Abstract
In the yeast Saccharomyces cerevisiae, Cdc13, Yku, and telomerase define three parallel pathways for telomere end protection that prevent chromosome instability and death by senescence. We report here that cdc13-1 yku70Δ mutants generated telomere deprotection-resistant cells that, in contrast with telomerase-negative senescent cells, did not display classical crisis events. cdc13-1 yku70Δ cells survived telomere deprotection by exclusively amplifying TG1-3 repeats (type II recombination). In a background lacking telomerase (tlc1Δ), this process predominated over type I recombination (amplification of subtelomeric Y′ sequences). Strikingly, inactivation of the Rad50/Rad59 pathway (which is normally required for type II recombination) in cdc13-1 yku70Δ or yku70Δ tlc1Δ mutants, but also in cdc13-1 YKU70+ tlc1Δ mutants, still permitted type II recombination, but this process was now entirely dependent on the Rad51 pathway. In addition, delayed senescence was observed in cdc13-1 yku70Δ rad51Δ and cdc13-1 tlc1Δ rad51Δ cells. These results demonstrate that in wild-type cells, masking by Cdc13 and Yku prevents the Rad51 pathway from amplifying telomeric TG1-3 sequences. They also suggest that Rad51 is more efficient than Rad50 in amplifying the sequences left uncovered by the absence of Cdc13 or Yku70.
Telomeres represent the ends of eukaryotic chromosomes and are absolutely required for the overall stability of the genome. More specifically, telomeres are needed to prevent chromosome ends from degrading and fusing together (for recent reviews, see references 2, 3, and 8). In addition, telomeres have particular features that accommodate telomerase recruitment and allow them to achieve complete replication of chromosomal ends (reviewed in references 2, 11, 13, and 44). Telomeres also play an important role in organizing particular chromatin structures and positioning chromosomes within the nucleus, thus potentially controlling transcriptional programs (reviewed in reference 11).
Previous studies have defined three components of telomere end protection in the budding yeast Saccharomyces cerevisiae, namely, telomerase, the Yku70/Yku80 heterodimer, and Cdc13 (10, 14, 15, 17-19, 32, 33, 36, 38, 41, 42, 49, 51). Some of these components are interconnected; for instance, overexpression of the catalytic subunit of telomerase, EST2, or of the telomerase RNA gene, TLC1, suppressed the temperature-sensitive growth defect of a yku80Δ mutant at 37°C (38, 49). However, these three components appear to act in distinct pathways, as the cdc13-1 yku70Δ, cdc13-1 tlc1Δ, and yku80Δ est2Δ pairs all exhibit severe synthetic defects (38, 42). Telomerase appears to exert telomere protection by preventing telomere shortening and, hence, chromosome instability and death by senescence (33), while Cdc13 appears to maintain a different aspect of telomere structure independent of telomere length (17). Loss of Yku function induces some recombinational events but does not provoke senescence (14, 42). The Yku complex has also been implicated in the process of telomerase access to telomeres in cooperation with telomerase itself and Cdc13 (reviewed in reference 11), in the subnuclear organization of telomeres (28), and in telomeric silencing (37). Similarly, Cdc13 has been implicated in both telomere end protection and telomere replication (reviewed in references 11 and 34).
The prominent roles of Cdc13, the Yku complex, and telomerase in telomere end protection result from their strategic physical position at telomere ends. In most of the eukaryotic species studied to date, double-stranded telomeric sequences end in 3′ single-stranded overhangs of differing lengths, the function of which is to bind specialized proteins (reviewed in references 2 and 40). Both Cdc13 and telomerase bind the 3′ overhang, while the Yku70/Yku80 heterodimer binds double-stranded telomeric sequences contiguous to the 3′ overhang (reviewed in references 11, 34, and 44). In telomerase-negative cells (33) and checkpoint-negative cdc13-1 cells (17), loss of telomere structure provokes senescence. Using Rad52-dependent homologous recombination mechanisms, a very small percentage of telomerase-negative cells can survive senescence by maintaining telomere structure (32). Two telomerase-independent telomere maintenance pathways have been identified (reviewed in reference 27). The Rad51 pathway, which also relies on Rad52, Rad54, Rad55, and Rad57, yields telomeres that display recombination events within the subtelomeric Y′ sequences (referred to as type I recombination) (29, 32, 48). The Rad50/Rad59 pathway, which amplifies the telomeric TG1-3 sequences (so-called type II recombination), relies on Rad52, Rad50, Mre11, Xrs2, Rad59, and Sgs1 (4, 6, 23, 26, 29, 32, 47, 48). Analysis of these pathways in a well-studied genetic system such as yeast has particular relevance to our comprehension of immortalization of human cells that maintain their telomeres via the alternative lengthening of telomeres recombination pathway (reviewed in reference 21).
In the present study, we report that postsenescence survivors emerge from senescing cdc13-1 yku70Δ double mutants grown at a temperature permissive for growth for both of the single mutants. While telomerase-negative cells (tlc1Δ) generated over 90% type I survivors, cdc13-1 yku70Δ mutants exclusively generated type II survivors. Type II recombination also predominated (100%) over type I in yku70Δ tlc1Δ and cdc13-1 tlc1Δ cells. Most importantly for our understanding of telomerase-independent telomere maintenance mechanisms, we find that, atypically, type II recombination on TG1-3 sequences can be accomplished by the Rad51 pathway in cdc13-1 and yku70Δ mutants. In addition, a rad51Δ-induced delay in the generation of type II survivors was observed in cdc13-1 yku70Δ and cdc13-1 tlc1Δ mutants but not in tlc1Δ mutants. The present data support the view that Yku, like Cdc13, protects the 3′ overhang from degradation, thus preventing recombination on the telomeric TG1-3 sequences. Moreover, the damaged substrate left after loss of Yku70 or Cdc13 function (presumably single-stranded TG1-3 sequences) may exhibit more affinity for Rad51 than for Rad50.
MATERIALS AND METHODS
Strains.
The rad59::KanMX4 strain (Euroscarf, Frankfurt, Germany) and all other strains used in this study (17) were backcrossed five times against the genetic background used in our laboratory prior to experimentation. The strain background was 15Daub, a ura3Δns derivative of BF264-15D, which was also leu2-3,112, trp1-1a, ade1, and his2 (43). For each experiment, the temperature at which cells were grown is indicated in the legend for the corresponding figure.
Analysis of telomere structure.
Genomic DNA from each culture was digested with either XhoI or a mixture of four restriction enzymes (AluI, HaeIII, HinfI, and MspI) that used a 4-bp recognition sequence (48), and the resulting Southern blots were analyzed with a 270-bp TG1-3 32P-labeled probe (17). In some cases (see Fig. 4C), a 32P-labeled Y′ probe was used (17). In some Southern blots, the amplified TG1-3 sequences resulting from type II homologous recombination appeared more smeary in the yku70Δ and cdc13-1 mutants than in the tlc1Δ mutants. This was possibly due to the presence of high amounts of single-stranded DNA in these mutant cells (15, 18, 42). To ascertain that digestion with the four-base cutter enzymes had been performed to completion, some gels were stained with ethidium bromide prior to being transferred to a nitrocellulose membrane and photographed (see Results). Such controls also served to confirm approximately equal loading in all lanes.
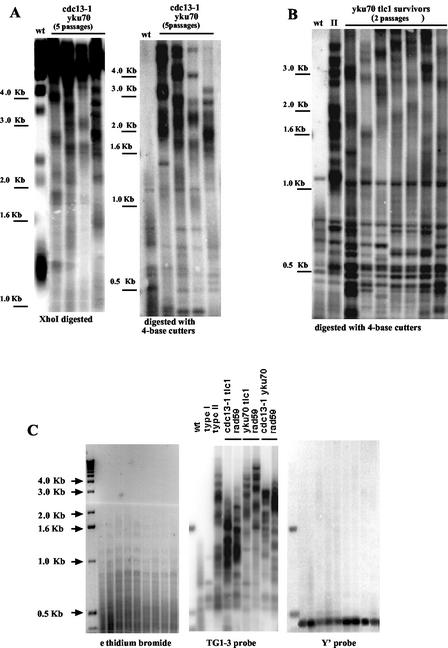
FIG. 4.
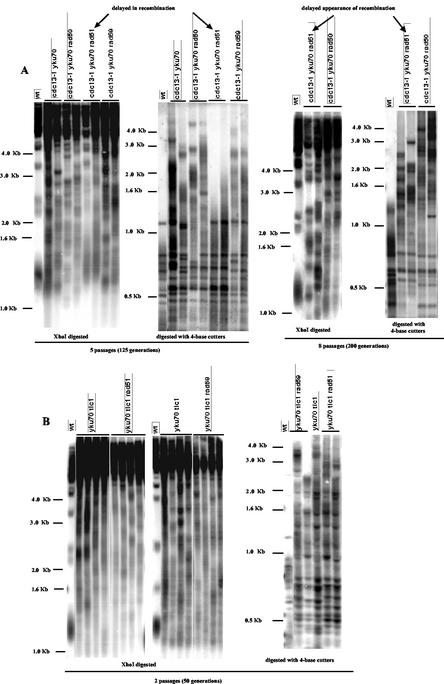
Removal of Yku70 exclusively triggers type II recombination that predominates over tlc1Δ-induced type I recombination. (A) The structure of telomeres of recombining cdc13-1 yku70Δ mutants grown at 25°C (at the fifth passage) following digestion of genomic DNAs with XhoI (left panel) or a mixture of AluI, HaeIII, HinfI, and MspI (4-base cutters, right panel) reveals that type II represents the exclusive pathway for survival by recombination in these cells, as explained for Fig. 1B. Four independent clones are shown here, but all 20 tested exhibited a similar pattern (data not shown). The lanes labeled “wt” represent telomere structure in nonrecombining wild-type colonies. A TG1-3 32P-labeled probe was used. (B) Survivors from senescing yku70Δ tlc1Δ cells are exclusively type II recombinants. The telomere structures of seven independent clones representative of a total of 18 survivors (all collected after the second passage) are shown here in comparison with that in a typical tlc1Δ type II survivor (fifth passage) (lane II) and that in a nonrecombining wild type (wt). Strains were grown at 25°C. The results of digestion with AluI, HaeIII, HinfI, and MspI and hybridization to a TG1-3 32P-labeled probe are shown. (C) Control experiments attest that the high-molecular-weight bands in samples from cdc13-1- or yku70Δ-induced type II recombinants represent recombining TG1-3 sequences. Left panel: a gel loaded with genomic DNA samples from the various strains indicated at the corresponding lanes in the middle panel was stained with ethidium bromide prior to transfer to a nitrocellulose membrane and photographed. Right panel: the same gel as that shown in left panel was then transferred to a nitrocellulose membrane and probed with a Y′ 32P-labeled probe. Middle panel: the same gel was then dehybridized and reprobed with a TG1-3 32P-labeled probe. All survivors were picked out as single colonies from the agar plates during the restreak following the appearance of the first survivors, a time that differed depending on the strains analyzed (Fig. 2). All strains were grown at 25°C.
Analysis of the kinetics of senescence and survival.
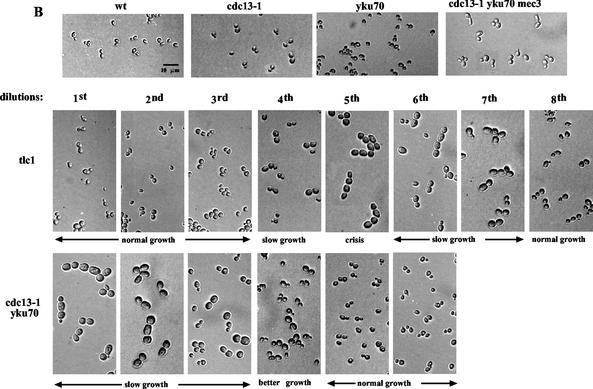
Streak assays for analysis of telomeric recombination events (performed on agar-based plates) were based on protocols described in reference 48 and also described in reference 17. Briefly, restreaking of single colonies on a yeast extract-peptone-dextrose (YEPD) plate was repeated every 48 h (typically, cells underwent ∼25 generations per “streakout,” or passage) to allow loss of viability and the appearance of survivors. To analyze recombination events, a single colony was picked out of the agar plate and grown overnight in liquid YEPD medium to obtain enough material for genomic DNA preparation and Southern blot analysis (see above). The possibility that the observed type II survivors were previous type I survivors that had evolved with time, as sometimes observed in liquid cultures, is very unlikely. Indeed, such an evolution has been recorded only for liquid cultures that, from the beginning of an experiment, contained both types of survivors and is due to the better growth of type II survivors (48). On the other hand, the presence of both types of survivors in a colony grown on agar-based medium and issued from a single cell has never been observed. In some experiments (Fig. 1B), cells were withdrawn from liquid cultures at various intervals during senescence and fixed with formaldehyde. Cells were then observed in a BX50 Olympus light microscope (using a 40× lens and Nomarski optics), and photographs were taken using 400 ASA Ilford films. Measurements of cell viability in liquid cultures (see Fig. 6B) were done as described in references 4 and 29.
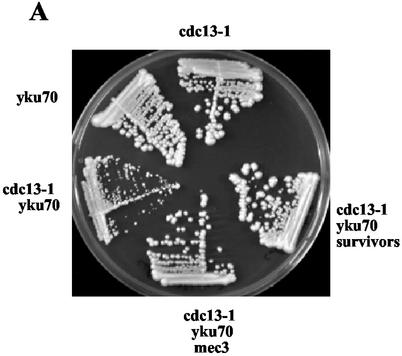
FIG. 1.
A cdc13-1 yku70Δ double mutant experiences senescence or survival at 25°C. (A) Growth of haploid cells containing the cdc13-1ts, yku70, and/or mec3 null mutations. A CDC13+/cdc13-1 YKU70+/yku70Δ MEC3+/mec3Δ heterozygous diploid was sporulated, and after selection among the progeny of the appropriate phenotypes, cdc13-1 and yku70 single mutants, cdc13-1 yku70 double mutants, and cdc13-1 yku70 mec3 triple mutants were propagated for 3 days at 25°C (∼40 generations) on YEPD plates and photographed. Because of the time for selection of the mutations, an estimated ∼80 generations elapsed between sporulation and photograph. The cdc13-1 yku70 survivors shown here originate from a fast-growing colony that appeared on the fourth restreak plate (following postsporulation selection) and were grown for 3 more days at 25°C. (B) Morphology of the cdc13-1 yku70Δ (bottom row) and tlc1Δ (middle row) cells during the process of senescence or survival. The top row shows the morphologies of cdc13-1 and yku70Δ single-mutant cells as well as of wild-type (wt) and cdc13-1 yku70Δ mec3Δ cells. Immediately after germination of the cdc13-1 yku70Δ double mutant or introduction of the tlc1Δ mutation, cells were cultivated at 25°C in YEPD medium and liquid cultures were diluted to 105 cells/ml every 24 h. Cells were fixed at various intervals from the time of genetic introduction of the mutations until the generation of survivors, observed in the light microscope, and photographed.
FIG. 6.
cdc13-1-induced type II recombination can be independent of the Rad50/Rad59 pathway. (A) cdc13-1 tlc1Δ can use the Rad51 pathway to accomplish recombination on TG1-3 sequences (type II) and associated survival. Two representative early survivors among cdc13-1 tlc1Δ (second and third lanes from the left in both panels) and cdc13-1 tlc1Δ rad59Δ (fourth and fifth lanes in both panels) strains, all taken at the second passage, are shown here, following digestion of genomic DNAs with XhoI (left panel) or a mixture of AluI, HaeIII, HinfI, and MspI (4-base cutters, right panel). Such a pattern of telomere structure attests to the presence of type II recombination in both strains. Nonrecombining wild-type cells are shown for comparison (lanes 1). Strains were grown at 25°C. A TG1-3 32P-labeled probe was used. (B) Operation of the Rad50/Rad59 pathway is delayed in cdc13-1 mutants. Top graph: cdc13-1 tlc1Δ rad51Δ mutant cells (full and empty squares) generate postsenescence survivors with a delay compared with cdc13-1 tlc1Δ rad59Δ cells (full and empty circles). All survivors were of type II (data not shown). cdc13-1 tlc1Δ rad50Δ mutant cells (full and empty triangles), which did not generate any survivor (see text), are shown for comparison. Cell viability was measured as described previously (4, 29). Briefly, cells were grown to saturation (108 cells/ml) in liquid YEPD medium and were then counted using a Malassez chamber every 24 h and diluted to 105 cells/ml with fresh medium. The results for two independent clones for each strain are shown. Bottom graph: tlc1Δ rad51Δ mutant cells (full and empty squares) generated postsenescence survivors at the same time as tlc1Δ RAD51+ cells (full and empty circles). Two independent clones for each strain are shown. Growth was at 25°C.
RESULTS
cdc13-1 yku70Δ mutant cells generate postsenescence survivors at 25°C in the presence of functional telomerase.
We decided to reinvestigate the phenotype of a cdc13-1 yku70Δ double-mutant strain. As reported previously (38, 42), this strain exhibited severe growth defects at 25°C, a permissive temperature for growth of both single mutants (Fig. 1A). However, a novel observation was that a small percentage of cdc13-1 yku70Δ cells escaped slow growth and generated fast-growing colonies reminiscent of survival from senescence (33) (Fig. 1A; Fig. 2, row 3). As expected from previous studies of telomerase-negative senescent cells (32), survival within the cdc13-1 yku70Δ strain took place through homologous recombination pathways (see below), as cdc13-1 yku70Δ rad52Δ triple-mutant cells did not generate fast-growing survivors (data not shown). These data therefore establish the important point that loss of Yku function, in combination with a partial loss of Cdc13 function, induces damage that can be rescued by recombination. This takes place in the presence of functional telomerase, a characteristic also observed following loss of Cdc13 function alone, at least in cdc13-1 mutant cells (17).
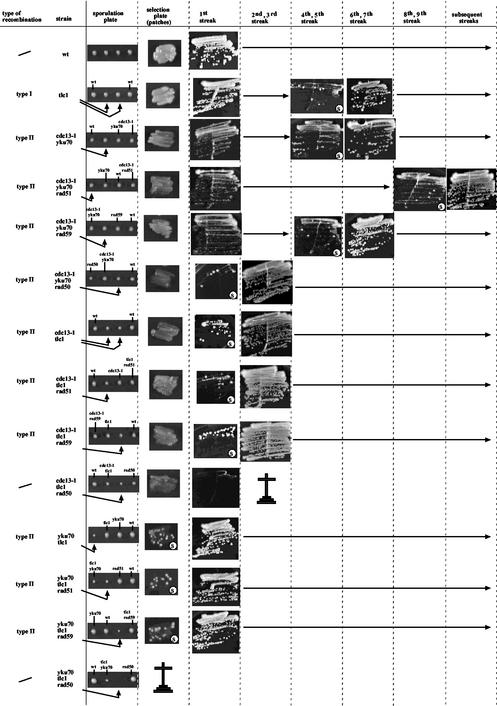
FIG. 2.
Summary of the fates of the strains analyzed in the present study. The relevant genotype of each strain is indicated in the second column. The first column indicates the type of the survivors for each strain: type I (recombination of the subtelomeric Y′ sequences) or type II (recombination of the TG1-3 sequences). Absence of indication of the survival type indicates that the strain either never generated survivors (such as the wild type [wt]) or died without generating survivors. In the latter situation, the Calvary cross indicates the time of death following creation of the strain. The third column shows, for each strain, a representative progeny from the sporulation plate. Based on phenotypic characteristics, the genotype of each spore is indicated. The fourth column represents, for the indicated strain, growth of cells taken off the sporulation plate and repatched on the selection plate for further propagation. Columns 5 to 10 represent the successive streakouts (as indicated) that were done every 48 h (as explained in Materials and Methods) to follow the fate of each strain and, in particular, the occurrence of survivors (for each strain, the first appearance of survivors is marked by the letter “s” on the concerned streak), which are easily distinguishable by their ability to form fast-growing colonies on the background of senescing cells. All strains were grown at 25°C.
While proceeding to a detailed analysis of the recombinational events taking place in the cdc13-1 yku70Δ mutant strain, we also wished to understand the reasons for senescence and the generation of survivors in this strain. Both mutations, when expressed at 37°C, have been associated with the generation of abnormally high levels of single-stranded DNA in telomeric regions of the chromosomes (15, 18, 19, 36, 42, 49). This single-stranded DNA has been proposed to represent the signal that activates the DNA damage checkpoint, thus halting cell cycle progression (15, 19, 36). Although at 25°C, a permissive temperature for growth for both mutations, the presence of single-stranded telomeres was not detected in cdc13-1 cells (15) or at very low levels in yku70Δ cells (18), we nevertheless knew that at 25°C, the cdc13-1 mutation conferred a defect in a tlc1Δ background (17). A mutation in one of the genes (such as MEC3) involved in the DNA damage checkpoint (54) has been previously used to monitor activation of the checkpoint network. Thus, cdc13-1 rad9Δ and cdc13-1 mec3Δ mutant cells displayed improved growth at 29°C, a semipermissive temperature for growth for cdc13-1 (15, 17, 54). This has been interpreted as being due to the absence of cell cycle slowdown resulting from the inactivation of the checkpoint; such cells continued to proliferate because the level of DNA damage inflicted to the cells was only moderate (15, 54). In the present study, we observed that at 25°C, cdc13-1 mec3Δ and yku70Δ mec3Δ cells grew at the same rate as cdc13-1 MEC3+ and yku70Δ MEC3+ cells, respectively (data not shown). In contrast, growth of cdc13-1 yku70Δ mec3Δ cells at 25°C was much improved compared to that of cdc13-1 yku70Δ MEC3+ cells (Fig. 1). These observations suggest that the combination of the cdc13-1 and yku70Δ mutations results in an increase in DNA damage, most probably in the form of abnormally high levels of single-stranded DNA (42). Evidence that this DNA damage exists and that the cdc13-1 yku70Δ cells respond to it comes from the experiments in which the DNA damage checkpoint had been inactivated, as explained above.
Next, we set out to describe the slow-growth phenotype of the cdc13-1 yku70Δ strain and the morphological events associated with entry into senescence and survival. In addition, because survivors of the cdc13-1 yku70Δ strain were generated in the presence of functional telomerase, we wished to compare these events with those taking place in telomerase-negative cells. To help analyze the morphological defects of these strains, wild-type, cdc13-1, yku70Δ, and cdc13-1 yku70Δ mec3Δ cells were also observed (Fig. 1B). Immediately after germination, cells of the cdc13-1 yku70Δ double mutant grew slowly. This was due to a slowing down of the cell cycle rather than to a high rate of death (Fig. 1B, bottom row, leftmost panel). The morphology of the cdc13-1 yku70Δ cells during the presenescence period (i.e., very big and large-budded cells) was typical of that of cells delayed in G2/M phase due to the activation of the DNA damage checkpoint. In cdc13-1 yku70Δ mec3Δ cells, as explained above, slowing down of the cell cycle in these cells was indeed suppressed following inactivation of the checkpoint (Fig. 1B, top row, rightmost panel). The cdc13-1 yku70Δ cells retained the same morphology during the first two or three passages. However, after the third and fourth dilutions, smaller cells emerged and (because they were dividing faster) started to overcome the entire population of slow-growing cells (Fig. 1B, bottom row). After the fifth or sixth dilution, cells of cdc13-1 yku70Δ that had adopted this normal growth displayed events of homologous recombination at their telomeres (see below). Meanwhile, from the time the mutation was introduced until after the third dilution, tlc1Δ cells exhibited morphology close to that of wild-type cells (Fig. 1B, middle row). This was in sharp contrast with the situation in senescing cdc13-1 yku70Δ cells, as seen above. After the fourth dilution, tlc1Δ cells started to enlarge (Fig. 1B, middle row), concomitant with the acquisition of slow growth. These defects became even more severe after the sixth dilution (a period during senescence known as the crisis) (Fig. 1B, middle row), in agreement with the previously described events of senescence in telomerase-negative cells (32, 33). As expected, during a subsequent period of slow growth after the sixth and seventh dilutions, fast-growing cells (survivors) emerged and overcame the population of slow-growing and arrested cells (Fig. 1B, middle row). This latter situation corresponded to the emergence of colonies growing faster than the bulk of senescing cells on agar plates (Fig. 2, rows 2 and 3). These survivors now exhibited events of telomeric homologous recombination (see below).
Importantly, telomere length in cdc13-1 yku70Δ cells was comparable with that in yku70Δ cells (Fig. 3, left panel). This observation eliminates the possibility that a further reduction in telomere length (yku70Δ cells have shorter telomeres than wild-type cells), like that taking place in telomerase-negative cells, is responsible for the senescence phenotype of cdc13-1 yku70Δ cells.
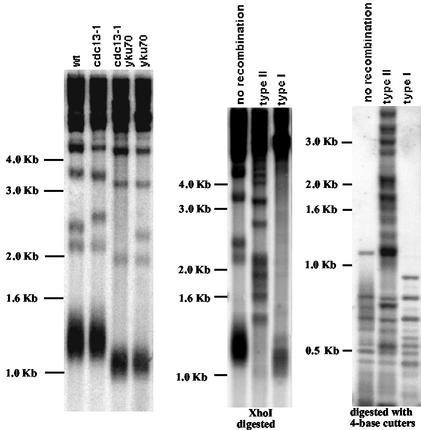
FIG. 3.
Telomere structure in cdc13-1 yku70Δ cells. Left panel: Southern blot of cdc13-1, yku70Δ, cdc13-1 yku70Δ, and wild-type (wt) cells, showing the mean size of telomeres following growth at 25°C (prior to recombination for cdc13-1 yku70Δ). Genomic DNA from each culture was digested with XhoI, and the resulting Southern blot was analyzed with a 270-bp TG1-3 32P-labeled probe. Telomere tracts of wild-type cells appear as a broad band of 1.1 to 1.3 kb. For non-Y′ chromosomes, XhoI cutting typically generates fragments migrating at ∼2.1, 2.3, 3.4, and 4.2 kb. Middle and right panels: telomeres of telomerase-negative (tlc1Δ) survivors show two distinct patterns (see, for instance, reference 48). Genomic DNAs from wild-type (no recombination), type I, and type II senescing tlc1Δ cells were digested either with XhoI (middle panel) or with a mixture of four restriction enzymes (AluI, HaeIII, HinfI, and MspI) that used a 4-bp recognition sequence (right panel), and the Southern blots were revealed with a TG1-3 probe. Since the four-base cutters cut within telomeric Y′ sequences but not within the TG1-3 sequences (48), the patterns they generated allowed us to distinguish between type II survivors, which amplify the TG1-3 sequences (right panel, lane 2), and type I survivors, which amplify the subtelomeric Y′ sequences (right panel, lane 3). Moreover, XhoI cutting evidences the erosion of telomeres in type I survivors (middle panel, lane 3) compared to wild-type cells (middle panel, lane 1) as well as the very long and heterogeneous TG1-3 sequences in type II survivors (middle panel, lane 2) compared to the shorter TG1-3 sequences in wild-type cells and type I survivors (middle panel, lanes 1 and 3). Individual colonies were picked out from agar plates (incubated at 29°C) during the sixth passage following introduction of the tlc1Δ mutation.
Previous data have established that although they exhibited no morphological defect at 25°C, cdc13-1 cells nevertheless already harbored a defect at this temperature. At 25°C, for instance, the cdc13-1 mutation effected the selection of survivor type in a tlc1Δ background (17). In our strain background, a growth temperature of 25°C for cdc13-1 cells appears to be equivalent to 23°C for other strain backgrounds (38, 42). To better document the properties of the cdc13-1 allele, we grew the cdc13-1 yku70Δ strain at 20°C; in contrast to the situation at 25°C (Fig. 1A), colonies formed at this temperature were the same size as those from each of the single-mutant and wild-type strains (data not shown). To determine the presence or absence of postsenescence survivors in the cdc13-1 yku70Δ strain at 20°C, several colonies were picked out at 2-day intervals during 3 weeks and analyzed for telomere structure. In parallel, liquid cultures of the cdc13-1 yku70Δ strain, grown at 20°C, were harvested every 2 days for 3 weeks for telomere structure analysis. In accordance with classical criteria (Fig. 3), we established that after growth at 20°C, no cdc13-1 yku70Δ cell exhibited recombining telomeres (data not shown). Therefore, cdc13-1 yku70Δ cells undergo senescence or survival at 25°C but not at 20°C (Table 1). To confirm that cdc13-1 was functional at 20°C, cdc13-1 tlc1Δ double-mutant cells were grown on plates at 20°C until senescence took place and analyzed for determination of the survival type. As expected, all eight analyzed survivor clones were of type I (data not shown). Since in cdc13-1 tlc1Δ cells grown at 25°C cdc13-1 causes 100% of the survivors to become type II (instead of the ∼90% type I in CDC13+ tlc1Δ cells) (17), these data further establish that cdc13-1 cells have no apparent telomeric defect at 20°C (Table 1).
TABLE 1.
Senescence and survival of cdc13-1 cells at various temperatures
| Strain | Type of clone produced or outcome at:
|
|||
|---|---|---|---|---|
| 20°C | 25°C | 29°C | 32°C | |
| cdc13-1 | Wild-type telomeres | Wild-type telomeres | Death | |
| cdc13-1 yku70 | Wild-type telomeres | Type II recombination | Death | |
| cdc13-1 mec3 | Wild-type telomeres | Wild-type telomeres | Type II recombination | Death |
| cdc13-1 tlc 1 | Type I recombination | Type II recombination | Death | |
In conclusion, cdc13-1 yku70Δ cells enter senescence and generate postsenescence survival (at 25 but not at 20°C) in the presence of functional telomerase, just as telomerase-negative cells do. However, analysis of the morphology and kinetics of senescence in both situations suggests that senescence in cdc13-1 yku70Δ cells is due to a different kind of DNA damage than that generated in telomerase-negative cells (see Discussion).
Survivors among cdc13-1 yku70Δ and yku70Δ tlc1Δ cells exclusively amplify the telomeric TG1-3 repeats (type II recombination).
Fast-growing colonies of senescing cdc13-1 yku70Δ cells (like those shown in Fig. 2, row 3) were picked out and grown on plates for additional generations. According to classical criteria of telomere structure analysis (Fig. 3), all of the 20 tested survivors were found to generate type II recombination (Fig. 4A), a Rad50/Rad59-dependent mechanism known to amplify the telomeric TG1-3 repeats (4, 29, 32, 48). The presence of high-molecular-weight bands following digestion with the four-base cutters (Fig. 3, right panel, and Fig. 4A, right panel) undoubtedly attests to the occurrence of type II recombination (48, 51). Since each of our DNA samples resulted from a single clone obtained from an agar plate, the type II survivors observed here could not have resulted from a conversion from type I such as is sometimes observed in heterogeneous liquid cultures (see Materials and Methods).
We noticed that in the presence of the cdc13-1 or yku70Δ mutations these bands tended to form heterogeneous smears (Fig. 4A and B) rather than a series of discrete bands as in type II tlc1Δ cells (Fig. 3, right panel). Since abnormally high levels of single-stranded DNA can be detected in cdc13-1 and yku70Δ mutants (15, 18, 36, 42, 49), we therefore asked ourselves whether this might affect digestion and/or migration of our DNA samples. To control for such possible problems, gels containing DNA samples were stained with ethidium bromide prior to transfer to a nitrocellulose membrane. The obtained images clearly showed that DNA digestion with the four-base cutters was complete and, in addition, that gels had been consistently loaded with similar amounts of DNA (Fig. 4C, left panel). As an additional control, DNA samples digested with the four-base cutters were probed with a Y′ probe. Under such conditions, the high-molecular-weight bands were no longer detectable (Fig. 4C, right panel), thus clearly showing that the bands that were detected with the TG1-3 probe indeed represent TG1-3 sequences (Fig. 4C, middle panel). We conclude that the heterogeneous smears sometimes visible in the Southern blotting analysis did not result from incomplete digestion due to the presence of single-stranded DNA in telomeric regions. However, it is very probable that cdc13-1 yku70Δ cells contain higher amounts of single-stranded DNA than wild-type cells, even at 25°C, as explained above. We propose that this renders recombining telomeres more unstable, thus generating recombination at a higher frequency than in tlc1Δ cells. Such telomeres would tend to frequently change size, thus explaining the tendency to form heterogeneous smears. This hypothesis might also explain the variation in the patterns of recombination from one clone to the other (Fig. 4A and B).
The results above demonstrated that cdc13-1 yku70Δ cells generated 100% type II survivors. However, the fact that senescing checkpoint-deficient cdc13-1 cells also generate 100% type II survivors (17) prevented us from drawing further conclusions at this point concerning the nature of the yku70Δ-induced defect. Since in most strain backgrounds studied to date (4, 6, 23, 32, 47), including ours (17), telomerase-negative cells (est1Δ, est2Δ, or tlc1Δ) generate a majority of type I survivors (one of which is shown in Fig. 3) relying on amplification by a Rad51-based pathway of the subtelomeric Y′ sequences (29, 32, 48), we next analyzed the behavior of senescing yku70Δ tlc1Δ cells. As reported previously (38), these cells exhibited accelerated senescence. Senescence was so rapid that sporulation of a yku70Δ/YKU70+ tlc1Δ/TLC1+ diploid generated yku70Δ tlc1Δ colonies (Fig. 2, row 11) containing cells with already recombinant telomeres (Fig. 4B). Most importantly, these surviving cells exhibited 100% (18 out of the 18 tested survivors) type II telomeric recombination (Fig. 4B). For comparison, YKU70+ tlc1Δ senescing cells generated a large majority of type I recombinants (over 90%; 24 out of 26 tested survivors), as previously reported concerning the genetic background used in the laboratory of Grandin et al. (17). Such a proportion was similar to that seen in other strain backgrounds (29, 32, 48), thus demonstrating that our strain background did not exhibit an unusual preference for type II recombination.
We conclude that the absence of Yku70 in telomerase-negative cells leads to a complete reversal of the proportions of type I and type II recombination among survivors, type II being shifted from less than 10% in YKU70+ tlc1Δ cells to 100% in yku70Δ tlc1Δ cells. The yku70Δ-induced predominance of type II recombination over type I recombination is reminiscent of that previously observed in the presence of the cdc13-1 mutation (17).
Type II recombination in cdc13-1 yku70Δ, yku70Δ tlc1Δ, or cdc13-1 tlc1Δ mutants can become Rad50/Rad59 independent.
To check whether type II recombination in cdc13-1 yku70Δ cells utilized the usual Rad50/Rad59-dependent pathway (4, 29, 47, 48), we constructed the cdc13-1 yku70Δ rad50Δ and cdc13-1 yku70Δ rad59Δ triple mutants and, as a control, the cdc13-1 yku70Δ rad51Δ strain. Much to our surprise, type II recombination still took place in cdc13-1 yku70Δ rad50Δ and cdc13-1 yku70Δ rad59Δ cells (Fig. 5A, left two panels, and Fig. 2, rows 4 to 6). These results demonstrate the possibility of Rad50/Rad59-independent type II telomeric recombination. As a control, we determined the survivor type in various tlc1Δ mutants. In agreement with previous experiments (4, 29, 47, 48), we found that tlc1Δ rad50Δ and tlc1Δ rad59Δ cells generated 100% type I survivors (20 out of 20 and 10 out of 10 tested survivors, respectively), while tlc1Δ rad51Δ cells generated 100% type II survivors (20 out of the 20 tested survivors). We therefore conclude that, atypically, type II recombination (in the presence of the cdc13-1 and yku70Δ mutations) can proceed even when the Rad50/Rad59 pathway, previously reported to be essential for this type of recombination, has been genetically inactivated.
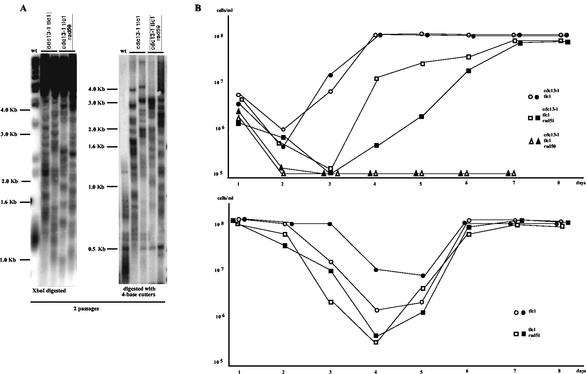
FIG. 5.
yku70Δ-induced type II recombination can be independent of the Rad50/Rad59 pathway. (A) cdc13-1 yku70Δ can either use the Rad50/Rad59 or Rad51 pathway to accomplish recombination on TG1-3 sequences and associated survival. However, operation of the Rad50/Rad59 pathway is delayed in this strain. First and second panels: two representative early survivors among cdc13-1 yku70Δ (second and third lanes from the left in both panels), cdc13-1 yku70Δ rad50Δ (fourth and fifth lanes in both panels), cdc13-1 yku70Δ rad51Δ (sixth and seventh lanes in both panels), and cdc13-1 yku70Δ rad59Δ (eighth and ninth lanes in both panels) strains are shown here, following digestion of genomic DNAs with XhoI (first panel) or a mixture of AluI, HaeIII, HinfI, and MspI (4-base cutters; second panel). Note that all of these strains had generated survivors by ∼100 generations following mixing of the mutations, with the noticeable exception of the cdc13-1 yku70Δ rad51Δ strain (all samples shown here are from ∼125-generation-old survivors). For that strain, cells had not generated survivors at that time but rather continued to proliferate slowly without showing signs of recombination, as attested by their pattern of telomere structure (sixth and seventh lanes [labeled “delayed in recombination”]) resembling that in nonrecombining wild-type cells (lanes 1). Third and fourth panels: however, after eight passages (∼100 generations later than cdc13-1 yku70Δ rad59Δ and a total of ∼200 generations following mixing of the mutations), cells of the cdc13-1 yku70Δ rad51Δ strain recombined (second and third lanes from left in both panels [labeled “delayed appearance of recombination”], representing the same samples as those shown in the sixth and seventh lanes in the first and second panels) and generated type II survivors also, just like the cdc13-1 yku70Δ rad50Δ cells taken at the same time (fourth and fifth lanes in both panels, representing the same samples as those shown in the fourth and fifth lanes in the first and second panels). Nonrecombining wild-type cells are shown for comparison (wt). All strains were grown at 25°C. Genomic DNAs were digested with XhoI (third panel) or a mixture of AluI, HaeIII, HinfI, and MspI (4-base cutters; fourth panel). A TG1-3 32P-labeled probe was used. (B) yku70Δ tlc1Δ can also undergo Rad59- or Rad51-dependent type II recombination. However, operation of the Rad50/Rad59 pathway is not delayed in the absence of Yku70. Left and middle panels: four representative early survivors among yku70Δ tlc1Δ (second to fifth lanes from the left in both panels), yku70Δ tlc1Δ rad51Δ (left panel, sixth to ninth lanes), and yku70Δ tlc1Δ rad59Δ (right panel, sixth to ninth lanes) strains are shown here, following digestion of genomic DNAs with XhoI. Right panel: Southern blot of the same strains as shown in the left and middle panels as indicated, following digestion with a mixture of AluI, HaeIII, HinfI, and MspI (4-base cutters). A TG1-3 32P-labeled probe was used. Survivors from the three strains shown here (growth at 25°C) were picked out during the second passage.
Similar observations were made using a yku70Δ tlc1Δ rad59Δ strain (yku70Δ tlc1Δ rad50Δ was totally inviable) (Fig. 5B; Fig. 2, rows 13 and 14), thus demonstrating that the yku70Δ mutation alone can induce this atypical Rad50/Rad59-independent type II recombination. Previous work has shown that type II survivors are generated at a low frequency in tlc1Δ rad50Δ (but not in tlc1Δ rad59Δ) but cannot be maintained during subsequent growth (4). We therefore determined the status of recombination in cdc13-1 yku70Δ rad50Δ cells after 200 generations. Southern analysis showed that, contrary to that in tlc1Δ rad50Δ cells (4), Rad50-independent type II recombination in cdc13-1 yku70Δ rad50Δ cells is stable over time (Fig. 5A, right two panels). Since in our strain background (data not shown), as in other strain backgrounds (4, 47), tlc1Δ rad50Δ rad51Δ cells were unable to generate any survivors, the present data suggest that this atypical Rad50/Rad59-independent type II recombination relies on the Rad51 pathway.
Next, we wished to determine whether the exclusive type II survivors encountered in cdc13-1 tlc1Δ (17) could, like the yku70Δ-induced type II survivors, be generated when the Rad50/Rad59 pathway was inactivated. To this end, we constructed cdc13-1 tlc1Δ rad59Δ mutants (cdc13-1 tlc1Δ rad50Δ was totally inviable), grew them at 25°C (a permissive temperature of growth for cdc13-1) until senescence took place, and looked for possible postsenescence survivors. Interestingly, this strain did generate survivors (Fig. 2, rows 9 and 10), which were all of type II and appeared at a rate similar to that found in cdc13-1 tlc1Δ RAD59+ cells (Fig. 6A). The absence of postsenescence survivors in cdc13-1 tlc1Δ rad50Δ mutants as noted previously (17) is presumably due to a presenescence lethal genetic interaction between cdc13-1 and tlc1Δ in the presence of the rad50Δ mutation, similar to that observed between yku70Δ and tlc1Δ in yku70Δ tlc1Δ rad50Δ cells, as seen above. The rad50Δ mutation generates a considerable amount of DNA damage (7) which, when combined with the cdc13-1 or yku70Δ mutation, appears to provoke presenescence lethality. Although Rad50 and Rad59 are in the same pathway of recombination, Rad50 has a role in loading telomerase and Cdc13 on nonrecombining telomeres (references 9 and 52 and references therein) that Rad59 does not have. This probably explains the viability of cdc13-1 (yku70Δ) tlc1Δ rad59Δ and, in contrast, the absence of viability of cdc13-1 (yku70Δ) tlc1Δ rad50Δ.
In conclusion, the data shown above establish the important finding that even in YKU70+ cells at a temperature permissive for growth, recombination on TG1-3 sequences can be independent of the Rad50/Rad59 pathway when the cdc13-1 mutation is present. The fact that tlc1Δ rad50Δ rad51Δ cells are unable to generate any survivor at all, as seen above, strongly supports the assumption that the type II recombination taking place in the cdc13-1 and yku70Δ mutants in which the Rad50/Rad59 pathway has been inactivated relies on the Rad51 pathway.
The absence of Yku70 as well as the cdc13-1 mutation introduces a delay in Rad50/Rad59-based recombination on TG1-3 sequences.
Next, we analyzed the phenotype of the cdc13-1 yku70Δ rad51Δ strain. In this strain, type II recombination (100% of the events) took place, as expected. Most surprisingly, however, postsenescence survivors arising from the cdc13-1 yku70Δ rad51Δ strain appeared with a delay compared with those from the cdc13-1 yku70Δ rad50Δ and cdc13-1 yku70Δ rad59Δ strains (Fig. 5A, first and second panels, lanes 6 and 7, and right two panels; Fig. 2, row 4). This rad51Δ-induced delay was substantial, as survivors from cdc13-1 yku70Δ rad51Δ cells emerged at the 8th to 10th passages (200 to 250 generations, depending on the tested clones) following germination, while for cdc13-1 yku70Δ RAD51+ cells, survivors emerged between the 3rd and 4th passages (Fig. 2, rows 3 and 4). During the delay in the appearance of postsenescence survivors observed in cdc13-1 yku70Δ rad51Δ, cells grew slowly, like the cdc13-1 yku70Δ cells (data not shown), but displayed substantially elongated telomeres (Fig. 5A, first panel; compare lanes 6 and 7 to lane 1 with respect to wild-type telomere size). The significance of this event is presently unknown. Eventually, survivors (100% type II) emerged from the cdc13-1 yku70Δ rad51Δ population. Once reached, type II recombination in cdc13-1 yku70Δ rad51Δ was indistinguishable from that in cdc13-1 yku70Δ rad50Δ (Fig. 5A, right two panels).
To better document the rad51Δ-induced delay in recombination described above, we then measured the occurrence of postsenescence survival for various cdc13-1 mutants. As reported previously, 100% of the survivors were of type II (data not shown), due to the predominance of the cdc13-1 mutation over the tlc1Δ mutation during amplification of the TG1-3 sequences (17). Postsenescence survivors arising from the cdc13-1 tlc1Δ rad51Δ strain appeared with a delay compared with those in the cdc13-1 tlc1Δ RAD51+ strain. The delay, smaller than that for the cdc13-1 yku70Δ rad51Δ strain (Fig. 5A), was seen best in liquid cultures (Fig. 6B, upper panel). Meanwhile, cdc13-1 yku70Δ rad50Δ cells, which were used as the negative control, did not generate any postsenescence survivors, as reported previously (17). As an important control for the two sets of experiments involving the rad51Δ-induced delay in the generation of postsenescence survivors in cdc13-1 and yku70Δ mutants, we monitored the kinetics of survival onset in tlc1Δ rad51Δ mutants. Interestingly, we found that the tlc1Δ rad51Δ and tlc1Δ strains entered senescence at the same time and also generated postsenescence survivors at the same time (Fig. 6B, lower panel). These data therefore suggest that Rad51 has no important role in initiating Rad50/Rad59-dependent recombination, at least in CDC13+ YKU70+ cells, and that the rad51Δ-induced delay described above is encountered only in the cdc13-1 and yku70Δ mutants.
Unlike cells of the cdc13-1 yku70Δ rad51Δ strain, which generated survivors later than those of the cdc13-1 yku70Δ RAD51+ strain (as seen above), cells of the yku70Δ tlc1Δ rad51Δ and yku70Δ tlc1Δ RAD51+ strains recombined at the same time (Fig. 5B). Absence of a rad51Δ-induced delay in the tlc1Δ background might be due to tlc1Δ-induced telomere uncapping being more drastic than that induced by cdc13-1. Indeed, yku70Δ tlc1Δ cells entered senescence immediately (in ∼5 to 10 generations versus ∼100 generations in cdc13-1 yku70Δ cells) after mixing of the two mutations (Fig. 2, rows 11 to 13). Under such conditions, cells either rapidly enter recombination and survival prior to lethal telomere erosion, thus masking the rad51Δ-induced delay observed when telomere uncapping is less drastic (e.g., in cdc13-1), or die.
DISCUSSION
Loss of telomerase activity in yeast cells leads to a phenotype of slow death known as senescence (33). Telomerase-negative cells can escape death from senescence by amplifying (using Rad50/Rad59-dependent homologous recombination) TG1-3 sequences or by amplifying (using a Rad51-dependent pathway of recombination) subtelomeric Y′ sequences (4, 29, 32, 47, 48). For the present study, we report that postsenescence survivors can be generated in cdc13-1 yku70Δ cells, even in the presence of functional telomerase. The present work also demonstrated that the Rad51 pathway can operate recombination on TG1-3 sequences in cdc13-1 and yku70Δ mutants when the Rad50/Rad59 pathway has been inactivated.
The emergence of survivors in cdc13-1 yku70Δ does not involve a classical series of crisis-senescence events.
Increased DNA damage following a combination of cdc13-1 and yku70Δ mutations has been suspected on the basis of a synthetic interaction between the two (38, 42). Here, we also observed improved growth of the double mutant, but not of any of the single mutants, in a mec3Δ background (Fig. 1A). Such behavior (as already observed for cdc13-1 cells in a rad9Δ background [15]) attests that removal of the recognition of the damage by the DNA damage checkpoint also relieves the associated slowing down of the cell cycle. Rad17 and Rad24 have been implicated in generating single-stranded DNA in cdc13-1 cells (35). It is therefore possible that inactivation of Mec3 (which functions in cooperation with Rad17 and Rad24) in cdc13-1 yku70Δ cells acts by diminishing the damage, thus amplifying the effect on cell cycle progression.
In cdc13-1 mec3Δ cells, which also underwent senescence at 29°C (17), genetic inactivation of the DNA damage checkpoint (and thus the absence of an appropriate G2/M delay) may have prevented repair of the damage, as postulated long ago (53) and verified since on many occasions. Because the DNA damage checkpoint was intact in the senescing cdc13-1 yku70Δ cells, we assume that less damage accumulated in these cells as they divided than in cdc13-1 mec3Δ cells. Indeed, the presence of functional checkpoint proteins, including Mec3, has been shown to activate DNA repair (1). The constant morphological defect of the cdc13-1 yku70Δ cells from the moment the mutations are mixed together until survivors are generated (Fig. 1B) does not appear to us to be a sensitive enough criterion for the monitoring of DNA damage accumulation. Indeed, cdc13-1 mec3Δ cells retained a wild-type morphology during senescence but nevertheless did accumulate DNA damage (17). In addition, we rule out the possibility that these events reflect a sort of adaptation to the damage present from the beginning. Indeed, adaptation to DNA damage corresponds to a modulation of cell cycle events downstream of the arrest caused by the damage (30, 50). Therefore, adaptation does not appear to augment DNA repair but rather changes the sensitivity of one or several cell cycle engines to the inflicted damage (30, 50). In contrast, what we see here in presenescence cdc13-1 yku70Δ cells is the initiation of mechanisms of homologous recombination that repair the telomeric DNA damage. Presumably, the appearance of fast-growing cells is a consequence of this repair. Although the exact reason for the delay existing before the damage is actually repaired has not been determined with certainty yet, it is probable that DNA damage must accumulate to some extent before it becomes the signal for recombinational repair.
Finally, it is to be noted that there was an absence of a detectable crisis in senescing cdc13-1 yku70Δ cells (Fig. 1B). This was unlike the classical crisis of telomerase-negative cells (32, 33). This difference is probably due to the kinetics of accumulation of DNA damage during senescence being different in these strains. It would be most interesting to compare the implications of the various levels of checkpoint machinery in these two different pathways of senescence.
Both Yku70 and Cdc13 play a role in telomere end protection and telomere replication.
What are the known properties of Yku70 and Cdc13 at the molecular level that are relevant to the behavior of the yku70Δ and cdc13-1 mutants revealed in the present study? Both Cdc13 and the Yku complex occupy a key position near the telomere ends (18, 31, 39). Cdc13 was found to bind the single-stranded telomeric DNA extension (31, 39). Cdc13's DNA binding domain was found to be essential (24). Although the Cdc13-1 mutant protein had no apparent defect in binding single-strand telomeric DNA at restrictive temperatures (24), the cdc13-1 mutation had a dramatic impact on the structure of telomeric DNA nevertheless (15). In addition, the Cdc13-1 mutant protein was defective in associating with Stn1 and Ten1, its partners in telomere end protection, at least in a two-hybrid system (16). On the other hand, at the junction between single-strand and double-strand telomeric DNA, Yku binding was not essential (18). The generation of abnormally high levels of single-stranded DNA in cdc13-1 and yku cells is thought to represent the signal for cell cycle arrest (15, 18). Intriguingly, the presence of single-stranded DNA in yku70Δ or yku80Δ did not always correlate with their temperature-sensitive growth defects (36, 49). In addition, the size of the double-stranded telomeric DNA tracts in yku70 mutants appeared to be a determinant for the response to single-stranded DNA as representing damage (19).
In addition to its role in telomere end protection, Cdc13 has a distinct role in recruiting telomerase (12, 39; reviewed in reference 34). Est1, another single-strand telomeric DNA binding protein, plays an essential role in mediating binding between telomerase and Cdc13 (12). Moreover, cooperation between Cdc13 and Est1 after the latter has attached to the telomeres in late S phase appears to be essential for the conversion of inactive telomere-bound telomerase into its active form (46). Cdc13 appears to alternate its function of protection with that of recruitment of telomerase by alternating physical interactions with Est1 and Stn1 (41). Although less documented than that of Cdc13, Yku70 also has an apparent role in telomerase recruitment (reviewed in reference 11).
Obviously, the roles of Cdc13 and Yku70 in telomere end protection seem to be more relevant to the properties of the mutants described here than their roles in telomere replication. Globally, on the basis of the phenotypes at restrictive temperatures of the various cdc13 and yku mutants, the functions of Cdc13 are more crucial for telomere maintenance than those of Yku70.
The role of Yku70 and Cdc13 in preventing telomere recombination.
More specifically, both Yku70 (this study) and Cdc13 (17; this study) protect against the inappropriate intervention of homologous recombination at the telomeres. Events of recombination at the telomeres have been described for ykuΔ mutants; however, none of these events appeared to be drastic enough to generate postsenescence survivors (14, 42). Moreover, senescence (but not recombination-based survival) can be generated by a ykuΔ mutation associated with the cdc13-1, rad50Δ, or mre11Δ mutation (10, 36, 38, 42). The generation of postsenescence survivors in cdc13-1 yku70Δ cells described here took place in the presence of functional telomerase, thus confirming the previous finding that telomere end protection not only depends on telomere length but also on properties independent of telomere length (17). Several other conclusions derived from the present work. There are probably several models that can be proposed to interpret these conclusions, among which our preferred one (Fig. 7) is explained below.
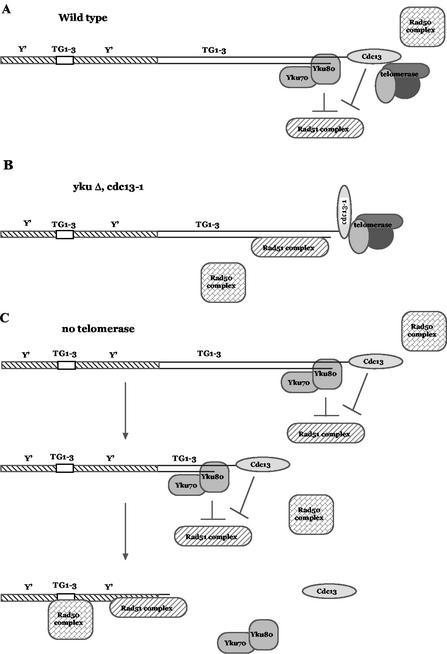
FIG. 7.
Model for the role of Yku and Cdc13 in selecting the type of telomere repair by homologous recombination. (A) In wild-type cells, protection of the substrate for recombination-dependent telomere replication involves the actors schematically represented here. Cdc13, which binds the 3′ overhang, both protects telomere ends and participates in telomerase recruitment, while the Yku70/Yku80 complex, which binds double-stranded DNA near the junction with the 3′ overhang, protects telomere ends and also appears to play a role in telomerase-mediated telomere replication. Telomerase (composed of Est2, the catalytic subunit, and TLC1, the RNA template) is represented here together with Est1, a telomerase-regulatory protein (reviewed in references 2, 11, and 34). The two pathways of recombination-dependent maintenance of telomeres, the Rad50- and Rad51-dependent machineries (reviewed in reference 27), are also schematically represented. Subtelomeric DNA, on the left, comprises the Y′ elements (hatched zone) with dispersed short stretches of TG1-3 sequences (small open rectangle). Telomeric DNA (the two parallel solid lines), which is exclusively composed of TG1-3 sequences, ends with a single-stranded 3′ overhang. (B) In the absence of protection by either Yku (yku70Δ cells) or Cdc13 (cdc13-1ts cells grown at 25°C), unmasking of telomere sequences creates unprotected single-stranded TG1-3 sequences (15, 18, 36) which become a substrate for type II recombination. Atypically, this type II recombination can now be performed by the Rad51-dependent machinery but also by the Rad50-dependent machinery in a less efficient way. The rad51Δ-induced delay in recombination observed in these mutants suggests either a role for Yku and Cdc13 in recruiting Rad50 or a higher affinity of the unmasked TG1-3 sequences for Rad51 than for Rad50. Such events can occur whether telomerase is present or absent. (C) Top and middle panels: in telomerase-negative CDC13+ YKU+ cells, Yku and Cdc13 remain attached to the terminal TG1-3 sequences and protect them (during telomere shortening resulting from the absence of telomerase) until telomere erosion reaches the subtelomeric Y′ sequences. At this time (bottom panel), the Rad51-dependent machinery operates recombination on the Y′ sequences (∼90% of recombination events) but no longer has the (missing) terminal TG1-3 sequences for the substrate. Meanwhile, the Rad50-dependent machinery operates recombination on the more internal, shorter TG1-3 sequences (∼10% of recombination events) which exhibit too little homology between them to be processed by the Rad51-dependent machinery (25).
First, based on the occurrence of 100% type II recombination in cdc13-1 yku70Δ and yku70Δ tlc1Δ cells, one can conclude that Yku70 plays a role in the protection of the terminal TG1-3 sequences (Fig. 7A). Previous data (confirmed here) have established that Cdc13 also plays such a role (17). Using a different strategy, a recent study has shown that type I survivors relied more on Yku70 for their viability than type II survivors (51). As both the cdc13-1 (15, 36) and ykuΔ (18, 36, 42, 49) mutants generate higher levels of single-stranded TG1-3 sequences than wild-type cells, we infer that these sequences represent the likely substrate for type II recombination. We further infer that in yku70Δ tlc1Δ as well as in cdc13-1 tlc1Δ cells, a terminal TG1-3 substrate is exposed before telomeres have been shortened enough to expose the subtelomeric Y′ sequences (Fig. 7A). This might explain the exclusive occurrence of type II recombination in these cells.
Second, atypically, the Rad51 pathway amplified the telomeric TG1-3 sequences (type II recombination) in cdc13-1 and yku70Δ mutants (Fig. 5 and 6A). This was atypical because telomerase-negative rad50Δ (or rad59Δ) cells have been shown to generate 100% type I survivors (4, 29, 47, 48; this study). Since Rad50/Rad59-independent type II recombination was encountered in both telomerase-negative and -positive cells (Fig. 5), it was most likely associated with the presence of either the cdc13-1 or the yku70Δ mutation. This strongly argues that in wild-type cells, both Cdc13 and Yku70 determine the selection of the recombinational pathway. They might do so by hindering access of Rad51 to these sequences (Fig. 7A). Removal of the protective action of Yku or of Cdc13 might thus allow Rad51 to reach the unprotected telomeres (Fig. 7B). If the Rad50/Rad59 pathway were then inactivated, as in the experiments shown here, the Rad51 pathway would remain the only way to amplify the damaged sequences. In fact, type II recombination in cdc13-1 and yku70Δ mutants can be either Rad51 or Rad50/Rad59 based (Fig. 5 and 6A), a conclusion based on genetic data establishing that tlc1Δ rad50Δ rad51Δ triple mutants were unable to generate any survivors (29, 47; this study).
The third important conclusion was that the Rad51 pathway was more efficient than the Rad50/Rad59 pathway in amplifying the TG1-3 sequences in cdc13-1 yku70Δ and cdc13-1 tlc1Δ cells (Fig. 5A and 6B). On the other hand, in tlc1Δ rad51Δ CDC13+ YKU70+ cells, type II recombination operating with the Rad50/Rad59 pathway alone appeared to be as effective as the bulk of recombination (type I and type II) operating with both pathways (Fig. 6B). Two possibilities arise to explain the rad51Δ-induced delay in the appearance of postsenescence survivors in cdc13-1 and yku70Δ mutants. First, this delay can be interpreted as reflecting a role for Yku and Cdc13 in efficiently recruiting Rad50/Rad59 at damaged telomeres. In support for this hypothesis, recent in vitro assays have demonstrated that the binding of Yku to a double-strand break not only protected it from further degradation but also stimulated intermolecular joining by the Rad50 complex (5). Recombinational repair of broken telomeres during telomerase-independent telomere maintenance might also use mechanisms of Rad50 recruitment similar to those used at other places in the genome. Second, this rad51Δ-induced delay might indicate a potential role for Rad51 in an initial step of Rad50/Rad59-dependent type II recombination. Clearly, Rad51 has no such role in tlc1Δ CDC13+ YKU70+ cells, as seen above. However, single-stranded DNA sequences becoming exposed in cdc13-1 or yku70Δ cells might represent a better recombination substrate for Rad51 than for Rad50. This assumption is supported by the previous observation that at chromosomal double-strand breaks other than telomeric, the Rad51 pathway was more efficient at performing break-induced replication (the supposed repair pathway at telomeres) than the Rad50/Rad59 pathway (45).
An important, still unanswered, question is that of which one of these two pathways operates when both are present in the cell. We propose that in cdc13-1 and yku70Δ mutant cells, Rad51 represents the predominant pathway of recombination on the TG1-3 sequences, as suggested by its higher efficiency (Fig. 7B). In telomerase-negative cells, on the other hand, the terminal TG1-3 sequences would remain capped, mainly by Cdc13 and Yku, until the telomeres become eroded so as to reach the subtelomeric Y′ sequences (Fig. 7C). The exposed Y′ sequences would then become a majority substrate for the Rad51 machinery, as has been known for a long time (32, 48). Meanwhile, protection of the 3′ overhang by Yku and Cdc13 would be maintained during telomere erosion and only the TG1-3 sequences located more internally (within the subtelomeric Y′ sequences) would become a substrate for the Rad50/Rad59 machinery (Fig. 7C). These rarer events would correspond to the minority of type II recombination recorded in telomerase-negative cells (4, 29, 32, 47, 48). Very recently, it was proposed that in telomerase-negative cells, Rad51 is probably very inefficient in amplifying the short and irregular TG1-3 sequences (25). However, the large amounts of single-stranded TG1-3 sequences generated in cdc13-1 and yku70Δ mutants (15, 18, 36, 42) might allow sequences to reach around 100 bp, the minimal level of homology required for Rad51 invasion (25). Since the much smaller internal TG1-3 sequences located within the Y′ sequences were too short to be amplified by the Rad51 machinery, they were processed only by the Rad50 machinery, which requires only 30 bp of homology (25).
The present data on Yku and Cdc13 may be of importance not only for our basic knowledge of DNA repair (20, 22) but also for the understanding of telomere functioning in yeast as well as in other eukaryotes, including mammals, in which the Ku complex and homologues of Cdc13 play similar roles in telomere end protection (2).
Acknowledgments
We thank Thomas Petes, James Haber, Dan Gottschling, Leland Hartwell, and Maria-Pia Longhese for the gifts of strains and plasmids as well as Matthias Rose and his colleagues at Euroscarf for efficiently conveying strains.
This work was supported by grants from the Association pour la Recherche contre le Cancer and the Comité Départemental de la Savoie de la Ligue Nationale contre le Cancer.
REFERENCES
- 1.Bashkirov, V. I., J. S. King, E. V. Bashkirova, J. Schmuckli-Maurer, and W.-D. Heyer. 2000. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol. Cell. Biol. 20:4393-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 3.Cervantes, R. B., and V. Lundblad. 2002. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 14:351-356. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, H., and D. A. Sinclair. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 98:3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Amours, D., and S. P. Jackson. 2002. The MRE11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3:317-327. [DOI] [PubMed] [Google Scholar]
- 8.de Lange, T. 2002. Protection of mammalian telomeres. Oncogene 21:532-540. [DOI] [PubMed] [Google Scholar]
- 9.Diede, S. J., and D. E. Gottschling. 2001. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 11:1336-1340. [DOI] [PubMed] [Google Scholar]
- 10.DuBois, M. L., Z. W. Haimberger, M. W. McIntosh, and D. E. Gottschling. 2002. A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161:995-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubrana, K., S. Perrod, and S. M. Gasser. 2001. Turning telomeres on and off. Curr. Opin. Cell Biol. 13:281-289. [DOI] [PubMed] [Google Scholar]
- 12.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 13.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 14.Fellerhoff, B., F. Eckardt-Schupp, and A. A. Friedl. 2000. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics 154:1039-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandin, N., C. Damon, and M. Charbonneau. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20:1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandin, N., C. Damon, and M. Charbonneau. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 20:6127-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gravel, S., M. Larrivée, P. Labrecque, and R. J. Wellinger. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280:741-744. [DOI] [PubMed] [Google Scholar]
- 19.Gravel, S., and R. J. Wellinger. 2002. Maintenance of double-stranded telomeric repeats as the critical determinant for cell viability in yeast cells lacking Ku. Mol. Cell. Biol. 22:2182-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haber, J. E. 2000. Partners and pathways: repairing a double-strand break. Trends Genet. 16:259-264. [DOI] [PubMed] [Google Scholar]
- 21.Henson, J. D., A. A. Neumann, T. R. Yeager, and R. R. Reddel. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21:598-610. [DOI] [PubMed] [Google Scholar]
- 22.Hopfner, K. P., C. D. Putnam, and J. A. Tainer. 2002. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12:115-122. [DOI] [PubMed] [Google Scholar]
- 23.Huang, P. H., F. E. Pryde, D. Lester, R. L. Maddison, R. H. Borts, I. D. Hickson, and E. J. Louis. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11:125-129. [DOI] [PubMed] [Google Scholar]
- 24.Hughes, T. R., R. G. Weilbaecher, M. Walterscheid, and V. Lundblad. 2000. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. USA 97:6457-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ira, G., and J. E. Haber. 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22:6384-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, F. B., R. A. Marciniak, M. McVey, S. A. Stewart, W. C. Hahn, and L. Guarente. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kass-Eisler, A., and C. W. Greider. 2000. Recombination in telomere-length maintenance. Trends Biol. Sci. 25:200-204. [DOI] [PubMed] [Google Scholar]
- 28.Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde, E. J. Louis, and S. M. Gasser. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653-656. [DOI] [PubMed] [Google Scholar]
- 29.Le, S., J. K. Moore, J. E. Haber, and C. W. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 31.Lin, J. J., and V. A. Zakian. 1996. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. USA 93:13760-13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 33.Lundblad, V., and J. W. Szostak. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57:633-643. [DOI] [PubMed] [Google Scholar]
- 34.Lustig, A. J. 2001. Cdc13 subcomplexes regulate multiple telomere functions. Nat. Struct. Biol. 8:297-299. [DOI] [PubMed] [Google Scholar]
- 35.Lydall, D., and T. Weinert. 1995. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270:1488-1491. [DOI] [PubMed] [Google Scholar]
- 36.Maringele, L., and D. Lydall. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16:1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra, K., and D. Shore. 1999. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by Rif proteins. Curr. Biol. 9:1123-1126. [DOI] [PubMed] [Google Scholar]
- 38.Nugent, C. I., G. Bosco, L. O. Ross, S. K. Evans, A. P. Salinger, J. K. Moore, J. E. Haber, and V. Lundblad. 1998. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8:657-660. [DOI] [PubMed] [Google Scholar]
- 39.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 40.Pardue, M. L., and P. G. DeBaryshe. 1999. Telomeres and telomerase: more than the end of the line. Chromosoma 108:73-82. [DOI] [PubMed] [Google Scholar]
- 41.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 42.Polotnianka, R. M., J. Li, and A. J. Lustig. 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8:831-834. [DOI] [PubMed] [Google Scholar]
- 43.Reed, S. I., J. A. Hadwiger, and A. T. Lorincz. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA 82:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shore, D. 2001. Telomeric chromatin: replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11:189-198. [DOI] [PubMed] [Google Scholar]
- 45.Signon, L., A. Malkova, M. L. Naylor, H. Klein, and J. E. Haber. 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taggart, A. K. P., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 47.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif1-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 48.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teo, S. H., and S. P. Jackson. 2001. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 51.Tsai, Y.-L., S.-F. Tseng, S.-H. Chang, C.-C. Lin, and S.-C. Teng. 2002. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 22:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsukamoto, Y., A. K. P. Taggart, and V. A. Zakian. 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11:1328-1335. [DOI] [PubMed] [Google Scholar]
- 53.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 54.Weinert, T. A., G. L. Kiser, and L. H. Hartwell. 1994. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication. Genes Dev. 8:652-665. [DOI] [PubMed] [Google Scholar]