Abstract
It is well established that steroid receptor function requires interaction with coactivators. However, the mechanisms through which steroid receptors elicit precise assembly of coactivator complexes and the way the steroid activation signal is transduced remain elusive. Using a T47D cell line stably integrated with a mouse mammary tumor virus-chloramphenicol acetyltransferase (MMTV-CAT) reporter, we demonstrate that specific steroid receptors exhibit preferential recruitment of SRC-1 family coactivators, which determines the subsequent recruitment of specific downstream coregulator molecules. Upon ligand treatment, progesterone receptor (PR) interacted preferentially with SRC-1, which recruited CBP and significantly enhanced acetylation at K5 of histone H4. In contrast, activated glucocorticoid receptor (GR) preferentially associated with SRC-2 (TIF-2/GRIP-1), which subsequently recruited pCAF and led to specific modification of histone H3, suggesting that specific coactivators recruit distinct histone acetyltransferases to modulate the transcription of steroid-responsive genes. Loss-of-function experiments further support the predicted roles of SRC-1 and SRC-2 in, respectively, PR- and GR-mediated transcription on the MMTV promoter. This study indicates that differential recruitment of coactivators by nuclear receptors determines the assembly of coactivator complexes on target promoters to mediate specific transcription signals.
Steroid receptors regulate transcription by recruiting coactivator complexes to target gene promoters. Coactivators recruited by ligand-bound nuclear receptors (NR) include members of the SRC family of coactivators, such as SRC-1, SRC-2 (TIF2/GRIP1), and SRC-3 (pCIP/RAC3/ACTR/AIB1/TRAM1) (32). These proteins serve as adaptors that potentiate the transcriptional activity of different NRs through conserved motifs termed NR boxes (15). Motifs within the receptor-interacting domain of SRCs have been demonstrated to determine coactivator preferences for specific NRs, while the transcriptional activation domains of SRCs mediate interactions with histone acetyltransferases (HATs) (1, 46, 48). Biochemical studies and protein-protein interaction screens suggest that these proteins function as components of large multiprotein complexes (37), indicating a mechanism for the integration of inputs from multiple signaling pathways.
Although members of the homologous SRC family possess limited functional redundancy, accumulating evidence suggests that SRCs also play distinct roles in biological processes (4, 13, 25, 32, 49). These functional differences could result in part from the preferential recruitment of coactivators induced by ligands (6, 34). SRC-1 and SRC-2 have been reported to exhibit similar, but not identical, binding preferences to nine different NRs. For example, the androgen receptor binds well to SRC-2 but poorly to SRC-1 (11). The molecular basis of NR preferences for coactivators is linked to the structure of the ligand-binding domain of NRs and their concomitant specificity for individual NR boxes (9, 11, 31, 35, 48). For instance, the vitamin D receptor and estrogen receptor beta interact with different alpha-helical NR boxes, as in SRC-3 (27). The differences in affinity for NR box 2 or 3 of SRC-2 by glucocorticoid receptor (GR) and thyroid receptor (TR) ligand-binding domains provide additional evidence for underlying specific receptor-coactivator interactions (9). Furthermore, these contacts are sensitive to conformational changes induced by distinct ligands or DNA binding (26).
Despite all these reports, the precise assembly of coactivators and components of coactivator complexes required for physiological nuclear receptor function are not well understood. Since the functions of components of coactivator complexes appear to be distinct (21), it is likely that transcription factor-specific differences in configuration and content within the coactivator complex dictate acetyltransferase activities that bring about histone modification and further transcriptional regulation. An intriguing question involves the physiologic contexts under which coactivator utilization is determined by different NRs. We have approached this question with the hypothesis that specific NR-coactivator interactions exist in cells and generate distinct regulatory patterns in differing signaling contexts. We explored the hypothesis by studying coregulator assembly on a mouse mammary tumor virus (MMTV) promoter that is both progesterone and glucocorticoid responsive.
The actions of GR and PR have been extensively studied in an effort to understand how ligands for either receptor can elicit distinct biological activities when both steroid receptors act on the same common hormone responsive element (HRE) (2, 10, 23). One of the mechanisms by which GR or PR activity could be achieved is differential interactions with transcriptional coregulators, which further direct downstream events during transcription. A T47D cell line with stably integrated MMTV-chloramphenicol acetyltransferase (CAT) reporter (24) was used to test our hypothesis. Since all these analyses have been performed on a single promoter in a single cell line, cell-specific and promoter-specific effects can be minimized.
In this work, we demonstrate coactivator specificity for PR or GR as well as the effect that initial receptor-coactivator interactions have on the assembly of subsequent cofactors on the MMTV promoter. Upon steroid treatment, both PR and GR were recruited to the MMTV promoter and induced transcription of MMTV-CAT. PR selectively recruited SRC-1 and CBP, resulting in acetylation of K5 on histone H4. GR preferentially associated with SRC-2, which subsequently recruited pCAF, leading to specific histone modification of histone H3. Studies using RNA interference further substantiate an important role of SRC-1 in PR function and of SRC-2 in GR function. Taken together, this study elucidates differential mechanisms by which distinct nuclear receptor-coactivator interactions mediate diverse signaling events on the identical gene under physiological conditions in nontransfected cells.
MATERIALS AND METHODS
Reagents.
Antibodies used in this study anti-PR (H-190), anti-GR (H-300), anti-SRC-1 (M-341), anti-RAC3 (C-20), CBP (A-22), and p300 (N-15) (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.); anti-TIF2 (T73620) (BD Biosciences, San Diego, Calif.); anti-acetyl-histone H4 (K5), anti-phospho-Histone H3 (Ser 10), anti-dimethyl-histone H3 (K9), and anti-acetyl-histone H3 (K14) (Upstate Biotechnology, Lake Placid, N.Y.); anti-polymerase II (MMS-126R; Covance, Richmond, Va.); and anti-TRAP220 (generated in this lab), anti-BRG-1 (Weidong Wang, National Institute on Aging, National Institutes of Health, Baltimore, Md.), and anti-pCAF (41).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) analyses were performed by following a modified procedure based on previously described protocols (3, 5, 41). The T47D/CAT0 cell line was grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% charcoal-dextran stripped serum for 3 days. Cells with 90% confluence (∼1 × 108) were treated with dexamethasone (10−8 M), progesterone (10−8 M), or R1881 (10−9 M; NEN Life Sciences Products, Boston, Mass.) for 1 h. Cells were washed once with phosphate-buffered saline (PBS), pH 7.4, and fixed with formaldehyde (1% final concentration) for 10 min at room temperature; cross-linking was terminated upon addition of glycine (0.125 M final concentration). Cells were then rinsed twice with cold PBS, collected, and swollen on ice in 20 volumes of nuclei preparation buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 8.0)-85 mM KCl-0.5% Nonidet P-40, supplemented with 1× protease inhibitor cocktail (Roche, Mannheim, Germany)] for 30 min. Nuclei were collected by microcentrifugation and resuspended in 300 μl of ChIP lysis buffer (50 mM Tris-HCl [pH 8.1], 1% sodium dodecyl sulfate [SDS], 10 mM EDTA, 1× protease inhibitor cocktail), followed by incubation on ice for 20 min. Samples were then sonicated on ice four to six times for 10 s each (duty output, 30%; duty cycle, 90%), followed by centrifugation for 10 min. The chromatin solution was diluted 10-fold with dilution buffer (20 mM Tris-HCl [pH 8.1], 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 1× protease inhibitor cocktail). The chromatin preparation was precleared with 80 μl of salmon sperm DNA-protein A/G-agarose (Upstate Biotechnology, Lake Placid, N.Y.) and 20 μl of preimmune serum. Chromatin complexes were then incubated with 5 to 10 μl of specific antibodies and rotated at 4°C overnight. Immune complexes were collected with 40 μl of protein A/G-agarose with agitation for 1 h at 4°C. A supernatant fraction (20 μl) from reaction mixtures lacking primary antibodies was saved as an input control and processed in parallel with the eluted immunoprecipitates beginning at the cross-link reversal step. Immunoprecipitates were sequentially washed for 5 to 10 min in wash buffer I (20 mM Tris-HCl [pH 8.1], 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 150 mM NaCl), wash buffer II (20 mM Tris-HCl [pH 8.1], 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 500 mM NaCl), wash buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and TE buffer (three times). Washed beads were extracted with 50 μl of elution buffer (1% SDS, 100 mM NaHCO3) three times. The elution was combined in one tube, and the protein-DNA cross-linking was reversed by incubation at 65°C for 4 h. Each sample was treated with 15 μg of proteinase K (Gibco BRL, Grand island, N.Y.) in proteinase K buffer (50 mM Tris-HCl [pH 8.5], 1% SDS, 10 mM EDTA) for 2 h at 45°C. The DNA was purified with the QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and eluted in 50 μl of elution buffer. Total input samples were eluted in 100 μl of elution buffer and diluted 1:10 before PCR analysis. Each PCR mixture contained 6 μl of immunoprecipitate or input, 0.5 μM each primer, 0.4 mM deoxynucleoside triphosphate mixture, 1× Titanium Taq PCR buffer (Clontech, Palo Alto, Calif.), and 1× Titanium TaqDNA polymerase (Clontech) in a total volume of 25 μl. The primers for the MMTV promoter were as follows: forward, 5′-TAT GGT TAC AAA CTG TTC TTA AAA CGA GGA TG-3′; reverse, 5′-GCA AGT TTA CTC AAA AAT CAG CAC TCT TT-3′. PCR was performed for 26 to 29 cycles with 1 min of denaturing at 94°C, annealing at 62°C, and extension at 68°C. PCR results were analyzed by agarose gel (1.5%) electrophoresis.
Cell culture and transient transfection of siRNA.
T47D-CAT0 (human breast cancer) cells were routinely maintained in DMEM supplemented with 10% fetal bovine serum and 0.2 mg of Geneticin/ml (G418; Sigma). Prior to transfections, 3 × 105 cells per well of a six-well multiplate were grown overnight until reaching 50 to 60% confluence. Cells were then washed with phenol red-free DMEM three times and supplemented with 1 ml of phenol red-free DMEM containing 5% dextran-coated charcoal-stripped fetal bovine serum. For each well, 15 μl of TransIT-TKO transfection reagent (Mirus, Madison, Wis.) was premixed with 200 μl of Opti-MEMI (Gibco BRL) serum-free medium for 15 min. Synthesized small interfering RNA (siRNA) (Dharmacon Research, Inc., Lafayette, Colo.) duplexes sufficient for 100 nM final concentrations per well were added to the diluted TransIT-TKO reagent and incubated at room temperature for 15 min. The transfection reagent-siRNA mixture was added dropwise to the cells with gentle rocking and incubated for 48 to 72 h. For optimal results, a combination of two SRC-1 siRNAs at 50 nM each was utilized, namely, siRNA 1a (5′-AAC ACG ACG AAA UAG CCA UAC-3′) and siRNA 1b (5′-AAG UGA UGA CUC GUG GCA CUG-3′). The siRNA sequence for SRC-2 is 5′-AAG UCA GAU GUA UCC UCU ACA-3′. The siRNA sequence for SRC-3 is the same as previously described (40). Transfection efficiency was monitored by fluorescence-labeled luciferase siRNA (Dharmacon Research, Inc.), which also served as a negative control.
In vitro mRNA synthesis and microinjection of Xenopus oocytes.
The Flag-tagged full-length SRC-1, SRC-2, and SRC-3 cDNAs cloned in pSP64 [poly(A)] vector (28) were linearized with a unique restriction enzyme cutting 3′ to the poly(A) site. The digested DNA was purified with phenol-chloroform extraction and ethanol precipitation. The in vitro synthesis of mRNA was then carried out with an SP6 Message Machine kit from Ambion (Austin, Tex.), following the manufacturer's instructions. The preparation and microinjection of Xenopus stage VI oocytes were performed essentially as previously described (28). Diluted mRNA solutions containing approximately 100 ng/μl were injected into the cytoplasm of stage VI oocytes (∼25 nl/oocyte). The injected oocytes were incubated for 24 h to allow the synthesis of proteins. An equivalent amount of injected or uninjected oocytes was harvested for subsequent studies.
Immunoprecipitation and Western blotting analysis.
Cells were washed once with PBS and lysed in ice cold immunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, and 1× protease inhibitor cocktail) for 30 min, and the debris was cleared by centrifugation at 13,400 × g for 10 min at 4°C. The in vitro translated proteins or lysate was incubated with 0.5 μg of desired antibodies overnight at 4°C. After incubation with 40 μl of protein A/G-agarose for 1 h, the immune complexes were collected and washed extensively with immunoprecipitation assay buffer. For Western blot analysis, the samples were resolved by SDS-7% polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Bio-Rad, Hercules, Calif.). The primary antibodies were diluted in blocking buffer (50 mM Tris-HCl, 150 mM NaCl [pH 7.5], 0.1% Tween 20, 5% skim milk) and added to the membranes for overnight rotation at 4°C. The membrane was washed four times in 1× PBS (Gibco BRL) and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. All blots were visualized with SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, Ill.).
RNA preparation and RT-PCR.
Total RNA was extracted with TRI reagent (Molecular Research Center, Inc., Cincinnati, Ohio) following the manufacturer's instruction. The reverse transcriptase (RT)-PCR analysis was performed with the Access RT-PCR system (Promega, Madison, Wis.). The primers for the CAT reporter gene were as follows: forward, 5′-GTG AGC TGG TGA TAT GGG ATA GTG TT-3′; reverse, 5′-CAT ATT GGC CAC GTT TAA ATC AAA A-3′. The PCR conditions were the same as described in the previous section except that 24 cycles of reaction were carried out.
CAT assay.
The reporter activity of MMTV-CAT was quantitatively measured by enzyme-linked immunosorbent assay. In accordance with the instructions with the CAT enzyme-linked immunosorbent assay kit (Roche), diluted cell lysates were added to the anti-CAT-coated microtiter plates and incubated at 37°C for 1 h. After extensive washes, incubation of digoxigenin-conjugated anti-CAT was followed by anti-digoxigenin-peroxidase and substrate. The photometric detection was carried out with a microtiter plate reader at 405 nm with a reference wavelength of approximately 490 nm. The results were normalized with respect to total protein concentration. Total protein concentration was measured with Bio-Rad protein assay dye reagent.
Statistical analysis and quantitative analysis.
Statistical analysis of multiple comparisons was performed with SPSS (Statistical Package for the Social Sciences software (SPSS, Inc.). Analysis of variance was carried out by both Tukey and Dunnett methods. The significance level was set at 0.05. The results described in the text are consistent with both Tukey and Dunnett analyses. Data quantitation was performed with NIH Image software version 1.62.
RESULTS
Selection of assay system.
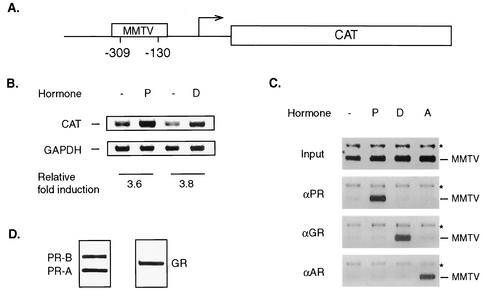
To address whether coactivators could be preferentially recruited by different steroid receptors, we chose the MMTV promoter as a template for the analysis of coactivator assembly. The MMTV promoter has previously been used as an in vivo model for studying the role of chromatin structure in transcription (10, 44). A cell line with a stably integrated MMTV-reporter construct is an appropriate system in which the utilization of cofactors on a single promoter in response to two different steroid hormones could be compared without the complication of cell background differences. To test if the T47D/CAT0 cell line containing the stably integrated MMTV-CAT was suitable for our study, RT-PCR was performed using primers specific for the CAT gene (Fig. 1A) to measure CAT mRNA expression after steroid hormone treatment. At a 10 nM concentration, both progesterone and dexamethasone significantly increased CAT mRNA levels (3.6- and 3.8-fold, respectively) after a 3-h treatment (Fig. 1B). More-than-10-fold increases in CAT enzyme activity were observed when cells were treated with either hormone for 24 h (data not shown and Fig. 5), indicating that either ligand can promote transcription from the MMTV promoter. To ensure that PR or GR was specifically responsible for the elevated transcriptional activity of the MMTV promoter, ChIP was performed with specific antibodies against PR, GR, and AR (androgen receptor) 1 h after steroid treatment. To ensure the quality of the ChIP analysis, we evaluated the sensitivity of the PCR amplification on serial dilutions of total DNA collected after sonication (input DNA) with specific primers. The results indicated that the signal is proportional to the amount of input DNA within 100 ng of DNA up to 31 cycles (date not shown). The DNA precipitated with antibodies against PR, GR, and AR was amplified by PCR using primers that were targeted to the region containing the HRE in the MMTV promoter (Fig. 1A). Antibodies directed against PR or GR sufficiently precipitated the HRE region of the MMTV only when progesterone or dexamethasone, respectively, was present (Fig. 1C). However, PR or GR was not recruited in the presence of dexamethasone or progesterone, respectively. Nor were they recruited after stimulation with R1881, a synthetic AR ligand. Similarly, AR, which bound to the same HRE on the MMTV in the presence of R1881, did not associate with the promoter DNA in the presence of either progesterone or dexamethasone (Fig. 1C). Importantly, PCR analysis performed in the same reaction did not detect any significant changes in PR, GR, or AR occupancy of a region encoding part of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA (Fig. 1C). This result indicated that recruitment of PR or GR to the MMTV-CAT gene was hormone specific. Furthermore, significant amounts of PR or GR were observed in this T47D cell line (Fig. 1D), consistent with the results of gene activation and nuclear receptor recruitment (Fig. 1B and C). Taken together, the T47D/CAT0 cell satisfied our requirements for further studies.
FIG. 1.
Properties of T47D/MMTV-CAT as a system for in vivo studies. (A) Schematic structure of MMTV-CAT integrated into the T47D cells. The locations of PCR primers specific for the MMTV promoter region containing the HRE are indicated. (B) MMTV-CAT gene expression induced by progesterone (lane P) or dexamethasone (lane D) was analyzed by RT-PCR after 3 h of treatment. The levels of GAPDH serve as a reference for quantitation as well as control for loading. The semiquantitative results represent the relative levels of CAT mRNA expression and GAPDH levels relative to the those of uninduced control. Lanes −, controls without hormone. (C) Ligand-dependent recruitment of nuclear receptors to the MMTV promoter examined by ChIP. Soluble chromatin was prepared from T47D cells treated with progesterone (lane P), dexamethasone (lane D), or synthetic androgen (lane A) R1881 for 1 h. Receptor-bound DNA immunoprecipitated with antibody against PR, GR, or AR was amplified by the MMTV primers illustrated in Fig. 1A. The PCR products are labeled −MMTV. Primers specific for the GAPDH coding region were included in the same PCRs to amplify nonspecifically immunoprecipitated DNA as a loading control, indicated by asterisks. The material in the Input panel was amplified by using DNA extracted from diluted chromatin that was not immunoprecipitated. (D) Significant amounts of PR and GR are present in the T47D/CAT0 cells. Endogenous PR and GR were immunoprecipitated from the T47D/CAT0 cells and detected by Western blotting, as described in Materials and Methods.
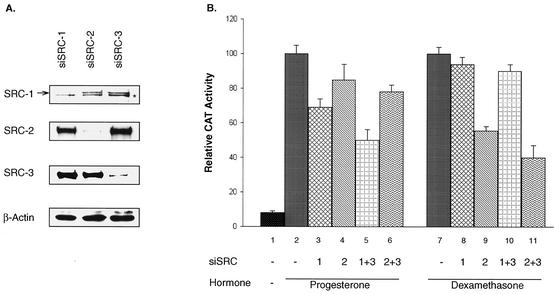
FIG. 5.
Modulation of PR or GR activity by loss of SRC coactivator function. (A) RNA interference was carried out by introducing siRNA against individual SRCs (siSRC-1, -2, and -3) into the T47D/CAT0 cells for 3 days. Total protein was extracted with TRI reagent (Molecular Research Center, Inc). Western blotting analysis was performed to monitor the reduction of specific proteins with the indicated antibodies. The asterisk indicates a nonspecific band, used as a loading control in place of β-actin. (B) CAT activity was measured after RNA interference with the indicated siSRCs and treatment with progesterone or dexamethasone for 24 h. The uninduced or induced controls (lanes 1, 2, and 7) were treated with an unrelated siRNA against luciferase. The siSRCs labeled 1+3 and 2+3 represent the combinations of siRNA against SRC-1 and SRC-3 and SRC-2 and SRC-3, respectively. Triplicate results were normalized against results for hormone-treated controls (lanes 2 and 7), which are arbitrarily set as 100 for comparison. Analysis of variance of each individual group was done. Statistical results are mentioned in the text.
Steroid receptor preference for coactivators.
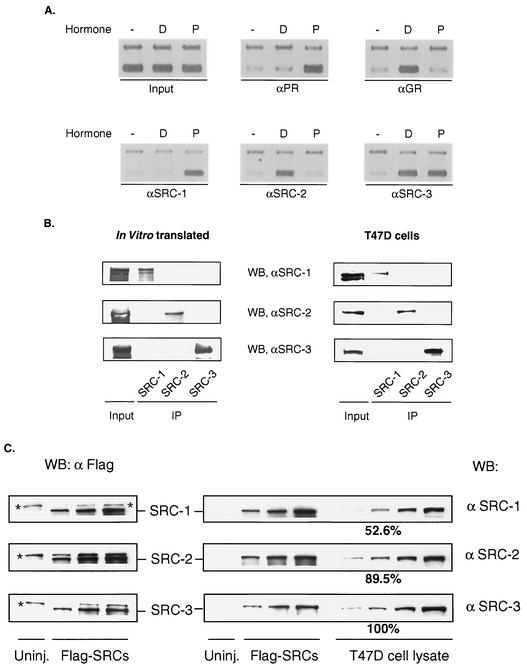
To explore the recruitment of SRC-1 family coactivators to the MMTV promoter in vivo, PR- or GR-mediated assembly of SRC coactivators was compared by using ChIP assays. As expected, selective recruitment of PR or GR was observed only when progesterone or dexamethasone was present (Fig. 1C and 2A). Interestingly, progesterone treatment resulted in a dramatic increase in the occupancy of SRC-1 on the MMTV promoter; dexamethasone failed to do so. In contrast, treatment with dexamethasone induced significant occupancy of the same promoter by SRC-2 (Fig. 2A). Similar studies detected recruitment of SRC-3 on the promoter after either progesterone or dexamethasone stimulation (Fig. 2A). The simultaneous recruitment of PR/SRC-1 and GR/SRC-2 on the MMTV promoter were detected as early as 15 min after hormone stimulation and were persistent during a time course of 160 min (data not shown). We did not observe clear cycling patterns of receptor or cofactors on the MMTV promoter in this particular cell line. These studies indicate that activated PR prefers SRC-1 recruitment while liganded GR preferentially recruits SRC-2 on the MMTV promoter; both also appear to recruit SRC-3.
FIG. 2.
Detection of PR/GR and SRC proteins on the MMTV promoter by ChIP. (A) Ligand- and receptor-mediated recruitment of SRC-1, SRC-2, and SRC-3 was determined by ChIP assays using the same batch of T47D/CAT0 cells treated with progesterone (lanes P) or dexamethasone (lanes D) or untreated cells (lanes −). The lower bands were amplified by using primers for the MMTV promoter, and the slower-migrating bands represent GAPDH control. (B) SRC antibody specificities were tested by using in vitro-translated SRC-1, -2, or -3 or T47D whole-cell lysate by immunoprecipitation-Western blotting with the indicated antibodies. About 20% of each sample was used for inputs. (C) Xenopus oocytes lysate expressed Flag-tagged SRC proteins were prepared in serial twofold dilutions for Western blot analysis using anti-Flag antibody (left panel). Equivalent amounts of each Xenopus lysate dilution were used in Western blotting with anti-SRC-1, -2, or -3. Similar serial dilutions of T47D cell lysate were prepared for Western blotting together with the blots containing Flag-tagged SRC proteins. The bands labeled with asterisks represent nonspecific reactions. Results of Western blotting were quantitated with NIH Image version 1.62. The relative levels of SRC proteins were calculated by normalization with quantitated Flag-SRC references. The relative level for SRC-3 was arbitrarily set as 100%.
Considering the high homology among members of the SRC family, one obvious uncertainty is the specificity of the antibodies used in the ChIP studies. To rule out the potential cross-reaction among these antibodies, in vitro-translated SRC proteins (Fig. 2B, left panel) or cell extracts (Fig. 2B, right panel) containing endogenous SRCs were immunoprecipitated by SRC antibodies. Triplicate blots containing precipitated SRC proteins resolved by SDS-polyacrylamide gel electrophoresis were probed with antibodies directed against SRC-1, SRC-2, or SRC-3. Antibody specificities were clearly demonstrated by these studies. Furthermore, PR, GR, and SRC antibodies were tested for immunoprecipitation under ChIP conditions followed by Western blotting (data not shown). These control experiments verified our observation of coactivator specificity for different NRs at the same promoter.
To confirm the expression of SRCs in T47D cells and to understand whether the preferential recruitment of SRC-1 and SRC-2 by activated NR is a consequence of dramatically different levels of these proteins, we estimated endogenous SRC levels by comparison with Flag-tagged SRC proteins expressed in Xenopus oocytes. Serial dilutions of equivalent amounts of Flag-tagged SRC-1, -2, and -3 normalized according to Western blotting with anti-Flag antibody were probed with individual SRC antibodies to provide references for the estimation of SRC antibody efficiency (Fig. 2C). Western blotting analyses of endogenous SRC proteins were performed simultaneously with the reference Flag-tagged SRC proteins. The normalized results shown in Fig. 2C demonstrated that SRC-2 and SRC-1 levels were 89.5 and 52.6% of that of SRC-3, respectively. These results suggest that endogenous SRC-1, -2, and -3 levels are comparable. It seems highly unlikely that the slight differences between SRC-1 and SRC-2 levels could contribute to NR selectivity.
Differential assembly of HATs mediated by disparate receptor-SRC interactions.
Multiple components of transcription coactivator complexes are important for transcriptional activation mediated by nuclear receptors (32, 37, 41). However, the composition of coactivator complexes in each NR signaling may not be the same due to relative affinity between the receptor and cofactors, competition between cofactors or coactivator complexes for common interaction motifs (47), or cell-specific cofactor levels (40). Having demonstrated a distinct pattern of SRC coactivator recruitment by two different NRs, we then examined the outcome of this differential utilization of SRCs on the recruitment of other coregulators. In a proof-of-principle experiment, we compared PR- and GR-mediated recruitment of additional coactivators on the MMTV promoter by using antibodies against several HATs (CBP, p300, and pCAF), TRAP220 (an important component of mediator complex), and BRG-1. Remarkably, CBP was significantly recruited in the presence of progesterone while minimally recruited by dexamethasone (Fig. 3). The observation that PR, SRC-1, and CBP were recruited concomitantly to the promoter coincides with a report where a large multicomponent complex was identified in T47D cells, the members of which include PR, SRC-1, and CBP (50). To our surprise, pCAF appeared to be recruited to the MMTV promoter in the presence of dexamethasone but not in the presence of progesterone. There were no significant differences between PR- and GR-mediated recruitment of TRAP220, polymerase II (Fig. 3), and BRG-1 (data not shown). However, p300 was recruited in response to stimulation with both steroids, but slightly higher levels of occupancy were repeatedly observed after dexamethasone induction. Furthermore, SRCs and HAT cofactors were recruited to the MMTV promoter within a similar time frame (data not shown). Based on the data in Fig. 2 and 3, it seems that PR, SRC-1, SRC-3, and CBP are major components in progesterone-dependent transcription, while GR, SRC-2, SRC-3, and pCAF constitute complexes for dexamethasone induction on the MMTV promoter. Thus, hormone stimulations of different NRs induce differential coactivator utilization within the same MMTV promoter context, and a different subset of HATs are recruited in each case for transcriptional activation, endowing potential downstream specificity to different ligand signals.
FIG. 3.
Differential assembly of HATs in disparate NR signaling. ChIP analysis was performed to detect the occupancy of HATs, TRAP220, or polymerase II on the MMTV promoter after a 1-h treatment with progesterone (lanes P) or dexamethasone (lanes D). The faster-migrating bands represent products amplified from the MMTV promoter.
Diverse histone modification pattern in different NR-signaling pathways.
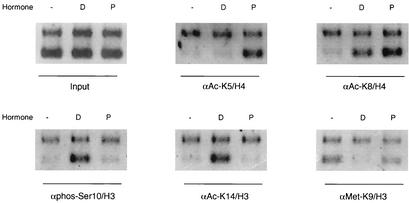
Histone modification has been shown to affect chromatin remodeling and correlates with promoter activation (17). In particular, acetylation of selective lysine residues by specific HATs has been linked to transcription (22). The preferential recruitment of HAT activities by PR or GR led us to ask whether a distinct pattern of histone acetylation on the MMTV promoter could be generated. ChIP experiments were performed using antibodies against site-specific histone modifications. Distinct patterns of histone modification were noticed after 1 h of hormone treatment. Strikingly, the MMTV promoter-associated histone H4 underwent significant acetylation at lysine 5, a known target site for CBP (20), only with progesterone stimulation (Fig. 4). Acetylation at lysine 8 (Fig. 4) and methylation at arginine 17 of H3 (not shown) were equally enhanced by both progesterone and dexamethasone treatments. In agreement with the GR/SRC-2-induced recruitment of pCAF, acetylation at lysine 14 of histone H3 was increased in the presence of dexamethasone. It was reported previously that pCAF catalyzes selective acetylation of lysine 14 of histone H3 in both free histones and nucleosomes (39). Since phosphorylation of serine 10 or methylation at lysine 9 are functionally linked to acetylation at lysine 14 (17, 30), we further examined modification of serine 10 and lysine 9. Intriguingly, significant phosphorylation of serine 10 and hypo-methylation of lysine 9 in histone H3 were associated with the acetylation at lysine 14 after dexamethasone stimulation, representing an active or accessible state of the chromatin. In contrast, progesterone did not induce significant changes at serine 10, lysine 9, and lysine 14 modifications (Fig. 4). Methylation at arginine 17 in histone H3 increased similarly in the presence of either progesterone or dexamethasone (data not shown). These findings suggest that the differential recruitment of coactivators by PR and GR results in distinct downstream patterns of histone modification on the MMTV promoter.
FIG. 4.
Diverse histone modifications associated with distinct hormone pathways. Hormone-dependent acetylation, phosphorylation, or methylation-demethylation of selective histone residues on the MMTV promoter was examined by ChIP assays with the indicated antibodies. The symbols are as described in the legends to Fig. 2A and 3.
Modulation of PR and GR activities by loss of coactivator function.
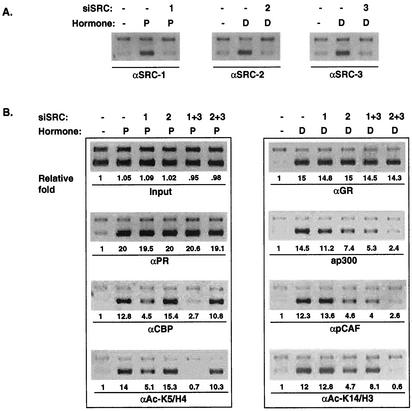
Given that the major SRC proteins recruited to the MMTV promoter were SRC-1 and SRC-3 for PR and SRC-2 and SRC-3 for the GR pathway, we sought to further substantiate that SRC proteins indeed contribute to the PR- or GR-mediated MMTV-CAT activity patterns in the T47D cells. To achieve this, we used an RNA interference approach (12) to eliminate the expression of SRC-1 or SRC-2 alone or in combination with SRC-3. The results in Fig. 5A demonstrated the efficiency and specificity of siRNA in T47D cells. Corresponding siRNA resulted in a reduction of 80 to 90% in levels of endogenous SRC proteins. Specific siRNAs had no cross-reactivity with other SRC counterparts. As a control, siRNA directed against luciferase had no effect on any of the SRC proteins (data not shown). To analyze CAT activity, the cells were transfected with siRNA for 2 days and then treated with the indicated hormones for 24 h. Reduction of SRC-1 levels in T47D cells reduced progesterone-stimulated activity by 31%, while siSRC-2 had only slight effect (Fig. 5B, lanes 3 [P < 0.01] and 4 [P = 0.1], compared to the control level in lane 2). Simultaneous knockdown of SRC-1 and SRC-3 resulted in a further reduction of PR-dependent transactivation (Fig. 5B, lane 5), while a combination of SRC-2 and SRC-3 siRNA had a greatly reduced effect (Fig. 5B, lane 6). In contrast, decreasing SRC-2 levels significantly diminished dexamethasone-stimulated CAT activity (Fig. 5B, lane 9 [P < 0.01]), while reducing SRC-1 had only a minimal effect (Fig. 5B, lane 8 [P = 0.39], compared to the control in lane 7). Interfering with both SRC-2 and SRC-3 expression together (Fig. 5B, lane 11), but not SRC-1 and SRC-3 together (Fig. 5B, lane 10), further restricted GR-mediated activation. A reduction of SRC-3 alone had only a minimal effect on PR- or GR-mediated CAT expression (data not shown). These data are consistent with our ChIP analysis, which shows that SRC-1 recruitment was preferred by PR and that SRC-2 was preferred by GR. Taken together, our results indicate that SRC-1, in combination with other coregulators, plays an important role in progesterone-mediated gene transcription, while SRC-2 is required for full glucocorticoid-stimulated activation of the MMTV promoter in T47D cells.
Transcription signal cascades determined by specific SRC-NR interactions.
We next wished to determine whether the differential recruitment of CBP versus pCAF by PR versus GR and distinct patterns of histone modification are the downstream effect of differential recruitment of SRC-1 or SRC-2. Therefore, we performed RNA interference of SRCs followed by ChIP to examine the potential changes that knockdown of either coactivator would produce. To eliminate the nonspecific effects of transfection reagent on cell growth, the control groups (Fig. 6A and B) were treated with siRNA against luciferase, which is absent in the T47D/CAT0 cells. Reduction of SRC-1, -2, or -3 levels in T47D cells by siRNA appeared to eliminate the recruitment of these coactivators to the MMTV promoter (Fig. 6A), while these siRNA had no effect on PR or GR recruitment (Fig. 6B), confirming the expected specificity of individual siRNA. The recruitment of CBP and acetylation of histone H4 lysine 5 with progesterone stimulation decreased significantly in the absence of SRC-1 and were even further decreased when both SRC-1 and SRC-3 were reduced by RNA interference (Fig. 6B). As expected, recruitment of CBP and acetylation at lysine 5 of histone H4 were not affected by the reduction of the SRC-2 level and were only marginally reduced by loss of both SRC-2 and SRC-3. In contrast, reduction of pCAF recruitment and acetylation of lysine 14 on histone H3 after dexamethasone treatment were associated with reduction of SRC-2 expression and were further decreased by reducing both SRC-2 and SRC-3. In controls, reduction of SRC-1 had little effect on either pCAF recruitment or acetylation of histone H3 at lysine 14. When both SRC-1 and SRC-3 expression were inhibited, reduction of pCAF recruitment was not proportional to the decrease in acetylation at lysine 14 on histone H3, indicating involvement of another HAT, possibly GCN5. GCN5 has been shown to acetylate lysine 14 of histone H3 as well. These observations suggest that initial NR-SRC interactions influence the way by which transcription signal is transmitted. Our results also substantiated an order of sequential recruitment: PR/GR to SRCs to other HATs, in agreement with previous reports (29, 42).
FIG. 6.
Effects of SRC silencing on cofactor recruitment and histone modification. (A) Reduction of SRC-1, -2, and -3 specifically eliminated recruitment of these proteins on the MMTV promoter. (B) The recruitment of receptors or HATs or acetylation of selective histone residues on the MMTV promoter was monitored by ChIP assays after a 1-h hormone treatment following RNA silencing of the indicated SRCs, as described in the legends to Fig. 2A, 3, and 4. Relative changes of PCR products were calculated based on the quantitated results from original images. Normalization was performed against GAPDH first and then against the adjusted input, followed by normalization against the untreated sample (−) that was arbitrarily set to 1.
DISCUSSION
Steroid receptors exhibit ligand-dependent interactions with multiple coactivators. However, the mechanisms for in vivo assembly of multiple coactivator components and coactivator specificity remain obscure. In this study, we show that SRCs are differentially utilized by NRs on the MMTV promoter, and more importantly, the preferential recruitment of SRCs by steroid receptors contributes to the determination of downstream events in NR-mediated transcription. The data suggest that members of the SRC family play an important and specific role in the construction of downstream events at target promoters and do not serve as functionally redundant platform proteins in the transcription processes.
Although SRC-1 and SRC-2 have been reported to interact with both PR and GR in overexpression transfection experiments (11, 14, 19, 32, 45), several indirect lines of evidence have suggested the existence of coactivator preferences. First, on the basis of chromatography and coimmunoprecipitation approaches, a large multicomponent complex containing PR, SRC-1, and CBP was identified in T47D cells (50). Second, in an attempt to isolate steady-state coregulator complexes, both SRC-1 and SRC-2 were identified in a preformed complex. However, upon progesterone induction, more SRC-1 than SRC-2 appeared to comigrate with PR (33). Third, an SRC-1 mutant devoid of activation domains but containing HAT and receptor interaction domains displayed a strong dominant-negative repressor activity for certain nuclear receptors (18, 36), but the dominant-negative activity of this mutant SRC-1 is less for GR (14). Finally, a preferential interaction between GR and SRC-2 has been suggested in several studies (9, 14, 16, 25, 48). In this study, we have provided in vivo evidence that PR prefers SRC-1 and GR prefers SRC-2 on MMTV promoter. Our data substantiate the observations mentioned above. Furthermore, our results suggest a potential mechanism by which different NRs mediate transcription signaling by preferential recruitment of a subset of cofactors.
The molecular details of PR and GR preferences for different coactivators in vivo are not clear. One possibility is NR box binding specificity, as suggested above. A small region of SRC-2 (amino acids 730 to 1121) retains steroid receptor binding, transactivation, and coactivator activities. However, this region of SRC-2 exhibits relatively low homology with SRC-1 (16), suggesting a potential molecular basis for functional differences between SRC-1 and SRC-2. In addition, allosteric effects upon DNA binding may trigger different receptor conformations (26, 51), which in turn could be a determinant for the binding of coactivators. In an effort to determine whether or not the coactivator preferences are observed in solution or evolve only on the MMTV promoter, we have performed coimmunoprecipitation studies of PR and GR with SRCs in the presence or absence of ligands. Since only GR/SRC-2 interaction was observed in solution (data not shown), it is possible that allosteric regulation of coactivator interactions on the MMTV promoter may influence the PR/SRC-1 preference.
Importantly, we have observed differential recruitment of CBP and pCAF on the MMTV promoter in response to progesterone or dexamethasone stimulation, whereas p300 is recruited in both cases. The differential recruitment of CBP or pCAF appears to correlate with the preferential targeting by SRC-1 or SRC-2. Although the function of SRC coactivators' recruitment of CBP/p300 is well documented, such specificity has not been shown before. Only specificity of HAT requirement for different transcription factors has been indicated in previous studies. For example, inactivation of the HAT domains of CBP has no influence on the coactivation of RAR. On the other hand, CREB (CRE-binding protein) function needs CBP-HAT activity and not P/CAF-HAT activity (21). Although SRC-1 has been shown to interact with p300, CBP, and pCAF (18, 19, 43, 45), only the recruitment of CBP/p300 was clearly observed upon progesterone treatment. It is interesting that pCAF associates with the GR/SRC-2 complex specifically in the presence of dexamethasone. The ability of pCAF to mediate GR function was recently tested by examining the effects of cotransfected pCAF on GR transactivation properties (14). The requirement of pCAF in RAR-mediated activation was also established (21). We show here that SRC-2 is required for pCAF recruitment, either directly or through an interaction with p300, as reduction of SRC-2 by RNA interference limited the recruitment of pCAF (Fig. 6). It is unlikely that CBP/p300 interacts directly with GR/PR (19), since reports from different groups reveal little or no direct binding to these receptors (7, 43).
The state of chromatin modification has long been recognized to influence gene expression. Our results indicate that differentially assembled coactivator complexes lead to specific patterns of targeted histone modification. The acetylation of specific lysine residues on histone H3 or H4 was correlated with the recruitment of distinct HAT activity that could be altered by knockdown of either SRC-1 or SRC-2 (Fig. 6). To date, the manner by which different HATs discriminate lysines in vivo is not fully understood. CBP and p300 have been shown to acetylate histone H4 at lysine 5, 8, 12, and 16 in vitro (20). However, we have observed significant acetylation of lysine 5 and 8 on histone H4 by CBP/p300 only after progesterone stimulation. This suggests that the actual target of p300/CBP acetylation could be redirected in a different context. It may also reflect the diversity of histone modification in vivo, since acetylation at lysine 12 on histone H4 was considered to be an inactivating modification in certain cellular contexts (17). The acetylation of K14 on histone H3 after dexamethasone induction is consistent with the recruitment of pCAF (39). However, GCN5, which bears homology to human pCAF, also displays a preference for K14 of histone H3. It is possible that GCN5 also could be recruited to the MMTV promoter after GR recruitment, as demonstrated for pCAF. Histone modification associated with acetylation of K14 (phosphorylation of serine 10 and hypomethylation of lysine 9) is consistent with the histone code pattern observed by others (17). Histone H3 phosphorylation at Ser10 has recently been elucidated as a novel regulatory mechanism involving histone kinases such as Rsk-2 and Msk-1 (8). These histone codes and the recently reported in vitro binding of a HAT bromodomain with acetylated lysines within H3 and H4 peptides have been implicated in a targeting step for events following histone modification.
In addition to coactivator specificities, our studies also suggest that SRC-3 is utilized similarly by either receptor. Elimination of SRC-3 along with either SRC-1 or SRC-2 resulted in a further reduction of PR- or GR-mediated CAT expression compared to knockdown of SRC-1 or SRC-2 alone. This suggests a role for SRC-3 in both pathways. ChIP assays indicate that SRC-3 also contributes to the recruitment of HAT and histone modification by SRC-1 or SRC-2. In addition, SRC-2 knockdown has a slight but reproducible influence on PR-mediated CAT expression (Fig. 6), indicating that it plays a minor role in PR function. However, such a contribution involving a limited amount of SRC-2 recruitment was not detectable by ChIP assays. Therefore, the reason for the moderate attenuation of the CAT activity after SRC-1 silencing (Fig. 6) could be explained by recruitment of SRC-3 and a small amount of SRC-2 to the MMTV promoter or to SRC coactivator-independent activation. Our observations are consistent with previous reports, which demonstrate the redundant functions of SRCs (38, 49).
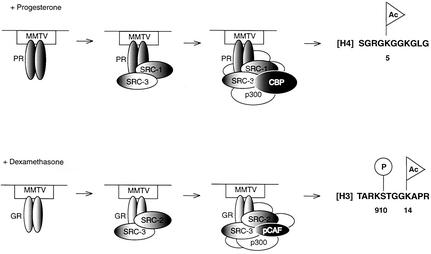
Finally, we have revealed the roles of SRCs in mediating transcription signal cascades by RNA interference. Our suggestion that SRC-1 could be important in the progesterone pathway has been supported by the study of SRC-1 knockout mice in our lab (49). Therefore, we propose a model in which preferential SRC family coactivator-steroid receptor interactions influence differential recruitment of downstream components by each receptor (Fig. 7). Progesterone signaling is transduced from PR/SRC-1/CBP along with other coactivators to induce specific histone modification, including acetylation at lysine 5 of histone H4 on the MMTV promoter. The cascade originating from dexamethasone induction is conveyed primarily through a complex containing GR/SRC-2/pCAF and leads to specific posttranslational modification on histone H3. Our study provides conceptual proof of principle for a mechanism by which coactivator preferences for nuclear receptors can regulate the fates of downstream events in transcription signaling.
FIG. 7.
Models of stepwise events in NR signaling. Progesterone activation signal is transmitted from PR recruitment of SRC-1/SRC-3, which further recruits CBP along with other coactivators (not included in the diagram) to induce specific histone modification, including acetylation at lysine 5 of histone H4. The cascade originating from dexamethasone induction is conveyed through GR's interaction with SRC-2 and SRC-3, which dictates recruitment of pCAF and other cofactors with various enzyme activities, leading to specific posttranslational modification of histone H3.
Acknowledgments
We thank Jack-Michel Renoir for providing the T47D/CAT0 cell line, Yoshihiro Nakatani for anti-pCAF, and Weidong Wang for antibody against BRG-1.
This work was supported by National Institutes of Health grants to B.W.O. (HD08818 and Atlas program).
REFERENCES
- 1.Alen, P., F. Claessens, E. Schoenmakers, J. V. Swinnen, G. Verhoeven, W. Rombauts, and B. Peeters. 1999. Interaction of the putative androgen receptor-specific coactivator ARA70/ELE1alpha with multiple steroid receptors and identification of an internally deleted ELE1beta isoform. Mol. Endocrinol. 13:117-128. [DOI] [PubMed] [Google Scholar]
- 2.Allan, G. F., N. H. Ing, S. Y. Tsai, G. Srinivasan, N. L. Weigel, E. B. Thompson, M. J. Tsai, and B. W. O'Malley. 1991. Synergism between steroid response and promoter elements during cell-free transcription. J. Biol. Chem. 266:5905-5910. [PubMed] [Google Scholar]
- 3.Bhakat, K. K., and S. Mitra. 2000. Regulation of the human O(6)-methylguanine-DNA methyltransferase gene by transcriptional coactivators cAMP response element-binding protein-binding protein and p300. J. Biol. Chem. 275:34197-34204. [DOI] [PubMed] [Google Scholar]
- 4.Borud, B., T. Hoang, M. Bakke, A. L. Jacob, J. Lund, and G. Mellgren. 2002. The nuclear receptor coactivators p300/CBP/cointegrator-associated protein (p/CIP) and transcription intermediary factor 2 (TIF2) differentially regulate PKA-stimulated transcriptional activity of steroidogenic factor 1. Mol. Endocrinol. 16:757-773. [DOI] [PubMed] [Google Scholar]
- 5.Boyd, K. E., and P. J. Farnham. 1999. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 19:8393-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bramlett, K. S., and T. P. Burris. 2002. Effects of selective estrogen receptor modulators (SERMs) on coactivator nuclear receptor (NR) box binding to estrogen receptors. Mol. Genet. Metab. 76:225-233. [DOI] [PubMed] [Google Scholar]
- 7.Chen, D., S. M. Huang, and M. R. Stallcup. 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 275:40810-40816. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., M. Tini, and R. M. Evans. 2001. HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13:218-224. [DOI] [PubMed] [Google Scholar]
- 9.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deroo, B. J., and T. K. Archer. 2001. Glucocorticoid receptor-mediated chromatin remodeling in vivo. Oncogene 20:3039-3046. [DOI] [PubMed] [Google Scholar]
- 11.Ding, X. F., C. M. Anderson, H. Ma, H. Hong, R. M. Uht, P. J. Kushner, and M. R. Stallcup. 1998. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC- 1): multiple motifs with different binding specificities. Mol. Endocrinol. 12:302-313. [DOI] [PubMed] [Google Scholar]
- 12.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 13.Gehin, M., M. Mark, C. Dennefeld, A. Dierich, H. Gronemeyer, and P. Chambon. 2002. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol. Cell. Biol. 22:5923-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He, Y., D. Szapary, and S. S. Simons, Jr. 2002. Modulation of induction properties of glucocorticoid receptor-agonist and -antagonist complexes by coactivators involves binding to receptors but is independent of ability of coactivators to augment transactivation. J. Biol. Chem. 277:49256-49266. [DOI] [PubMed] [Google Scholar]
- 15.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 16.Hong, H., K. Kohli, M. J. Garabedian, and M. R. Stallcup. 1997. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol. Cell. Biol. 17:2735-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 18.Kalkhoven, E., J. E. Valentine, D. M. Heery, and M. G. Parker. 1998. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 17:232-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamei, Y., L. Xu, T. Heinzel, J. Torchia, R. Kurokawa, B. Gloss, S. C. Lin, R. A. Heyman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1996. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85:403-414. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, A., and M. Horikoshi. 1998. How do histone acetyltransferases select lysine residues in core histones? FEBS Lett. 431:131-133. [DOI] [PubMed] [Google Scholar]
- 21.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 22.Kuo, M. H., J. E. Brownell, R. E. Sobel, T. A. Ranalli, R. G. Cook, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383:269-272. [DOI] [PubMed] [Google Scholar]
- 23.Kusk, P., S. John, G. Fragoso, J. Michelotti, and G. L. Hager. 1996. Characterization of an NF-1/CTF family member as a functional activator of the mouse mammary tumor virus long terminal repeat 5′ enhancer. J. Biol. Chem. 271:31269-31276. [DOI] [PubMed] [Google Scholar]
- 24.Le Bihan, S., V. Marsaud, C. Mercier-Bodard, E. E. Baulieu, S. Mader, J. H. White, and J. M. Renoir. 1998. Calcium/calmodulin kinase inhibitors and immunosuppressant macrolides rapamycin and FK506 inhibit progestin- and glucocorticosteroid receptor-mediated transcription in human breast cancer T47D cells. Mol. Endocrinol. 12:986-1001. [DOI] [PubMed] [Google Scholar]
- 25.Leers, J., E. Treuter, and J. A. Gustafsson. 1998. Mechanistic principles in NR box-dependent interaction between nuclear hormone receptors and the coactivator TIF2. Mol. Cell. Biol. 18:6001-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefstin, J. A., and K. R. Yamamoto. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392:885-888. [DOI] [PubMed] [Google Scholar]
- 27.Leo, C., H. Li, and J. D. Chen. 2000. Differential mechanisms of nuclear receptor regulation by receptor-associated coactivator 3. J. Biol. Chem. 275:5976-5982. [DOI] [PubMed] [Google Scholar]
- 28.Li, J., B. W. O'Malley, and J. Wong. 2000. p300 requires its histone acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol. Cell. Biol. 20:2031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, Z., J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2001. Sequential recruitment of steroid receptor coactivator-1 (SRC-1) and p300 enhances progesterone receptor-dependent initiation and reinitiation of transcription from chromatin. Proc. Natl. Acad. Sci. USA 98:12426-12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 31.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 33.McKenna, N. J., Z. Nawaz, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1998. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc. Natl. Acad. Sci. USA 95:11697-11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee, R., S. Sun, L. Santomenna, B. Miao, H. Walton, B. Liao, K. Locke, J. H. Zhang, S. H. Nguyen, L. T. Zhang, K. Murphy, H. O. Ross, M. X. Xia, C. Teleha, S. Y. Chen, B. Selling, R. Wynn, T. Burn, and P. R. Young. 2002. Ligand and coactivator recruitment preferences of peroxisome proliferator activated receptor alpha. J. Steroid Biochem. Mol. Biol. 81:217-225. [DOI] [PubMed] [Google Scholar]
- 35.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 36.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 37.Rosenfeld, M. G., and C. K. Glass. 2001. Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 276:36865-36868. [DOI] [PubMed] [Google Scholar]
- 38.Sadow, P. M., O. Chassande, K. Gauthier, J. Samarut, J. Xu, B. W. O'Malley, and R. E. Weiss. 2003. Specificity of thyroid hormone receptor subtype and steroid receptor coactivator (SRC)-1 on thyroid hormone action. Am. J. Physiol. Endocrinol. Metab. 284:E36-E46. [DOI] [PubMed] [Google Scholar]
- 39.Schiltz, R. L., C. A. Mizzen, A. Vassilev, R. G. Cook, C. D. Allis, and Y. Nakatani. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189-1192. [DOI] [PubMed] [Google Scholar]
- 40.Shang, Y., and M. Brown. 2002. Molecular determinants for the tissue specificity of SERMs. Science 295:2465-2468. [DOI] [PubMed] [Google Scholar]
- 41.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 42.Sharma, D., and J. D. Fondell. 2002. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl. Acad. Sci. USA 99:7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheppard, H. M., J. C. Harries, S. Hussain, C. Bevan, and D. M. Heery. 2001. Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol. Cell. Biol. 21:39-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, C. L., H. Htun, R. G. Wolford, and G. L. Hager. 1997. Differential activity of progesterone and glucocorticoid receptors on mouse mammary tumor virus templates differing in chromatin structure. J. Biol. Chem. 272:14227-14235. [DOI] [PubMed] [Google Scholar]
- 45.Smith, C. L., S. A. Onate, M. J. Tsai, and B. W. O'Malley. 1996. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc. Natl. Acad. Sci. USA 93:8884-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 47.Treuter, E., L. Johansson, J. S. Thomsen, A. Warnmark, J. Leers, M. Pelto-Huikko, M. Sjoberg, A. P. Wright, G. Spyrou, and J. A. Gustafsson. 1999. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J. Biol. Chem. 274:6667-6677. [DOI] [PubMed] [Google Scholar]
- 48.Voegel, J. J., M. J. Heine, M. Tini, V. Vivat, P. Chambon, and H. Gronemeyer. 1998. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 17:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, J., Y. Qiu, F. J. DeMayo, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1998. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922-1925. [DOI] [PubMed] [Google Scholar]
- 50.Xu, Y., L. Klein-Hitpass, and M. K. Bagchi. 2000. E1A-mediated repression of progesterone receptor-dependent transactivation involves inhibition of the assembly of a multisubunit coactivation complex. Mol. Cell. Biol. 20:2138-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi, P., M. D. Driscoll, J. Huang, S. Bhagat, R. Hilf, R. A. Bambara, and M. Muyan. 2002. The effects of estrogen-responsive element- and ligand-induced structural changes on the recruitment of cofactors and transcriptional responses by ER alpha and ER beta. Mol. Endocrinol. 16:674-693. [DOI] [PubMed] [Google Scholar]