Amplification Kit (Clontech), according to the instructions of the manufacturer. The procedure was performed with the RNA sample used for the SSH experiment with the exception that the total RNA was treated with DNase (Boehringer) to remove any remaining genomic DNA. The treated RNA was used to generate full-length cDNAs. 5′- and 3′-RACE PCRs were performed with relevant specific primers and the universal primers from Clontech. PCR products obtained were cloned into pCR2.1 vector (TOPO TA cloning system; Invitrogen).
Sequence analysis.
The Genetics Computer Group (Madison, Wis.) package was used for sequence assembly. The Blastn and Blastx programs from the National Center for Biotechnology Information were used to identify similar sequences, and specific conserved domains were searched with the different programs available through the Infobiogen DEAMBULUM Web page (http://www.infobiogen.fr/services/deambulum/fr/). PF (pfam) motifs are available at: http://pfam.wustl.edu/textsearch.shtml, and PD (PRODOM) domains are available at http://prodes.toulouse.inra.fr/prodom/doc/form2001.2.html.
Nucleotide sequence accession numbers.
The sequences of differential clones have been deposited in the GenBank database under accession no. AF483527 to AF48547.
RESULTS
Identification of candidate genes.
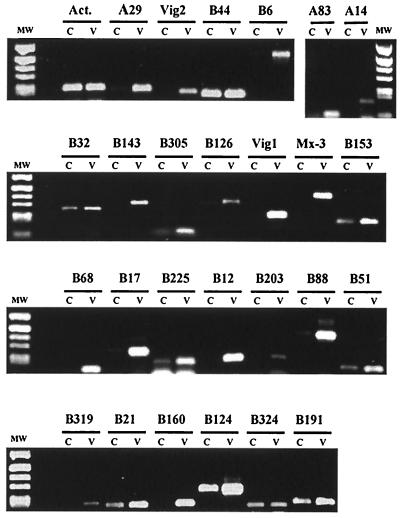
In order to identify unique viral induced gene transcripts represented in fish leukocytes, we used the method of SSH to generate a subtracted cDNA library. Leukocytes were isolated from a single fish so that genetic variances that occur between different fish would not affect the SSH procedure. Subtraction of cDNA from virus-treated cells (tester) with excess cDNA from control cells (driver) was used to enrich for genes induced by the virus. After several rounds of subtraction, the equalized cDNA was ligated to plasmid DNA and used in transformations with competent E. coli. Following transformation, 350 bacterial clones were isolated and subjected to the differential screening method. Based on hybridization results using forward-subtracted, reverse-subtracted, and unsubtracted tester and unsubtracted driver probes, one hundred clones were shown to be induced by virus exposure and were then subjected to DNA sequencing. Several of these clones represented fragments of the same gene. Consequently, the total number of differential genes represented by these clones was 28 (Table 2). Among differentially expressed sequences identified by SSH and hybridization screening, we found three genes, which were previously characterized as VHSV-induced genes: Mx-3 (44), vig-1 (8), and vig-2 (9). This finding was a good indication of the relevance of the methodology. To further confirm the differential expression of the candidate genes, we performed semiquantitative RT-PCR assays on the original RNA samples used for the library construction. By this method, we tested all identified genes except clones A27 and B322, which corresponded to Alu repeats and an immunoglobulin M constant domain, and therefore were not investigated further. We used sets of specific primers derived from the sequence of each candidate, and rainbow trout actin expression was used to normalize the amount of cDNA present in each PCR experiment. We reproducibly observed a clear difference between virus-treated cDNA and untreated control samples for 16 candidate genes (clones A29, vig-2, B6, A83, A14, B143, B126, vig-1, Mx-3, B68, B17, B12, B203, B88, B319, and B160). For eight candidate genes (clones B32, B305, B153, B225, B51, B21, B124, and B191), a basal expression was observed in the control sample, but an increase of expression was promoted by virus treatment. Clones B44 and B324 showed no difference between virus-treated cDNA and untreated control samples and were considered as artifacts of the SSH method. Thus, 21 new virus-induced sequences were identified in addition to vig-1, vig-2, and Mx-3 (Fig. 2). Similarity searches with the Blast programs resulted in significant scores for 12 of these new virus-induced genes (Table 2). No significant similarity was found for the nine remaining sequences, which were considered to be expressed sequence tags (ESTs). Not surprisingly, the two sequences that were not differentially amplified by the semiquantitative assay did have similarities to trout globin and to a ribosomal protein. Also, the number of clones observed for each gene is shown in Table 2. These numbers should be considered as a representative proportion of mRNA abundance for these genes and therefore reflect their expression levels. The number of clones corresponding to a given gene varied from 21 for the most-represented to 1 for the less-represented transcripts. It appeared that vig-2 and clones similar to genes encoding the ubiquitin-like family of proteins were the most represented in the subtracted library. However, the composition of the library may have been biased by a specific feature of the genes, such as the RsaI site distribution in their sequence.
TABLE 2.
Sequence analysis of SSH clones selected by hybridization screening
| Identification (GenBank, accession no.) | No. of clones | Other similar sequence(s) | RT-PCR assay result |
|---|---|---|---|
| A29 (AF483529) | 21 | Ubiquitin-like (O. mykissAF206323, E = 2e − 40; H. sapiensAB003730E = 6e − 25)a | + |
| vig-2 (AF290477) | 14 | Vig-2 (O. mykissAF290477, E = 0) | + |
| B44 | 11 | Globin (O. mykissD82926, E = 3c − 24, blastn) | − |
| B6 (AF483530) | 7 | Retinoic acid and interferon-induced protein (H. sapiens U34605, E = 7e − 11) | + |
| A83 (AF483533) | 6 | Galectin-9 (H. sapiensAJ288083, E = 0.0034; O. mykissAB027462, E = 5e − 03) | + |
| A14 (AF483527) | 5 | CXC chemokine (H. sapiens P19875, E = 7e − 5) | + |
| B32 (AF483536) | 5 | Estrogen responsive protein (H. sapiens D21205, E = 7e − 7) | + |
| B143 (AF483539) | 4 | + | |
| B305 (AF483542) | 3 | + | |
| B126 (AF483532) | 3 | Hypothetical protein (Drosophila T13747, E = 2e − 12) | + |
| vig-1 (AF076620) | 2 | vig1 (O. mykissAF076620, E = 0.0) | + |
| mx-3 (U47946) | 2 | mx3 (O. mykissU47946, E = 0.0) | + |
| B153 (AF483547) | 2 | + | |
| A27 | 1 | Alu repeat | ND |
| B322 | 1 | ImmunoglobulinM (Salmo salar, Y12391E = 9e − 10, blastn) | ND |
| B68 (AF483528) | 1 | CXC chemokine (S. scrofulX77935, E = 0.072; H. sapiensU59286, E = 0.8) | + |
| B17 (AF483531) | 1 | Interferon-responsive protein (H. sapiensAJ312776, E = 0.007) | + |
| B225 (AF483540) | 1 | + | |
| B12 (AF483534) | 1 | Hypothetical protein (H. sapiensAK022542, E = 5e − 13) | + |
| B203 (AF483538) | 1 | uvrD-helicase domain containing protein (H. sapiens emb/CAC15527, E = 4e − 13) | + |
| B88 (AF483541) | 1 | + | |
| B51 (AF483537) | 1 | Coatomer protein complex (H. sapiensAK003458, E = 2e − 41) | + |
| B319 (AF483543) | 1 | + | |
| B21 (AF483544) | 1 | + | |
| B160 (AF483545) | 1 | + | |
| B124 (AF483546) | 1 | + | |
| B324 | 1 | Ribosomal protein (H. sapiensXM_096067, E = 1 − 13) | − |
| B191 (AF483535) | 1 | C-lectin (O. mykissAF281349, E = 6e − 04) | + |
Accession numbers, name of the species, and E values of Blast hits are indicated. E values correspond to the BlastX program, except when “Blastn” is specified.
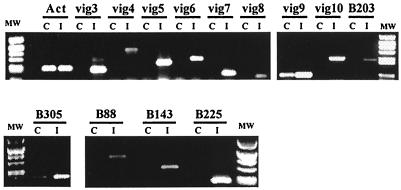
FIG. 2.
RT-PCR screen for differential expression of candidate genes. Semiquantitative PCR assay was performed on cDNA from rainbow trout leukocytes, after incubation with VHSV (V) or with culture medium as a control (C). The cDNA samples (V and C) were first normalized on the basis of actin expression and then subjected to PCR amplification with primers specific for each candidate gene.
Validation of differential expression.
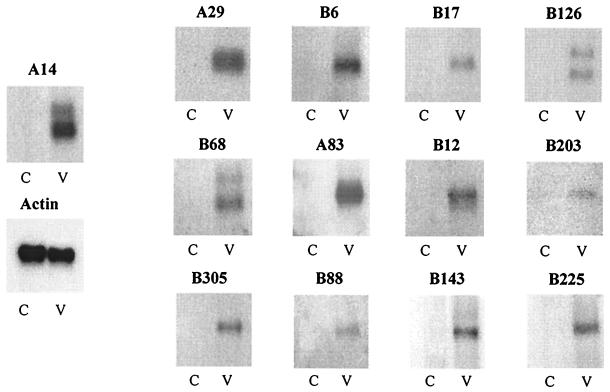
We selected the nine most-promising candidate genes on the basis of their similarity to known proteins, and we performed Northern blot experiments to authenticate their differential expression. We also performed Northern blots with four ESTs (clones B305, B88, B143, and B225) randomly selected among those appearing differentially expressed in the RT-PCR assay. After hybridization with specific probes, the membranes were rehybridized with an actin probe as a control for the mRNA inputs. Specific radioactive signals were quantified using a phosphorimager, and a stimulation index was calculated for each candidate gene on the basis of the ratio between its radioactive signal and the signal for actin mRNA (Table 3). As shown in Fig. 3 and Table 3, these experiments confirmed that all of the genes that showed a clear virus induction in the RT-PCR experiments were also clearly positive in Northern blots. Most of the hybridizations revealed a unique mRNA species at a size compatible with the size expected from the corresponding full-length cDNA. However, two discrete bands were consistently observed with the CXC chemokines and the B126 specific probes. This mRNA heterogeneity could result from the presence of different copies of the same gene, transcribed into mRNAs of different length. Alternatively, double bands could correspond to alternative polyadenylation or alternative splicing (13).
TABLE 3.
Genes modulated by VHSV
| Gene | Stimulation indexa | Protein family/ most significant hits and E values for BlastX program | Potential function(s) |
|---|---|---|---|
| vig-3 (clone A29) | 7.2 | Ubiquitin-like protein (Carassius auratus AF206323, E = 2e − 40; H. sapiens ISG15 NM_0051017, E = 7e − 18) | Interferon induced (ISG15); human homolog blocked by influenza virus |
| vig-4 (clone B6) | 1.9 | Retinoic acid- and interferon-inducible protein with tetratricopepetide domain (H. sapiens NM_012420 (E = 7e − 655)) | Interferon induced (p58/p56); myeloid differenciation marker, interaction with eukaryotic initiation factor (translation regulation), glucocorticoid attenuated response |
| vig-5 (clone B17) | 2.4 | Transmembrane protein 7 (H. sapiens NM_031440 E = 2e − 22) | Interferon induced (p28) Unknown function |
| vig-6 (clone B126) | 2.6 | Protein with six transmembrane regions (Mus musculus AK015888, E = 5e − 44; H. sapiens XM_059238, E = 2e − 41, Drosophila melanogaster AAF45721, E = 2e − 29); Hs and Dm promoters have ISRE | Interferon induced (?) Unknown function |
| vig-7 (clone A14) | 2 | CXC chemokine (L. fluviatilis IL-8 AJ231072 E = 1e − 11 and other CXC) | Chemoattractant |
| vig-8 (clone B68) | 1.6 | CXC chemokine (S. scrofa PDPP X77935, E = 6e − 12 and other CXC) | Chemoattractant |
| vig-9 (cloneA83) | 2.6 | Galectin-like protein (O. mykiss AB027452, E = 2e − 92) galectin-9 Mus musculus U55061, E = 2e − 59) | Chemoattractant, apoptosis |
| vig-10 (clone B12) | 1.6 | Unknown protein with poly ADP-ribose polymerase catalytic domain (H. sapiens AK022542, E = 2e − 59) | Transcription repression, apoptosis |
| clone B203 (partial cDNA) | 2.1 | Unknown nucleic acid binding protein with ribonuclease and uvr-helicase domains (H. sapiens XM_028918 (KIAA1769), E = 1e − 104) | RNA metabolism |
Stimulation index (S) was calculated as follows: from quantification of Northern blots using a PhosphorImager: S = (Vs/ Vact)/(Cs/Cact), where Vs and Vact correspond to the signal obtained for the virus-treated RNA hybridized with specific and actin probe, respectively; Cs and Cact correspond to the signal obtained for the control RNA hybridized with specific and actin probe, respectively.
FIG. 3.
Differential RNA expression of selected candidate genes. The RNA expression of selected genes was analyzed by Northern blotting, comparing RNAs from virus-treated cells (V) and untreated cells (C). Hybridizations were first performed with 32P-labeled specific probes, and the blots were stripped and rehybridized with an actin probe as a control for the total amounts of RNA. For all genes, specific signals and corresponding actin expression (only shown for A14) were quantified and were used to calculate a stimulation index (see Table 3). Northern blot experiments were also performed for VHSV-induced ESTs B305, B88, B143, and B225.
Description of selected genes.
To obtain full-length cDNA clones for particular genes of interest, we used the 5′ -and 3′-RACE methodology. Full-length sequences with a complete open reading frame (ORF) and a polyadenylation signal were obtained for eight candidate genes (A29, B6, B17, B126, A14, B68, A83, and B12). Consequently the complete genes corresponding to clones A29, B6, B17, B126, A14, B68, A83, and B12 were named VHSV-induced genes (vig-3 to -10), respectively. Due to technical difficulties, the sequence of B203 was not expanded further than 1,320 bp. In fact it was highly similar (BlastX E value, 1e-104) to the human KIAA1769 sequence, which is 7,482 bp long. Sequence homology searches within GenBank and EST, were then refined by using the longest sequence available for each candidate gene. The sequences were also subjected to a systematic search of characteristic motifs using the Expasy and ProDom tools to investigate their potential functions. The VHSV-induced genes were then divided into three groups on the basis of their similarities with genes belonging to different functional pathways (Table 3).
Interferon responsive genes.
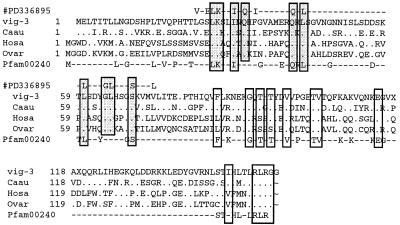
vig-3 (clone A29) corresponded to a 762-bp cDNA, which encoded a 156-amino-acid (-aa) protein that was highly similar to the ubiquitin family of proteins. It displayed the conserved motifs pfam PF00240 (ubiquitin family) and ProDom PD336895 (interferon induction repeat, Fig. 4). The ubiquitin-like protein encoded by vig-3 was highly similar to the mammalian interferon-responsive protein ISG15.
FIG. 4.
Multiple sequence alignment of the vig-3 encoded protein. ClustalW alignment of vig-3 and the ubiquitin-like proteins from Carassius auratus (Caau; GenBank accession no. AF206323), human ISG15 (Hosa; GenBank accession no. M21786), and sheep ISG17 (Ovar; GenBank accession no. AF152103). Similar residues are indicated, a dash corresponds to a gap, and a dot corresponds to a different residue. The interferon induction motif (PRODOM #PD336895) is indicated on the top line, with conserved residues boxed in grey. The ubiquitin-like motif (pfam00240) is shown in the bottom line, with conserved residues boxed in white.
vig-4 (clone B6) corresponded to a cDNA sequence of 1824 bp. It contained a 1443 bp ORF encoding a 481 AA protein with tetratricopeptide repeats. Tetratricopeptide repeats motifs are involved in protein-protein interactions and were identified in various organisms from bacteria to mammals (34). The most-similar human protein was an interferon-induced protein, which is expressed during myeloid differentiation (36). The vig-4 encoded protein was also highly similar to the interferon-induced protein P56, which binds to the P48 subunit of eukaryotic initiation factor-3 and inhibits translation (20).
vig-5 (clone B17) corresponded to a full-length cDNA sequence of 1,213 bp. It encoded a 168-aa protein with a predicted trans-membrane domain. It showed a high sequence similarity and similar organization to several interferon responsive proteins from mouse and human. There are no published data about the biological function of these proteins sharing the ProDom motif PD358088. The identification of the trout sequence clearly showed that the proteins of this family were conserved throughout vertebrates as virus- and interferon-induced proteins.
vig-6 (clone B126) corresponded to a full-length cDNA of 992 bp. It contained a 714-bp ORF, encoding a 238-aa protein. This protein was similar to several proteins from mouse, human, drosophila, and Caenorhabditis elegans (Fig. 5). A significant similarity was also observed with the rat TM6P1, which was described as a fasting-induced protein (49). This family of proteins displayed six trans-membrane domains predicted on the basis of the hydrophobicity profile. Moreover, TM6P1 was shown to be an integral membrane protein (49). Since vig-6 protein was the first in the family to be connected to the virus-induced response, we scanned the genomic sequence of the human gene with the TFSEARCH program, and we identified an interferon regulatory factor 1-interferon regulatory factor 2 binding site at position −1060 from the ATG of the human gene. Together with our observations from this work, the presence of this potential binding site for transcription factor in the promoter strongly suggested that proteins of this family probably belong to the interferon-induced pathway.
FIG. 5.
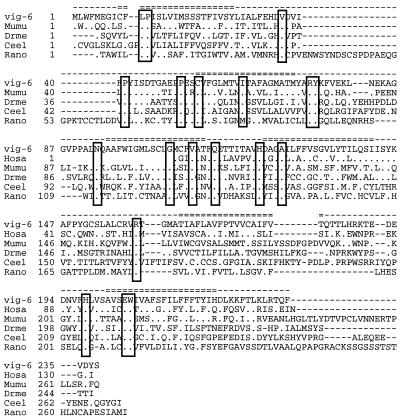
Multiple alignment of the vig-6 encoded protein. ClustalW alignment of vig-6 with similar proteins from human (Hosa; GenBank accession no. AK002121), mouse (Mumu; GenBank accession no. AK015888), drosophila hypothetical protein 22E5.9 (Drme; GenBank accession no. T1374), C. elegans (Ceel; GenBank accession no. T19654), and rat (Rano; GenBank accession no. AF186469). Residues similar to the trout sequence are noted by a dot; a dash corresponds to a gap. Conserved positions are boxed. Predicted trans-membrane domains are noted (=) on the top line.
Chemo-attractants.
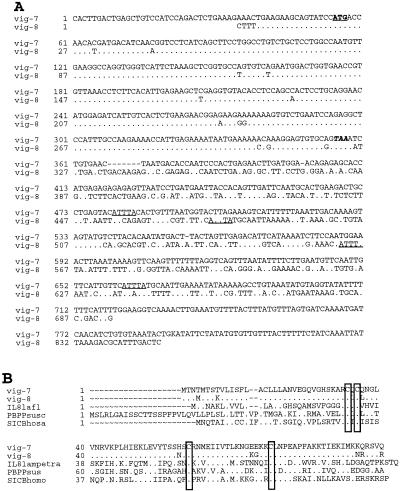
vig-7 and vig-8 (clones A14 and B68) corresponded to two different transcripts 850 and 694 bp in length, respectively. Their sequences were highly similar within the 300-bp ORF region but were clearly divergent in the 3′ untranslated region (Fig. 6A). The 100-aa encoded proteins had typical characteristics of CXC chemokines. They contained the interleukin-8 (IL-8) motif PF00048 typical of different secreted growth factors or interferon-induced chemokines involved in mitogenic, chemotactic, and inflammatory activities (reviewed in reference 32). The vig-7 and vig-8 encoded proteins displayed 90% identity in their amino acid sequence (Fig. 6B) and were therefore closer to each other than to any sequence described so far. However, considering they had ten divergent amino acids, they may perform different functions. It is interesting that both rainbow trout CXC sequences are related to the interferon-inducible protein IP10. Mouse IP10 is a chemo-attractant for T cells and was described as a danger signal-molecule in host defense against several viruses (29).
FIG. 6.
Nucleotide and deduced amino acid sequence of vig-7 and vig-8. (A) Alignment of vig-7 and vig-8 nucleotide sequences. A dot indicate residues similar in both sequences. The start codon is in boldface type and is underlined, and stop codon in boldface italic type. The destabilization motifs are underlined. Dots indicate similar nucleotides, and dashes correspond to gaps. (B) Multiple alignment of amino acid sequence of vig-7 and vig-8, with the most similar CXC chemokines: (IL-8 from Lampetra fluviatilis, GenBank accession no. AJ231072; platelet basic protein precursor from Sus scrofa, GenBank accession no. P43030; and small inducible cytokine IP10 from Homo sapiens, GenBank accession no. 225751). Conserved cysteine residues are boxed. Dots indicate similar residues, and dashes correspond to gaps.
The sequence of vig-9 (clone A83) was 1,373 bp. It contained a 1,044-bp ORF, encoding a 348-aa protein, which had a high similarity index with mammalian galectins. The vig-9 encoded protein was most similar to galectin-9 from human, rat, and mouse. In human specimens, galectin-9 was described as a potent chemoattractant for eosinophils (33) and mediated apoptosis of activated T cells (45).
DNA-RNA binding proteins.
vig-10 (clone B12) and clone B203 did not show any significant homology to proteins with defined functions. However, they had regions significantly similar to conserved nucleic acid binding domains, and thus we classified them as putative DNA-RNA binding proteins. The sequence from vig-10 was 1,438 bp, with an 894-bp ORF encoding a 298-aa protein. B12 had a region (residues 146 to 220) corresponding to a poly ADP-ribose polymerase catalytic domain (PF00644). In mammals, the poly ADP-ribose polymerase is a DNA-binding protein involved in apoptosis, transcriptional repression (42), and maintenance of the genomic integrity. Clone B203 was extended to 1,320 bp and contained a partial ORF of 440 aa. The protein encoded by this ORF was highly similar to KIAA-1769, a large human protein (GenBank accession no. XP_028918) probably involved in RNA degradation, due to the presence of RNase (PF00773) and helicase UV-resistant (UVR) domains (PF00580). The B203 partial sequence had an uvr-helicase domain and may perform similar functions.
Interferon induction.
Several studies have stressed the importance of the alpha/beta interferon pathway in the host response to viral infection (19, 27). A high proportion of the virus-induced genes retrieved in this study had sequence characteristics that clearly assigned them to the group of interferon-induced genes. The rainbow trout epithelial cell line (RTG cell line) subjected to vesicular stomatitis virus releases into the culture medium a protein component with typical characteristics of alpha/beta interferon, i.e., acid resistance and antiviral activity. It was therefore interesting to test this rainbow trout interferon for direct induction of the proven virus-induced genes and ESTs. The supernatant of the rainbow trout epithelial cell line RTG-2 treated with vesicular stomatitis virus was extensively centrifuged to eliminate viral particles (40,000 × g for 2 h) and subjected to acid treatment (pH 2.2). The antiviral activity of this preparation was 10−4 in a 50% plaque reduction test. To test this rainbow trout interferon for direct induction of the proven virus-induced genes and ESTs, rainbow trout leukocytes were treated with the interferon preparation (1/100 dilution) or with supernatant from untreated RTG cells for 14 h at 14°C. The expression of validated virus-induced genes and ESTs was investigated with a semiquantitative RT-PCR assay. As shown in Fig. 7, all of the candidate genes and ESTs tested in this experiment were clearly induced following an 8-h interferon treatment, except for vig-3 (A83), which showed a moderate increase over some basal expression. These results were a clear indication that the leukocyte response to VHSV was for the most part orchestrated through the alpha/beta interferon inducible pathway.
FIG. 7.
Induction of selected genes and ESTs by rainbow trout interferon. The expression of candidates was assessed using an RT-PCR assay in rainbow trout leukocytes treated by a conditioned medium with interferon activity at a 1/100 dilution (I), or treated with medium from normal culture of RTG cells as a control (C). Specific primers were used for each candidate. cDNAs samples (I and C) were normalized on the basis of actin expression.
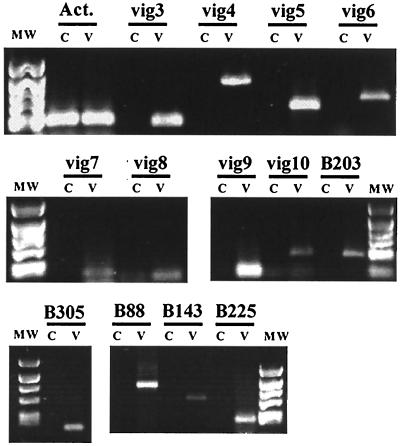
In vivo virus induction.
The VHSV induced-genes identified by SSH in leukocyte primary cultures treated in vitro with the virus may not represent genes activated during natural infection. To explore the relevance of this set of genes in natural infection, juvenile rainbow trout were infected with 5 × 106 PFU of VHSV (strain 07-71) by the intramuscular route. Fish were sacrificed on day 6 postinfection and tissue was sampled from the site of injection, where the virus is known to first replicate. The samples were then processed for RNA extraction. We used a semiquantitative PCR assay to search for the expression of the candidate genes. All of the genes tested were expressed in sample tissue from infected fish, while they were not detected in tissue from the noninfected controls (Fig. 8). These results demonstrated that the virus-induced genes identified by SSH experiments in leukocytes were also up-regulated during experimental infection in vivo and are likely to play a role in the course of natural infection.
FIG. 8.
Expression of selected genes and ESTs in experimental VHSV infection. For each candidate, a specific RT-PCR analysis was performed with samples taken at the inoculation site from VHSV-infected (V) or phosphate-buffered saline-injected control fish (C). Total amounts of cDNA were normalized on the basis of actin expression in the two samples (V and C).
DISCUSSION
The identification of host genes induced by viruses is one of the critical steps leading to the elucidation of the host defense pathways. We used several methodologies to search for such genes induced by a fish rhabdovirus in a lower vertebrate, the rainbow trout. In our approach, we focused on genes induced in primary culture of rainbow trout leukocytes infected in vitro with a well-characterized strain of VHSV (8, 9). In doing so, we have deliberately reduced the scope of the analysis to a selected population of fish cells. We reasoned that fish leukocytes should maintain a high reactivity to virus stimuli and concurrently, they are not totally permissive to viral replication. In the present work, we used the SSH methodology with RNA from infected leukocytes as the tester sample, and RNA from noninfected cells as the driver. We have identified 24 virus-induced sequences and further characterized the nine most-promising sequences. These transcripts provide a framework for future functional studies and pave the way for comparative studies of vertebrate's host cell response to viruses.
Among this set of genes, six were assigned to the interferon pathway on the basis of their sequence characteristics. Three had been already described in rainbow trout: Mx-3, vig-1, and vig-2. Mx-3 and vig-1 did have mammalian homologs with a documented antiviral activity (12, 38), which emphasizes the relevance of the methodology. The three others—vig-3 (A29), vig-4 (B6), and vig-5 (B17)—are new fish genes with a high similarity to mammalian interferon responsive genes. The ubiquitin-like protein encoded by vig-3 was highly similar to the mammalian interferon-responsive protein ISG15. As previously described for proteins of the ubiquitin-like family, ISG15 is probably able to bind to target proteins through its C terminal glycine residues (30), and this conjugation could result in the degradation of the target protein (25). The protein encoded by vig-3 had the critical C-terminal glycine residues and could therefore have a similar function to ISG15 in mammals. Interestingly, influenza A and B viruses were shown to have developed strategies to block ISG15. The NS1 protein of influenza B blocked ISG15 conjugation with the target protein, and influenza A virus inhibited the synthesis of ISG15 (48). The development of different viral mechanisms to block the ISG15 protein highlights its antiviral potential. Further studies are needed to determine if the trout ubiquitin-like protein identified in the present study is a true functional homolog of the mammalian ISG15. Most importantly, all the genes and ESTs shown by Northern blotting to be unequivocally induced by the virus showed also direct induction by a rainbow trout interferon preparation. This strongly suggests that the rainbow trout cell response to the virus is mainly mediated through the interferon responsive pathways. In fact, interferon-inducible genes were found to be systematically up-regulated in recent global studies of transcriptional changes induced by viruses of different families: human immunodeficiency virus type 1 (17), hepatitis C virus (5, 37), cytomegalovirus (51), rabies virus (39), and Hirame rhabdovirus (1). Human papillomavirus constitutes an exception to this general scheme. Several interferon-inducible genes were shown to be down-regulated in human keratinocytes infected by Human papillomavirus (11).
The role of chemokines and chemo-attractants in innate immunity to pathogens is already well documented, and our results confirmed the likely importance of this class of molecules in the rainbow trout response to a virus infection. Galectins were clearly linked to innate and specific immunity in mammals. Our results showed this feature could be conserved through vertebrates. Proteins from this family were also reported in nematodes (22), suggesting that members of this family probably belong to an ancient pathway of innate defense. In contrast, we have found no virus- or interferon-induced homologs in the data banks for vig-10 (B12) and B203. Semiquantitative PCR and Northern blot analyses confirmed their induction by the virus in leukocyte primary cultures. These genes were also up-regulated by rainbow trout interferon in the same cells, and in vivo, at the site of virus replication. In fact, it was not surprising to find genes encoding for nucleic acid binding proteins among the VHSV-induced genes and these could be involved in RNA regulation.
The set of genes induced by VHSV described in this study is most probably far from exhaustive. Considering the large number of genes modulated during viral infections, as revealed by DNA micro array methods (23, 37, 51), the SSH method probably identified only the most differentially expressed transcripts. Moreover, it was surprising that interferon was not found among the VHSV-induced genes. It is likely that the interferon transcripts are rare, unstable, or down-regulated by the time of our SSH experiment. Indeed, it is noteworthy that interferon transcripts were rarely retrieved in global surveys of virus-induced genes. Although inductions that bypass interferon were described for several interferon-activated genes in mammals (21, 35), and for vig-1 in rainbow trout (8), the hypothesis that the interferon pathway may have not been activated in our experiment is much less likely. Indeed, trout leukocyte primary cultures were shown to produce interferon when incubated with VHSV (40). It was also surprising that neither MHC nor the interferon-inducible genes of the proteasome were retrieved in our SSH screening. More experiments are required to determine if this lack was due to the nature of the cells, to the kinetics of MHC and proteasome induction, or to technical limitations. In fact, both differential display-PCR (28) and SSH (16) were first considered exhaustive. However, it became evident that these methods suffer some limitations in the description of differential transcriptomes. Methods based on cDNA arrays are far more efficient for global analysis of transcriptome modifications, and several teams have already used them for the study of virus-induced genes (5, 10, 11, 24, 51). Unfortunately, cDNA arrays are not yet available for all animal species. Therefore, SSH remains likely to be the most efficiently optimized alternative method.
Acknowledgments
C.O. and N.V. contributed equally to this work.
This work was supported by the Institut National de la Recherche Agronomique (INRA) and the France national program on microbiology (PRFMMIP, Ministère de la Recherche). Caroline O'Farrell and Nikta Vaghefi were recipients of postdoctoral fellowships from the Division of Animal Health, INRA.
We acknowledge René L'haridon for making the rainbow trout interferon preparation.
REFERENCES
- 1.Aoki, T., I. Hirono, M. G. Kim, T. Katagiri, Y. Tokuda, H. Toyohara, and E. Yamamoto. 2000. Identification of viral induced genes in Ig+ leucocytes of Japanese flounder Paralichthys olivaceus, by differential hybridisation with subtracted and un-subtracted cDNA probes. Fish Shellfish Immunol. 10:623-630. [DOI] [PubMed] [Google Scholar]
- 2.Basurco, B., P. Vende, A. F. Monnier, J. R. Winton, P. de Kinkelin, and A. Benmansour. 1995. Genetic diversity and phylogenetic classification of viral hemorrhagic septicemia virus (VHSV). Vet. Res. 26:460-463. [PubMed] [Google Scholar]
- 3.Benmansour, A., G. Paubert, J. Bernard, and P. De Kinkelin. 1994. The polymerase-associated protein (M1) and the matrix protein (M2) from a virulent and an avirulent strain of viral hemorrhagic septicemia virus (VHSV), a fish rhabdovirus. Virology 198:602-612. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, J., F. Lecocq-Xhonneux, M. Rossius, M. E. Thiry, and P. de Kinkelin. 1990. Cloning and sequencing the messenger RNA of the N gene of viral haemorrhagic septicaemia virus. J. Gene. Virol. 71:1669-1674. [DOI] [PubMed] [Google Scholar]
- 5.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boudinot, P., M. Blanco, P. de Kinkelin, and A. Benmansour. 1998. Combined DNA immunization with the glycoprotein gene of viral hemorrhagic septicemia virus and infectious hematopoietic necrosis virus induces double-specific protective immunity and nonspecific response in rainbow trout. Virology 249:297-306. [DOI] [PubMed] [Google Scholar]
- 7.Boudinot, P., S. Boubekeur, and A. Benmansour. 2001. Rhabdovirus infection induces public and private T cell responses in teleost fish. J. Immunol. 167:6202-6209. [DOI] [PubMed] [Google Scholar]
- 8.Boudinot, P., P. Massin, M. Blanco, S. Riffault, and A. Benmansour. 1999. vig-1, a new fish gene induced by the rhabdovirus glycoprotein, has a virus-induced homologue in humans and shares conserved motifs with the MoaA family. J. Virol. 73:1846-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudinot, P., S. Sahli, M. Blanco, and A. Benmansour. 2001. Viral haemorrhagic septicaemia virus induces vig-2, a new interferon responsive gene of rainbow trout. Fish Shellfish Immunol. 11:383-397. [DOI] [PubMed] [Google Scholar]
- 10.Browne, E., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y. E., and L. A. Laimins. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 74:4174-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chin, K. C., and P. Cresswell. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 98:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claverie, J. 2001. What if there are only 30000 human genes. Science 291:1255-1257. [DOI] [PubMed] [Google Scholar]
- 14.Collet, B., and C. J. Secombes. 2001. The rainbow trout (Oncorhynchus mykiss) Mx1 promoter. Structural and functional characterization. Eur. J. Biochem. 268:1577-1584. [PubMed] [Google Scholar]
- 15.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Nat. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diatchenko, L., Y. F. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiss, G., R. Bumgarner, M. An, M. Agy, A. van 't Wout, E. Hammersmark, V. Carter, D. Upchurch, J. Mullins, and M. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 18.Graf, L., and S. B. Torok. 1995. Identification of a novel DNA sequence differentially expressed between normal human CD34+CD38hi and CD34+CD38lo marrow cells. Blood 86:548-556. [PubMed] [Google Scholar]
- 19.Guidotti, L., and F. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 20.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 22.Hirabayashi, J., T. Ubukata, and K. Kasai. 1996. Purification and molecular characterization of a novel 16-kDa galectin from the nematode Caenorhabditis elegans. J. Biol. Chem. 271:2497-2505. [DOI] [PubMed] [Google Scholar]
- 23.Huang, Q., D. Liu, P. Majewski, L. C. Schulte, J. M. Korn, R. A. Young, E. S. Lander, and N. Hacohen. 2001. The plasticity of dendritic cell responses to pathogens and their components. Science 294:870-875. [DOI] [PubMed] [Google Scholar]
- 24.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jentsch, S., and G. Pyrowolakis. 2000. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10:335-342. [DOI] [PubMed] [Google Scholar]
- 26.Kim, C. H., M. C. Johnson, J. D. Drennan, B. E. Simon, E. Thomann, and J. A. Leong. 2000. DNA vaccines encoding viral glycoproteins induce nonspecific immunity and Mx protein synthesis in fish. J. Virol. 74:7048-7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy, D., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:245-257. [DOI] [PubMed] [Google Scholar]
- 28.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 29.Liu, M. T., B. P. Chen, P. Oertel, M. J. Buchmeier, D. Armstrong, T. A. Hamilton, and T. E. Lane. 2000. The T cell chemoattractant IFN-inducible protein 10 is essential in host defense against viral-induced neurologic disease. J. Immunol. 165:2327-2330. [DOI] [PubMed] [Google Scholar]
- 30.Loeb, K. R., and A. L. Haas. 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 267:7806-7813. [PubMed] [Google Scholar]
- 31.Lorenzen, N., E. Lorenzen, K. Einer-Jensen, J. Heppell, and H. L. Davis. 1999. Genetic vaccination of rainbow trout against viral haemorrhagic septicaemia virus: small amounts of plasmid DNA protect against a heterologous serotype. Virus Res. 63:19-25. [DOI] [PubMed] [Google Scholar]
- 32.Luster, A. D. 1998. Chemokines-chemotactic cytokines that mediate inflammation. N. Engl. J. Med. 338:436-445. [DOI] [PubMed] [Google Scholar]
- 33.Matsushita, N., N. Nishi, M. Seki, R. Matsumoto, I. Kuwabara, F. T. Liu, Y. Hata, T. Nakamura, and M. Hirashima. 2000. Requirement of divalent galactoside-binding activity of ecalectin/galectin-9 for eosinophil chemoattraction. J. Biol. Chem. 275:8355-8360. [DOI] [PubMed] [Google Scholar]
- 34.Melville, M. W., M. G. Katze, and S. L. Tan. 2000. P58IPK, a novel cochaperone containing tetratricopeptide repeats and a J-domain with oncogenic potential. Cell. Mol. Life Sci. 57:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Memet, S., F. Besancon, M. F. Bourgeade, and M. N. Thang. 1991. Direct induction of interferon-gamma- and interferon-alpha/beta-inducible genes by double-stranded RNA. J. Interferon Res. 11:131-141. [DOI] [PubMed] [Google Scholar]
- 36.Niikura, T., R. Hirata, and S. C. Weil. 1997. A novel interferon-inducible gene expressed during myeloid differentiation. Blood Cells Mol. Dis. 23:337-349. [DOI] [PubMed] [Google Scholar]
- 37.Patzwahl, R., V. Meier, G. Ramadori, and S. Mihm. 2001. Enhanced expression of interferon-regulated genes in the liver of patients with chronic hepatitis C virus infection: detection by suppression-subtractive hybridization. J. Virol. 75:1332-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavlovic, J., O. Haller, and P. Staeheli. 1992. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J. Virol. 66:2564-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prosniak, M., D. C. Hooper, B. Dietzschold, and H. Koprowski. 2001. Effect of rabies virus infection on gene expression in mouse brain. Proc. Natl. Acad. Sci. USA 98:2758-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogel-Gaillard, C., S. Chilmonczyk, and P. de Kinkelin. 1993. In vitro induction of interferon like activity from rainbow trout leucocytes stimulated by Egtved virus. Fish Shellfish Immunol. 3:383-394. [Google Scholar]
- 41.Schutze, H., E. Mundt, and T. C. Mettenleiter. 1999. Complete genomic sequence of viral hemorrhagic septicemia virus, a fish rhabdovirus. Virus Genes 19:59-65. [DOI] [PubMed] [Google Scholar]
- 42.Soldatenkov, V. A., S. Chasovskikh, V. N. Potaman, I. Trofimova, M. S. Smulson, and A. Dritschilo. 2002. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J. Biol. Chem. 277:665-670. [DOI] [PubMed] [Google Scholar]
- 43.Thiry, M., X. F. Lecoq, I. Dheur, A. Renard, and K. P. De. 1991. Sequence of a cDNA carrying the glycoprotein gene and part of the matrix protein M2 gene of viral haemorrhagic scepticaemia virus, a fish rhabdovirus. Biochim. Biophys. Acta 1090:345-347. [DOI] [PubMed] [Google Scholar]
- 44.Trobridge, G. D., P. P. Chiou, and J. A. Leong. 1997. Cloning of the rainbow trout (Oncorhynchus mykiss) Mx2 and Mx3 cDNAs and characterization of trout Mx protein expression in salmon cells. J. Virol. 71:5304-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wada, J., and Y. S. Kanwar. 1997. Identification and characterization of galectin-9, a novel beta-galactoside-binding mammalian lectin. J. Biol. Chem. 272:6078-6086. [DOI] [PubMed] [Google Scholar]
- 46.Welsh, R. M., and G. C. Sen. 1997. Nonspecific host responses to viral infections, p. 109-141. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven, Philadelphia, Pa.
- 47.Yabu, T., H. Hirose, I. Hirono, T. Katagiri, T. Aoki, and E. Yamamoto. 1998. Molecular cloning of a novel interferon regulatory factor in Japanese flounder, Paralichthys olivaceus. Mol. Mar. Biol. Biotechnol. 7:138-144. [PubMed] [Google Scholar]
- 48.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, J., A. J. D'Ercole, and L. E. Underwood. 2000. Identification of a new gene (rat TM6P1) encoding a fasting-inducible, integral membrane protein with six transmembrane domains. Biochim. Biophys. Acta 1492:280-284. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]
- 51.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]