Abstract
The transcription of neuron-specific genes must be repressed in nonneuronal cells. REST/NRSF is a transcription factor that restricts the expression of many neuronal genes through interaction with the neuron-restrictive silencer element at the promoter level. PAHX-AP1 is a neuronal gene that is developmentally up-regulated in the adult mouse brain but that has no functional NRSE motif in its 5′ upstream sequence. Here, we report that the transcription factor AP4 and the corepressor geminin form a functional complex in which SMRT and histone deacetylase 3 are recruited. The functional complex represses PAHX-AP1 expression in nonneuronal cells and participates in regulating the developmental expression of PAHX-AP1 in the brain. This complex also serves as a transcriptional repressor of DYRK1A, a candidate gene for Down’s syndrome. Furthermore, compared with that in normal fetal brain, the expression of AP4 and geminin is reduced in Down’s syndrome fetal brain at 20 weeks of gestation age, at which time premature overexpression of dual-specificity tyrosine-phosphorylated and regulated kinase 1A (DYRK1A) is observed. Our findings indicate that AP4 and geminin act as a previously undescribed repressor complex distinct from REST/NRSF to negatively regulate the expression of target genes in nonneuronal cells and suggest that the AP4–geminin complex may contribute to suppressing the precocious expression of target genes in fetal brain.
The expression of neuronal genes in neural tissues is regulated by activator and repressor systems that provide the proper transcriptional pattern (1, 2). One of these systems, interaction of the neuron-restrictive silencer element (NRSE) with repressor element 1-silencing transcription factor (REST, also known as neuron-restrictive silencer factor or NRSF), mediates the repression of several neuronal genes in nonneuronal cells, such as type II sodium channel, SCG10, and synapsin I. REST acts on promoters that carry the NRSE sequence to repress transcription, which is thought to be a general mechanism for the control of neuron-specific gene expression (3–5). However, the transcriptional regulatory mechanisms required to direct the temporal expression of brain-specific genes are not fully understood.
Phytanoyl-CoA α-hydroxylase-associated protein 1 (PAHX-AP1) was isolated as a novel neuron-specific protein that interacts with Refsum disease gene product (PAHX) (6) and the cytoplasmic region of brain-specific angiogenesis inhibitor 1, a seven-span transmembrane protein (7). Refsum disease is an autosomal recessive disorder of lipid metabolism; retinitis pigmentosa and peripheral neuropathy are major clinical findings (8). PAHX-AP1 is involved in the developmental regulation of the photoreceptor’s function (9) and is weakly expressed in most embryonic tissues, but its expression pattern changes dramatically after birth when it is specifically expressed in the brain in a developmentally up-regulated pattern (6). Studies in transgenic (TG) mice showed that the 5-kb region of 5′ PAHX-AP1 gene is sufficient to direct the developmental expression of a reporter gene in the brain only, especially neuronal cells, in a pattern similar to that of endogenous PAHX-AP1 (10), which indicates that the 5-kb region contains the sequences required to direct temporal brain-specific expression in vivo. However, no functional NRSE motif was found within 5 kb of the PAHX-AP1 promoter (Fig. 7, which is published as supporting information on the PNAS web site), which indicates that elements distinct from the NRSE must be present for neuron-specific transcriptional up-regulation of PAHX-AP1 in the adult brain.
Identification of NRSE-like sequences that repress transcription and binding repressor proteins is important because these sequences may represent common regulatory elements involved in the transcriptional control of other neuronal genes. In this regard, the PAHX-AP1 gene can provide an attractive model for revealing other NRSE-like sequences. Previously, we identified a putative repressive element (PRE) in the PAHX-AP1 promoter sequence that is suspected to be required for directing brain-specific expression (10), and we hypothesized that PRE-binding proteins may participate in the negative regulation of PAHX-AP1 expression in brain at the promoter level.
In the present study, we screened for PRE-binding proteins to elucidate the responsible molecular mechanisms and potential implications of this PRE-binding transcription factor in the regulatory control of brain-specific gene expression. Here, we report that activator protein 4 (AP4) is a PRE-binding transcription factor and that AP4 and the corepressor geminin (Gem) form a previously uncharacterized functional complex to repress target gene expression in nonneuronal cells. The AP4–Gem complex may provide a second mechanism for silencing neuronal genes in nonneuronal cells that operates in parallel to the previously reported REST/neuron-restrictive silencer factor (NRSF).
Results
AP4 Is a Transcriptional Repressor of PAHX-AP1, and Gem Interacts with AP4 to Form a Complex.
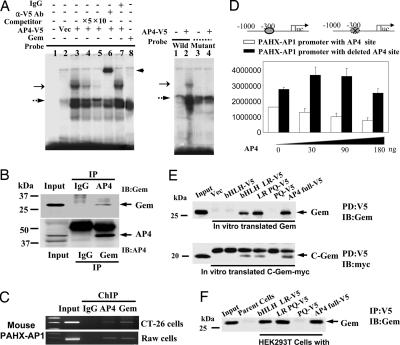
To search for PRE-binding proteins, we used the yeast one-hybrid assay and obtained several positive clones, two of which simultaneously expressed the transcription factor AP4 and Gem; the others contained AP4 only. AP4 is a ubiquitously expressed transcription factor that binds to the DNA consensus sequence 5′-CAGCTG-3′ and a helix–loop–helix protein that contains two distinctive leucine-repeat elements, LR1 and LR2 (11). Gem is a bifunctional protein that acts in embryonic neuralization (12) and in replication licensing in the cell cycle (13). We found that in vitro translated AP4-V5 (Fig. 1A Left) or native AP4 in nuclear extract (Fig. 8, which is published as supporting information on the PNAS web site) binds specifically to WT PAHX-AP1 PRE but not to mutant PRE oligonucleotide (Fig. 1A Right). Unexpectedly, however, Gem did not bind to the PRE (Fig. 1A and 8) but interacted with AP4 (Fig. 1B). Binding of endogenous AP4 to the AP4-binding site on PAHX-AP1 promoter sequence was confirmed by ChIP (Fig. 1C). The positive result of the ChIP assay with anti-Gem antibody indicated that AP4 and Gem form a complex. In HEK293T cells, cotransfection of AP4 dose-dependently repressed the luciferase activity of the PAHX-AP1 promoter reporter, whereas site-directed mutation of the AP4 binding site resulted in a loss of repression by AP4 (Fig. 1D).
Fig. 1.
AP4 is a transcriptional repressor of the PAHX-AP1 promoter, and the C-terminal domain of Gem binds to the leucine-repeat elements (LR1 and LR2) on AP4. (A) AP4 binds to the PAHX-AP1 PRE, but Gem does not. (Left) Labeled PAHX-AP1 PRE oligonucleotide was incubated with in vitro-translated vector (lane 2), in vitro-translated AP4-V5 (lanes 3–7), or Gem (lane 8). Lane 1 is probe only. AP4-V5 was incubated with probe alone (lane 3), excess cold probe (lanes 4 and 5), anti-V5 antibody (lane 6), or IgG (lane 7). The shifted band due to AP4 binding with PRE (thin arrow) and the supershifted band (thick arrow) are indicated. The dashed arrow indicates nonspecific bands. (Right) Labeled WT (lanes 1 and 2) or mutant PAHX-AP1 PRE oligonucleotide (lanes 3 and 4) was incubated with AP4-V5 (lanes 2 and 4). (B) Interaction of AP4 with Gem. HEK293T cells were immunoprecipitated with anti-AP4 antibody and then blotted with anti-Gem antibody (Upper) or reciprocally immunoprecipitated and probed (Lower). Input is HEK293T cells. (C) ChIP assay showing that AP4 and Gem bind to the AP4-binding site on the murine PAHX-AP1 promoter. (D) Increasing doses of AP4 were transiently cotransfected with WT PAHX-AP1 promoter reporter (open bars) or AP4-binding site-mutated PAHX-AP1 promoter reporter (filled bars) in HEK293T cells. After 48 h, cell extracts were prepared and assayed for luciferase activity. Note that the basal value of mutated PAHX-AP1 promoter reporter was larger than that of WT PAHX-AP1 promoter reporter. Values are mean ± SEM (n = 3). (E) Various deletion constructs of AP4 were pulled down by whole Gem (Upper) or the C-terminal domain of Gem (Lower) and analyzed by immunoblotting. Input is in vitro-translated Gem. (F) HEK293T cells were transiently transfected with various AP4 deletion constructs, including bHLH LR-V5, LR PQ-V5, PQ V5, and AP4 full-V5. V5 immunoprecipitation (IP) was performed, and Gem was analyzed by immunoblotting. Input is HEK293T cells.
The transcription factors function cooperatively by forming large complexes with one another at their promoter sequences. Protein–protein interactions between DNA-binding transcription factors and non-DNA-binding factors have been shown to be responsible for the transcriptional regulation of gene expression (14, 15). Although no evidence suggests that Gem binds to promoter DNA directly, it is possible that Gem interacts with transcription factors or other proteins to alter their activity. AP4 contains multiple protein–protein interfaces that function to promote homodimer formation and restrict heterocomplexes (14). Although Gem’s neuralizing activity is localized to an N-terminal domain, a C-terminal domain contains the predicted coiled-coil region that inhibits the initiation of DNA replication (12). We thus prepared various AP4 and Gem deletion constructs to map the interacting domain (Figs. 9 A and B, which is published as supporting information on the PNAS web site) and found that the C-terminal domain of Gem binds to the LR1- and LR2-containing region of AP4 (Fig. 1 E and F).
AP4–Gem Functional Complex Works as a Transcriptional Repressor of PAHX-AP1.
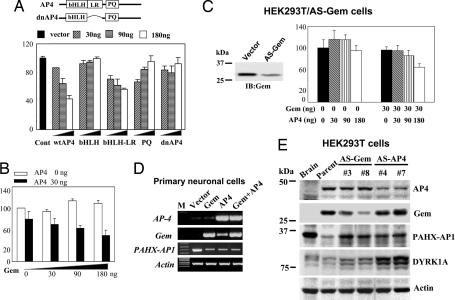
Cotransfection with increasing doses of various AP4 constructs harboring deletions of LR1 and LR2, such as bHLH-V5, PQ-V5, and dominant-negative AP4 (dnAP4), with a PAHX-AP1 promoter reporter did not repress luciferase activity in HEK293T cells compared with cotransfection with WT AP4 or basic helix–loop–helix LR-V5 (Fig. 2A). This result indicates that the LR-containing Gem-binding domain of AP4 is important for the transcriptional repressor activity of AP4 on the PAHX-AP1 promoter. In HEK293T cells, cotransfection with increasing doses of Gem with a PAHX-AP1 promoter reporter did not substantially affect luciferase activity, whereas addition of AP4 (30 ng) dose-dependently repressed luciferase activity (Fig. 2B). However, compared with that in parent cells, repression of the PAHX-AP1 promoter by increasing doses of AP4 alone was not observed in stably anti-sense Gem-transfected HEK293T cells that expressed very low Gem levels (Fig. 2C Left). However, the addition of Gem (30 ng) to the increasing doses of AP4 restored the dose-dependent repression activity of AP4 on the PAHX-AP1 promoter (Fig. 2C Right). These results indicate that AP4 and Gem work as transcriptional repressor and corepressor, respectively, for the exogenous PAHX-AP1 promoter.
Fig. 2.
AP4–Gem complex works as a transcriptional repressor of PAHX-AP1 expression. (A) Schematic of the dominant-negative AP4 (dnAP4). HEK293T cells were cotransfected with increasing doses of wild-type AP4 or various AP4 deletion constructs, including bHLH-V5, bHLH LR-V5, PQ-V5, and dnAP4, and PAHX-AP1 promoter reporter. (A–C) After 48 h, luciferase activity was measured as in Fig. 1D. Values are mean ± SEM (n = 3). (B) Increasing doses of Gem were transiently cotransfected with (filled bars) or without (open bars) 30 ng of AP4 along with WT PAHX-AP1 promoter reporter in HEK293T cells. (C Left) Stably anti-sense Gem-transfected HEK293T cells express less Gem than vector-transfected cells. (C Right) Increasing doses of AP4 were transiently cotransfected without or with Gem (30 ng) along with PAHX-AP1 promoter reporter in anti-sense Gem-transfected cells. (D) Primary mouse neuronal cells were transiently transfected with Gem or AP4 or both 10 days after seeding. After 2 days, total RNA was extracted and PAHX-AP1 expression was examined by RT-PCR. (E) Western blot analyses of PAHX-AP1 and DYRK1A in HEK293T cells stably transfected with anti-sense AP4 or anti-sense Gem construct.
To determine whether AP4 and Gem are necessary for the transcriptional repression of PAHX-AP1 in vivo, we attempted to overexpress them in primary neuronal cells. In cultured mouse neuronal cells, transfection of AP4 alone, Gem alone, or both weakly decreased the expression of PAHX-AP1 (Fig. 2D), which indicates that AP4 and Gem also work as repressors for the endogenous PAHX-AP1 promoter in neuronal cells. Collectively, these data show that AP4 can form a specific, functional complex with its partner Gem to repress the transcription of PAHX-AP1.
AP4 and Gem Work as a Functional Complex in Suppressing Neuronal Gene Expression in Nonneuronal Cells.
We next asked whether suppression of AP4 or Gem derepresses target gene expression in nonneuronal cells. We studied the activation of PAHX-AP1 expression in embryonic HEK293T cells that were stably transfected with an anti-sense AP4 or anti-sense Gem construct. The depressed PAHX-AP1 expression in HEK293T cells was relieved by suppression of AP4 or Gem expression (Fig. 2E), indicating that AP4 and Gem act as a transcription complex distinct from REST in suppressing neuronal gene expression in nonneuronal cells.
AP4–Gem Complex Interacts with Corepressor SMRT.
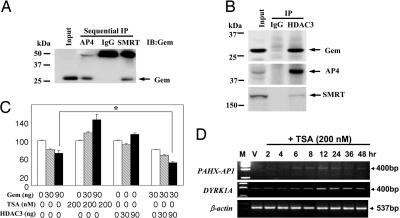
To explore whether AP4 and Gem depend on other corepressors to recruit or deacetylate proteins, we searched for Gem-interacting proteins with the yeast one-to-one assay and found that the corepressor SMRT, but not mSin3 or N-CoR, partners with Gem (Figs. 9C and 10A, which are published as supporting information on the PNAS web site). Through in vitro pull-down assays, we confirmed that SMRT binds to Gem but not to AP4 (Fig. 10 B and C). Thus, these results indicate that AP4–Gem complex recruits SMRT.
AP4–Gem–SMRT Complex Recruits HDAC to Establish and Maintain Trichostatin A (TSA)-Dependent Repression of Its Target Neuronal Gene in Nonneuronal Cells.
To demonstrate that an AP4–Gem–SMRT complex recruits histone deacetylase 3 (HDAC3), which interacts specifically with SMRT (16), we performed sequential immunoprecipitation (IP). First, the AP4–Gem–SMRT complex was immunoprecipitated in HEK293T cells with an anti-AP4 antibody and eluted away from the antibody. The eluant was immunoprecipitated again with SMRT. Finally, the IP product was probed for Gem (Fig. 3A) but not for HDAC3, because the size of HDAC3 is ≈53 kDa, which overlaps with IgG. Instead, HDAC3 was immunoprecipitated in HEK293T cells and then probed subsequently for AP4, Gem, and SMRT (Fig. 3B), indicating that the AP4–Gem–SMRT complex binds to HDAC3. Next, we used a GAL4-containing reporter construct to test whether functional HDAC activity is involved in the AP4-Gem repressor system. A synthetic GAL4-Gem construct was found to repress the target gene activity of GAL4 luciferase in HEK293T cells with increasing doses of Gem (Fig. 3C), and this repression was blocked by pretreatment with 200 nM TSA, an HDAC inhibitor (17). Moreover, cotransfection of Gem with HDAC3 repressed reporter activity more than transfection did with Gem alone (Fig. 3C, P < 0.05). We then assessed the expression of PAHX-AP1 in TSA-pretreated HEK293T cells to investigate whether the target genes of AP4–Gem become activated through blocking of histone deacetylation. Maximal activation of PAHX-AP1 expression was observed 12 h after treatment with 200 nM TSA (Fig. 3D), which indicates that transcriptional repression of AP4–Gem target genes in nonneuronal cells is relieved when HDAC activity is inhibited by TSA, similar to NRSE-containing genes (2). Thus, our results showed that AP4 and Gem recruit SMRT and HDAC3 and, therefore, that histone deacetylation is essential for transcriptional silencing via the AP4–Gem system. In contrast, REST represses transcription by recruiting mSin3 and HDAC (17, 18). Transcriptional repression via the recruitment of HDAC has been characterized for many transcription factors, most of which recruit HDAC by binding the corepressor mSin3 or NCor/SMRT (19, 20).
Fig. 3.
AP4–Gem–SMRT complex interacts with HDAC3 to establish and maintain TSA-dependent repression of its target gene in nonneuronal cells. (A) AP4–Gem–SMRT complex was immunoprecipitated with anti-AP4 antibody in HEK293T cells, eluted away from the antibody, and then immunoprecipitated again with an anti-SMRT antibody. The final precipitate was probed for Gem. (B) HDAC3 IP was performed in HEK293T cells, and Gem, AP4, and SMRT subsequently were analyzed by immunoblotting. (C) Increasing doses of Gem were transiently cotransfected with synthetic GAL4-Gem construct in vehicle- or TSA (200 nM)-pretreated HEK293T cells, and increasing doses of HDAC3 were transiently cotransfected with or without 30 ng of Gem along with synthetic GAL4-Gem construct in HEK293T cells. Values are mean ± SEM (n = 3). ∗, P < 0.05. (D) Time course for derepression of the PAHX-AP1 and DYRK1A genes in HEK293T cells after exponentially proliferating cells were incubated in the presence of TSA.
AP4 and Gem Participate in the Developmental Expression of PAHX-AP1 in the Brain.
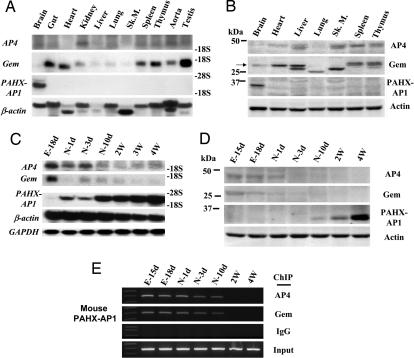
Because REST does not work as a transcriptional repressor for PAHX-AP1, we investigated whether the brain-specific temporal expression of PAHX-AP1 also is regulated by AP4 and Gem. We examined in vivo expression patterns of AP4, Gem, and PAHX-AP1 during brain development. Northern (Fig. 4A) and Western blot (Fig. 4B) analyses showed that AP4 and Gem are widely expressed in most adult tissues but very weakly in an adult brain. Also, AP4 and Gem exhibited similar patterns of down-regulation during a temporal window from embryonic to adult brain, in contrast with PAHX-AP1, which was developmentally up-regulated during this window (Fig. 4 C and D). To examine whether the AP4–Gem complex has an in vivo regulatory role in the developmental expression of target genes in brain, ChIP was performed with an anti-AP4 or anti-Gem antibody at the PAHX-AP1 promoter in brain tissues at several developmental time points. We found a gradual reduction in AP4 and Gem occupancies at the PAHX-AP1 promoter from the embryonic to adult brain (Fig. 4E). This finding suggests that differences in availability of the AP4–Gem complex during the developmental period may determine the temporal expression of its target protein in the brain.
Fig. 4.
AP4 and Gem work as negative regulators of the developmental expression of PAHX-AP1 in brain. (A and B) Tissue distribution of AP4, Gem, and PAHX-AP1 by Northern (A) and Western (B) blot analyses. (C and D) Temporal expressions of AP4, Gem, and PAHX-AP1 in the mouse brain by Northern (C) and Western (D) blot analyses. RNAs or proteins prepared from seven different stages of brain development (embryonic, neonatal, and adult) were used. For each RNA or protein blot, fidelity was confirmed by reblotting with a GAPDH or actin probe. (E) ChIP was performed with anti-AP4 and anti-Gem antibodies at PAHX-AP1 promoter in brain tissue during several developmental time points. Supernatant fluid from sonicated tissue of the corresponding time point was divided into four aliquots. One aliquot was the same protein amounts among each time point and was used for ChIP assay (IgG, anti-AP4, or anti-Gem antibody) or preparation of input DNA.
AP4 and Gem also Work as Transcriptional Repressors of Dual-Specificity Tyrosine-Phosphorylated and Regulated Kinase 1A (DYRK1A), a Candidate Gene for Down’s Syndrome (DS).
We hypothesized that abnormal expression of AP4 or Gem in fetal brain causes premature overexpression of target genes and results in neurologic disease. DYRK1A is a candidate gene for DS and is overexpressed in a DS fetal brain (21). The DYRK1A gene promoter has three potential AP4-binding sites. Moreover, the C-terminal domain of DYRK1A interacts with PAHX-AP1, and both genes have similar expression and distribution patterns in brain. Also, PAHX-AP1 was reported to induce a relocalization of DYRK1A from the nucleus to the cytoplasm, where PAHX-AP1 was localized and, thereby, may contribute to new cellular functions of DYRK1A, suggesting possible involvement of the interaction of PAHX-AP1 with DYRK1A in the neurological pathogenesis of DS (22). Therefore, we chose DYRK1A as a candidate target gene for testing our AP4–Gem neurologic hypothesis that altered expression of AP4 and Gem in the fetal brain may result in neuropathologic disease.
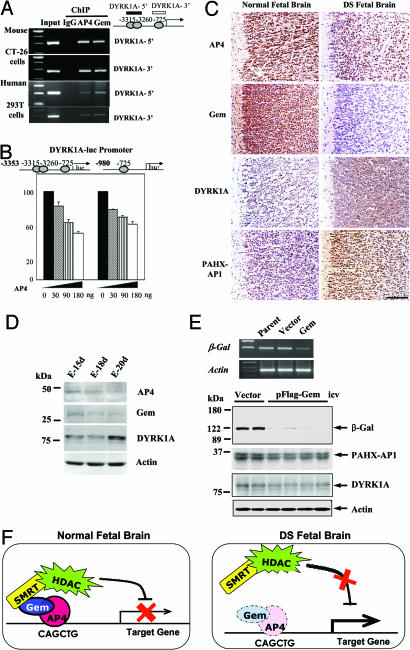
Binding of AP4–Gem to the three cis elements on the DYRK1A promoter sequence was confirmed by ChIP in mouse and human cells (Fig. 5A). Cotransfection of increasing doses of AP4 with DYRK1A promoter luciferase decreased reporter activity dose-dependently in HEK293T cells (Fig. 5B, left side). However, deletion of two 5′ AP4 binding sites (−3318 ≈ −3313 and −3263 ≈ −3258) did not result in a loss of transcriptional repression in response to AP4 (Fig. 5B, right side), which indicates that the remaining 3′ site (−728 ≈ −723) is also working as a cis repressor element. In addition, suppression of AP4 or Gem through an anti-sense strategy resulted in elevated DYRK1A expression in HEK293T cells (Fig. 2E), and DYRK1A expression is activated in HEK293T cells after TSA treatment (Fig. 3D), just as for PAHX-AP1. These data indicate that the AP4–Gem complex also works as a transcriptional repressor of DYRK1A.
Fig. 5.
Expression of AP4 and Gem is decreased in cerebral cortex of the DS fetal brain but DYRK1A is overexpressed. (A) ChIP assays showing that AP4–Gem binds to the AP4-binding sites on murine and human DYRK1A promoter sequences. DYRK1A-5′ and DYRK1A-3′ contain two 5′ and one 3′ AP4-binding site, respectively. (B) Increasing doses of AP4 were transiently cotransfected with three (−3315, −3260, −725; left side) or one (−725; right side) AP4 binding site-containing DYRK1A promoter luciferase in HEK293T cells. Values are mean ± SEM (n = 3). (C) Immunohistochemical analyses of AP4, Gem, DYRK1A, and PAHX-AP1 expression in cerebral cortex of age-matched (20 weeks of gestation, n = 3) normal and DS fetal brain. The data show one of three representative experiments. (Scale bar: 30 μm.) (D) Western blot analyses of expressions of AP4, Gem, and DYRK1A in embryonic mouse brain. (E) Primary neuronal cells from PAHX-AP1 promoter-LacZ TG brain were transiently transfected with Gem or vector, and reporter expression was examined by RT-PCR (Upper). PAHX-AP1 promoter-LacZ TG mice were given i.c.v. Gem or empty vector mixed with FuGENE 6. Five days after injection, proteins from whole brain were prepared and analyzed by immunoblotting (Lower). (F) Schematic of the transcriptional repression of the target gene by AP4–Gem–SMRT complex in normal or DS fetal brain.
Decreased AP4 and Gem Expression and DYRK1A Overexpression in the Cerebral Cortex of a DS Fetal Brain.
Next, we examined the in vivo expression patterns of AP4, Gem, PAHX-AP1, and DYRK1A in healthy and DS fetal cortex at 20 weeks of gestational age by immunohistochemistry. We found that the expression of AP4 and Gem is markedly lower in the DS fetal cortex, especially Gem, than in an age-matched healthy cortex, whereas the expression of DYRK1A and PAHX-AP1 is higher (Fig. 5C). Also, DYRK1A expression was developmentally up-regulated in mouse embryonic brain from embryonic days 15 (E-15d) to E-20d, in contrast with AP4 and Gem, which were developmentally down-regulated (Fig. 5D). These in vivo expression patterns indicate that down-regulation of AP4 and Gem in the DS fetal brain correlates with the premature overexpression of DYRK1A and suggest the reciprocal regulation of DYRK1A expression in the DS fetal brain by the AP4–Gem complex.
Restoring Gem Suppresses Reporter Expression in PAHX-AP1 Promoter-LacZ TG Brain.
Considering that primary neuronal cells have poor transfection efficiency and are mixed with glial cells, which do not express PAHX-AP1 (6), the repressive effects of AP4–Gem transfer on endogenous PAHX-AP1 expression in neuronal cells are relatively small (Fig. 2D). Thus, we tried gene delivery in TG mice that express the lacZ reporter gene overdriven by exogenous PAHX-AP1 promoter (10). In primary cultured neuronal cells isolated from TG brain, which barely expresses endogenous Gem, transfection of Gem modestly decreased the expression of β-gal mRNA (Fig. 5E). However, i.c.v. administration of the Gem construct markedly repressed reporter expression but only weakly suppressed endogenous PAHX-AP1 and DYRK1A in brain tissue (Fig. 5E). This finding indicates that centrally administered Gem efficiently suppressed the elevated transgene expression in the brains of adult TG mice, which expressed little Gem just as in the DS fetal brain (Figs. 4D and 5C). Considering that most neuronal cells in the adult brain are postmitotic cells that have poor transfection efficiency, our results suggest that delivery of Gem cDNA into the lateral ventricle may work in that compartment of neuronal cells, which expressed reporter transgene driven by exogenous PAHX-AP1 promoter, rather than in other neuronal cells driven by endogenous PAHX-AP1 promoter.
Discussion
AP4 is a helix–loop–helix protein and dimerization through the helix–loop–helix domain is a prerequisite for specific DNA binding via a short region of basic residues (11). AP4 interacts with specific sites in both promoter and enhancer regions of viral and cellular enhancers (23), and mutation of these sites inactivates enhancer activity and abolishes binding of AP4 in vitro (24). In this study, we found that two leucine-repeat elements are important for binding with the C-terminal domain of Gem and for repressor activity of AP4, which indicates that interaction with Gem did not affect the specific DNA-binding activity of AP4. Gem acts as a replication licensing protein in the cell cycle (25), where it inhibits cycle progression by sequestering Cdt1 (13), a key component for the assembly of prereplication complex (26). Gem–Cdt1 interaction blocks binding of the minichromosome maintenance complex to origins of replication (13). Recently, Gem was reported to bind to the transcription factors Hox (27) and Six3 (28), and this complex controls cell proliferation by directly inhibiting the interaction of Gem with Cdt1. In the present study, the interaction of Gem with AP4 did not affect cell proliferation (data not shown); rather, AP4 acts cooperatively with Gem to restrict target neuronal gene expression in nonneuronal cells by recruiting SMRT and HDAC at the transcriptional level.
The mechanisms by which the developmental expression of neuronal genes is regulated in the brain are not fully understood. Zinc finger proteins are essential components of developmental gene regulation and transcriptional activation. Modulation of zinc finger proteins, such as Sp1 and zif268, has been shown to alter developmental genes expression in the brain (29, 30). REST, also a zinc finger protein, is widely expressed in nonneuronal tissues and in neural precursors of the brain during fetal life (3, 4), and REST mRNA levels decrease during rat brain development (31). This report suggests that REST acts to prevent premature expression of terminally differentiated genes in neural precursors and to repress neuronal genes in nonneural tissues. Chen et al. (5) provided in vivo genetic evidence that REST is required to control the proper spatial and temporal expression of neuronal genes. In this study, we found that AP4 and Gem are widely expressed in most adult tissues, but very weakly in adult brain, and that the temporal expression of PAHX-AP1 in brain is increased as development progresses, whereas that of AP4 and Gem gradually decreases. This result suggests an inverse relation between the developmental expression patterns of AP4–Gem and PAHX-AP1 in brain. Considering that the AP4–Gem functional complex acts as a transcriptional repressor of PAHX–AP1, which is not under regulation by REST (Fig. 7), our results of a gradual reduction in occupancy of this complex at the PAHX-AP1 promoter during the temporal window suggest that AP4–Gem may work to regulate the developmental expression of PAHX-AP1 in the brain.
REST-dependent neuronal βIII tubulin is derepressed in nonneuronal cells of REST−/− mice, whereas SCG10 and synapsin I are not (5). This report indicates that complex mechanisms control their neuron-specific expression and may reflect differences in their complement of cis-regulatory elements or in lacking positive trans-acting factors needed in addition to relief from REST-imposed silencing. The former explanation suggests the presence of additional cis-regulatory repressor elements that may determine derepression when silencing by REST is relieved. Thus, we hypothesize that the AP4 site may be an additional cis-repressor element that may determine the derepression of neuronal genes in REST−/− mice.
Identification of function of candidate genes and characterization of their regulation are essential steps toward understanding the complex neurodevelopmental abnormalities in DS. DYRK1A was reported to be involved in the development of neurological deficits in DS (21), but regulatory mechanisms in the fetal brain are not understood. Impairment of REST and REST-dependent genes, such as SCG10, in DS neuronal progenitor cells has been shown at the RNA level (32), but REST-dependent proteins, such as SCG10, synapsin I, and BDNF, were comparably expressed in the cortex of DS and control fetal brains (33). Thus, the REST hypothesis of DS, a positive link between dysregulation of REST and some of the neurologic deficits seen in DS, has not been confirmed at the 18th to 19th week of gestation (33). In this study, we found that AP4–Gem represses the transcription of DYRK1A. Moreover, expression of AP4 and Gem, especially Gem, is markedly depressed at 20 weeks of gestation in the DS fetal cortex, the major target tissue in neurologic abnormalities in DS, which suggests that the specific regulatory pathways of AP4–Gem in the DS fetal brain are disrupted. Gem, one of the potential regulators of transcriptional events during early neural development (12), is down-regulated before neuronal differentiation and acts as a differentiation timer controlling the transition from undifferentiated neural progenitors to differentiated neurons by suppressing Brg1 and proneural basic helix–loop–helix factors. In addition, reduced Gem expression was reported to result in precocious neurogenesis in both Xenopus embryos and P19 cells (34). Furthermore, we observed that delivery of Gem into adult TG brain tissue markedly decreased the reporter transgene expression, suggesting again that certain actions of Gem are interrupted in the DS fetal brain because of marked depression. Overall, our present results indicate that down-regulation of AP4 and Gem in the DS fetal brain correlates with the premature overexpression of DYRK1A and suggest that the AP4–Gem complex contributes to suppressing the inappropriate temporal expression of target genes in the fetal brain (Fig. 5F).
Materials and Methods
The investigation conformed to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH publication no. 85-23, revised 1996). The Ethics Committee of Chonnam National University Medical School and Chonnam National University Hospital approved all experimental protocols.
Plasmids and Oligonucleotides.
PAHX-AP1 reporter constructs (-833-luciferase) were derived from a 5-kb fragment of the mouse PAHX-AP1 promoter (10). CAGCTG was substituted for GAATTC in the E box of the PAHX-AP1 promoter by PCR-based site-directed mutagenesis. Various AP4 or Gem cDNAs were obtained from RT-PCR of mouse testis RNA and were cloned into pcDNA3-V5-His mammalian expression vector (Invitrogen, Carlsbad, CA). A mammalian expression vector for Gal4 fusion to Gem was constructed by inserting the appropriate Gem fragment into pCMXGal4/N. The Gal4-tk-luc reporter plasmid contains five tandem repeats of GAL4-binding sites, followed by herpes simplex virus-thymidine kinase minimal promoter. A series of corepressors, SMRT (a gift from Ronald M. Evans, The Salk Institute, San Diego, CA), mSin3A, and N-CoR, were constructed by inserting appropriate fragments into pcDNA3.1-V5-His plasmid (Invitrogen). Human and murine DYRK1A promoters (5-kb) were obtained by PCR of genomic DNA.
Immunohistochemistry.
Three human fetal brain tissue specimens from DS fetuses and three age-matched fetal brain tissue specimens from spontaneously aborted fetuses (20 weeks of gestation) were obstetrically obtained through therapeutic abortion at Chonnam University Hospital (Kwangju, Korea) and used for immunohistochemistry. The brain was excised and immersed in a 4% paraformaldehyde fixative overnight at 4°C. The tissue blocks were washed, dehydrated, embedded in paraffin, cut into 6-μm sections, and mounted. Immunohistochemistry was performed by using an immunoperoxidase procedure as described in ref. 10. The sections were incubated for 12 h with antibodies to AP4 (Chemicon, Temecula, CA), Gem (Santa Cruz Biotechnology, Santa Cruz, CA), PAHX-AP1 (6), and DYRK1A (a gift from W. J. Song, Inje University, Daejeon, Korea) diluted in PBS with 0.3% BSA.
Supporting Information.
Additional details can be found in Supporting Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank S. A. Hwang for assistance in neuronal culture. This work was supported by the Korea Science and Engineering Foundation through the Medical Research Center for Gene Regulation at Chonnam National University Grant R13-2002-013-02000-0 and partly by Korea Research Foundation Grant KRF-2001-015-DP0421.
Abbreviations:
- AP4
activator protein 4
- DS
Down’s syndrome
- DYRK1A
dual-specificity tyrosine-phosphorylated and regulated kinase 1A
- Gem
geminin
- HDAC3
histone deacetylase 3
- NRSE
neuron-restrictive silencer element
- PAHX-AP1
phytanoyl-CoA α-hydroxylase-associated protein 1
- PRE
putative repressive element
- REST
repressor element 1-silencing transcription factor
- TG
transgenic
- TSA
trichostatin A.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Goodman R. H., Mandel G. Curr. Opin. Neurobiol. 1998;8:413–417. doi: 10.1016/s0959-4388(98)80069-x. [DOI] [PubMed] [Google Scholar]
- 2.Roopra A., Huang Y., Dingledine R. Mol. Interv. 2001;1:219–228. [PubMed] [Google Scholar]
- 3.Chong J. A., Tapia-Ramirez J., Kim S., Toledo-Aral J. J., Zheng Y., Boutros M. C., Altshuller Y. M., Frohman M. A., Kraner S. D., Mandel G. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 4.Schoenherr C. J., Anderson D. J. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z. F., Paquette A. J., Anderson D. J. Nat. Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 6.Lee Z. H., Kim H, Ahn K. Y., Seo K. H., Kim J. K., Bae C. S., Kim K. K. Mol. Brain Res. 2000;75:237–247. doi: 10.1016/s0169-328x(99)00304-6. [DOI] [PubMed] [Google Scholar]
- 7.Koh J. T., Lee Z. H., Ahn K. Y., Kim J. K., Bae C. S., Kim H. H., Kee H. J., Kim K. K. Mol. Brain Res. 2001;87:223–237. doi: 10.1016/s0169-328x(01)00004-3. [DOI] [PubMed] [Google Scholar]
- 8.Mihalik S. J., Morrell J. C., Kim D., Sacksteder K. A., Watkins P. A., Gould S. J. Nat. Genet. 1997;17:185–189. doi: 10.1038/ng1097-185. [DOI] [PubMed] [Google Scholar]
- 9.Ahn K. Y., Nam K. I., Kim B. Y., Cho C. W., Jeong S. K., Yang K. J., Kim K. K. Int. J. Dev. Neurosci. 2002;20:93–102. doi: 10.1016/s0736-5748(02)00017-5. [DOI] [PubMed] [Google Scholar]
- 10.Kim M. Y., Ahn K. Y., Lee S. M., Koh J. T., Chun B. J., Bae C. S., Lee K. S., Kim K. K. FEBS Lett. 2004;566:87–94. doi: 10.1016/j.febslet.2004.03.106. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y. F., Luscher B, Admon A., Mermod N., Tjian R. Genes Dev. 1990;4:1741–1752. doi: 10.1101/gad.4.10.1741. [DOI] [PubMed] [Google Scholar]
- 12.Kroll K. L., Salic A. N., Evans L. M., Kirschner M. W. Development (Cambridge, U.K.) 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- 13.Wohlschlegel J. A., Dwyer B. T., Dhar S. K., Cvetic C., Walter J. C., Dutta A. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 14.Vandromme M., Gauthier-Rouviere C., Lamb N., Fernandez A. Trends Biochem. Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 15.Grave B. J. Science. 1998;279:1000–1002. doi: 10.1126/science.279.5353.1000. [DOI] [PubMed] [Google Scholar]
- 16.Guenther M. G., Barak O., Lazar M. A. Mol. Cell. Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naruse Y., Aoki T., Kojima T., Mori N. Proc. Natl. Acad. Sci. USA. 1999;96:13691–13696. doi: 10.1073/pnas.96.24.13691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roopra A., Sharling L., Wood I. C., Briggs T., Bachfischer U., Paquette A. J., Buckley N. J. Mol. Cell. Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassig C. A., Fleischer T. C., Billin A. N., Schreiber S. L., Ayer D. E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 20.Laherty C. D., Yang W. M., Sun J. M., Davie J. R., Seto E., Eisenman R. N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 21.Guimera J., Casas C., Estivill X., Pritchard M. Genomics. 1999;57:407–418. doi: 10.1006/geno.1999.5775. [DOI] [PubMed] [Google Scholar]
- 22.Bescond M., Rahmani Z. Int. J. Biochem. Cell Biol. 2005;37:775–783. doi: 10.1016/j.biocel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Mermod N., Williams T. J., Tjian R. Nature. 1988;332:557–561. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- 24.Fodor E., Weinrich S. L., Meister A., Mermod N., Rutter W. J. Biochemistry. 1991;30:8102–8108. doi: 10.1021/bi00247a002. [DOI] [PubMed] [Google Scholar]
- 25.McGarry T. J., Kirschner M. W. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 26.Maiorano D., Moreau J., Mechali M. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 27.Luo L., Yang X., Takihara Y., Knoetgen H., Kessel M. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 28.Del Bene F., Tessmar-Raible K., Wittbrodt J. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- 29.Thiel G., Schoch S., Peterson D. J. Biol. Chem. 1994;269:15294–15301. [PubMed] [Google Scholar]
- 30.Zawia N. H., Sharan R., Brydie M., Oyama T., Crumpton T. Brain Res. Dev. 1998;107:291–298. doi: 10.1016/s0165-3806(98)00023-6. [DOI] [PubMed] [Google Scholar]
- 31.Palm K., Belluardo N., Metsis M., Timmusk T. J. Neurosci. 1998;18:1280–1296. doi: 10.1523/JNEUROSCI.18-04-01280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bahn S., Mimmack M., Ryan M., Caldwell M. A., Jauniaux E., Starkey M., Svendsen C. N., Emson P. Lancet. 2002;359:310–315. doi: 10.1016/S0140-6736(02)07497-4. [DOI] [PubMed] [Google Scholar]
- 33.Sohn S. Y., Weitzdoerfer R., Mori N., Lubec G. J. Neural. Transm. Suppl. 2003;67:59–66. doi: 10.1007/978-3-7091-6721-2_5. [DOI] [PubMed] [Google Scholar]
- 34.Seo S., Herr A., Lim J. W., Richardson H., Kroll K. L. Genes Dev. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.