Abstract
Receptor tyrosine kinases (RTKs) direct diverse cellular and developmental responses by stimulating a relatively small number of overlapping signaling pathways. Specificity may be determined by RTK expression patterns or by differential activation of individual signaling pathways. To address this issue we generated knock-in mice in which the extracellular domain of the mouse platelet-derived growth factor alpha receptor (PDGFαR) is fused to the cytosolic domain of Drosophila Torso (αTor) or the mouse fibroblast growth factor receptor 1 (αFR). αTor homozygous embryos exhibit significant rescue of neural crest and angiogenesis defects normally found in PDGFαR-null embryos yet fail to rescue skeletal or extraembryonic defects. This phenotype was associated with the ability of αTor to stimulate the mitogen-activated protein (MAP) kinase pathway to near wild-type levels but failure to completely activate other pathways, such as phosphatidylinositol (PI) 3-kinase. The αFR chimeric receptor fails to rescue any aspect of the PDGFαR-null phenotype. Instead, αFR expression leads to a gain-of-function phenotype highlighted by ectopic bone development. The αFR phenotype was associated with a failure to limit MAP kinase signaling and to engage significant PI3-kinase response. These results suggest that precise regulation of divergent downstream signaling pathways is critical for specification of RTK function.
Evolutionary conservation of a gene requires that it contributes specific functions that enhance the fitness of the organism in which it is expressed. Receptor tyrosine kinases (RTKs) represent a large family of genes that have been conserved due to their ability to control multiple fundamental cellular processes. Upon binding to specific extracellular ligands, activated RTKs regulate cell proliferation, survival, migration, differentiation, and metabolism. The conservation of over 50 RTKs in mammals suggests that each executes critical specific cellular functions. How functional specificity is generated remains a matter of controversy. A vast amount of research has focused on understanding the molecular mechanisms that underlie the ability of these receptors to mediate diverse cellular functions. Surprisingly, studies have shown that even divergent RTKs activate highly redundant array of downstream signaling proteins, such as mitogen-activated protein (MAP) kinases, Src family kinases, phospholipase C (PLC)-γ, and phosphatidylinositol (PI) 3-kinase. Several models have been proposed to explain how RTKs achieve functional specificity at the cellular level while at the molecular level appearing to activate redundant signaling pathways (12, 27).
One model proposes that specific responses to RTKs are a result of differential activation of downstream pathways or biochemical differences in RTK signaling. A great deal of evidence based on experiments performed in cultured cell lines to assess individual contributions of downstream signaling pathways supports this idea (14, 26). Differences in the strength and/or duration of a redundant signaling pathway by two distinct RTKs can translate into specific cellular responses. For example, in PC12 cells, both epidermal growth factor and nerve growth factor (NGF) stimulation results in MAP kinase activation. However, transient and strong activation by epidermal growth factor results in cell proliferation, whereas prolonged and less intense activation by NGF results in cell differentiation (33). More recently, studies assessing the relative contributions of individual downstream signaling pathways in the context of development have also provided support for this model. For example, studies on the development of the Drosophila melanogaster eye have illustrated that in a single population of cells, the decision to differentiate into R1 to R7 cells or R8 cells or to promote cell survival can be controlled by altering the duration of MAP kinase signaling (9). Previous work demonstrated that in the mouse, removal of the platelet-derived growth factor beta receptor (PDGFβR) signaling domain and replacement with the PDGFαR signaling domain resulted in substantial yet incomplete rescue of normal PDGFβR function. The inability of PDGFαR signaling to compensate for loss of PDGFβR signaling correlated with an inability to mediate sustained MAP kinase activation in response to PDGF (17). These studies, taken together, support the idea that precise regulation of specific downstream signaling pathways is critical to ensure the proper cellular outcome.
Another model suggests that RTK signaling is essentially generic and that functional specificity has evolved due to distinct ligand binding capacities and specific patterns of spatiotemporal expression of ligand and receptor. Several lines of evidence support this idea, including the observation that activation of wild-type or mutant PDGFβ receptor as well as fibroblast growth factor receptor (FGFR) results in similar transcriptional response of immediate-early genes (5). In addition, mice harboring point mutations in the PDGFβ receptor that prevent activation of specific downstream signaling pathways display only subtle phenotypic differences (32). Further support for this model comes from several studies that have utilized a strategy of creating chimeric receptors which fuse the intracellular signaling domain of one receptor to the extracellular ligand binding domain of another. In Drosophila, it was demonstrated that the Torso receptor intracellular domain was able to functionally replace the signaling domain of the FGFR (Breathless) by inducing appropriate cell migration and activation of cell fate determinants during tracheal development (3). A chimeric FGFR3 receptor in which the cytoplasmic domain is replaced with the corresponding domain from FGFR1 is still a negative regulator of chondrocyte proliferation when expressed in the growth plate, although FGFR1 has potent mitogenic activity in cell culture (34). Thus, specificity appears to result from the ability of individual cell types to differentially interpret redundant signals. Furthermore, studies from our laboratory have demonstrated that while PDGFαR signaling cannot fully compensate for loss of PDGFβR signaling, replacement of the PDGFαR with a chimeric receptor that transmits a PDGFβR-type signal fully rescues normal development (17). These studies clearly support the idea that RTK signaling can have a generic component, and the cell type in which the RTK is expressed is a defining factor in controlling developmental outcome.
The focus of this study was to determine whether the biochemical nature of the RTK signal or the cell-type-specific expression is the critical determinant of RTK function in development. We have analyzed the signaling capacity of chimeric receptors in which the extracellular domain of the mouse PDGFαR (αR) is fused to the intracellular signaling domain of divergent RTKs, the Drosophila Torso (αTor), and the mouse FGFR1 (αFR). These three RTKs share similarities in structure and in mechanisms of activation. All three RTKs are known to activate an overlapping set of intracellular signaling proteins, such as Ras, MAP kinase, PLC-γ, and Src family kinases. However, these three RTKs differ in the activation of certain downstream pathways. The PDGFαR binds and activates signaling molecules directly, including PI3-kinase, which is the dominant downstream effector of αR signaling (16). Previous studies of the Drosophila Torso protein have demonstrated its ability to directly bind corkscrew, a SHP-2-like phosphatase which activates the Ras MAP kinase pathway through its recruitment of additional factors, such as Grb2 (1, 2). FGFR1 directly binds PLC-γ (21) and the large adaptor protein FRS2 (36). Upon phosphorylation, FRS2 brings to the receptor complex other signaling proteins, including Grb2, Gab1, and SHP-2, which ultimately stimulate MAP kinase and PI3-kinase (7, 8, 18). Perhaps as a result of this indirect recruitment, the kinetics of activation of these pathways is quite different. For example, PDGFR stimulates MAP kinase more transiently than FGFR, while PI3-kinase stimulation is more robust by PDGFR (37).
To obviate issues related to overexpression and to ensure proper comparative analyses, we have generated mice harboring these chimeric receptors at the PDGFαR locus. Chimeric receptors were unable to fully replace αR function in embryonic development, but the well-characterized phenotype of homozygous PDGFαR-null mice made it possible to assess partial rescue of PDGFαR signaling. Further analysis supports the idea that the temporal control as well as proper kinetic modulation of MAP kinase and PI3-kinase activation by RTKs is critical for proper embryonic development.
MATERIALS AND METHODS
Derivation of knock-in mice.
The targeting constructs used to generate the knock-in mouse lines were essentially the same as those previously described for the targeted mutation of the PDGFαR genomic locus (17, 28). cDNA inserts encoded chimeric PDGFα receptors. The αTor cDNA encodes the extracellular domain of the PDGFαR fused to the transmembrane and intracellular domain of the αFR receptor. The fusion point in the PDGFαR is at amino acid 524 and amino acid 400 in the Torso receptor. The αFR cDNA encodes the extracellular and transmembrane domains of the PDGFαR fused to the intracellular domain of the mouse FGFR1. The fusion point for the αFR chimeric receptor in the PDGFαR is at amino acid 549 and at amino acid 398 for FGFR1. The chimeric αTor cDNA was generated by using PCR splicing of overlapping ends of the following primer pairs: for αR1, (5′-TGACCACCATGGCTCTGGCGGGGGACAGACT-3′), and for αTor1, (5′-GATAATAAAGAGTACCAATTCAGATCGCAGAGTGGG-3′); αTor2, (5′-CCCACTCTGCGATCTGAATTGGTACTCTTTATTATC-3′), and Tor1, (5′-TAAGTCCAATTGCATTTTGCTCCTGCCTGA-3′). Mouse PDGFαR and αFR cDNAs were used as templates. The products from the first PCRs were used as templates for the second set of reactions with the outside primers from the first reaction. The final PCR product contained a unique 5′ NheI site in the extracellular portion of the PDGFαR and a unique 3′ MfeI site in the intracellular region of Torso. These sites were used to subclone this fragment into the corresponding sites in the PDGFαR and Torso cDNAs, thus creating the chimeric αTor cDNA. The same technique was used to create the αFR chimeric cDNA. The primer pairs used were αR1 (5′-TGACCACCATGGCTCTGGCGGGGGACAGACT-3′) and αFR1 (5′-GTCCTGGTGGTCATTTGGAAGATGAAGAGCGGCACC-3′) and αFR2 (5′-GGTGCCGCTCTTCATCTTCCAAATGACCACCAGGAC-3′) and αFR1 (5′-GCCTAAGACCAGTCTGTCTCGTGG-3′). The final αFR PCR product again contained the 5′ NheI site from the PDGFαR and the 3′ BamHI in the FGFR1 cDNA for subcloning to create the full-length αFR cDNA.
The chimeric cDNAs or the H2Be-GFP construct (13) (generous gift from S. Fraser) was inserted in targeting vectors between a splice acceptor (SA) from the adenovirus late transcript (6) and a trimerized simian virus 40 polyadenylation (3pA) (20). The knock-in vectors replace a 6.5-kb BamHI-SmaI fragment in the PDGFαR that corresponds to the signal peptide and the first two immunoglobulin domains with SA chimeric cDNA-3pA-lox-PGKneo-lox sequence. The flanking arms for the targeting vector were exactly the same as those previously described for the PDGFαR locus (28). The targeting vectors were linearized with SacII and were electroporated into 129S4-derived AK7 embryonic stem (ES) cells, and colonies were selected in 300 μg of G418 (total powder)/ml. Correct targeting was verified by Southern blotting by using cDNA probes as previously described (28). PCR genotyping was performed on tail or yolk sac DNAs as previously described for the PDGFαR locus (28). Both mixed background (C57BL/6 × 129) and congenic 129S4 mice were analyzed in this study and displayed indistinguishable phenotypes.
FACS analysis.
Cells suspensions were obtained from mouse embryonic fibroblasts or from embryos that were homogenized to single-cell suspension in 0.25% trypsin-1 mM EDTA pelleted and resuspended in ice-cold phosphate-buffered saline. Cells were incubated in anti-PDGFαR antibody (1:500; Research Diagnostics) for 30 min and then were incubated with secondary antibody conjugated to phycoerythrin (PE). Fluorescence-activated cell sorter analysis was performed on a Becton Dickinson FACScan.
Derivation of cell lines.
Primary mouse embryonic fibroblast cell lines were derived by dissecting embryos at embryonic day 12.5 (E12.5) to E14.5 in phosphate-buffered saline and then incubating them in 0.25% trypsin-1 mM EDTA at 37°C for 5 min. Embryos were disassociated with a transfer pipette and were plated on gelatin-coated dishes. Cells were expanded to passage 2 before freezing and analysis. The expression of the αTor chimeric receptor in primary mouse embryonic fibroblast lines was assayed by FACS analysis with an antibody that recognizes the extracellular domain of the PDGFαR.
The signaling activity of αFR chimeric receptor was assayed by stable transfection of a fibroblast cell line from Patch (Ph) mice, which harbor a genomic deletion of the PDGFαR locus (29), with a pLXSH expression vector containing the chimeric αFR or wild-type cDNA. The expression levels of the αFR chimeric and wild-type receptors was assayed by FACS sorting, and two cell lines were isolated that expressed nearly identical levels of αFR or wild-type receptor.
Immunoprecipitations and Western blot analysis.
Western blotting was performed as previously described (15), with the exception that secondary antibodies were conjugated to horseradish peroxidase. The phospho-MAP kinase (91-10) and phospho-Akt (Ser473) antibodies were purchased from Cell Signaling Technology. The anti-phosphotyrosine antibody was purchased from Upstate Biotechnology. Immunoprecipitations were performed as previously described (18) by using an antibody to the C-terminal portion of FRS2 generously provided by I. Lax and J. Schlessinger.
Skeletal preparations.
E18 embryos were soaked in water for 24 h and then were skinned and eviscerated before being fixed in 95% ethanol overnight. Embryos were then stained at 37°C in 0.015% alcian blue-0.005% alizarin red-5% acetic acid in 70% ethanol and then were cleared in 1% KOH and transferred to decreasing strengths of KOH and increasing concentrations of glycerol.
Histology.
Tissues were fixed in 4% paraformaldehyde overnight at 4°C, embedded in paraffin, sectioned at 7 μM, and stained with hematoxylin and eosin. Embryos for whole-mount PECAM (CD31 adhesion molecule) analysis were fixed with 4% paraformaldehyde, dehydrated in methanol, and bleached in 5% hydrogen peroxide for 3 h. Embryos were rehydrated and blocked in 5% rabbit serum for 2 h. Immunohistochemical detection of PECAM was performed by using a monoclonal antibody purchased from Pharmingen (1951D) and used at a dilution of 1:200. Secondary antibody detection was done by using the VectaStain ABC kit and the DAB detection kit from Vector Technology.
RESULTS
Knock-in at the PDGFαR locus faithfully recapitulates expression pattern.
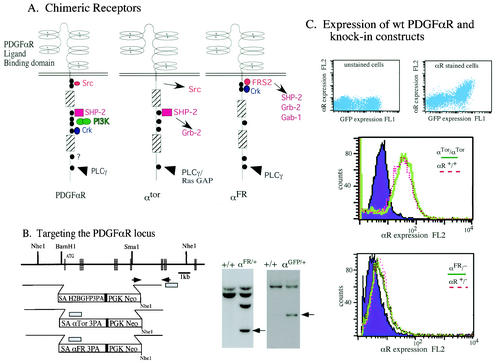
We have generated chimeric cDNAs that encode the extracellular ligand binding domain of the PDGFαR fused to the cytoplasmic domain of the αTor or αFR (Fig. 1A). We have previously described a knock-in vector that will simultaneously disrupt the expression of the endogenous pdgfαr gene while allowing for the expression of the chimeric receptor cDNA under the control of the endogenous PDGFαR promoter (Fig. 1B) (17). By using this vector we have demonstrated that a knock-in of wild-type PDGFαR cDNA is able to completely rescue the loss of the endogenous pdgfαr gene as measured by the normal embryonic and adult development of mice harboring this knock-in (17). ES cells were screened for proper targeting of the chimeric αTor or αFR cDNA to the PDGFαR locus by Southern blot analysis, as was previously described (Fig. 1B) (28). The expression level of chimeric receptors was evaluated by FACS analysis by using an antibody that recognizes the extracellular domain of PDGFαR. The nearly identical FACS profiles from αTor and αFR embryonic fibroblasts demonstrate that chimeric receptors are expressed at the cell surface at levels similar to those of wild-type controls (Fig. 1C).
FIG. 1.
Targeted knock-ins of αGFP, αFR, and αTor at the PDGFαR locus. (A) Schematic representation of the chimeric receptors and relevant binding sites for downstream effector molecules. (B) Schematic of the region of the wild-type PDGFαR genomic locus and the cDNA knock-in constructs. Hatched boxes represent the positions of exons. SA, splice acceptor; 3PA, triple polyadenylation sequence. Inset boxes are Southern blot analyses of wild-type (+/+) or targeted (αFR/+ or αGFP/+) ES clones with a probe corresponding to the shaded box and NheI-digested DNA. The probe hybridizes to a 4-kb fragment which indicates correct targeting (arrow) and an additional 8.7-kb fragment due to a duplication of this area in the αFR and αTor cDNA. (C) Top left panel, analysis of GFP expression in unstained cells derived from an αGFP/+ E9.5 embryo. Top right panel, cells from same embryo stained for PDGFαR expression with an antibody conjugated to PE. FACS analysis of cell lines expressing chimeric receptors (middle panel, αTor/αTor; lower panel, αFR/−) by using an antibody that recognizes the extracellular domain of the PDGFαR conjugated to PE. The purple filled peak represents signal from αR−/− cells, the solid green trace represents signal from cells expressing the chimeric receptors (αTor/αTor or αFR/−), and the dotted red line is the trace of cells expressing the wild-type PDGFαR.
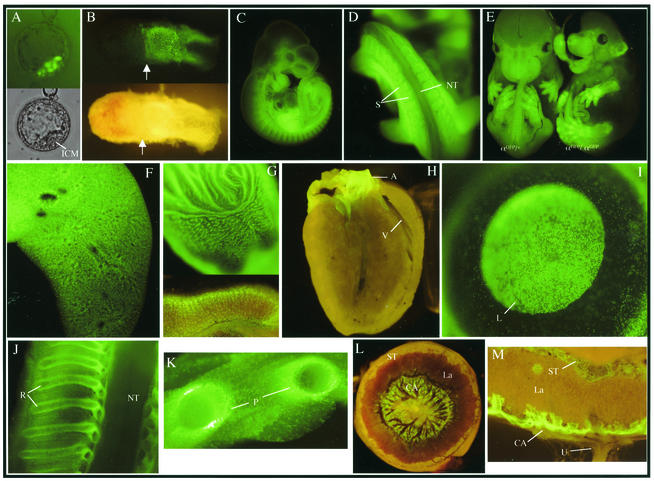
To confirm that cDNAs knocked into the PDGF locus are expressed in a manner identical to that of the PDGFαR, the histone H2B-eGFP fusion protein reporter construct was knocked into the PDGFαR locus (αGFP). According to previous expression studies, heterozygous αGFP mice demonstrate that green fluorescent protein (GFP) expression faithfully reproduces the PDGFαR expression pattern (11, 23-25, 30). Nuclear αGFP expression is clearly visible in cells of the polar trophectoderm of the E4.5 blastocyst and the extraembryonic ectoderm at E6.5 (Fig. 2A and B). Later, expression is noted in the cardiac neural crest derivatives of the heart, somites, and branchial arches at E10.5 as well as in the eye lens, lungs, stomach, and the perichondrium (Fig. 2C, D, F, G, H, I, J, and K). Analysis of extraembryonic structures demonstrate αGFP expression in the parietal endoderm, the chorioallantoic plate, and spongiotrophoblast layer of the placenta (Fig. 2L and M). Furthermore, at E13.5 homozygous αGFP mice display a phenotype identical to that of the PDGFαR homozygous null mice (Fig. 2E). FACS analysis of cells from αGFP/+ embryos demonstrate that H2BGFP and the wild-type PDGFαR are expressed in the same population of cells (Fig. 1C), further supporting the idea that αGFP faithfully reproduces the native wild-type PDGFαR expression pattern.
FIG. 2.
Expression patterns of αGFP in embryonic and adult tissues. (A) Expression in polar trophectoderm of E4.5 blastocysts. Bottom panel, visible light; top panel, UV. (B) Expression in extraembryonic ectoderm of an E6.5 embryo. Top panel, UV; bottom panel, visible light. Arrows denote the boundary of embryonic and extraembryonic tissues, and Reichert's membrane was not removed. (C) Whole-mount E10.5 embryo αGFP/+. (D) Magnification of the caudal somite. (E) E13.5 littermates. Left embryo, αGFP/+; right embryo, αGFP/αGFP. Expression in whole-mount E18.5 organs, lung (F), stomach (G, top portion), and transverse section of adult stomach in the glandular region (G, lower portion). (H) Sagittal section of adult heart. (I) Whole-mount adult eye. (J) Sagittal section of ribs and spinal cord of an E16.5 αGFP/+ embryo. (K) E18.5 αGFP/+ transverse section through ribs. Whole-mount (L) and transverse section (M) of an E14.5 αGFP/+ placenta visualized under fluorescence. ICM, inner cell mass; NT, neural tube; S, somites; V, ventricle; A, aorta; L, lens; R, ribs; P, perichondrium; La, labyrinth; ST, spongiotrophoblast layer; CA, chorioallantoic layer; U, umbilical cord.
Partial rescue by Torso.
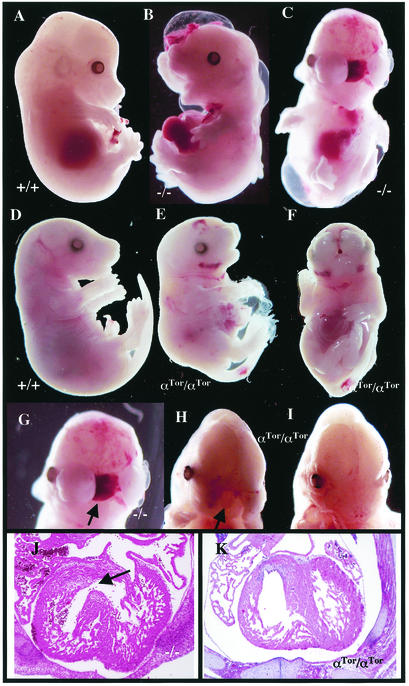
High-percentage chimeras (>80%) derived from two ES clones transmitted the αTor chimeric cDNA through the germ line. Crosses of heterozygous αTor mice revealed that no αTor homozygous pups survived to birth (n = 132; data not shown), demonstrating that αTor is not able to completely replace homozygous null PDGFαR (αR) function in development. However, embryos were then analyzed to determine if Torso signaling was able to partially compensate for loss of the PDGFαR. Previous studies have characterized the pleiotropic defects in αR embryos (28, 31). Embryonic lethality in PDGFαR-null mice occurs in two phases: an early phase in which greater than 50% of homozygous embryos die before E12.5 and a late phase in which the remainder die by E15.5. Genotyping of embryos from heterozygous crosses at stages E12 to E14 revealed that the expected Mendelian proportion of homozygous αTor embryos survived to this stage (40 αTor/αTor out of 149 offspring [27%]), which is significantly higher than the 10% recovery rate of αR-null embryos (31 and M. Tallquist and P. Soriano, unpublished data). Furthermore, a small percentage of live αTor/αTor embryos were recovered at late stages of development (5 αTor/αTor embryos out of 58 offspring at E17.5 to E18.5 [9%]). In contrast, PDGFαR-null embryos have never been recovered at this stage of development. Late-stage αTor/αTor embryos display typical αR phenotypes, including a cleft face or palate, failed ventral closure, and spina bifida, although the defects are significantly less severe compared to those of PDGFαR-null mutants (Fig. 3E and F). These results suggest that the αTor chimeric receptor is at least partially able to replace the function of the αR in mouse development.
FIG. 3.
Partial rescue of embryonic defects by αTor. (A to C) Wild-type (A) and PDGFαR−/− (B and C) E14.5 whole-mount embryos. (D to F) Wild-type (D) and αTor/αTor (E and F) E17.5 embryos. (G to I) Cleft face phenotype of PDGFαR−/− (G) and αTor/αTor (H) E14.5 embryos and E15.5 αTor/αTor embryos (I). (J and K) Transverse sections through E14.5 embryos at heart level for PDGFαR−/− (J) and αTor/αTor (K) embryos. Arrows indicate incomplete septum formation.
Rescue of neural crest defects and vascular development by αTor.
To determine if the rescue by the αTor chimeric receptor is general or tissue specific, homozygous mutant embryos were analyzed for different developmental defects. Loss of the PDGFαR affects multiple tissues. Some αR-null phenotypes, such as rib fusions, have variable penetrance (28). The cleft face phenotype, however, is one of the most dominant features of αR-null embryos and is 100% penetrant (Fig. 3B and C). Studies that used conditional mutagenesis have demonstrated that the cleft face phenotype is due to the specific loss of αR function in the neural crest cells (31). Examination of over 50 homozygous αTor embryos at E13 to E15 shows that while almost all exhibit some degree of facial clefting, this clefting is far less severe in the vast majority of the αTor homozygotes than in αR-null counterparts. In fact, only 10% of these embryos look phenotypically identical to the αR homozygous null. In most cases the clefting does not extend into the anterior forebrain, as occurs in αR-null embryos (Fig. 3G and H). Perhaps the most striking rescue was observed in a E15.5 embryo that exhibits a complete rescue of the facial clefting (Fig. 3I). This degree of closure at the midline of the face is never seen in PDGFαR-null embryos.
Conditional mutagenesis studies disrupting αR function in neural crest cells also demonstrated that formation of the membranous portion of the heart septa is dependent upon αR function. Heart septation defect in αR-null mice is also a completely penetrant phenotype (Fig. 3J) (31). Serial transverse sections through the heart were done on E14 to E16 αTor homozygous embryos to examine the formation of the septa. All of the αTor/αTor embryos that exhibited severe facial clefting also had incomplete septa formation (n = 3). However, embryos in which facial clefting was partially rescued had complete rescue of the heart septation defect (n = 4; Fig. 3K). These results illustrate the ability of αTor to functionally replace PDGFαR in both cranial and cardiac neural crest lineages.
Another prominent phenotype that is completely penetrant in PDGFαR-null E13.5 embryos is a vascular defect evidenced by the presence of hemorrhaging and edema (Fig. 3). It is possible that these phenotypes are the result of defective angiogenesis or vasculogenesis, which is known to require PDGFαR activity (4, 28). Examination of homozygous E13 to E15 αTor embryos reveals that hemorrhaging is rarely present, and edema can only be detected in very-late-stage (E17) embryos. To determine whether αTor is able to rescue the vascular phenotype due to its ability to function in angiogenesis, whole-mount PECAM staining was performed on E9 to E11 embryos. At E9.5 vascularization of the midbrain and forebrain is easily detected as newly formed vessels migrate towards the midline (Fig. 4A). By E11.5 the vascular network has become much more intricate at the midline of the midbrain through a complex process of angiogenesis which involves branching and remodeling of the early vascular network (Fig. 4D). Angiogenesis appears to be fairly normal at E9.5 in PDGFαR-null embryos, although the migration of vessels to the midline appears slightly retarded (Fig. 4B). Defects in angiogenesis become prominent in PDGFαR-null embryos by E11.5 as fewer vessels are found in the midbrain (Fig. 4E). The presence of larger vessels is likely due to a decrease in the process of branching and remodeling of the vascular network. In contrast, this process appears to be fairly normal in αTor/αTor embryos, as evidenced by vascularization in the midbrain at E9.5 and the extensively branched vascular network in the midbrain at E11.5 (Fig. 4C and F). It is worth noting that rescue of the vascular defect appears to be independent of the neural crest rescue, as αTor/αTor embryos with cleft face phenotypes as severe as those of the PDGFαR null still display rescue of the vascular defects. Therefore, this represents the second specific tissue in which αTor is able to replace the function of the αR.
FIG. 4.
Rescue of vascular development by αTor. Whole-mount PECAM staining in the mid- and forebrain region of E9.5 wild-type (A), PDGFαR −/− (B), and αTor/αTor (C) embryos is shown. Also shown is PECAM staining in the midbrain of E11.5 wild-type (D), PDGFαR −/− (E), and αTor/αTor (F) embryos.
Failed rescue of skeletal and extraembryonic defects.
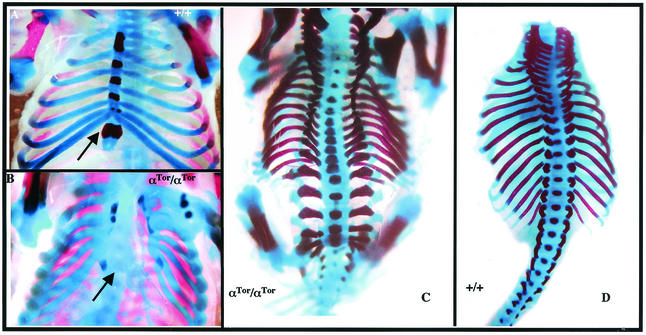
Previous studies have demonstrated that PDGFαR is required for normal development of the axial skeleton (28). Late-stage αR-null embryos display severe defects in skeletal development, including rib fusions, split sternum, spina bifida, and fusions of cervical vertebrae. Several of these defects are recapitulated in homozygous αTor embryos. The inability of ribs to fuse at the midline and to form the sternum is completely penetrant, even in embryos that exhibit almost complete rescue of neural crest defects (Fig. 5B). Furthermore, all αTor homozygous embryos have dorsal closure defects, including spina bifida (Fig. 5C). Lack of rescue in skeletal development while exhibiting partial rescue of neural crest defects by αTor is reminiscent of previous studies on mice harboring point mutant PDGFαR that selectively eliminated capacity to bind to PI3-kinase (αPI3K) or Src (αSrc) (16). As in the case of αTor, αPI3K mice displayed a less severe neural crest defect (cleft palate versus the cleft face presented by α-receptor-null mice) while exhibiting extensive spina bifida.
FIG. 5.
Skeletal defects in αTor mice. Skeletons of E16.5 embryos. (A) Wild-type ventral view of ribs and sternum. (B) αTor/αTor ventral view of rib cage. (C) αTor/αTor dorsal view of spinal column. (D) Wild-type dorsal view of spinal column with limbs removed. Arrows denote the positions of sternum.
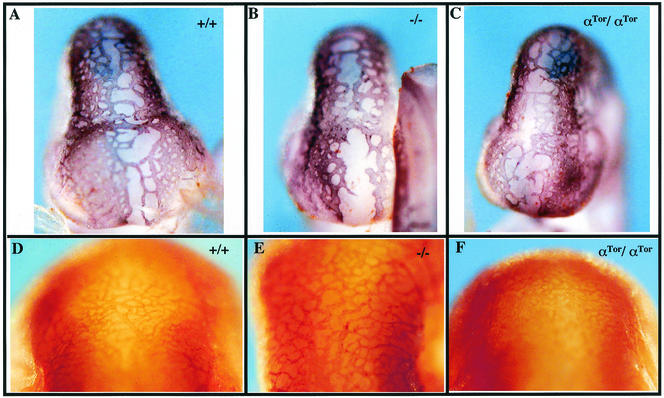
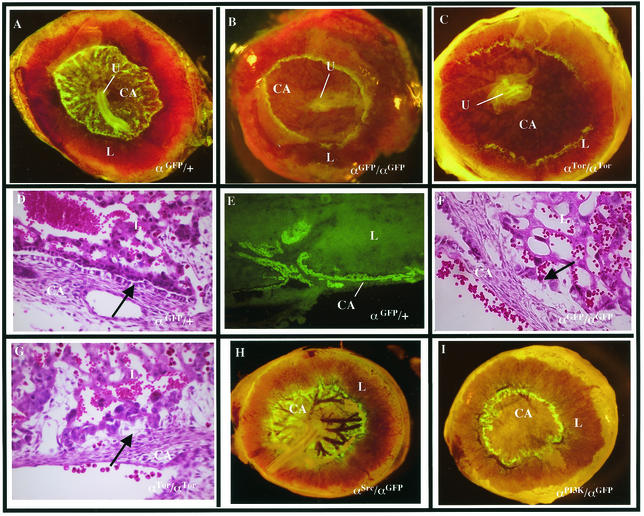
The extraembryonic defects associated with PDGFαR-null mice have not been well characterized. Previous studies have demonstrated that αR is expressed in a subset of the trophoblast lineages in the placenta (22), particularly in the spongiotrophoblasts. Analysis of the αGFP/+ mice has shown that αR is also highly expressed in the parietal endoderm from E8 to E10 (data not shown). As the allantois fuses to the chorion to form the chorioallantoic plate (E9 to E10), αGFP-expressing cells are found at the edges of the chorioallantoic plate, the junction of the yolk sac and placenta. In whole-mount αGFP/+ placentas from E10 to E13, αGFP-expressing cells are easily identified and will populate the entire chorioallantoic plate from the periphery to the forming umbilical cord (Fig. 6A). Transverse sections of E12 to E13 αGFP/+ placentas reveal that αGFP is expressed in an epithelial cell layer between the chorioallantoic plate and the labyrinth (Fig. 6E). Vascular development in the chorioallantoic plate of αGFP/αGFP embryos is significantly retarded, and αGFP-expressing cells fail to populate the chorioallantoic plate, instead remaining at the yolk sac junction (Fig. 6B). The epithelial cell layer between the chorioallantoic plate and labyrinth is also completely absent (Fig. 6F). Although this defect does not prevent fusion of the allantois, it is likely that the decreased vascularization of the placenta affects the development of αGFP/αGFP embryos and contributes to the lethality. Analysis of hemizygous αGFP/αTor placentas revealed that the PDGFαR-null defect is not rescued by the αTor chimeric receptor (7 out of 7 embryos), as evidenced by the lack of the αGFP-expressing cells in the chorioallantoic plate and the loss of the epithelial cells between the labyrinth and the chorioallantoic plate (Fig. 6C and G).
FIG. 6.
Placental defects in αTor mice. (A to C) αGFP/+ (A), αGFP/αGFP (B), and αGFP/αTor (C) E13.5 whole-mount placentas visualized under fluorescence. (D, F, and G) Hematoxylin and eosin staining of E13.5 transverse placental sections of αGFP/+ (D), αGFP/αGFP (F), and αGFP/αTor (G) embryos. (E) Transverse frozen section at E13.5 placenta. (H and I) αGFP/αSrc (H) and αGFP/αPI3K (I) whole-mount placentas at E14.5. CA, chorioallantoic place; U, umbilical cord; L, labyrinth. Arrows denote positions of the epithelial layer between chorioallantoic layer and labyrinth, which is absent in mutants.
To investigate if the placental defect correlated with the loss of a specific signaling pathway, we analyzed placentas from hemizygous mice carrying the previously described point mutations αPI3K or αSrc (16) in combination with the αGFP allele. αPI3K/αGFP cells failed to populate the chorioallantoic plate (Fig. 6I), while the αSrc/αGFP placentas show no defects (Fig. 6H), illustrating a specific requirement for PI3-kinase signaling. This provides a possible explanation for the failure of αTor to functionally replace PDGFαR in extraembryonic tissues. The inability of αTor homozygous embryos to undergo proper development of the axial skeleton and development of extraembryonic tissues may be due to failure of the Torso RTK to activate a subset of specific signaling pathways normally triggered by the PDGFαR which are critical to the development of these tissues.
αFR dominant phenotype.
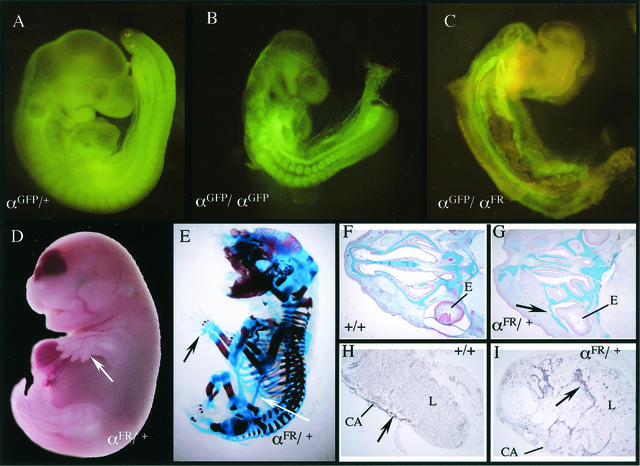
High-percentage chimeras derived from four different αFR ES clones exhibited a variety of defects, including subepidermal growths and skeletal defects, and were much smaller than nonchimeric or low-contribution chimeric littermates (data not shown). Furthermore, higher percentage chimeras were unable to mate. In the course of this study two chimeras were generated that had a low degree of chimerism, allowing normal development and the ability to transmit the αFR allele. Genotyping of agouti pups revealed that none carried the αFR knock-in allele. Heterozygous αFR/+ embryos could be recovered at E16.5 but displayed a variety of severe defects. The most dramatic are the skeletal malformations which are completely penetrant, including polydactyly (Fig. 7D), spina bifida, failed ventral closure, rib fusions, and abnormal cranial bone development which results in bone growth in front of the developing eye (Fig. 7G). Perhaps most surprising is an ectopic bone growth extending from the pubic bone and fusing, in most cases, to the elbow (Fig. 7E). Extraembryonic defects could also be detected in αFR heterozygous mice. Placentas from αFR heterozygous mice were significantly larger (data not shown), and trophoblast lineages were disorganized, as evidenced by an expansion of the chorioallantoic layer into the surrounding labyrinth trophoblast layer (Fig. 7I).
FIG. 7.
αFR dominant phenotypes. (A to C) αGFP/+ (A), αGFP/αGFP (B), and αGFP/αFR (C) E9.5 whole-mount embryos visualized under fluorescence. (D and E) E17.5 αFR/+ embryo (D) and skeleton (E). Arrows denote extra digits and ectopic bone. (F and G) Alcian blue-stained transverse sections through nasal cartilage of E16.5 wild-type (F) and αFR/+ (G) embryos. Arrows indicate cartilage growth in front of the eye. (H and I) ASMA staining of transverse sections of E16.5 wild-type (H) or αFR/+ (I) placentas. Arrows denote positively stained chorioallantoic trophoblasts. L, labyrinth; CA, chorioallantoic plate; E, eye.
Interestingly, αFR heterozygous mice display a combination of αR-null phenotypes (spina bifida, failed ventral closure, cleft palate) and novel, possibly gain-of-function phenotypes (ectopic bone growths), which could be due to the interaction of the αFR receptor with the wild-type αR to produce dominant-negative heterodimers. To examine the effect of the αFR receptor in the absence of the wild-type αR, germ line chimeras were crossed to αGFP heterozygous mice. Hemizygous αFR (αFR/αGFP) embryos isolated at E9.5 display much more severe defects than αR-null embryos, including failure of the embryo to turn and inhibition of somitogenesis (Fig. 7C). In fact, no hemizygous αFR embryos were ever recovered after E10.5, in contrast to homozygous αR-null embryos, which can survive to E15.5. These results clearly illustrate that FGFR1 signaling cannot substitute for PDGFαR in mouse development and further show that simply having a functional kinase domain is insufficient to rescue αR function.
Activation of signaling pathways by chimeric receptors.
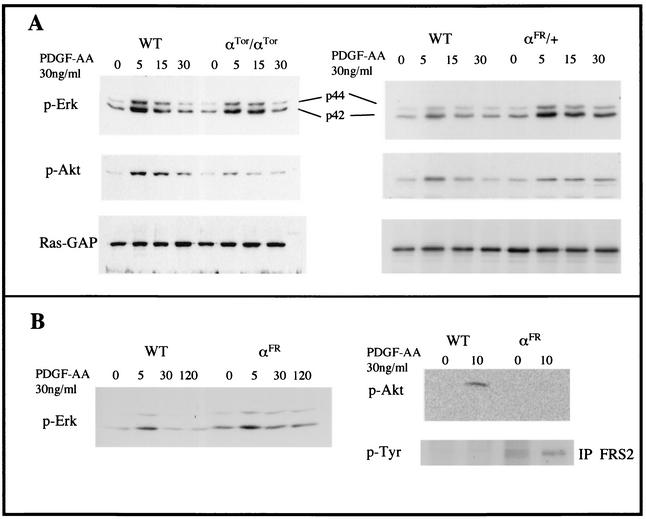
To determine whether the failure of chimeric receptors to fully rescue PDGFαR function was due to inherent differences in their signaling capacity, embryonic fibroblast cell lines were derived and analyzed. Cell lines were treated with PDGF-AA, the exclusive ligand for the PDGFαR, and the ability of chimeric receptors to activate MAP kinase and PI3-kinase signaling was assayed by Western blot analysis of whole-cell lysates. The kinetics of MAP kinase phosphorylation by αTor is very similar to that of wild-type αR (Fig. 8A, left panels), which demonstrated that αTor is activated by PDGF-AA and suggests that the function of the Torso receptor has been conserved well enough to activate this signaling pathway, most likely through SHP-2 and Grb2. Interestingly, we were able to detect signaling via PI3-kinase, although it was much weaker than that with the αR, as evidenced by the phosphorylation of Akt, a downstream target of PI3-kinase. It is not clear at this time whether this occurs via direct binding of PI3-kinase, since no consensus binding sites are found in the Torso receptor, or through a secondary pathway.
FIG. 8.
Activation of MAP kinase and PI3-kinase signaling pathways. (A) Western blot analysis of whole-cell lysates from wild-type (WT) or αTor/αTor (left panels) or αFR/+ (right panels) primary embryonic fibroblasts stimulated with 30 ng of PDGF-AA/ml harvested at the time points indicated above the lanes (in minutes). Blots were probed with antibodies to phosphorylated MAP kinase (p-Erk), phosphorylated Akt (downstream of PI3-kinase), or Ras-GAP as a loading control. (B) Western blot analysis of cell lysates from Ph fibroblasts transfected with wild-type PDGFαR or αFR stimulated with PDGF-AA for the indicated times. The expression levels of receptors were normalized by FACS sorting, and protein level loading controls were quantified by Ponceau S staining (not shown). Blots were probed with antibodies to phosphorylated Erk or phosphorylated Akt. Bottom right panel, Western blot analysis of FRS2 immunoprecipitation of lysates from indicated cell types. Blots were probed with antibodies to phosphorylated tyrosine.
FGFRs have the ability to stimulate MAP kinase and PI3-kinase signaling through an interaction with the adaptor molecule, FRS2 (7). The ability of the αFR chimeric receptor to be activated by PDGF-AA was assayed by using a Ph cell line, which contains a genomic deletion of the pdgfαr locus (29), stably transfected with the αFR receptor or the wild-type αR. Cell lines isolated by FACS sorting that expressed equal levels of wild-type or αFR receptor (data not shown) were stimulated with PDGF-AA. The kinetics of MAP kinase activation shows increased and sustained stimulation in αFR-expressing cells compared to that of wild-type αR, which is consistent with previous studies of FGFR (7, 37) (Fig. 8B, left panel). We were unable to detect any αFR-stimulated PI3-kinase signaling as assayed by phosphorylation of Akt (Fig. 8B, right panels), but it is possible that the signaling threshold was below the detection levels of Western blotting. It has already been shown that FGFR stimulation of PI3-kinase is significantly weaker than that of PDGFR (37).
We directly assayed the ability of αFR to activate FGFR-specific signaling pathways by determining whether FRS2 is tyrosine phosphorylated by αFR. FRS2 was immunoprecipitated from αFR and wild-type lysates, and then tyrosine phosphorylation was assayed by Western blot analysis (Fig. 8B, right panels). FRS2 phosphorylation is constitutive as a result of αFR overexpression and is possibly slightly increased with PDGF-AA. Importantly, cells expressing the wild-type αR do not activate FRS2, even when stimulated by PDGF-AA. These results demonstrate that αFR receptors are responsive to PDGF-AA and transmit a distinct FGFR intracellular signal. Taken together, differences in the ability of both chimeric receptors to activate PDGF-AA-driven pathways or in the kinetics of activation could be the reason for the inability to completely rescue PDGFαR function in development.
The ability of αFR to exert the dominant phenotype seen in αFR/+ embryos was analyzed in primary embryonic fibroblast cell lines. These cell lines were stimulated by the addition of PDGF-AA, and activation of MAP kinase and PI3-kinase signaling were assayed by Western blot analysis (Fig. 8A, right panels). The kinetics of MAP kinase signaling are significantly altered in αFR/+ fibroblasts, demonstrating increased and sustained activation similar to what was observed with αFR overexpression in Ph cells. PI3-kinase signaling is slightly lower, while the duration is longer. Since we were unable to detect PI3-kinase stimulation by the αFR receptor alone in Ph cells, the signaling observed in αFR/+ cells is likely generated by the wild-type αR. The slight decrease in activation compared to that of the wild type could be due to the fact that the cells are heterozygous for the PDGFαR, and the increased duration of PI3-kinase signaling could be due to the formation of αFR-wild-type heterodimeric receptors. The increased duration of MAP kinase activation by αFR demonstrated the increased stability of this signaling complex. It is possible that the stability of heterodimers is also increased, thereby allowing for greater PI3-kinase signaling.
DISCUSSION
Studies on the molecular mechanisms of RTK signaling have revealed that when activated by ligand binding, the intracellular domains of diverse RTK families stimulate an overlapping set of signaling pathways. A fundamental question is how RTKs are able to generate distinct cellular outcomes while initiating these seemingly redundant signaling pathways. The interpretation of RTK signals may depend on several factors, such as the cell type in which it is present and the kinetics of activation of specific downstream pathways. We have utilized two general approaches to determine how functional specificity of RTK signaling is generated during development. The first approach involves the generation of an allelic series of point mutations abrogating specific signaling pathways. Whereas the analysis of PDGFβR mutants has demonstrated only subtle differences in the requirements for specific signaling pathways (10, 32, and M. Tallquist and P. Soriano, unpublished), more dramatic differences between the contributions of individual signaling pathways are observed with the PDGFαR (16). The second approach, adopted in this work, involves replacing the signaling moiety of each PDGFR with that of a related (17) or more divergent receptor. Our results illustrate that the signaling domains of divergent RTKs are not interchangeable and further support the idea that cellular outcomes of RTK signaling are the result of specific regulation of activated downstream effectors. Thus, functional specificity and consequent evolutionary conservation of individual RTKs is not simply due to differences in spatiotemporal expression and/or ligand binding capacity.
Analysis of the homozygous αTor/αTor mice illustrates the ability of the Torso signaling domain to functionally replace αR function in tissues derived from neural crest lineages and in the vasculature but not in development of the axial skeleton and the placenta. The ability of the αTor receptor to rescue αR function in a tissue-specific manner could be interpreted in different ways. One possibility is that the αTor is able to produce a weakened generic RTK signal due to a decreased capacity to bind the mammalian homologues of the signaling molecules it would normally activate. The tissue specificity could result from the presence of another activated RTK in the neural crest or vascular progenitors, which may be able to partially compensate for the loss of PDGFαR signaling. A potential candidate for compensation might be the PDGFβR, which has been shown to work together with the PDGFαR in controlling neural crest and vascular development (17). Failed rescue of certain tissues would then suggest that PDGFαR is more critical in these tissues and thus is unable to be compensated by other RTKs.
A second possibility it that the αTor receptor is able to partially rescue αR function due to its ability to activate a subset of the specific signaling pathways normally activated by the PDGFαR. Previous studies of Torso signaling have demonstrated its ability to activate the MAP kinase pathway via interaction with corkscrew, a SHP-2 homologue (1, 2). Our studies show that αTor is able to activate MAP kinase with kinetics very similar to those of the native PDGFαR, suggesting that Torso is able to bind the mammalian components of the MAP kinase pathway. This is a possible reason for the partial rescue of αR function in neural crest-derived and vascular lineages. The finding that αTor is able to weakly stimulate the PI3-kinase pathway is surprising, since Torso has not been demonstrated to activate this pathway in Drosophila and lacks direct PI3-kinase binding sites. It is possible that binding of a SHP-2/Grb2 complex could recruit Gab1, which is known to activate PI3-kinase signaling (7). However, the suboptimal activation of PI3-kinase by αTor likely contributes to the failure of this chimeric receptor to fully rescue embryonic development. Previous results have indeed indicated that elimination of PI3-kinase signaling by the PDGFαR (αPI3K point mutant) results in tissue-specific developmental defects affecting mostly the axial skeleton and placental development, while the neural crest-derived tissues are more modestly affected (16). The overlapping tissue-specific defects of αTor and αPI3K mutants suggest that PI3-kinase signaling is not rescued by the αTor chimeric receptor. This provides further compelling evidence that the ability to trigger a precise blend of signaling pathways is critical for the function of RTKs, at least in certain cell types, in the context of development.
In light of the ability of the αTor receptor to partially rescue αR function, it was surprising that the αFR receptor was unable to functionally substitute for αR in any significant manner. The inability of hemizygous αFR/αGFP mice to survive past E10.5 illustrated the failure of the αFR receptor to produce a PDGFαR-like intracellular signal. This result is significant and somewhat unexpected in light of an earlier study comparing the downstream transcriptional responses of activated PDGFβR and FGFR in cultured cells. In that study, microarray analysis of fibroblasts that overexpressed the PDGFβR or FGFR1 demonstrated that the transcriptional responses to stimulation of these RTKs were highly redundant (5). Clearly our observation that an activated αFR chimeric receptor cannot rescue any aspect of the PDGFαR-null phenotype demonstrates that significant differences do exist between FGF and PDGF signaling in the context of development which are not detected by studies of transcriptional responses of immediate-early genes.
The gain-of-function phenotypes observed in αFR/+ heterozygous mice illustrate even more dramatically the differences in the signaling capacity between PDGFRs and FGFRs. These defects may also reflect inherent differences in the signaling capacity of these two receptor families. Previous biochemical analysis has demonstrated that the duration of MAP kinase activation is greater by FGFRs than by PDGFRs (37), and this was further demonstrated by our analysis of cells derived from the mutant αFR/+ embryos. Additionally, the biochemical data from heterozygous αFR/+ cells suggest that the αFR receptor does not negatively interfere with the wild-type αR by creating inactive heterodimers. Instead, it supports the idea that αFR receptors result in overstimulation of downstream signaling pathways, thus behaving like a gain-of-function mutation. The kinetics of MAP kinase activation has been shown to be a determining factor in the development of the Drosophila eye, controlling differentiation, survival, and proliferation (9). Levels of MAP kinase activation have also been shown to be critical in mediating NGF responses (19, 33). This provides a plausible explanation for the gross malformations seen in the chondrocyte lineages and chorioallantoic trophoblasts of αFR/+ embryos. Another interpretation is that many of the defects could be explained by the presence of excess FGF signaling in tissues where αFR receptor expression overlaps with FGFRs. Mutations in FGFRs that result in increased or ectopic signaling indeed account for a number of human skeletal dysplasia disorders (35). PDGFαR and FGFR1 are both expressed in the perichondrium of the developing axial skeleton. It is possible that the αFR receptor behaves much like an activating FGFR1 mutation, increasing the level of FGF signaling produced in the perichondrium, which results in the skeletal malformations exhibited by αFR/+ heterozygous mice. Overall, our data clearly show that PDGFRs and FGFRs produce distinct signals which result in unique developmental outcomes.
The experimental approach adopted here provides a testable model to help address why a large number of RTKs, which appear to transmit highly redundant intracellular signals, have been retained over evolution in mammals. We now have generated a series of knock-in mice expressing chimeric receptors which demonstrate that the highly conserved signaling domain of the PDGFβR can functionally replace the PDGFαR in mouse embryonic development, while the signaling domains from divergent RTK families either partially replace or severely hamper PDGFαR function in vivo. These observations as well as the analysis of an allelic series of signaling mutants (16) illustrate that several factors contribute to the functional specificity of the PDGFαR in mammalian development. Therefore, not only are correct expression of the RTK and ligand important determinants, but also the precise regulation of divergent downstream signaling pathways is critical for optimization of RTK function. This in turn is likely to promote organismal fitness and in part accounts for the maintenance of a richly diverse gene family throughout evolution.
Acknowledgments
We thank Jason Frazier and Peter Mueting-Nelsen for excellent technical assistance; Alice Davy, Josée Aubin, and Weisheng Chen for continuing interest and many helpful discussions; Brad Olwin for providing the FGFR1 cDNA; Deborah Morrison for providing the Torso cDNA; I. Lax and J. Schlessinger for the anti-FRS2 antibody; and our laboratory colleagues for critical review of the manuscript.
During the course of this study T.G.H. was supported by National Institutes of Health (NIH) postdoctoral fellowship HD08741-01. This work was supported by NIH grants HD25326 and HD24875 to P.S.
REFERENCES
- 1.Cleghon, V., P. Feldmann, C. Ghiglione, T. D. Copeland, N. Perrimon, D. A. Hughes, and D. K. Morrison. 1998. Opposing actions of CSW and RasGAP modulate the strength of Torso RTK signaling in the Drosophila terminal pathway. Mol. Cell 2:719-727. [DOI] [PubMed] [Google Scholar]
- 2.Cleghon, V., U. Gayko, T. D. Copeland, L. A. Perkins, N. Perrimon, and D. K. Morrison. 1996. Drosophila terminal structure development is regulated by the compensatory activities of positive and negative phosphotyrosine signaling sites on the Torso RTK. Genes Dev. 10:566-577. [DOI] [PubMed] [Google Scholar]
- 3.Dossenbach, C., S. Rock, and M. Affolter. 2001. Specificity of FGF signaling in cell migration in Drosophila. Development 128:4563-4572. [DOI] [PubMed] [Google Scholar]
- 4.Edelberg, J. M., W. C. Aird, W. Wu, H. Rayburn, W. S. Mamuya, M. Mercola, and R. D. Rosenberg. 1998. PDGF mediates cardiac microvascular communication. J. Clin. Investig. 102:837-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fambrough, D., K. McClure, A. Kazlauskas, and E. S. Lander. 1999. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell 97:727-741. [DOI] [PubMed] [Google Scholar]
- 6.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513-1523. [DOI] [PubMed] [Google Scholar]
- 7.Hadari, Y. R., N. Gotoh, H. Kouhara, I. Lax, and J. Schlessinger. 2001. Critical role for the docking-protein FRS2 alpha in FGF receptor-mediated signal transduction pathways. Proc. Natl. Acad. Sci. USA 98:8578-8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadari, Y. R., H. Kouhara, I. Lax, and J. Schlessinger. 1998. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol. Cell. Biol. 18:3966-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halfar, K., C. Rommel, H. Stocker, and E. Hafen. 2001. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development 128:1687-1696. [DOI] [PubMed] [Google Scholar]
- 10.Heuchel, R., A. Berg, M. Tallquist, K. Ahlen, R. K. Reed, K. Rubin, L. Claesson-Welsh, C. H. Heldin, and P. Soriano. 1999. Platelet-derived growth factor beta receptor regulates interstitial fluid homeostasis through phosphatidylinositol-3′ kinase signaling. Proc. Natl. Acad. Sci. USA 96:11410-11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmgren, L., A. Glaser, S. Pfeifer-Ohlsson, and R. Ohlsson. 1991. Angiogenesis during human extraembryonic development involves the spatiotemporal control of PDGF ligand and receptor gene expression. Development 113:749-754. [DOI] [PubMed] [Google Scholar]
- 12.Hunter, T. 2000. Signaling-2000 and beyond. Cell 100:113-127. [DOI] [PubMed] [Google Scholar]
- 13.Kanda, T., K. F. Sullivan, and G. M. Wahl. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377-385. [DOI] [PubMed] [Google Scholar]
- 14.Kazlauskas, A. 1994. Receptor tyrosine kinases and their targets. Curr. Opin. Genet. Dev. 4:5-14. [DOI] [PubMed] [Google Scholar]
- 15.Kazlauskas, A., A. Kashishian, J. A. Cooper, and M. Valius. 1992. GTPase-activating protein and phosphatidylinositol 3-kinase bind to distinct regions of the platelet-derived growth factor receptor beta subunit. Mol. Cell. Biol. 12:2534-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klinghoffer, R. A., T. G. Hamilton, R. Hoch, and P. Soriano. 2002. An allelic series at the PDGFαR locus indicates unequal contributions of distinct signaling pathways during development. Dev. Cell. 2:103-113. [DOI] [PubMed] [Google Scholar]
- 17.Klinghoffer, R. A., P. F. Mueting-Nelsen, A. Faerman, M. Shani, and P. Soriano. 2001. The two PDGF receptors maintain conserved signaling in vivo despite divergent embryological functions. Mol. Cell 7:343-354. [DOI] [PubMed] [Google Scholar]
- 18.Kouhara, H., Y. R. Hadari, T. Spivak-Kroizman, J. Schilling, D. Bar-Sagi, I. Lax, and J. Schlessinger. 1997. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell 89:693-702. [DOI] [PubMed] [Google Scholar]
- 19.Marshall, C. J. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80:179-185. [DOI] [PubMed] [Google Scholar]
- 20.Maxwell, I. H., G. S. Harrison, W. M. Wood, and F. Maxwell. 1989. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. BioTechniques 7:276-280. [PubMed] [Google Scholar]
- 21.Mohammadi, M., D. C. Li, W. Li, N. Spivak, T. Honegger, A. M. Jaye, and M. J. Schlessinger. 1992. Point mutation in FGF receptor eliminates phosphatidylinositol hydrolysis without affecting mitogenesis. Nature 358:681-684. [DOI] [PubMed] [Google Scholar]
- 22.Ohlsson, R., P. Falck, M. Hellstrom, P. Lindahl, H. Bostrom, G. Franklin, L. Ahrlund-Richter, J. Pollard, P. Soriano, and C. Betsholtz. 1999. PDGFB regulates the development of the labyrinthine layer of the mouse fetal placenta. Dev. Biol. 212:124-136. [DOI] [PubMed] [Google Scholar]
- 23.Orr-Urtreger, A., M. T. Bedford, M. S. Do, L. Eisenbach, and P. Lonai. 1992. Developmental expression of the alpha receptor for platelet-derived growth factor, which is deleted in the embryonic lethal Patch mutation. Development 115:289-303. [DOI] [PubMed] [Google Scholar]
- 24.Pringle, N. P., and W. D. Richardson. 1993. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development 117:525-533. [DOI] [PubMed] [Google Scholar]
- 25.Schatteman, G. C., K. Morrison-Graham, A. van Koppen, J. A. Weston, and D. F. Bowen-Pope. 1992. Regulation and role of PDGF receptor alpha-subunit expression during embryogenesis. Development 115:123-131. [DOI] [PubMed] [Google Scholar]
- 26.Schlessinger, J., and A. Ullrich. 1992. Growth factor signaling by receptor tyrosine kinases. Neuron 9:383-391. [DOI] [PubMed] [Google Scholar]
- 27.Simon, M. A. 2000. Receptor tyrosine kinases: specific outcomes from general signals. Cell 103:13-15. [DOI] [PubMed] [Google Scholar]
- 28.Soriano, P. 1997. The PDGFα receptor is required for neural crest cell development and normal patterning of the somites. Development 124:2691-2700. [DOI] [PubMed] [Google Scholar]
- 29.Stephenson, D. A., M. Mercola, E. Anderson, C. Y. Wang, C. D. Stiles, D. F. Bowen-Pope, and V. M. Chapman. 1991. Platelet-derived growth factor receptor alpha-subunit gene (Pdgfra) is deleted in the mouse patch (Ph) mutation. Proc. Natl. Acad. Sci. USA 88:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takakura, T., H. Yoshida, Y. Ogura, H. Kataoka, S. Nishikawa, and S. I. Nishikawa. 1997. PDGFR alpha expression during mouse embryogenesis: immunolocalization by whole-mount immunohistostaining using the monoclonal anti-mouse PDGFR alpha antibody APA5. J. Histochem. Cytohistochem. 45:883-889. [DOI] [PubMed] [Google Scholar]
- 31.Tallquist, M. D., and P. Soriano. 2002. Cell autonomous requirement for PDGFRα in populations of cranial and cardiac neural crest cells. Development 130:507-518. [DOI] [PubMed] [Google Scholar]
- 32.Tallquist, M. D., R. A. Klinghoffer, R. Heuchel, P. F. Mueting-Nelsen, P. D. Corrin, C. H. Heldin, R. J. Johnson, and P. Soriano. 2000. Retention of PDGFR-β function in mice in the absence of phosphatidylinositol 3′-kinase and phospholipase Cγ signaling pathways. Genes Dev. 14:3179-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traverse, S., N. Gomez, H. Paterson, C. Marshall, and P. Cohen. 1992. Sustained activation of the mitogen-activated protein (MAP) kinase cascade may be required for differentiation of PC12 cells. Comparison of the effects of nerve growth factor and epidermal growth factor. Biochem. J. 288:351-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, Q., R. P. Green, G. Zhao, and D. M. Ornitz. 2001. Differential regulation of endochondral bone growth and joint development by FGFR1 and FGFR3 tyrosine kinase domains. Development 128:3867-3876. [DOI] [PubMed] [Google Scholar]
- 35.Webster, M. K., and D. J. Donoghue. 1997. FGFR activation in skeletal disorders: too much of a good thing. Trends Genet. 13:178-182. [DOI] [PubMed] [Google Scholar]
- 36.Xu, H. L. K., and M. Goldfarb. 1998. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of SNT adapter proteins. J. Biol. Chem. 273:17987-17990. [DOI] [PubMed] [Google Scholar]
- 37.Yim, S. H., J. A. Hammer, and R. H. Quarles. 2001. Differences in signal transduction pathways by which platelet-derived and fibroblast growth factors activate extracellular signal-regulated kinase in differentiating oligodendrocytes. J. Neurochem. 76:1925-1934. [DOI] [PubMed] [Google Scholar]