Abstract
Chronic hepatitis C is a common cause of liver disease, the complications of which include cirrhosis and hepatocellular carcinoma. Treatment of chronic hepatitis C is based on the use of alpha interferon (IFN-α). Recently, indirect evidence based on mathematical modeling of hepatitis C virus (HCV) dynamics during human IFN-α therapy suggested that the major initial effect of IFN-α is to block HCV virion production or release. Here, we used primary cultures of healthy, uninfected human hepatocytes to show that: (i) healthy human hepatocytes can be infected in vitro and support HCV genome replication, (ii) hepatocyte treatment with IFN-α results in expression of IFN-α-induced genes, and (iii) IFN-α inhibits HCV replication in infected human hepatocytes. These results show that IFN-α acts primarily through its nonspecific antiviral effects and suggest that primary cultures of human hepatocytes may provide a good model to study intrinsic HCV resistance to IFN-α.
Hepatitis C has emerged in recent years as a common cause of liver disease, and an estimated 170 million people are thought to be infected worldwide. Hepatitis C virus (HCV) infection is characterized by viral persistence and chronic liver disease in approximately 80% of cases. The complications of chronic hepatitis C include cirrhosis in 20% of cases and hepatocellular carcinoma, which has an incidence of up to 4 to 5% per year in patients with cirrhosis. Hepatitis C-related end-stage liver disease is now the principal indication for liver transplantation in industrialized countries (2).
HCV is a single-strand positive-sense RNA virus belonging to the family Flaviviridae. Translation of its only open reading frame leads to the synthesis of a single polyprotein which is secondarily cleaved by both host and viral proteases, giving rise to structural and nonstructural proteins (22). The nonstructural protein 5B (NS5B) is an RNA-dependent RNA polymerase (RdRp). The mechanisms of HCV replication in host cells are poorly understood. It is thought that RdRp, along with other nonstructural proteins, the HCV RNA template, and host cell factors, forms a replication ribonucleoprotein complex associated with perinuclear membranous structures that would be the site of RNA replication (13, 43). By analogy with other members of the family Flaviviridae, the replication strategy within this complex would be the production of a negative-strand copy of the RNA genome, which would in turn serve as a template for the production of progeny positive-strand RNA. Indeed, negative-strand HCV RNA has been detected in various cells and tissues supporting HCV replication (1, 18, 20, 28, 39, 40).
HCV RdRp, like other viral RNA polymerases, has a high error rate, with misincorporation frequencies averaging about 10−4 to 10−5 per base site, in the absence of a proofreading mechanism. As a result, mutations accumulate in newly generated HCV genomes. Most mutant viral particles are replication deficient, but some propagate efficiently. The fittest infectious particles are selected continuously on the basis of their replication capacities and environmental selective pressures (mainly the host immune response). This explains why each infected individual harbors a pool of genetically distinct but closely related HCV variants referred to collectively as a quasispecies (24, 45).
Treatment of chronic hepatitis C is aimed at preventing complications, especially cirrhosis and hepatocellular carcinoma. It is currently based on subcutaneous injection of recombinant alpha interferon (IFN-α) three times a week or of its pegylated form (i.e., IFN-α combined with polyethylene glycol) once a week. The antiviral efficacy of IFN-α is potentiated by ribavirin, a nucleoside analog with an unknown mechanism of action (2, 10, 21, 25, 26, 36, 47). Combination therapy with pegylated IFN-α plus ribavirin for 24 to 48 weeks leads to permanent viral clearance in 42 to 82% of patients according to HCV genotype; other patients have ongoing viral replication and remain at risk of disease progression [23; M. W. Fried, M. L. Shiffman, R. K. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., et al., Gastroenterology 120(Suppl. A):55, 2001].
After subcutaneous administration, IFN-α specifically binds to high-affinity receptors at the surface of target cells. IFN-α binding to its receptor triggers a cascade of intracellular reactions, leading to activation of numerous IFN-induced genes (11, 31, 38, 41, 44). The products of these genes mediate the cellular actions of IFN-α. As IFN-α binds to surface receptors of immune cells, it has immunomodulatory effects (34, 42). IFN-α binding to various cells also induces numerous proteins and enzymatic pathways involved in establishing a non-virus-specific antiviral state through distinct but complementary mechanisms (3, 12, 27, 37, 49). Specific IFN-α binding to human hepatocytes and subsequent activation of IFN-α-induced genes leading to the establishment of an antiviral state have not yet been documented.
Recently, indirect evidence based on mathematical modeling of HCV dynamics during human IFN-α therapy suggested that the major initial effect of IFN-α is to block HCV virion production or release (30). Inhibition of HCV replication by both IFN-α and IFN-β has been observed in a human lymphocytic cell line supporting HCV genome replication (40). More recently, IFN-α was shown to inhibit subgenomic HCV RNA replication in HuH-7 human hepatoma cell lines (8, 9) and full-length HCV RNA replication in a binary expression system in CV-1 monkey kidney cell lines (4). Nevertheless, IFN-α blockade of HCV replication has never been demonstrated in healthy, uninfected human hepatocytes.
MATERIALS AND METHODS
Primary cultures of human hepatocytes.
Healthy human hepatocytes were isolated from surgical liver resection specimens from 15 liver donors (Table 1). The two-step collagenase perfusion method was used (35). Cell viability, assessed by the trypan blue exclusion test, was greater than 85%. Hepatocytes were plated at confluence (14 × 104 cells/cm2) in 60- or 35-mm-diameter culture dishes precoated with type I collagen (Iwaki Glass, Chiba, Japan) in a total volume of 3.0 or 1.5 ml of standard culture medium consisting of a mixture of Williams' E medium and Ham F12 medium (1:1 volume) supplemented as recommended elsewhere (14) and containing 5% fetal calf serum to favor cell attachment. After 4 h, the standard medium was replaced with 3.0 or 1.5 ml of a previously described modified serum-free long-term culture medium (6, 17). This medium was renewed after 24 h and then every 48 h. The cultures were maintained in humidified 95% air-5% carbon dioxide at 37°C.
TABLE 1.
Characteristics of the primary cultures of human hepatocytes and the corresponding HCV-positive sera used for inoculation and summary of experiments and results
| Culture | Hepatocyte characteristics
|
Infecting serum
|
IFN-α treatment | IRF-1 and PKR induction by IFN-α | IFN-α toxicity | (+) and (−) HCV RNA strandsf | Accumulation of (+) HCV RNAg | Effect of IFN-α on genetic evolution | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor sexa | Donor age (yr) | Origin | Serum | Genotype | Viral loadd | |||||||
| FT141 | F | 52 | Left lobe | S23 | 1b | 7.3 | Yes | NTe | NT | Present | NT | Reduced |
| FT143 | F | 64 | Right lobe | S34 | 1b | 5.6 | Yes | NT | NT | Present | NT | Unchanged |
| FT144 | M | 60 | Right lobe | S27 | 1b | >7.4 | Yes | NT | NT | Present | NT | NT |
| FT147 | M | 54 | Right lobe | S26 | 1b | 6.7 | Yes | NT | NT | Present | NT | Unchanged |
| FT154 | M | 61 | Segments VI-VII | S23 | 1b | 7.3 | Yes | NT | No toxicity | Present | NT | NT |
| FT155 | F | 55 | Left lobe | S20 | 1b | >7.4 | Yes | NT | No toxicity | Present | NT | Reduced |
| FT156 | M | 77 | Right lobe | S17 | 1b | 6.9 | Yes | NT | No toxicity | Present | NT | Reduced |
| FT161 | M | Unknown | Deceased organ donor | S42 | 1b | 5.2 | Yes | NT | NT | Present | NT | NT |
| FT164 | F | 47 | Right lobe | NIb | NAc | NA | Yes | Present | NT | NT | NT | NT |
| FT171 | M | 48 | Segment V-VII | NI | NA | NA | Yes | Present | NT | NT | NT | NT |
| FT172 | F | 57 | Left lobe | S42 | 1b | 5.2 | Yes | Present | NT | Present | Present | NT |
| FT168 | F | 75 | Left lobe | S34 | 1b | 5.6 | No | NT | NT | Present | Present | NT |
| FT187 | M | 66 | Segment VI-VII | S155 | 1b | 6.0 | Yes | NT | NT | Present | Present | NT |
| FT189 | M | 48 | Left lobe | S155 | 1a | 6.0 | No | NT | NT | Present | Present | NT |
| FT195 | M | 17 | Deceased organ donor | S155 | 1a | 6.0 | No | NT | NT | Present | Present | NT |
M, male; F, female.
NI, no infectious challenge.
NA, not applicable.
Viral load in log10 HCV RNA international units/milliliter
NT, not tested.
Positive-sense (+) and negative-sense (−) HCV RNA strands by the qualitative, nonquantitative, strand-specific rTth RT-PCR-based assay.
Accumulation of positive-sense (+) HCV RNA by the quantitative real-time PCR-based assay.
In vitro infection of primary cultures of human hepatocytes.
Serum samples from nine patients with chronic HCV genotype 1 infection were used for cell infection (the HCV subtypes and viral loads are shown in Table 1). Three days after plating (to permit cell recovery from isolation), the cells were infected in vitro by overnight incubation with 25 μl of HCV-positive serum in 3.0 ml of medium. The cells were then washed three times with 3.0 ml of fresh medium, and the cultures were continued under normal conditions in long-term culture medium. Cells and culture medium were collected at various times during culture and stored at −80°C. All experiments were carried out in duplicate.
Treatment of primary hepatocyte cultures with IFN-α.
Recombinant IFN-α 2a (Roferon-A; Hoffmann-La Roche, Basel, Switzerland) was used at final concentrations of 500 to 10,000 U/ml. IFN-α treatment was started at the time of HCV infection in most experiments and 3 days after infection in some cases. IFN-α was replaced on a daily basis as the culture medium was changed. All experiments were carried out in duplicate.
Western blot analysis of IRF-1 and PKR induction.
Total protein corresponding to 400,000 cells was extracted in Laemmli buffer, electrophoresed on 12.5% polyacrylamide gels, and then transferred electrophoretically to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Bedford, Mass.). Western blotting was performed using the Santa Cruz polyclonal antibodies against interferon-responsive factor 1 (IRF-1) (kindly provided by N. Mechti, INSERM U475, Montpellier, France) and a monoclonal antibody against human RNA-dependent protein kinase (PKR) (kindly provided by E. Meurs, Institut Pasteur, Paris, France). The proteins were visualized by using an enhanced chemiluminescence detection method (Amersham Pharmacia, Abingdon, United Kingdom), and the blots were analyzed with a National Institutes of Health image analyzer in order to measure the amounts of induced proteins.
Cellular RNA extraction and strand-specific rTth RT-PCR.
At the time of cell harvest, the medium was removed, and the cultures were washed three times with cold phosphate-buffered saline. RNA was purified from 4 × 106 or 2 × 106 cultured hepatocytes or from 100-μl samples of HCV-positive sera, using a guanidium isothiocyanate-acid phenol extraction procedure (RNABLE; Eurobio, Les Ulis, France). Precipitated RNA was dissolved in 50 μl of diethyl pyrocarbonate-treated water and quantified by UV spectrum analysis. Extracted RNA was analyzed with a modification of the previously described strand-specific rTth reverse transcription-PCR (RT-PCR) assay (7). Primers located in the HCV 5′ noncoding region, including antisense primer HCV-I (5′-TGG[A]TGCACGGTCTACGAGACCTC-3′, nucleotides [nt] 342 to 320) and sense primer HCV-II (5′-CACTCCCCTGTGAGGAACT-3′, nt 38 to 56) (19), were used. In the positive-strand HCV RNA assay, 1 μg of cellular RNA (corresponding to approximately 6 × 10 4 cells) in 10 μl of diethyl pyrocarbonate-treated water was layered with mineral oil and heated at 95°C for 1 min. The temperature was then lowered to 70°C, and a 20-μl reaction mixture containing 50 ng of primer HCV-I, 1× RT buffer (Applied Biosystems, Foster City, Calif.), 1 mM MnCl2, 200 μM (each) deoxynucleoside triphosphate, and 5 U of rTth enzyme (Applied Biosystems) was prepared for cDNA synthesis. Primer annealing was performed at 60°C for 2 min, followed by the RT reaction at 70°C for 20 min. In order to inactivate the RT activity of rTth, Mn2+ was chelated with 40 μl of a mixture containing 8 μl of 10× EGTA chelating buffer (Applied Biosystems). Forty microliters of the prewarmed (70°C) PCR mixture containing 50 ng of primer HCV-II and 3.75 mM MgCl2 was added. PCR was performed on the GeneAmp PCR-System 9600 apparatus (Applied Biosystems) and consisted of the following: (i) an initial denaturation step of 1 min at 94°C; (ii) 50 cycles, with 1 cycle consisting of 15 s at 94°C, 30 s at 58°C, and 30 s at 72°C; and (iii) a final extension step of 7 min at 72°C. PCR products were analyzed by agarose gel electrophoresis. The negative-strand HCV RNA assay was performed by the same procedure, except that the primers were used in reverse order.
Real-time PCR quantification of positive- and negative-strand HCV RNA.
Both positive- and negative-strand HCV RNAs were quantified by means of a real-time PCR assay using the LightCycler instrument and technology (Roche Applied Science, Indianapolis, Ind.) and SYBR green I dye for detection. The primer pair was located in the HCV 5′ noncoding region and included antisense primer KY78 (5′-CTCGCAAGCACCCTATCAGGCAGT-3′, nt 311 to 288) and sense primer KY80 (5′-GCAGAAAGCGTCTAGCCATGGCGT-3′, nt 68 to 91) (46). One microgram of cellular RNA was used for cDNA synthesis in a 20-μl reaction mixture containing 5 U of rTth polymerase and 1 μM RT primer. Primer HCV-I was used for positive-strand cDNA synthesis, and primer HCV-II was used for negative-strand cDNA synthesis. In addition, PCR amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was performed as an internal control for the quality of extracted cellular RNA, with RT using primer GAPDH-31 (5′-GCCTGCTTCACCACCTTCTTG-3′, nt 869 to 849). cDNA was synthesized at 70°C for 20 min in all instances, and generated cDNA was purified with the HighPure PCR product purification kit (Roche Applied Science) in a 50-μl volume. Positive- and negative-strand HCV PCR amplifications were performed with 3 μl of purified cDNA in a 10-μl reaction mixture containing 1 μl of LightCycler-FastStart DNA Master SYBR green (Roche Applied Science) and 0.5 μM (each) HCV primer KY78 and KY80. PCR consisted of an initial denaturation step of 6 min at 95°C, followed by 45 cycles, with 1 cycle consisting of 15 s at 95°C, 5 s at 70°C, and 15 s at 72°C. All the samples were analyzed in triplicate. PCR amplification of GAPDH mRNA used primers GAPDH-51 (5′-ACAGTCCATGCCATCACTGCC-3′, nt 603 to 624) and GAPDH-31. One microliter of purified cDNA was used in 10 μl of mixture containing 1 μl of LightCycler-FastStart DNA Master SYBR green and 0.5 μM (each) primer. DNA was quantified in real time during the PCR by measuring fluorescent dye incorporation into PCR products at 530 nm. At the end of each run, a DNA melting step was performed, and the fusion curve was recorded to control for the homogeneity and quality of amplified DNA. In each run, 10-fold serial dilutions of synthetic positive- and negative-strand RNAs were tested in duplicate to establish a standard curve to calculate the amount of positive- and negative-strand HCV RNA in each sample. Tenfold serial dilutions of purified GAPDH mRNA amplicons were tested in duplicate to quantify GAPDH mRNA in each sample. The measured amounts of HCV RNA were normalized to the amount of GAPDH mRNA in each sample, and the results were expressed per culture plate (4 × 106 or 2 × 106 cells).
Generation of HCV quasispecies sequences.
Extracted RNAs were reverse transcribed at 42°C for 60 min by using 70 ng of primer ASPR per μl (5′-AGCTCCGCCAAGGCAGAAGACAC-3′, nt 7347 to 7369) in the presence of 8 U of avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.). The first nested PCR round was performed using 5 pmol of degenerated sense primer HC378-1b (5′-TCCCRTGYGAGCCYGAACCG-3′, nt 6808 to 6828) and antisense primer MKed (5′-TTCCARGACTCTARCART-3′, nt 7193 to 7234) with Pwo high-fidelity DNA polymerase (Roche Molecular Biochemicals, Mannheim, Germany). The second-round PCR used sense primer NS5A-S (5′-CCCACATTACAGCAGAGACGGC-3′, nt 6865 to 6986) and antisense primer WARid (5′-GGRTTGTARTCCGGSCGYGCCCATA-3′, nt 7189 to 7213). After denaturation for 5 min at 94°C, the two PCR rounds comprised 30 cycles (1 cycle consisting of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C) and a final elongation step at 72°C for 5 min. Amplified products were analyzed by electrophoresis through 2% agarose gel and staining with ethidium bromide.
PCR products were cloned using the TOPO TA cloning kit for sequencing (Invitrogen BV, Gröningen, The Netherlands), according to the manufacturer's protocol. They were then ligated into the M13 vector, and recombinant plasmid DNA was transformed into competent Escherichia coli cells according to the manufacturer's protocol. Transformants were grown on plates containing brain heart infusion and ampicillin. Cloned DNA was reamplified by using both sense and antisense M13-specific primers in the presence of high-fidelity Pwo DNA polymerase. PCR products were purified with Microcon 100 before sequencing (Millipore, Dublin, Ireland).
After cloning and PCR amplification of 20 clones per time point, each clone was sequenced with the Cy5.0/Cy5.5 dye primer kit (Visible Genetics, Inc., Toronto, Canada) on a LongRead Tower automated DNA sequencer (Visible Genetics, Inc.) according to the manufacturer's instructions. The sequencing primers were the labeled upstream and downstream PCR primers.
Genetic and phylogenetic analyses of HCV quasispecies evolution.
Nucleotide sequences were aligned using the CLUSTAL X program. Distances between pairs of sequences were calculated by using the DNADIST module in the PHYLIP package version 3.572 (distributed by J. Felsenstein, Department of Genetics, University of Washington, Seattle). Distance calculation was based on a Kimura two-parameter distance matrix with a transition-to-transversion ratio of 4.0. The means ± standard errors of the means (SEMs) of within-sample genetic distances were calculated for the inocula, and the means ± SEMs for between-sample genetic distances were calculated on the basis of distances between pairs of inocula (day 0) and postculture (day 8 of infection) sequences. Accumulation of synonymous and nonsynonymous substitutions per synonymous and nonsynonymous site, respectively, was calculated with the Jukes-Cantor correction for multiple substitutions, using the MEGA program (15). Statistical comparisons were made using a t test.
The PHYLIP program, version 3.572, was used to construct phylogenetic trees by means of the neighbor-joining method with a sequence matrix determined by the two-parameter method of Kimura. Patient viral sequence trees were constructed with nucleotide sequences. Phylogenetic analyses of all viral sequences generated in this study showed distinct clusters of viral sequences corresponding to each patient (data not shown), indicating the absence of PCR cross-contamination.
RESULTS
HCV replicates in primary cultures of healthy human hepatocytes infected in vitro.
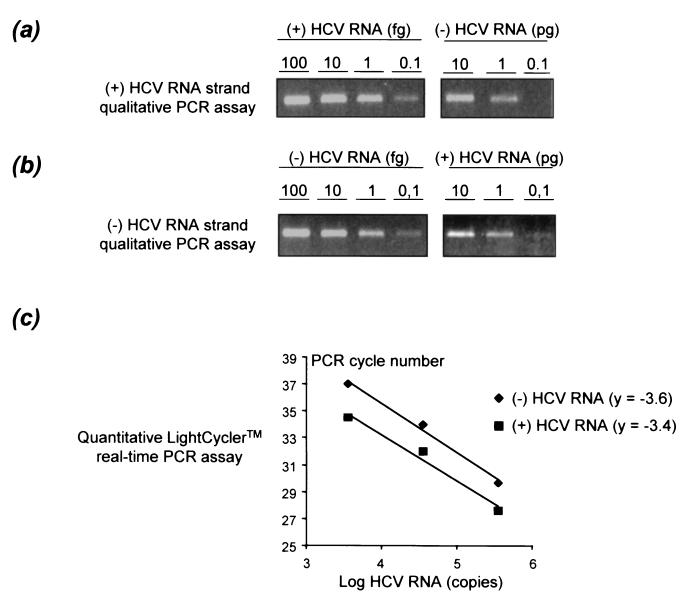
We prepared primary cultures of healthy human hepatocytes from 15 HCV-seronegative donors (Table 1). Ferrini et al. previously showed that healthy human hepatocytes retain a differentiated phenotype for at least 35 days under the conditions used here (6). Serum samples from nine patients chronically infected with HCV genotype 1 (subtype 1a or 1b) who had never been treated were used for in vitro infection of 13 of the 15 primary hepatocyte cultures (Table 1). Cultures were infected 3 days after plating and harvested at various times between 3 and 12 days of culture for extraction of total cellular RNA. Two complementary assays were used to study positive- and negative-sense HCV RNA strands in the inocula and infected hepatocyte cultures. The first assay is a highly sensitive qualitative (i.e., nonquantitative) detection assay, based on a modification of our previously described strand-specific rTth RT-PCR assay (7). As shown in Fig. 1a and b, the assay detects 0.1 fg of the correct RNA strand (i.e., 3 × 102 molecules), whereas at least 1 to 10 pg of the incorrect RNA strand is required to obtain a detectable signal. The second assay is a quantitative assay based on real-time PCR with the LightCycler technology allowing quantification of positive- and negative-sense HCV RNA strands in both the inocula and hepatocyte cultures (Fig. 1c). As shown in Fig. 1c, the tested interval from 3.5 to 5.5 log HCV RNA copies per capillary was within the dynamic range of quantification of the assay. The quantitative assay is less sensitive for HCV RNA strand detection than the qualitative assay, with a lower detection cutoff of 1 to 2 log HCV RNA copies per milliliter higher than the latter. The results are summarized in Table 1.
FIG. 1.
Characteristics of the strand-specific HCV RNA assays used in this study. (a) Strand specificity of the positive-strand-specific HCV RNA rTth RT-PCR assay. Decreasing amounts of positive-strand (+) HCV RNA (100, 10, 1, and 0.1 fg) and of negative-strand (-) HCV RNA (10, 1, and 0.1 pg) synthesized from an appropriate plasmid were subjected to the rTth RT-PCR assay. The products were analyzed by agarose gel electrophoresis. (b) Strand specificity of the negative-strand-specific HCV RNA rTth RT-PCR assay. Decreasing amounts of negative-strand HCV RNA (100, 10, 1, and 0.1 fg) and positive-strand HCV RNA (10, 1, and 0.1 pg) synthesized from the same plasmid as for panel a were analyzed by the same procedure. (c) Range of linear quantification of the quantitative assay based on real-time PCR using the LightCycler technology and SYBR green I dye for detection. The range of linear quantification of the assay was studied by testing 10-fold serial dilutions of synthetic positive- and negative-sense HCV RNA strands after RT at 70°C with the rTth polymerase. Each point is the mean of three experimental values for each dilution. y is the slope of the linear plots.
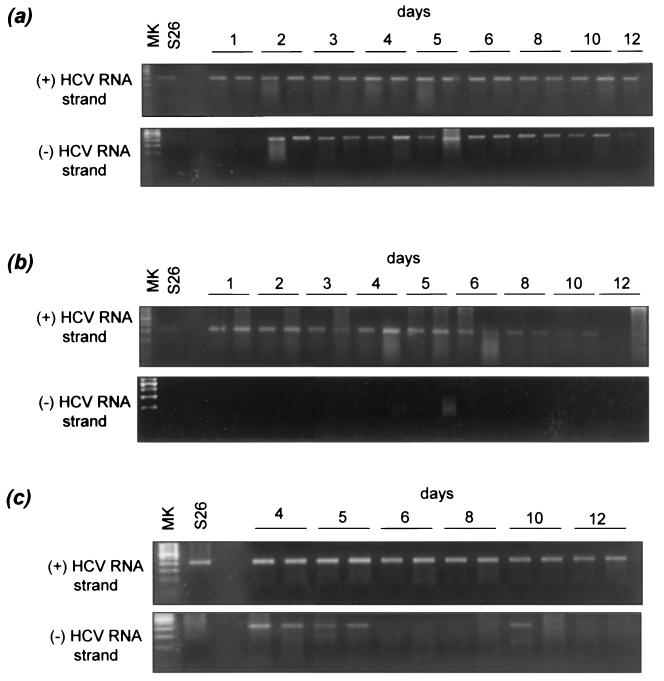
As expected, the positive-sense RNA strand was the only form of HCV RNA present in the inoculum. We thus used detection of the positive- and negative-sense RNA strands with the qualitative assay as a marker of HCV replication in hepatocyte cultures. Both positive- and negative-sense RNA strands were detected in the cultures. In culture FT147 infected with serum S26 (Fig. 2a), positive RNA strands were detected on day 1 postinfection and were still present in the last plate harvested. The negative-sense RNA strand was detected on day 2 postinfection and remained detectable up to day 10, proving viral replication in the culture. We observed the presence of positive-strand HCV RNA throughout the culture period and persistent expression of negative-strand HCV RNA in the other cultures infected with different sera (Table 1 and data not shown).
FIG. 2.
Qualitative assay detection of positive- and negative-strand HCV RNA in a primary culture of healthy human hepatocytes infected in vitro with an HCV-positive serum and effect of IFN-α. The hepatocyte culture FT147, infected 3 days after plating by HCV-positive serum S26, is shown. Positive-strand (+) RNA but not negative-strand (-) RNA was present in the inoculum. (a) Primary hepatocyte culture in the absence of IFN-α. Positive-strand HCV RNA was detected with the qualitative strand-specific rTth PCR assay from day 1 to the end of the culture (day 12), whereas negative-strand RNA was detected from days 2 to 10. (b) Culture in the presence of 5,000 U of IFN-α per ml. Positive-strand HCV RNA was detected from days 1 to 10, whereas negative-strand RNA was never detected. (c) Culture treated on day 3 with 5,000 U of IFN-α per ml. Positive-strand RNA was detected throughout the culture period, whereas negative-strand RNA was no longer detected after day 5. Similar patterns (not shown) were observed with the following cultures infected with the corresponding sera: FT141 and S23, FT143 and S34, FT144 and S27, FT154 and S23, FT155 and S20, and FT156 and S17. MK, molecular size standards.
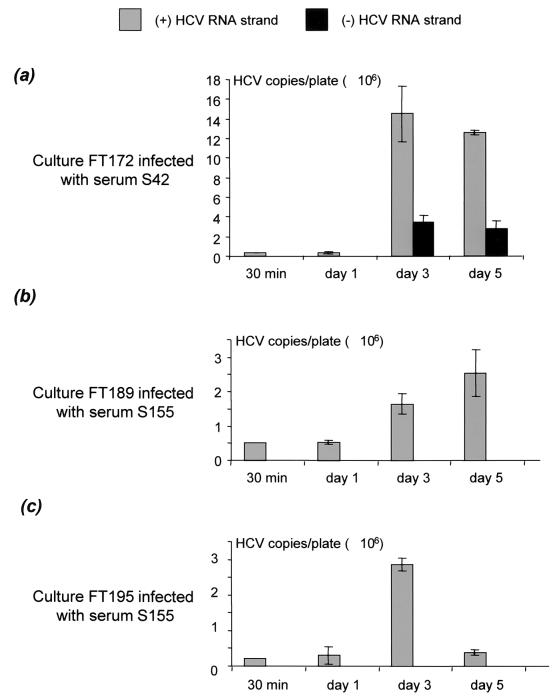
HCV replication in hepatocyte cultures was further supported by the accumulation of HCV RNA strands, as measured by real-time quantitative RT-PCR. Indeed, we observed a significant increase in the amount of both positive- and negative-sense HCV RNA strands in culture FT172 infected with serum S42 (Fig. 3a). A similar increase in the amount of positive-strand HCV RNA was observed in cultures FT189 and FT195, both infected with S155, but the total amount of positive-strand HCV RNA was smaller in these cultures at the various time points, suggesting less replication than that in culture FT172 (Fig. 3b and c). The negative-strand HCV RNA also accumulated in culture FT172, but the amount of negative-strand HCV RNA was consistently smaller than the amount of positive-strand HCV RNA on days 3 and 5 (Fig. 3a). This explains why, in cultures FT189 and FT195, negative-strand HCV RNA was not detected with the quantitative assay, whereas it was detected with the more sensitive qualitative assay; i.e., its amount was below the detection cutoff of the quantitative assay. Similar results were obtained with culture FT168 infected with serum S34 (data not shown). Finally, neither positive- nor negative-strand HCV RNA was detected in culture supernatants by the sensitive qualitative assay.
FIG. 3.
Accumulation of positive- and negative-strand HCV RNA in hepatocyte cultures FT172 (a), FT189 (b), and FT195 (c), infected with sera S42, S155, and S155, respectively, as measured by the quantitative LightCycler real-time RT-PCR assay. The hepatocyte cultures were infected 3 days after plating. The cells were harvested 30 min and 1, 3, and 5 days after infection for positive-strand (gray) and negative-strand (black) HCV RNA quantification. The amounts of HCV RNA strands are shown as means ± SEMs of three determinations, expressed in numbers of HCV RNA copies per 2 × 106 cells, normalized to GAPDH mRNA. Similar results (not shown) were obtained with culture FT168 infected with serum S34.
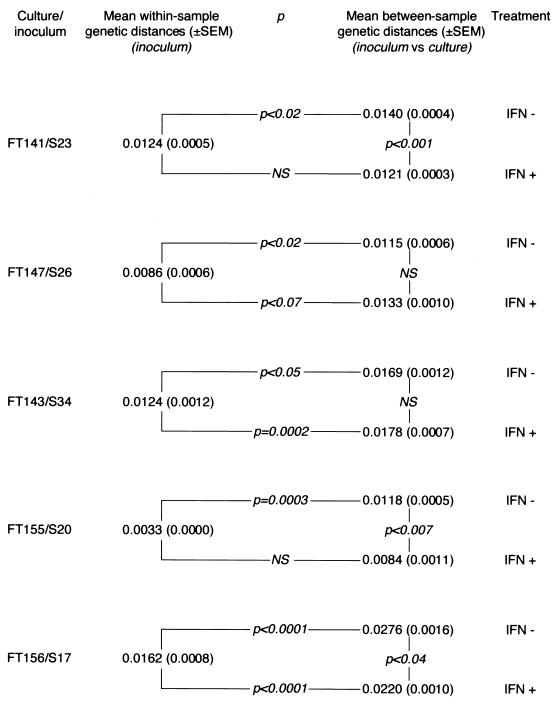
In order to prove that HCV replicated in primary hepatocyte cultures, i.e., that HCV RdRp synthesized both negative- and positive-sense HCV RNA strands, we examined the accumulation of mutations on HCV genomes in five cultures. A 300-bp fragment located within the NS5A gene was chosen for this study. In all instances, nucleotide mutations accumulated on positive-strand HCV genomes during the culture period. Comparison of NS5A quasispecies sequences in the inoculum and after 8 days of culture (20 clones per time point) showed significantly higher between-sample genetic distances (calculated by pairwise comparison of NS5A quasispecies sequences in the inoculum versus the culture) than within-sample genetic distances (calculated by pairwise comparison of NS5A quasispecies sequences in the inoculum) (Fig. 4). In all instances, accumulation of synonymous mutations per synonymous site was significantly greater than accumulation of nonsynonymous mutations per nonsynonymous site (data not shown), indicating that the accumulation of mutations on HCV genomes in culture resulted from random nucleotide misincorporations by RdRp, in the absence of positive selection forces driving genetic evolution.
FIG. 4.
Accumulation of nucleotide substitutions on HCV genomes during replication in five primary cultures of healthy human hepatocytes in the presence and absence of IFN-α. The accumulation of mutations on HCV genomes was assessed by comparing the mean ± SEM within-sample genetic distance (calculated by pairwise comparison of NS5A quasispecies sequences in the inoculum) with the mean ± SEM between-sample genetic distance (calculated by pairwise comparison of NS5A quasispecies sequences in the culture versus the inoculum). A significantly higher between-sample than within-sample genetic distance was interpreted as a significant accumulation of genomic mutations over time as a result of HCV replication in the culture; the lack of significant difference was interpreted as a lack of genetic evolution in the culture, reflecting inhibition of HCV replication. The between-sample genetic distances were also compared for each culture in the presence (+) and absence (-) of IFN-α. A significantly smaller between-sample genetic distance in the presence of IFN-α reflected reduced accumulation of mutations in the culture and was interpreted as an inhibition of HCV replication by IFN-α. NS, not significantly different (i.e., P > 0.05).
Together, these findings demonstrated unequivocally that HCV replicated in the primary cultures of healthy human hepatocytes as a result of viral RdRp function. Complete infectious virions did not appear to be secreted in the medium.
Primary cultures of healthy human hepatocytes exhibit a biological response to IFN-α.
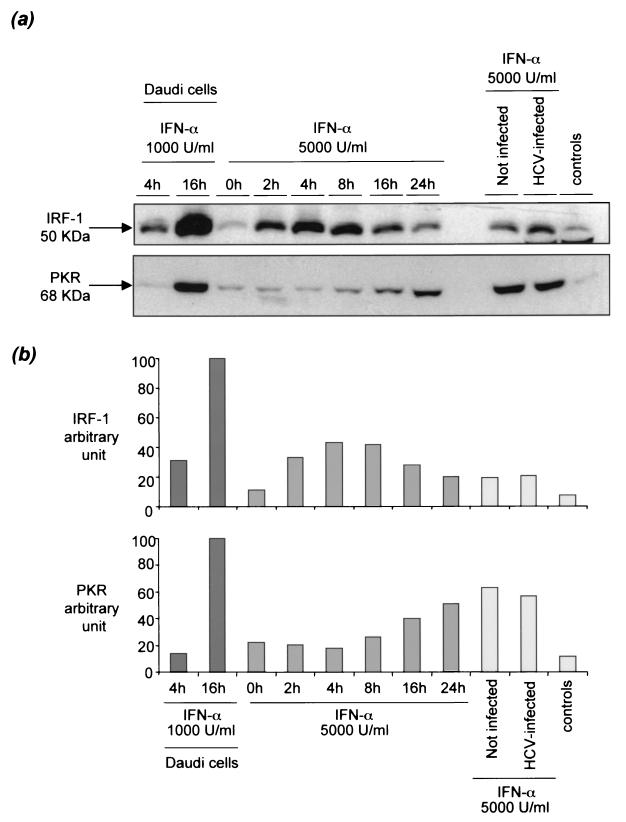
To determine whether primary hepatocyte cultures are equipped to respond appropriately to IFN-α stimulation, uninfected cultures were treated with 5,000 U of IFN-α per ml. We then extracted total cellular protein at various times from 0 to 24 h and analyzed the expression of IRF-1 and double-stranded PKR by Western blotting. These two proteins are encoded by two prototypic IFN-α-regulated genes: IRF-1 is a transcription factor induced as a primary response to IFN-α, while PKR induction is a secondary response, necessitating prior synthesis of IRF-1. IFN-α-stimulated Daudi cells were used as positive controls for these experiments. IFN-α induced the expression of both IRF-1 and PKR in cultured hepatocytes by factors of approximately 4 and 3, respectively (Fig. 5). As expected, IRF-1 expression preceded PKR expression by approximately 8 h. Similar experiments were carried out after 8 days of culture, with both uninfected and HCV-infected hepatocytes. Identical results were obtained, indicating that the response to IFN-α is maintained for more than a week and is not eliminated by HCV infection (Fig. 5).
FIG. 5.
Effects of IFN-α on IRF-1 and PKR expression in primary cultures of human hepatocytes. Immunoblot analysis was performed with anti-IRF-1 and anti-PKR antibodies after 8 days of culture in infected and noninfected primary hepatocytes treated with 5,000 U of IFN-α per ml and in Daudi cells treated with 1,000 U of IFN-α per ml used as positive controls. Cells not treated with IFN-α were used as controls. Immunoblot experimental results (a), together with their quantitative representation after National Institutes of Health image analysis (b) are shown. (b) (Left) Effect of 1,000 U of IFN-α per ml on Daudi cells harvested after 4 and 16 h of treatment. (Center) Effect of 5,000 U of IFN-α per ml on hepatocyte culture FT172 harvested after 0, 2, 4, 8, 16, and 24 h of treatment. (Right) Effect of HCV infection of the primary hepatocyte culture on the effect of IFN-α on IRF-1 and PKR expression. Similar results (not shown) were obtained in cultures FT164 and FT171.
IFN-α is not toxic for primary cultures of healthy human hepatocytes at the concentrations used in this study.
As an effect of IFN-α on markers of intracellular HCV replication might merely reflect cytotoxicity rather than inhibition of viral RdRp, we studied IFN-α toxicity in our primary hepatocyte culture system. Phase-contrast microscopy revealed no signs of cellular toxicity. Furthermore, culture of hepatocytes from three different donors (FT154, FT155, and FT156), treated for 5 days with 5,000 U of IFN-α per ml, revealed no significant reduction in total de novo protein synthesis (not shown), a sensitive marker of cytotoxic stress in cultured hepatocytes (6).
IFN-α inhibits the expression of positive- and negative-sense HCV RNA strands in primary cultures of healthy human hepatocytes infected in vitro.
We tested eight primary hepatocyte cultures infected in vitro for the effects of continuous incubation with 5,000 and 10,000 U of IFN-α per ml by means of the qualitative HCV RNA assay. In culture FT147 infected with serum S26 (Fig. 2b), the positive-sense RNA strand was detected from day 1 but disappeared after day 10 in the presence of IFN-α, whereas the negative-sense RNA strand remained undetectable throughout the culture period (i.e., until day 12). Similarly, the negative-sense RNA strand was never detected in any other IFN−α-treated culture (not shown). In contrast, when IFN-α treatment was started 3 days after HCV infection, the positive-sense RNA strand was detected throughout the culture period, whereas the negative-sense RNA strand was detected from infection through day 5 before disappearing (Fig. 2c).
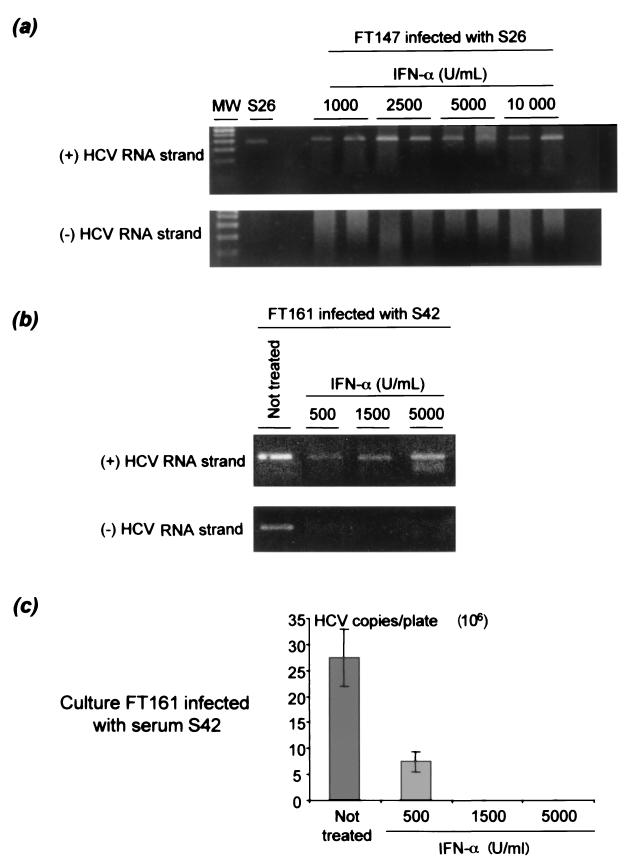
The effect of increasing IFN-α concentrations (500 to 10,000 U/ml) on the detection of positive- and negative-sense HCV RNA strands was studied 5 or 8 days after infection. In cultures FT147 and FT161 (Fig. 6a), the negative-sense HCV RNA strand was never detected, whatever the IFN-α concentration used, whereas the positive-sense HCV RNA strand was always detected, even at the maximum IFN-α concentration used, i.e., 10,000 U/ml. Inhibition of viral replication in the culture was confirmed by using the less sensitive, but quantitative, real-time RT-PCR assay. Indeed, this assay did not detect positive-strand HCV RNA at IFN-α concentrations higher than 500 U/ml, meaning that the intracellular amount of positive-strand HCV RNA was below the detection cutoff level (Fig. 6b). Altogether, these results suggested potent concentration-dependent inhibition of positive-sense HCV RNA strand accumulation in response to IFN-α treatment.
FIG. 6.
Effect of increasing concentrations of IFN-α on the accumulation of positive- and negative-strand HCV RNA in primary hepatocyte cultures infected in vitro. Cultures FT147 (infected with serum S26) and FT161 (infected with serum S42) were treated for 5 and 8 days with IFN-α concentrations ranging from 1,000 to 10,000 U/ml and 500 to 5,000 U/ml, respectively. Qualitative detection of positive-sense (+) and negative-sense (-) HCV RNA strands is shown in cultures FT147 (a) and FT161 (b). In both instances, positive-strand HCV RNA was detected at all concentrations used, whereas the negative strand was never detected. MW, molecular size standards. (c) In culture FT161, LightCycler real-time RT-PCR quantitative analysis of the same extracts showed a reduction in the amount of positive-sense HCV RNA strand in the culture when the IFN-α concentration increased, suggesting IFN-α concentration-dependent inhibition of HCV replication in the culture. Similar results (not shown) were obtained with culture FT187 infected with serum S155.
IFN-α inhibits the accumulation of mutations on the HCV genome during replication in primary cultures of healthy human hepatocytes infected in vitro.
In order to confirm that IFN-α inhibited HCV replication, we studied its effect on the accumulation of mutations on HCV genomes. In the absence of IFN-α, significant HCV genetic evolution was always observed, due to random accumulation of mutations (Fig. 4). In contrast, when 5,000 or 10,000 U of IFN-α per ml was added daily before testing for positive-strand RNA on day 8 of infection (i.e., several days before it otherwise became undetectable), no significant genetic evolution was observed in two of the five cultures tested (Fig. 4). In addition, the between-sample genetic distances (calculated by pairwise comparison of the quasispecies sequences in culture on day 8 versus that in the inoculum) were significantly lower in the presence of IFN-α than in the absence of IFN-α in three of the five cultures, again suggesting that IFN-α significantly inhibited the accumulation of HCV genome mutations. No significant difference was seen in the remaining two cultures. In one culture, phylogenetic analysis followed by phylogenetic tree plotting showed a trend toward distinctive clustering of postculture and inoculum quasispecies sequences, respectively, in the absence of IFN-α; this clustering was abolished in the presence of IFN-α (data not shown). No such trend was clearly visible in the remaining four cultures.
DISCUSSION
This study shows that HCV can replicate in primary cultures of human hepatocytes infected in vitro, as a result of viral RdRp function. As previously described (7), in vitro infection resulted in the production of negative-sense HCV RNA strands, an HCV replication intermediate. In addition, real-time RT-PCR quantification showed a significant accumulation of positive-sense HCV RNA strands and, when present in sufficient amounts, of negative-sense HCV RNA strands during hepatocyte culture. Significant accumulation of random mutations on the HCV genome showed that the viral RdRp, an error-prone RNA polymerase with no proofreading activity, was responsible for the accumulation of positive-strand HCV RNA genomes during culture. The random accumulation of mutations in the region studied, in the absence of positive pressure toward amino acid changes, was not surprising in this in vitro culture system. However, the same region of the NS5A gene displays a similar pattern of genetic evolution in HCV-infected patients (33). As NS5A is part of the replication complex and is most likely involved in regulating RdRp function (13, 43), the conservation constraints on NS5A evolution occurring in vivo might also be present in our hepatocyte culture system.
The principal finding of this study is the effect of IFN-α on HCV replication in primary hepatocyte culture. IFN-α was recently shown to inhibit the replication of dengue virus, another member of the Flaviviridae family, in hepatoma cell lines infected in vitro (5). Mathematical modeling of HCV dynamics during human IFN-α treatment recently suggested that IFN-α blocks HCV virion production or release as a result of its direct, nonspecific antiviral effect (29, 30). The capacity of IFN-α to directly inhibit HCV replication in healthy human hepatocytes had not previously been demonstrated.
We show that IFN-α blocks HCV genome synthesis by the HCV RdRp in cultured healthy human hepatocytes in a concentration-dependent manner. This effect is probably mediated by IFN-α-induced cellular pathways supporting nonspecific antiviral actions. Indeed, we observed the following. (i) IFN-α-induced genes were expressed in primary hepatocyte cultures treated with IFN-α, and their expression was not altered by HCV infection. (ii) Expression of negative-strand HCV RNA was always suppressed in IFN-α-treated cultures. (iii) IFN-α significantly inhibited the accumulation of mutations on the HCV genome in three of five hepatocyte cultures. The concentrations of IFN-α used here were relatively high (500 to 10,000 U/ml), but we showed that IFN-α toxicity could not explain the inhibitory effect on HCV replication. An earlier report suggested that IFN-α could act by inhibiting de novo infection of hepatocytes (48). If IFN-α effectively prevents HCV entry into hepatocytes in vivo, this effect would probably be mediated by IFN-α-induced humoral responses (neutralizing antibodies), which are not present in hepatocyte cultures. We did not examine whether IFN-α could affect virus entry in our model, in addition to inhibiting viral replication. It is conceivable that the reduction of positive-strand HCV RNA accumulation in IFN-α-treated cells could be enhanced by receptor down-regulation or by decreased internalization or membrane fusion.
Interestingly, despite the disappearance of negative-strand HCV RNA from all IFN-α-treated cultures, positive-strand RNA always persisted for several days, suggesting that the kinetics of HCV RNA strands in cell culture differ from those in the peripheral circulation. This finding was not surprising, because most of the mechanisms governing viral clearance are absent in vitro, especially when only intracellular HCV RNA is concerned, whereas the estimated mean half-life of free HCV virions is only 2.7 h in vivo (30). In contrast, the apparent lack of IFN-α inhibition of mutation accumulation on HCV genomes in two cultures, despite a clear effect on negative-strand HCV RNA production, was surprising. The inhibitory effect of IFN-α may have been weaker in these two cultures, permitting low-level viral replication, while negative-strand HCV RNA was undetectable, even with our sensitive qualitative strand-specific HCV RNA assay. Such variability in the effect of IFN-α might be explained by partial hepatocyte resistance to IFN-α stimulation or by partial viral resistance to IFN-α, possibly mediated by viral proteins inhibiting antiviral effectors induced by IFN-α (32). It is noteworthy that all the cultures were infected with HCV genotype 1, a genotype that displays various levels of IFN-α sensitivity based on initial IFN-α blocking efficacy (16, 30), possibly owing to differences in the sequences of viral proteins and, thus, in their structure and function. Unfortunately, data on early viral dynamics during IFN-α therapy in the patients whose blood samples were used for in vitro infection are not available to confirm this hypothesis.
In conclusion, we show that primary cultures of healthy human hepatocytes can be infected in vitro by HCV and support its sustained replication. We further show that IFN-α is able to block HCV replication in this culture model, which is close to the HCV-infected human liver. These results strengthen the hypothesis that IFN-α acts primarily through its nonspecific antiviral effects and suggest that primary cultures of human hepatocytes may provide a good model to study intrinsic HCV resistance to IFN-α. However, clearance of infected cells resulting from IFN-α-modulated immune responses probably plays a major role in permanent HCV RNA clearance during therapy (16, 30), emphasizing the need for both in vitro and in vivo studies to understand IFN-α treatment failure in HCV-infected patients.
Acknowledgments
Valerie Castet and Chantal Fournier contributed equally to this work.
We thank Jacques Domergue, Jean-Michel Fabre, and Henri Joyeux for providing surgical liver samples and Marie-France Saint-Marc-Girardin (Hoffmann-La Roche) for providing recombinant IFN-α 2a. We are grateful to Pierre-Olivier Frainais for excellent technical assistance in viral quasispecies assessment and Patricia Ponsoda for assessment of IFN-α toxicity. We thank Robert E. Lanford, Robert R. Purcell, and Avidan U. Neumann for helpful comments on the manuscript.
This work was supported in part by grant no. 2000/102 from the “Agence Nationale de Recherches sur le SIDA et l'Hépatite C” (ANRS) to J.-M.P., a grant from the “Réseau National Hépatites” to P.M., and a grant from Hoffmann-La Roche (Neuilly-sur-Seine, France). V.C. was the recipient of an ANRS predoctoral grant. P.M. also thanks the Lion's Club d'Uzès for financial support.
REFERENCES
- 1.Agnello, V., G. Abel, G. B. Knight, and E. Muchmore. 1998. Detection of widespread hepatocyte infection in chronic hepatitis C. Hepatology 28:573-584. [DOI] [PubMed] [Google Scholar]
- 2.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence, and sequelae in hepatitis C virus infection: a perspective on long-term outcome. Semin. Liver Dis. 20:17-35. [DOI] [PubMed] [Google Scholar]
- 3.Chieux, V., D. Hober, J. Harvey, G. Lion, D. Lucidarme, G. Forzy, M. Duhamel, J. Cousin, H. Ducoulombier, and P. Wattre. 1998. The MxA protein levels in whole blood lysates of patients with various viral infections. J. Virol. Methods 70:183-191. [DOI] [PubMed] [Google Scholar]
- 4.Chung, R. T., W. He, A. Saquib, A. M. Contreras, R. J. Xavier, A. Chawla, T. C. Wang, and E. V. Schmidt. 2001. Hepatitis C virus replication is directly inhibited by IFN-alpha in a full-length binary expression system. Proc. Natl. Acad. Sci. USA 98:9847-9852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrini, J. B., L. Pichard, J. Domergue, and P. Maurel. 1997. Long-term primary cultures of adult human hepatocytes. Chem. Biol. Interact. 107:31-45. [DOI] [PubMed] [Google Scholar]
- 7.Fournier, C., C. Sureau, J. Coste, J. Ducos, G. Pageaux, D. Larrey, J. Domergue, and P. Maurel. 1998. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J. Gen. Virol. 79:2367-2374. [DOI] [PubMed] [Google Scholar]
- 8.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 9.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heathcote, E. J., M. L. Shiffman, W. G. Cooksley, G. M. Dusheiko, S. S. Lee, L. Balart, R. Reindollar, R. K. Reddy, T. L. Wright, A. Lin, J. Hoffman, and J. De Pamphilis. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C and cirrhosis. N. Engl. J. Med. 343:1673-1680. [DOI] [PubMed] [Google Scholar]
- 11.Heim, M. H. 2000. Intracellular signalling and antiviral effects of interferons. Dig. Liver Dis. 32:257-263. [DOI] [PubMed] [Google Scholar]
- 12.Hovanessian, A. G. 1991. Interferon-induced and double-stranded RNA-activated enzymes: a specific protein kinase and 2′,5′-oligoadenylate synthetases. J. Interferon Res. 11:199-205. [DOI] [PubMed] [Google Scholar]
- 13.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 14.Isom, H. C., and I. Georgoff. 1984. Quantitative assay for albumin-producing liver cells after simian virus 40 transformation of rat hepatocytes maintained in chemically defined medium. Proc. Natl. Acad. Sci. USA 81:6378-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 16.Lam, N. P., A. U. Neumann, D. R. Gretch, T. E. Wiley, A. S. Perelson, and T. J. Layden. 1997. Dose-dependent acute clearance of hepatitis C genotype 1 virus with interferon alfa. Hepatology 26:226-231. [DOI] [PubMed] [Google Scholar]
- 17.Lanford, R. E., K. D. Carey, L. E. Estlack, G. C. Smith, and R. V. Hay. 1989. Analysis of plasma protein and lipoprotein synthesis in long-term primary cultures of baboon hepatocytes maintained in serum-free medium. In Vitro Cell. Dev. Biol. 25:174-182. [DOI] [PubMed] [Google Scholar]
- 18.Lanford, R. E., C. Sureau, J. R. Jacob, R. White, and T. R. Fuerst. 1994. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology 202:606-614. [DOI] [PubMed] [Google Scholar]
- 19.Laskus, T., M. Radkowski, L. F. Wang, J. Cianciara, H. Vargas, and J. Rakela. 1997. Hepatitis C virus negative strand RNA is not detected in peripheral blood mononuclear cells and viral sequences are identical to those in serum: a case against extrahepatic replication. J. Gen. Virol. 78:2747-2750. [DOI] [PubMed] [Google Scholar]
- 20.Lerat, H., S. Rumin, F. Habersetzer, F. Berby, M. A. Trabaud, C. Trepo, and G. Inchauspe. 1998. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood 91:3841-3849. [PubMed] [Google Scholar]
- 21.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 22.Major, M. E., and S. M. Feinstone. 1997. The molecular virology of hepatitis C. Hepatology 25:1527-1538. [DOI] [PubMed] [Google Scholar]
- 23.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 24.Martell, M., J. I. Esteban, J. Quer, J. Genesca, A. Weiner, R. Esteban, J. Guardia, and J. Gomez. 1992. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J. Virol. 66:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McHutchison, J. 1999. Hepatitis C therapy in treatment-naive patients. Am. J. Med. 107:56S-61S. [DOI] [PubMed] [Google Scholar]
- 26.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 27.Meurs, E., K. Chong, J. Galabru, N. S. Thomas, I. M. Kerr, B. R. Williams, and A. G. Hovanessian. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379-390. [DOI] [PubMed] [Google Scholar]
- 28.Negro, F., E. Giostra, K. Krawczynski, R. Quadri, L. Rubbia-Brandt, G. Mentha, G. Colucci, L. Perrin, and A. Hadengue. 1998. Detection of intrahepatic hepatitis C virus replication by strand-specific semi-quantitative RT-PCR: preliminary application to the liver transplantation model. J. Hepatol. 29:1-11. [DOI] [PubMed] [Google Scholar]
- 29.Neumann, A. U., N. P. Lam, H. Dahari, M. Davidian, T. E. Wiley, B. P. Mika, A. S. Perelson, and T. J. Layden. 2000. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. J. Infect. Dis. 182:28-35. [DOI] [PubMed] [Google Scholar]
- 30.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 31.Novick, D., B. Cohen, and M. Rubinstein. 1994. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell 77:391-400. [DOI] [PubMed] [Google Scholar]
- 32.Pawlotsky, J. M. 2000. Hepatitis C virus resistance to antiviral therapy. Hepatology 32:889-896. [DOI] [PubMed] [Google Scholar]
- 33.Pawlotsky, J. M., G. Germanidis, A. U. Neumann, M. Pellerin, P. O. Frainais, and D. Dhumeaux. 1998. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5A gene quasispecies mutations. J. Virol. 72:2795-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters, M. 1996. Actions of cytokines on the immune response and viral interactions: an overview. Hepatology 23:909-916. [DOI] [PubMed] [Google Scholar]
- 35.Pichard, L., I. Fabre, M. Daujat, J. Domergue, H. Joyeux, and P. Maurel. 1992. Effect of corticosteroids on the expression of cytochromes P450 and on cyclosporin A oxidase activity in primary cultures of human hepatocytes. Mol. Pharmacol. 41:1047-1055. [PubMed] [Google Scholar]
- 36.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, and J. Albrecht. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 37.Proud, C. G. 1995. PKR: a new name and new roles. Trends Biochem. Sci. 20:241-246. [DOI] [PubMed] [Google Scholar]
- 38.Reis, L. F., H. Harada, J. D. Wolchok, T. Taniguchi, and J. Vilcek. 1992. Critical role of a common transcription factor, IRF-1, in the regulation of IFN-beta and IFN-inducible genes. EMBO J. 11:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu, Y. K., A. Iwamoto, M. Hijikata, R. H. Purcell, and H. Yoshikura. 1992. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc. Natl. Acad. Sci. USA 89:5477-5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu, Y. K., and H. Yoshikura. 1994. Multicycle infection of hepatitis C virus in cell culture and inhibition by alpha and beta interferons. J. Virol. 68:8406-8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, N., T. Kawakami, and T. Taniguchi. 1993. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol. Cell. Biol. 13:4531-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tilg, H. 1997. New insights into the mechanisms of interferon alfa: an immunoregulatory and anti-inflammatory cytokine. Gastroenterology 112:1017-1021. [DOI] [PubMed] [Google Scholar]
- 43.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 44.Uze, G., G. Lutfalla, and I. Gresser. 1990. Genetic transfer of a functional human interferon alpha receptor into mouse cells: cloning and expression of its cDNA. Cell 60:225-234. [DOI] [PubMed] [Google Scholar]
- 45.Weiner, A. J., M. J. Brauer, J. Rosenblatt, K. H. Richman, J. Tung, K. Crawford, F. Bonino, G. Saracco, Q. L. Choo, and M. Houghton. 1991. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology 180:842-848. [DOI] [PubMed] [Google Scholar]
- 46.Young, K. K., R. M. Resnick, and T. W. Myers. 1993. Detection of hepatitis C virus RNA by a combined reverse transcription-polymerase chain reaction assay. J. Clin. Microbiol. 31:882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeuzem, S. 2000. Hepatitis C virus: kinetics and quasispecies evolution during anti-viral therapy. Forum 10:32-42. [PubMed] [Google Scholar]
- 48.Zeuzem, S., J. M. Schmidt, J. H. Lee, B. Ruster, and W. K. Roth. 1996. Effect of interferon alfa on the dynamics of hepatitis C virus turnover in vivo. Hepatology 23:366-371. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753-765. [DOI] [PubMed] [Google Scholar]