Abstract
Min (Multiple intestinal neoplasia) mice carry a dominant mutation in the adenomatous polyposis coli (Apc) gene and develop multiple adenomas throughout their intestinal tract (Moser et al. 1990; Su et al. 1992). Polyp multiplicity in Min mice is greatly influenced by genetic background. A modifier locus, Mom1 (Modifier of Min 1), was identified and localized to distal mouse chromosome 4 (Moser et al. 1992; Dietrich et al. 1993), and accounts for some of the genetic variance in polyp multiplicity. Mom1 is a semidominant modifier of polyp size and multiplicity in Min mice (Gould and Dove 1997), and encodes the secretory type II nonpancreatic phospholipase A2 (Pla2g2a) gene (MacPhee et al. 1995; Cormier et al. 1997, 2000). We now report the identification of a second Modifier of Min 2 (Mom2) locus that is the result of a spontaneous mutation. One resistant Mom2 allele can suppress 88%–95% of polyps detected in ApcMin/+ mice, indicating that Mom2 acts in a dominant fashion. Linkage analysis has localized Mom2 to distal mouse chromosome 18. The effects of the Mom2 locus on reducing polyp multiplicity are stronger than the effects of the Mom1 locus, in both the small and large intestines. Some ApcMin/+ mice that carried one resistant Mom2 allele were tumor-free at 21 weeks of age, even in the absence of a resistant Mom1 allele. Thus, the resistant Mom2 allele can, in some cases, completely suppress the penetrance of the ApcMin mutation.
Colorectal cancer is the third leading cause of cancer morbidity and mortality in the U.S. and other Western developed countries (Greenlee et al. 2000). Familial adenomatous polyposis (FAP) is a dominantly inherited disorder characterized by the development of hundreds to thousands of adenomatous polyps throughout the intestinal tract; although the polyps are benign, some adenomas progress to malignant adenocarcinomas by 30–40 years of age (for review, see Goss and Groden 2000; Lal and Gallinger 2000). Most individuals with FAP have been shown to inherit one defective adenomatous polyposis coli (APC) gene, making them highly susceptible to the development of colorectal adenomas. These adenomas result from the inactivation of the remaining wild-type allele through somatic mutation and/or loss of heterozygosity (LOH) (Groden et al. 1991; Nishisho et al. 1991; Powell et al. 1992; Miyaki et al. 1994). In addition to the APC mutations seen in FAP patients, somatic mutations in both alleles of the APC gene have been shown to occur in the majority of sporadic colorectal tumors (Groden et al. 1991; Kinzler et al. 1991; Nishisho et al. 1991; Miyoshi et al. 1992; Powell et al. 1992; Miyaki et al. 1994). Inactivation of the APC gene results in the upregulation of β-catenin (CTNNB1), which binds to the transcription factor TCF4 (Korinek et al. 1997; Morin et al. 1997). Hereditary nonpolyposis colorectal cancer (HNPCC) is another dominantly inherited disorder which predisposes patients to colorectal carcinomas at an early age (Bocker et al. 1999). Although the primary genetic defect is a mutation in either the MLH1 or MSH2 mismatch repair genes (Fishel et al. 1993; Bronner et al. 1994; Papadopoulos et al. 1994), more than half of HNPCC tumors have secondary mutations which are either regulatory mutations in the CTNNB1 gene or inactivating mutations in the APC gene, suggesting that the majority of carcinomas in HNPCC patients develop through alteration of the APC-CTNNB1 pathway (Miyaki et al. 1999a). This recent finding, coupled with the observation that >70% of sporadic colorectal tumors have mutations or LOH of the APC gene itself (Miyaki et al. 1994), highlights the central role of the APC pathway in colorectal carcinogenesis.
Mouse models of cancer have proven useful for discovering oncogenes and serve as powerful tools to dissect complex genetic factors influencing cancer susceptibility (Buchberg and Siracusa 1995; Bedell et al. 1997; Balmain and Nagase 1998; Klausner 1999). Several models for intestinal tumorigenesis have been generated by introducing specific mutations into the murine homolog of the APC gene (Apc), allowing investigation of its role in polyp initiation and neoplastic progression to carcinoma (Moser et al. 1995a; Shibata et al. 1997; Smits et al. 1998; Takaku et al. 1998; Heyer et al. 1999; Taketo 1999). One such model for intestinal tumorigenesis that has been extensively studied is the Min (multiple intestinal neoplasia) mouse model, which was discovered through phenotypic screening following ENU mutagenesis (Moser et al. 1990). Min mice carry a dominant, fully penetrant mutation in the Apc gene, which results in the development of multiple adenomas throughout the intestinal tract (Moser et al. 1990; Su et al. 1992). The Min mouse is heterozygous for a nonsense mutation at codon 850 in exon 15 of the Apc gene (called ApcMin), which is on mouse chromosome 18 (Su et al. 1992). The murine ApcMin mutation is analogous to mutations found in the APC gene of some FAP kindreds (Miyoshi et al. 1992; Su et al. 1992). Embryos homozygous for the ApcMin mutation exhibit abnormalities prior to gastrulation and die early during development (Moser et al. 1995b).
Polyp multiplicity in Min mice is greatly influenced by genetic background (Moser et al. 1992; Dietrich et al. 1993). B6 mice heterozygous for the ApcMin mutation were initially reported to develop an average of 29 polyps (when counting was performed on ∼1/3 of the small intestine) and rarely live past 150 days of age (Moser et al. 1990, 1992; Su et al. 1992). Hybrid F1 offspring generated from B6 ApcMin/+ mice crossed to AKR/J (AKR), MA/MyJ (MA), or Mus musculus castaneus (CAST) mice show a significant decrease in polyp number (Moser et al. 1992; Dietrich et al. 1993). This difference in polyp number suggests that certain inbred strains carry modifier genes that affect polyp multiplicity in ApcMin mice. A modifier locus, Mom1 (Modifier of Min 1), was identified and localized to mouse chromosome 4 by quantitative trait loci (QTL) analysis (Dietrich et al. 1993). Mom1 is a semidominant modifier of both polyp size and multiplicity in ApcMin/+ mice (Gould and Dove 1997), and encodes the secretory type II nonpancreatic phospholipase A2 (Pla2g2a) gene (MacPhee et al. 1995; Cormier et al. 1997, 2000). However, the Mom1 locus was estimated to account for only ∼50% of the genetic variance in intestinal polyp multiplicity in ApcMin/+ animals, suggesting the presence of additional modifier loci within the mouse genome (Dietrich et al. 1993).
A series of crosses between B6 ApcMin/+ mice and other inbred strains were established to confirm the relationship between inheritance of the different alleles of the Pla2g2a gene and the Mom1 phenotype (MacPhee et al. 1995). These studies led us to the identification of a second modifier locus, Mom2 (Modifier of Min 2), that significantly affects polyp multiplicity in both the small and large intestines of ApcMin/+ mice. We describe here for the first time the identification, characterization, and map location of the Mom2 locus. Our data suggest that the resistant Mom2 allele is the result of a spontaneous mutation that was present in a B6 ApcMin/+ male. The impact of spontaneous mutations on quantitative phenotypic studies is discussed.
RESULTS
A series of intercrosses between C57BL/6J (B6) ApcMin/+ and other inbred strains were established to confirm the relationship between the inheritance of the Pla2g2a gene and the Mom1 phenotype (MacPhee et al. 1995). Hybrid F1 offspring that carried the ApcMin mutation were aged and analyzed at specific times for polyp multiplicity along the intestinal tract. Most crosses confirmed the relationship between high polyp numbers (60–100 polyps/mouse) in the small intestines of ApcMin/+ F1 animals inheriting the null Pla2g2a allele and low polyp numbers (10–30 polyps/mouse) in the small intestines of ApcMin/+ F1 animals inheriting a wild-type Pla2g2a allele from the test parent (MacPhee et al. 1995; data not shown).
An Exceptional Mating Cage Reveals an Unusual Distribution in Apc-Induced Intestinal Polyp Multiplicity
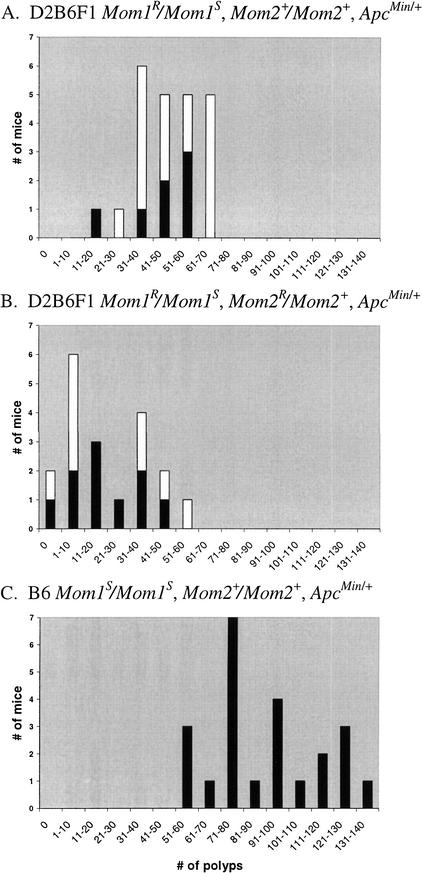
To test the effects of the DBA/2J (D2) inbred strain background on ApcMin-induced intestinal neoplasia, we performed an intercross by mating D2 females with B6 ApcMin/+ males. Offspring from most (4 of 5) mating cages gave a distribution where most ApcMin/+ F1 animals exhibited a moderate polyp multiplicity (31–70 polyps/mouse; Fig. 1A). However, one exceptional mating cage produced unusual results, in that their ApcMin/+ F1 offspring showed a wide range in polyp number (0–60 polyps/mouse; Fig. 1B). This pattern of small intestinal polyp multiplicity resembled a bimodal distribution, since 12 mice had low polyp numbers in the range of 0–30 polyps each and 7 mice had moderate polyp numbers in the range of 31–60 polyps each. In addition, this unusual distribution was clearly different from that exhibited by pure B6 ApcMin/+ mice, whose polyp numbers were in the range of 51–140 polyps/mouse (Fig. 1C).
Figure 1.
Polyp multiplicity of offspring from a DBA/2J X C57BL/6J − ApcMin/+ intercross. ApcMin/+ progeny from each mating cage were aged for 130–150 days and then analyzed for polyp number in the small intestines. Black bars represent male progeny, and white bars represent female progeny. (A) Polyp counts of the small intestine in D2B6F1 ApcMin/+ offspring from 4 of 5 mating cages showing the expected results. Average polyp multiplicity ± standard deviation was 48.2 ± 13.1. (B) Polyp counts of the small intestine in D2B6F1 ApcMin/+ offspring from the exceptional mating cage reveal an apparent bimodal distribution in polyp multiplicity. (C) Polyp counts of the small intestine in B6 ApcMin/+ offspring aged for 165–190 days; average polyp multiplicity ± standard deviation was 89.9 ± 25.0.
Since hybrid F1 animals are genetically identical, the apparent bimodal distribution could be due to distinct phenomena such as: (1) a difference in polyp multiplicity between males and females, (2) strain contamination (i.e., one of the inbred parents was heterozygous for a locus that conferred resistance to polyp multiplicity, such as the Mom1 locus), or (3) one parent harbored a spontaneous mutation that conferred resistance to intestinal polyp multiplicity. To investigate the first possibility, we examined the sex of each F1 offspring and found that the differences in polyp numbers were not sex-specific; the low polyp group was composed of roughly half males (58%) and half females (42%), as was the moderate polyp group (43% males and 57% females) (Fig. 1B). This finding eliminated the possibility that the B6 Y chromosome was responsible for the phenotype of low polyp multiplicity. To investigate the second possibility, we first examined the F1 offspring for the presence of the Mom1 locus by genotyping each mouse for the Pla2g2a gene (see Methods). Both parents were homozygous for their expected Pla2g2a alleles (the D2 parent was Pla2g2a+/Pla2g2a+ and the B6 parent was Pla2g2a−/Pla2g2a−), and all 19 F1 offspring were heterozygous (Pla2g2a+/Pla2g2a−) for the Pla2g2a gene (Fig. 1B). The F1 offspring were also typed for 22 microsatellite markers distributed on all autosomes and the X chromosome (see Methods). All F1 offspring were found to be heterozygous for D2 and B6 alleles at the loci tested. These results indicated that the apparent bimodal inheritance was not due to strain contamination. Support for the third possibility of spontaneous mutation was derived from further analysis of the distribution of F1 offspring with moderate (7 mice) and low (12 mice) polyp multiplicity. These mice showed no significant difference (χ2 = 1.32; p = 0.25) from a normal Mendelian ratio of 1:1, which would be expected if a single dominant modifier allele was segregating independently of the Apc gene. Therefore, the data suggested that the apparent bimodal distribution could be due to inheritance of a new mutation which was segregating in the D2B6F1 ApcMin/+ offspring.
The New Mutation Can Be Transmitted through the Germline
A new mutation that confers a dominant, resistant phenotype to polyp formation could potentially be invaluable in designing new strategies for prevention and/or treatment of APC-induced intestinal neoplasia. However, the offspring shown in Figure 1 were already euthanized. Therefore, we needed to confirm the existence of a new mutation by determining whether any of the surviving F1 offspring from the exceptional mating cage could transmit the new mutation. Seven D2B6F1 ApcMin/+ mice were the only remaining progeny from the exceptional mating cage that had yielded the apparent bimodal distribution of polyp multiplicity (Fig. 1B). We reasoned that the new mutation which was segregating in the F1 offspring must have been inherited from either the D2 female parent or the B6 male ApcMin/+ parent. Therefore, to eventually determine the chromosomal location of the new mutation as well as to make the new mutation congenic on the originating strain background, the seven D2B6F1 ApcMin/+ offspring were mated with B6 and/or D2 mice as shown in Table 1. We established a total of 11 mating cages; D2B6F1 ApcMin/+ males #1, 2, 5, and 6 were moved between a cage with B6 females and a cage with D2 females, D2B6F1 ApcMin/+ male #3 was mated with a B6 female, D2B6F1 ApcMin/+ female #4 was mated with a D2 male, and D2B6F1 ApcMin/+ female #7 was mated with a B6 male. The goal was to ensure that at least one F1 offspring that carried the mutation would be selected and pass the new mutation through the germline. This assessment was performed blind, since it was not possible to know a priori which surviving F1 animals might have inherited the new mutation.
Table 1.
N2 Offspring from Crosses of C57BL/6J and DBA/2J Mice to D2B6F1 ApcMin/+ Mice Reveal Germline Transmission of a New Modifier of Min 2 (Mom2) Locus
| D2B6F1 ApcMin parenta | Sex of F1 parent | # Polyps in F1 parentb | Age (days)c | D2B6F1 ApcMin × C57BL/6Ja | D2B6F1 ApcMin × DBA/2Ja | ||
|---|---|---|---|---|---|---|---|
| Highd | Lowd | Highd | Lowd | ||||
| 1 | male | 39 | 351 | 6 (70.2 ± 25.5) | 0 | 9 (58.1 ± 16.4) | 1 (10) |
| 2 | male | 37 | 331 | 6 (98.0 ± 24.9) | 0 | 9 (53.1 ± 22.4) | 0 |
| 3 | male | 61 | 351 | 5 (52.8 ± 14.7) | 0 | — | — |
| 4 | female | 100 | 315 | — | — | 9 (49.4 ± 19.4) | 1 (17) |
| Total mice | 17 | 0 | 27 | 2 | |||
| 5e | male | 6 | 472 | 2 (91.0 ± 48.1) | 17 (5.0 ± 3.3) | 4 (63.8 ± 18.2) | 8 (8.8 ± 6.9) |
| 6e | male | 7 | 405 | 3 (70.7 ± 34.0) | 9 (4.6 ± 4.8) | 1 (76) | 8 (7.3 ± 5.2) |
| 7e | female | 52 | 658 | 10 (100.5 ± 57.3) | 38 (6.3 ± 5.3) | — | — |
| Total mice | 15 | 64 | 5 | 16 | |||
Seven D2B6F1 ApcMin mice were crossed to C57BL/6J and/or DBA/2J mice as indicated in the last two column headings. The number of N2 offspring in the high and low polyp class is shown. N2 offspring were counted at 4–5 mo of age, with the exception of four mice from each backcross that were counted at >1 y of age. Average polyp numbers of resulting N2 offspring are shown ± standard deviation in parentheses.
The number of polyps present in the small intestine of each D2B6F1 ApcMin/+ parent.
The age in days of the D2B6F1 ApcMin/+ parent at the time polyps were counted.
Based on the distribution of polyps in the F1 animals (Fig. 1B), those animals with ≤24 polyps constitute the “low” polyp group and those animals with ≥25 polyps constitute the “high” polyp group.
Parents #5, #6, and #7 produced sufficient numbers of offspring with low polyp multiplicity and were therefore classified as the only D2B6F1 ApcMin parents that both carried and transmitted the resistant Mom2R allele.
Polyp number in the small intestine was used to determine which N2 offspring carried the new mutation. Based on the distribution of polyp numbers in the F1 offspring (Fig. 1), those N2 offspring with ≤24 polyps were placed in the “low” polyp class, and those N2 offspring with ≥25 polyps were placed in the “high” polyp class (Table 1). We also established the criteria that at least 20% of the N2 offspring from each mating cage had to fall in the low polyp class in order for the D2B6F1 ApcMin/+ parent to be considered a carrier of the new mutation. Table 1 shows that D2B6F1 ApcMin/+ parents #1, 2, 3, and 4 produced N2 progeny with mostly high polyp numbers (44 of 46 mice fell in the high polyp class), and therefore were not carriers of the new mutation. In contrast, D2B6F1 ApcMin/+ parents #5, 6, and 7 produced a total of 100 N2 progeny with both high (20 of 100 mice) and low (80 of 100 mice) polyp numbers (Table 1). Therefore, three of the seven D2B6F1 ApcMin/+ parents were able to transmit the new mutation through the germline. These results indicated that the low polyp phenotype was most likely due to an inherited mutation which acts in a dominant fashion to reduce polyp multiplicity. We named this new locus Mom2, for Modifier of Min 2.
Chromosomal Localization of the Mom2 Locus
The ratio of offspring with high polyp multiplicity to the total number of offspring in the F1 and N2 crosses was 11/26 (7/19 plus 4/7) and 20/100, respectively (Fig. 1, Table 1). Although the data from the F1 offspring (Fig. 1B) initially suggested that the new mutation was unlinked to the Apc gene (see above), the data from the N2 offspring, which constituted a four-fold larger sample size, suggested that the ApcMin mutation and the resistant Mom2 allele were indeed linked (in the cis configuration) on chromosome 18. The combined recombination frequency from both the F1 and N2 crosses (31/126) suggested that the Mom2 locus is 24.6 ± 3.8 cM away from the Apc gene on chromosome 18. Since the Apc gene had been localized 15 cM distal to the centromere of mouse chromosome 18 (Radice 2000), the most likely position for the Mom2 locus was distal to the Apc gene.
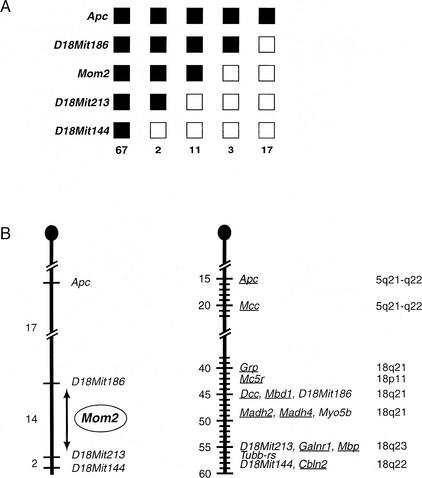
To test the hypothesis that the resistant Mom2 allele was located on the same chromosome as the ApcMin mutation, genomic DNA from all 100 N2 offspring was typed by PCR with microsatellite markers (Copeland et al. 1993; Dietrich et al. 1994, 1996) that spanned the length of chromosome 18. The expectation was that ApcMin/+ N2 offspring with low polyp numbers would carry a B6 allele at or near the location of the resistant Mom2 allele; these low-polyp offspring would have inherited a nonrecombinant chromosome 18 from their D2B6F1 ApcMin/+ parent. In contrast, ApcMin/+ N2 offspring with high polyp numbers would carry a D2 allele at or near the wild-type (susceptible) Mom2 allele; these high-polyp offspring would have inherited a recombinant chromosome 18 from their D2B6F1 ApcMin/+ parent. The haplotype results demonstrated a perfect concordance between polyp phenotype and allele genotype, in that all 80 N2 offspring with a low polyp number inherited the B6 allele at the D18Mit186 locus, and all 20 N2 offspring with a high polyp number inherited the D2 allele at the D18Mit213 locus (Fig. 2A). Therefore, the Mom2 locus must reside between the D18Mit186 and D18Mit213 loci (Fig. 2B). These data are consistent with the hypothesis that the resistant Mom2 allele was present in cis with the ApcMin mutation in D2B6F1 ApcMin/+ parents #5, 6, and 7. The resistant Mom2 allele could have arisen from a spontaneous mutation that occurred in the original B6 ApcMin/+ male, who was the father in the exceptional mating cage (Fig. 1B). Alternatively, the Mom2 mutation could have arisen in an ApcMin/+ predecessor of that same B6 ApcMin/+ male. Regardless of its origin, the mapping data confirm the position of the Mom2 locus distal to the Apc gene on mouse chromosome 18, and placed the Mom2 locus in a region of synteny with human chromosome 18q (Radice 2000).
Figure 2.
Mapping and chromosomal localization of the Mom2 locus in the mouse genome. (A) Summary of the results of the N2 backcross analysis. Loci mapped in the analysis are listed to the left. Black boxes represent the C57BL/6J (B6) allele, and white boxes represent the DBA/2J (D2) allele. The Mom2 allele was determined by phenotype: black boxes represent the resistant B6 allele where mice had ≤24 polyps, and white boxes represent the susceptible (wild-type) D2 allele where mice had ≥25 polyps in the small intestine. The numbers of each type of chromosome identified in the backcross progeny are listed at the bottom. (B) Genetic localization of the Mom2 locus. The region of chromosome 18 analyzed in our backcrosses (left) is shown adjacent to a partial consensus linkage map of mouse chromosome 18 (Radice 2000). The loci mapped are listed to the right, and the genetic distances (in centimorgans) between adjacent loci are listed to the left of the chromosomes. The two maps were arbitrarily aligned at the D18Mit186 and D18Mit213 loci. The data from our backcross analysis gives a genetic distance of 14 ± 3.5 cM between D18Mit186 and D18Mit213, whereas the genetic distance given in the consensus map shows the two loci as being 10 cM apart. The locations of the human homologues of mouse genes are listed to the right.
Effects of the Mom2 Locus on Polyp Multiplicity in the Small Intestine
Multiple modifier loci can significantly impact polyp multiplicity in ApcMin mice (Dietrich et al. 1993; Buchberg and Siracusa 1995; Dove et al. 1995; van der Houven van Oordt et al. 1999). The D2 strain was previously reported to contain only one modifier of ApcMin, which was the Mom1 locus (Moser et al. 1995a). Therefore, resistant and susceptible alleles of both the Mom1 and Mom2 loci were segregating in the N2 offspring of D2B6F1 ApcMin/+ parents #5, 6, and 7 (Table 1). To directly compare the effects of both modifier loci on polyp multiplicity, we classified each N2 offspring according to their genotype at both modifier loci. A resistant Mom1 allele (Mom1R) was determined by the presence of a wild-type Pla2g2a allele, whereas a susceptible Mom1 allele (Mom1S) was determined by the presence of a null Pla2g2a allele. A resistant Mom2 allele (Mom2R) was determined by a mouse having ≤24 polyps, whereas a susceptible Mom2 allele (Mom2+) was determined by a mouse having ≥25 polyps. The susceptible Mom2+ allele represents the wild-type (+) allele, since the resistant Mom2R allele resulted from a spontaneous mutation that decreased polyp multiplicity.
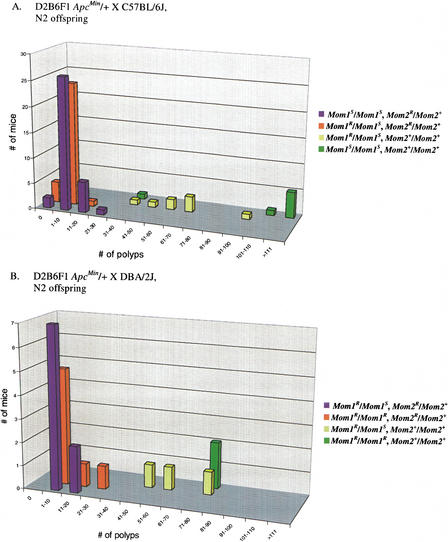
Figure 3 shows the distribution of the four classes of offspring from the B6 × D2B6F1 ApcMin backcross (Fig. 3A) and the D2 × D2B6F1 ApcMin backcross (Fig. 3B). In both the B6 and D2 backcrosses, the two classes of offspring that contain one resistant Mom2R allele (Mom1S/Mom1S, Mom2R/Mom2+ and Mom1R/Mom1S, Mom2R/Mom2+) have distinctly lower polyp numbers than the two classes of offspring that are homozygous for the wild-type Mom2 (Mom2+) allele (Mom1R/Mom1S, Mom2+/Mom2+ and Mom1S/Mom1S, Mom2+/Mom2+). In addition, 6 of 64 Mom2R/Mom2+ animals from the B6 backcross exhibited a complete suppression of polyp formation in the small intestines, regardless of the allele status at the Mom1 locus (Fig. 3A). These data show that even in a predominantly susceptible genetic background [where on average, 75% of the genome is of B6 origin in (B6 × D2B6F1)N2 ApcMin/+ offspring], one Mom2R allele can so effectively suppress the penetrance of the ApcMin mutation that no polyps are found in a subset of animals.
Figure 3.
Distribution of N2 progeny from the B6 and D2 backcrosses to D2B6F1 ApcMin/+ mice based on polyp number in the small intestine and genotypes at the Mom1 and Mom2 loci. N2 offspring (Table 1) were genotyped for the Pla2g2a gene (see Methods) to determine whether they carried the B6 susceptible Mom1S allele and/or the D2 resistant Mom1R allele. N2 offspring were phenotyped for polyp number to determine whether they were homozygous for susceptible Mom2 (Mom2+/Mom2+) alleles (≥25 polyps = high-polyp phenotype) or whether they were heterozygous for the resistant Mom2R (Mom2R/Mom2+) allele (≤24 polyps = low-polyp phenotype); molecular markers on mouse chromosome 18 were consistent with the determination of Mom2 allele status (Fig. 2). Genotypes at the Mom1 and Mom2 loci are shown to the right. (A) A total 79 N2 offspring were generated from the backcross to B6, and (B) a total of 21 N2 offspring were generated from the backcross to D2 (Table 1).
Table 2A shows the average polyp numbers in the small intestine for all four classes of offspring from both backcrosses. Mom1R/Mom1S offspring (that were Mom2+/Mom2+) from the backcross to B6 showed a 53.6% reduction in average polyp multiplicity compared to their Mom1S/Mom1S littermates (60.6 vs. 130.6); similarly, Mom1R/Mom1S offspring (that were Mom2R/Mom2+) showed a 59.0% reduction in average polyp multiplicity compared to their Mom1S/Mom1S littermates (3.2 vs. 7.8). Both comparisons were statistically significant at the p < 0.005 level. This finding is consistent with previous estimates that the presence of one resistant modifier allele at the Mom1 locus reduces polyp multiplicity by ∼50% (Dietrich et al. 1993; MacPhee et al. 1995; Gould et al. 1996).
Table 2.
Effects of the Mom1 and Mom2 Loci on Polyp Multiplicity in the Small Intestine of N2 Offspring from the B6 × D2B6F1 ApcMin and D2 × D2B6F1 ApcMin Backcrossesa
| Genotypes of B6 × D2B6F1 ApcMin N2 offspring | Mom1S/Mom1S | Mom1R/Mom1S | p values |
|---|---|---|---|
| Mom2+/Mom2+ | 130.6 ± 51.6 (7) | 60.6 ± 16.9 (8) | p < 0.005 |
| Mom2R/Mom2+ | 7.8 ± 5.1 (35) | 3.2 ± 2.9 (29) | p < 0.001 |
| p values | p < 0.001 | p < 0.001 |
| Genotypes of D2 × D2B6F1 ApcMin N2 offspring | Mom1R/Mom1S | Mom1R/Mom1R | Total |
|---|---|---|---|
| Mom2+/Mom2+ | 58.3 ± 17.9 (3) | 78.0 ± 2.8 (2) | 65.4 ± 16.1 (5) |
| Mom2R/Mom2+ | 7.8 ± 4.5 (9) | 8.3 ± 7.9 (7) | 8.0 ± 6.0 (16) |
| p values | p < 0.001 |
N2 progeny of D2B6F1 ApcMin/+ parents #5, #6, and #7 from Table 1 were classified according to strain background as well as genotype and phenotype at the Mom1 and Mom2 loci, respectively. Average polyp numbers are shown ± standard deviation. The number of offspring in each group is shown in parentheses. P values were determined by the two sample t-test.
N2 offspring from the backcross to D2 could be either homozygous for resistance or heterozygous at the Mom1 locus (Table 2). The differences in average polyp multiplicity between Mom1R/Mom1R and Mom1R/Mom1S offspring that were homozygous recessive for the susceptible Mom2 allele (Mom2+/Mom2+) from the D2 backcross could not be reliably assessed, due to the small numbers of animals in each group (two and three mice, respectively). Therefore, to assess the effects of one resistant Mom2 allele, we combined the Mom1R/Mom1R and Mom1R/Mom1Sclasses (Table 2). Mom2R/Mom2+ offspring showed an 88% reduction in average polyp multiplicity (8.0 polyps/mouse) compared to their Mom2+/Mom2+ littermates (65.4 polyps/mouse). Statistical analyses revealed that the average polyp number in Mom2R/Mom2+ offspring was significantly (p < 0.001) lower than the average polyp number in Mom2+/Mom2+ offspring.
One resistant Mom2 allele provided even more striking differences in the N2 offspring from the B6 backcross (Table 2). Mom2R/Mom2+ offspring showed a 94%–95% reduction in average polyp multiplicity compared to their Mom2+/Mom2+ littermates, regardless of whether their genotype at the Mom1 locus was Mom1S/Mom1S (7.8 vs. 130.6) or Mom1R/Mom1S (3.2 vs. 60.6). Statistical analyses revealed that the differences in average polyp multiplicity between Mom2R/Mom2+ and Mom2+/Mom2+ offspring was highly significant (p < 0.001)
These dramatic and highly significant (p < 0.001) reductions in average polyp number resulting from the presence of one Mom2R allele in offspring from both backcrosses was observed in Mom1R/Mom1R, Mom1R/Mom1S, and Mom1S/Mom1S mice. The fact that these highly significant reductions in polyp number are observed regardless of which Mom1 alleles are present suggests that the Mom1 and Mom2 loci act independently of each other. Furthermore, one Mom2R allele can reduce average polyp number by 88%–95%; in contrast, one Mom1R allele can reduce average polyp number by at most ∼50%. These data show that the Mom2 mutation has more dramatic effects on reducing polyp number than the Mom1 locus.
Effects of the Mom2 Locus on Polyp Multiplicity in the Colon
The incidence of colon polyps was also markedly reduced by one resistant Mom2 allele (Table 3). The data show that 71.4% of Mom1S/Mom1S and 37.5% of Mom1R/Mom1S offspring from the backcross to B6 had colon polyps in the absence of a Mom2R allele. However, in the presence of one resistant Mom2 allele, the incidence of colon polyps decreased more than eight-fold to 8.6% in Mom1S/Mom1S offspring and more than 11-fold to 3.4% in Mom1R/Mom1S offspring. These differences between the Mom2R/Mom2+ and Mom2+/Mom2+ classes of offspring were significant at the p < 0.03 level. Similarly, a dramatic decrease in colon polyp incidence was observed in N2 offspring from the backcross to D2 mice (Table 3). The incidence of colon polyps decreased more than nine-fold from 60.0% in Mom2+/Mom2+ offspring to 6.3% in Mom2R/Mom2+ offspring. This difference between the Mom2R/Mom2+ and Mom2+/Mom2+ classes of offspring was significant at the p < 0.03 level. Taken together, these data indicate that (1) Mom2 effectively suppresses polyp formation and/or development in both the small and large intestines of ApcMin animals, and (2) the suppression by Mom2R is detectable in both the B6 and D2 strain backgrounds.
Table 3.
Effects of the Mom1 and Mom2 Loci on the Incidence of Colon Polyps in N2 Offspring from the B6 × D2B6F1 ApcMin and D2 × D2B6F1 ApcMin Backcrossesa
| Genotypes of B6 × D2B6F1 ApcMin N2 offspring | Mom1S/Mom1S | Mom1R/Mom1S | p values |
|---|---|---|---|
| Mom2+/Mom2+ | 71.4% (5/7) | 37.5% (3/8) | p = 0.3147 |
| Mom2R/Mom2+ | 8.6% (3/35) | 3.4% (1/29) | p = 0.6198 |
| p values | p = 0.0012 | p = 0.0256 |
| Genotypes of D2 × D2B6F1 ApcMin N2 offspring | Mom1R/Mom1S | Mom1R/Mom1R | Total |
|---|---|---|---|
| Mom2+/Mom2+ | 66.7% (2/3) | 50.0% (1/2) | 60.0% (3/5) |
| Mom2R/Mom2+ | 0.0% (0/9) | 14.2% (1/7) | 6.3% (1/16) |
| p value | p = 0.0276 |
aN2 progeny of D2B6F1 ApcMin parents #5, #6, and #7 from Table 1 were classified according to strain background as well as genotype and phenotype at the Mom1 and Mom2 loci, respectively. The % of mice within each class is given. The numbers in parentheses show the actual number of mice with ≥1 colon polyp over the total number of mice in each class. P values were determined by the Fisher's exact test.
DISCUSSION
The results presented demonstrate that we have identified a new modifier locus, Mom2, which acts in a dominant fashion to markedly reduce intestinal polyp multiplicity in ApcMin/+ mice. The mutation was present in the original B6 ApcMin/+ male parent who was mated with a D2 female. This “founder” male lived to be more than one year old (he was euthanized at 14 months of age) and had only 33 polyps in his small intestine and no polyps in his large intestine. In contrast, B6 ApcMin/+ males that do not carry the resistant Mom2R mutation have an average of 89.9 ± 25.0 tumors in the small intestine and a 95.6% incidence of colon polyps by six months of age. In our laboratory, B6 ApcMin/+ males and females tend to live for 5–6 months; their death is usually the result of anemia due to bleeding caused by intestinal polyps and/or intestinal blockage caused by a high tumor burden. These findings suggest that the lifespan of ApcMin mice is increased by the presence of one resistant Mom2 allele.
Moreover, although the ApcMin mutation was reported to have 100% penetrance on a B6 background (Moser et al. 1990), our data show a complete absence of polyp formation in 6 of 64 N2 offspring from the B6 backcross that inherited one resistant Mom2R allele (Fig. 3). Although two males and two females carried one resistant Mom1 allele (which could contribute to the absence of polyp formation), the remaining two male mice with no polyps were homozygous susceptible (Mom1S/Mom1S) at the Mom1 locus (Fig. 3). Thus, the resistant Mom2R allele is capable of completely suppressing polyp formation in 9.4% of the progeny regardless of the presence or absence of a resistant Mom1 allele. In addition, two of the 12 low-polyp D2B6F1 ApcMin/+ offspring from the exceptional mating cage (Fig. 1B) actually had no polyps, whereas their non-Mom2R counterparts (Fig. 1A) had 11 or more polyps each. Our results clearly demonstrate that the presence of one resistant Mom2R allele is capable of significantly reducing the penetrance of the ApcMin mutation in both the small and large intestine (Tables 2 and 3).
Comparison of the Effects of the Mom2 and Mom1 Loci
We have observed that Mom2 acts in a dominant fashion, with one resistant Mom2 allele significantly suppressing polyp multiplicity in animals carrying the ApcMin/+ mutation. The results demonstrated that a single resistant Mom2 allele is a more potent modifier of polyp multiplicity than either one or two resistant Mom1 alleles. ApcMin mice that were heterozygous for Mom2R/Mom2+ showed an 88%–95% reduction in polyp number in the small intestine. These dramatic reductions in ApcMin-induced tumorigenesis caused by one resistant Mom2 allele are much greater than the ∼50% reduction in polyp multiplicity detected with one resistant Mom1 allele. In addition, the action of the resistant Mom2 allele is not restricted to either the B6 or D2 background, as its effects are detectable regardless of strain background.
Since the magnitude of the effects of the resistant alleles of Mom1 and Mom2 together are greater than the effects of either the Mom1R or Mom2R alleles alone (Tables 1, 2, and 3), the most likely hypothesis is that Mom1 and Mom2 act in different pathways. If Mom1 and Mom2 acted in the same pathway, we would not have detected the effects of the Mom1 resistant allele (∼50% reduction) in the presence of the much more potent Mom2 resistant allele (88%–95% reduction). Further analysis of Mom2 and how it affects polyp formation could reveal a distinct pathway with which polyp development can be potentially controlled in humans.
We also observed an 8–11 fold reduction in polyp incidence in the colon in the presence of a single resistant Mom2R allele. This difference was statistically significant (p < 0.03, Table 3), indicating that Mom2 functions in controlling polyp development in the colon as well as the small intestine. Interestingly, although the presence of one resistant Mom1R allele appeared to reduce colon polyp incidence by ∼50% (71.4% compared to 37.5% in Mom2+/Mom2+ offspring and 8.6% compared to 3.4% in Mom2R/Mom2+ offspring), these differences were not statistically significant (P < 0.3, Table 3). This finding suggests that the Mom1 locus does not affect the incidence of colon polyps in ApcMin mice. This finding is in contrast to previously published reports indicating that Mom1 (specifically Pla2g2a) significantly affects colon polyp multiplicity (Cormier et al. 2000). These differing results could be due to differences in sample size or strain background.
Chromosomal Location of the Mom2 Region
Molecular genetic linkage studies of the Mom2 phenotype showed that the Mom2 locus most likely resides within a 10–14 cM interval bounded by D18Mit186 and D18Mit213. The Mom2 region is syntenic with human chromosome 18q21 and 18q23 (Fig. 2; Radice 2000). Chromosome 18q21 is a region frequently found to undergo LOH in human colorectal tumors (Takagi et al. 1996; Thiagalingam et al. 1996). At least two genes in this region have been shown to result in intestinal neoplasia when cells undergo LOH, namely Smad2 and Smad4 (Koyama et al. 1999; Miyaki et al. 1999b; Tarafa et al. 2000; Xu et al. 2000). In addition, inheritance of an inactivating mutation in SMAD4 is responsible for a subset of the juvenile polyposis disorders (Howe et al. 1998). However, it is unclear as to how to reconcile the action of a Smad4 mutation acting as a dominant modifier that reduces the number of intestinal polyps unless it is a hypermorph. Alternatively, other genes within this region of mouse chromosome 18 may have been altered by the Mom2 mutation. Genes that influence cell division, differentiation, or survival of cells in the intestines may be candidates for the Mom2 locus. The mechanism by which the resistant Mom2R allele provides protection from polyp development is currently under study. It is not yet known whether the protective effects of the Mom2R mutation are limited to intestinal tissue or whether these effects extend to other Apc-induced tumors such as gastric, desmoid, and mammary tumors (Moser et al. 1993; Smits et al. 1998). Refinement of the map position of the Mom2 locus, cloning of the Mom2 gene, and characterization of the mutant Mom2 allele will ultimately answer these questions.
Importance of Mating Cage Comparisons
The fact that only one of five initial D2 × B6 ApcMin/+ mating cages produced offspring that exhibited this novel “low-polyp” phenotype suggests that the resistant Mom2R allele was the result of a spontaneous mutation. The linkage between the ApcMin mutation and the resistant Mom2R allele indicates that the new mutation was present in the original B6 ApcMin/+ male parent and most likely occurred on the B6 ApcMin chromosome. This finding highlights the importance of comparing results from individual mating cages, even if the parents originate from the same inbred strain(s).
The observation that the original B6 ApcMin/+ male parent appeared to transmit the resistant Mom2R allele to ∼50% of its ApcMin/+ offspring. 11 F1 offspring had moderate polyp numbers and 15 F1 offspring had low polyp numbers (χ2 = 0.62; 0.5 < p < 0.25) suggests that this male inherited the mutant ApcMin, Mom2R chromosome 18 from its ApcMin parent. Since the original B6 ApcMin/+ male parent had been purchased from The Jackson Laboratory (Bar Harbor), it is possible that siblings, cousins, or other relatives of this B6 ApcMin/+ male may also have inherited the Mom2 mutation. Therefore, results of studies with ApcMin/+ mice should be subjected to careful evaluation.
Spontaneous Mutations and Their Impact on QTL Studies
In any mating scheme designed to continuously select for a specific allele or chromosomal region, mutations that are linked to the selected allele or region are likely to be inherited along with the selected allele or region. In many cases, these linked mutations may be detrimental to the health of the animal; however, in the case of ApcMin/+ mice, any linked mutation that improves the health or extends the life of the animal would have an increased probability of being passed on to offspring.
The impact of these findings extends to many other types of experiments such as pharmacological drug testing, behavior studies, and disease phenotype-related studies. A spontaneous mutation with effects on the phenotype under study could distort the findings. Furthermore, crosses designed to detect quantitative trait loci usually require many mating cages to produce the large number of offspring required to detect the QTLs; the results of such studies could be skewed if one or several parents carried a new mutation that affected the phenotype under study. Spontaneous mutations will continue to occur in any set of crosses; these mutations can be detected because of diligence in examining phenotype and meticulous comparisons of results between individual mating cages.
The identification of a spontaneous dominant mutation which has dramatic effects on reducing polyp multiplicity in ApcMin/+ mice now opens the door for mutagenesis experiments designed to screen for additional modifier genes that influence Apc-induced tumorigenesis (Schimenti and Bucan 1998; Justice et al. 1999). Mutagenesis strategies can be designed to detect either dominant mutations that confer resistance to intestinal polyps on an otherwise susceptible background or recessive mutations that confer susceptibility to intestinal polyps on an otherwise resistant background in ApcMin/+ mice. The significance of designing studies to detect dominant mutations is that only a single allele is altered to produce the desired effect of polyp reduction and/or elimination. Understanding the molecular basis of such dominant mutations and the pathways they affect may directly lead to new methods for tumor prevention.
METHODS
Mice
All mice were bred at the Kimmel Cancer Center animal facility, except for the original C57BL/6J ApcMin/+ males and DBA/2J animals, which were purchased from The Jackson Laboratory (Bar Harbor).
DNA Isolation
Genomic DNAs were isolated from tail biopsies as described previously (Siracusa et al. 1987).
Genotyping for the Pla2g2a Locus
In the strains susceptible to polyp formation, a 1-bp thymidine insertion in exon 3 of the Pla2g2a gene abolishes the BamHI site present in the wild-type sequence (Kennedy et al. 1995; MacPhee et al. 1995). Using the primers (forward 5′ CAA TAC AGG TCC AAG GGA AC 3′ and reverse 5′ GTG ATT TGG CCC CCT TGG TG 3′), a fragment of ∼400 bp containing the BamHI site is amplified from the Pla2g2a locus. Each 20 μL PCR reaction contains 100 ng of genomic tail DNA, which is amplified along with 50 ng each of the oligomers, 0.5 mM of each nucleotide (dCTP, dGTP, dTTP, and dATP), in a buffer (final concentration 50 mM KCl, 10 mM Tris-HCl pH 8.0, 1.5 mM MgCl2 and 0.1 mg/mL gelatin) along with 5 U/μL of Taq DNA polymerase (Roche Molecular Biochemicals). Samples were amplified under the following conditions: one cycle at 94°C for 4 min, followed by 40 cycles at 94°C for 30 sec, 60°C for 45 sec, and 72°C for 30 sec, followed by one cycle at 72°C for 7 min. Five μL of the PCR product was then incubated with 10 U of BamHI overnight, which cleaves the wild-type sequence into two fragments (∼100 and 300 bp), without any effect on the mutant sequence in which the BamHI site has been abolished. Fragments were resolved on a 3% TBE agarose gel and visualized by staining with ethidium bromide (Sambrook et al. 1989).
Genotyping for the ApcMin Mutation
Mice were typed for the presence of the A-T transversion in codon 850 of the Apc gene by PCR analysis of genomic tail DNA (Dietrich et al. 1993).
Genotyping with SSLP Markers
SSLP markers were selected based on polymorphisms that were identified between the C57BL/6J and DBA/2J strains (Copeland et al. 1993; Dietrich et al. 1994, 1996). SSLP primer pairs were purchased from Research Genetics. Polymorphisms of 12 bp or higher were detected on 3% TBE agarose gels and visualized by staining with ethidium bromide (Sambrook et al. 1989). The following markers were used for genotyping: D1Mit416, D2Mit237, D3Mit6, D5Mit370, D6Mit101, D6Mit274, D7Mit301, D8Mit4, D9Mit191, D10Mit10, D10Mit180, D11Mit179, D11Mit333, D12Mit157, D13Mit248, D14Mit115, D15Mit111, D15Mit161, D16Mit114, D17Mit139, D19Mit19, and DXMit25.
Scoring for Polyps along the Intestinal Tract
Mice carrying the ApcMin/+ mutation were aged to 130–150 days and sacrificed by CO2 asphyxiation. The entire intestinal tract was dissected. The small intestine was divided into three sections (proximal, middle, and distal), gently scraped to remove fecal matter, and then cut open longitudinally. Each piece was washed with phosphate buffered saline (pH 7.0) to clear away residual fecal matter. The colon was also cleaned, cut longitudinally, and washed with PBS. The colon and each of the small intestinal segments were scored for polyps (≥0.13 mm in diameter) using a Nikon SMZ-U dissecting microscope (15× magnification). Polyps were counted by a single observer prior to genotyping of each mouse.
Acknowledgments
The symbol Mom2 was approved by the International Committee on Standardized Genetic Nomenclature for Mice. We thank Dr. Leslie Lock, Dr. Jay Rothstein, and Peter Wermuth for critical reading of the manuscript, and Dr. Walter Hauck and Ed Pequignot for statistical analyses. K.A.S. was supported by N.I.H. Training Grant T32-CA09678. R.K. was supported by N.I.H. Training Grant T32-CA09678. Research was supported in part by grants from the N.C.I. to A.M.B. and L.D.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL buchberg@mail.kimmelcancercenter.org; FAX (215) 923-4153.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.206002.
REFERENCES
- Balmain A, Nagase H. Cancer resistance genes in mice: Models for the study of tumour modifiers. Trends Genet. 1998;14:139–144. doi: 10.1016/s0168-9525(98)01422-x. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Largaespada DA, Jenkins NA, Copeland NG. Mouse models of human disease. Part II: Recent progress and future directions. Genes Dev. 1997;11:11–43. doi: 10.1101/gad.11.1.11. [DOI] [PubMed] [Google Scholar]
- Bocker T, Ruschoff J, Fishel R. Molecular diagnostics of cancer predisposition: Hereditary non-polyposis colorectal carcinoma and mismatch repair defects. Biochim Biophys Acta. 1999;1423:O1–O10. doi: 10.1016/s0304-419x(99)00008-6. [DOI] [PubMed] [Google Scholar]
- Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- Buchberg AM, Siracusa LD. The Genetics of Cancer Susceptibility. In: Fortner JG, Rhoads JE, editors. General Motors Cancer Research Foundation: Accomplishments in Cancer Research. Philadelphia, PA: J.B. Lippincott Company; 1995. [Google Scholar]
- Copeland NG, Gilbert DJ, Jenkins NA, Nadeau JH, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Steen RG, Lincoln SE, et al. Genome maps IV 1993. Wall chart. Science. 1993;262:67–82. doi: 10.1126/science.8211131. [DOI] [PubMed] [Google Scholar]
- Cormier RT, Bilger A, Lillich AJ, Halberg RB, Hong KH, Gould KA, Borenstein N, Lander ES, Dove WF. The Mom1 AKR intestinal tumor resistance region consists of Pla2g2a and a locus distal to D4Mit64. Oncogene. 2000;19:3182–3192. doi: 10.1038/sj.onc.1203646. [DOI] [PubMed] [Google Scholar]
- Cormier RT, Hong KH, Halberg RB, Hawkins TL, Richardson P, Mulherkar R, Dove WF, Lander ES. Secretory phospholipase Pla2g2a confers resistance to intestinal tumorigenesis [see comments] Nat Genet. 1997;17:88–91. doi: 10.1038/ng0997-88. [DOI] [PubMed] [Google Scholar]
- Dietrich WF, Lander ES, Smith JS, Moser AR, Gould KA, Luongo C, Borenstein N, Dove W. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell. 1993;75:631–639. doi: 10.1016/0092-8674(93)90484-8. [DOI] [PubMed] [Google Scholar]
- Dietrich WF, Miller J, Steen R, Merchant MA, Damron-Boles D, Husain Z, Dredge R, Daly MJ, Ingalls KA, O'Connor TJ, et al. A comprehensive genetic map of the mouse genome [see comments] [published erratum appears in Nature 1996 May 9;381(6578):172] Nature. 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Dietrich WF, Miller JC, Steen RG, Merchant M, Damron D, Nahf R, Gross A, Joyce DC, Wessel M, Dredge RD, et al. A genetic map of the mouse with 4,006 simple sequence length polymorphisms. Nat Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- Dove WF, Gould KA, Luongo C, Moser AR, Shoemaker AR. Emergent issues in the genetics of intestinal neoplasia. Cancer Surv. 1995;25:335–355. [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Goss KH, Groden J. Biology of the adenomatous polyposis coli tumor suppressor. J Clin Oncol. 2000;18:1967–1979. doi: 10.1200/JCO.2000.18.9.1967. [DOI] [PubMed] [Google Scholar]
- Gould KA, Dietrich WF, Borenstein N, Lander ES, Dove WF. Mom1 is a semi-dominant modifier of intestinal adenoma size and multiplicity in Min/+ mice. Genetics. 1996;144:1769–1776. doi: 10.1093/genetics/144.4.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KA, Dove WF. Localized gene action controlling intestinal neoplasia in mice. Proc Natl Acad Sci. 1997;94:5848–5853. doi: 10.1073/pnas.94.11.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50:7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- Heyer J, Yang K, Lipkin M, Edelmann W, Kucherlapati R. Mouse models for colorectal cancer. Oncogene. 1999;18:5325–5333. doi: 10.1038/sj.onc.1203036. [DOI] [PubMed] [Google Scholar]
- Howe JR, Ringold JC, Summers RW, Mitros FA, Nishimura DY, Stone EM. A gene for familial juvenile polyposis maps to chromosome 18q21.1. Am J Hum Genet. 1998;62:1129–1136. doi: 10.1086/301840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Hum Mol Genet. 1999;8:1955–1963. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- Kennedy BP, Payette P, Mudgett J, Vadas P, Pruzanski W, Kwan M, Tang C, Rancourt DE, Cromlish WA. A natural disruption of the secretory group II phospholipase A2 gene in inbred mouse strains. J Biol Chem. 1995;270:22378–22385. doi: 10.1074/jbc.270.38.22378. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Nilbert MC, Su LK, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hedge P, McKechnie D, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- Klausner RD. Studying cancer in the mouse. Oncogene. 1999;18:5249–5252. doi: 10.1038/sj.onc.1203089. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma [see comments] Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Koyama M, Ito M, Nagai H, Emi M, Moriyama Y. Inactivation of both alleles of the DPC4/SMAD4 gene in advanced colorectal cancers: Identification of seven novel somatic mutations in tumors from Japanese patients. Mutat Res. 1999;406:71–77. doi: 10.1016/s1383-5726(99)00003-5. [DOI] [PubMed] [Google Scholar]
- Lal G, Gallinger S. Familial adenomatous polyposis. Semin Surg Oncol. 2000;18:314–323. doi: 10.1002/(sici)1098-2388(200006)18:4<314::aid-ssu6>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- MacPhee M, Chepenik KP, Liddell RA, Nelson KK, Siracusa LD, Buchberg AM. The secretory phospholipase A2 gene is a candidate for the Mom1 locus, a major modifier of ApcMin-induced intestinal neoplasia. Cell. 1995;81:957–966. doi: 10.1016/0092-8674(95)90015-2. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Iijima T, Kimura J, Yasuno M, Mori T, Hayashi Y, Koike M, Shitara N, Iwama T, Kuroki T. Frequent mutation of beta-catenin and APC genes in primary colorectal tumors from patients with hereditary nonpolyposis colorectal cancer. Cancer Res. 1999a;59:4506–4509. [PubMed] [Google Scholar]
- Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, Utsunomiya J, Kuroki T, Mori T. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999b;18:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- Miyaki M, Konishi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, Tanaka K, Muraoka M, Takahashi H, Amada Y, Fukayama M, et al. Characteristics of somatic mutation of the adenomatous polyposis coli gene in colorectal tumors. Cancer Res. 1994;54:3011–3020. [PubMed] [Google Scholar]
- Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Somatic mutations of the APC gene in colorectal tumors: Mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC [see comments] Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Moser AR, Dove WF, Roth KA, Gordon JI. The Min (multiple intestinal neoplasia) mutation: Its effect on gut epithelial cell differentiation and interaction with a modifier system. J Cell Biol. 1992;116:1517–1526. doi: 10.1083/jcb.116.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser AR, Luongo C, Gould KA, McNeley MK, Shoemaker AR, Dove WF. ApcMin: A mouse model for intestinal and mammary tumorigenesis. Eur J Cancer. 1995a;31A:1061–1064. doi: 10.1016/0959-8049(95)00181-h. [DOI] [PubMed] [Google Scholar]
- Moser AR, Mattes EM, Dove WF, Lindstrom MJ, Haag JD, Gould MN. ApcMin, a mutation in the murine Apc gene, predisposes to mammary carcinomas and focal alveolar hyperplasias. Proc Natl Acad Sci. 1993;90:8977–8981. doi: 10.1073/pnas.90.19.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247:322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- Moser AR, Shoemaker AR, Connelly CS, Clipson L, Gould KA, Luongo C, Dove WF, Siggers PH, Gardner RL. Homozygosity for the Min allele of Apc results in disruption of mouse development prior to gastrulation. Dev Dyn. 1995b;203:422–433. doi: 10.1002/aja.1002030405. [DOI] [PubMed] [Google Scholar]
- Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsunomiya J, Baba S, Hedge P. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- Radice GL. Committee Reports, Mouse Genome Database (MGD). Bar Harbor, Maine: Mouse Genome Informatics, The Jackson Laboratory; 2000. Chromosome 18. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, a Laboratory Manual. New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schimenti J, Bucan M. Functional genomics in the mouse: Phenotype-based mutagenesis screens. Genome Res. 1998;8:698–710. doi: 10.1101/gr.8.7.698. [DOI] [PubMed] [Google Scholar]
- Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, Matsumoto H, Takano H, Akiyama T, Toyoshima K, Kanamaru R, Kanegae Y, Saito I, Nakamura Y, Shiba K, Noda T. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- Siracusa LD, Russell LB, Jenkins NA, Copeland NG. Allelic variation within the Emv-15 locus defines genomic sequences closely linked to the agouti locus on mouse chromosome 2. Genetics. 1987;117:85–92. doi: 10.1093/genetics/117.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits R, van der Houven van Oordt W, Luz A, Zurcher C, Jagmohan-Changur S, Breukel C, Khan PM, Fodde R. Apc1638N: A mouse model for familial adenomatous polyposis-associated desmoid tumors and cutaneous cysts. Gastroenterology. 1998;114:275–283. doi: 10.1016/s0016-5085(98)70478-0. [DOI] [PubMed] [Google Scholar]
- Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene [published erratum appears in Science 1992 May 22;256(5060):1114] Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- Takagi Y, Kohmura H, Futamura M, Kida H, Tanemura H, Shimokawa K, Saji S. Somatic alterations of the DPC4 gene in human colorectal cancers in vivo [see comments] Gastroenterology. 1996;111:1369–1372. doi: 10.1053/gast.1996.v111.pm8898652. [DOI] [PubMed] [Google Scholar]
- Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- Taketo MM. Apc gene knockout mice as a model for familial adenomatous polyposis. Prog Exp Tumor Res. 1999;35:109–119. doi: 10.1159/000062007. [DOI] [PubMed] [Google Scholar]
- Tarafa G, Villanueva A, Farre L, Rodriguez J, Musulen E, Reyes G, Seminago R, Olmedo E, Paules AB, Peinado MA, Bachs O, Capella G. DCC and SMAD4 alterations in human colorectal and pancreatic tumor dissemination. Oncogene. 2000;19:546–555. doi: 10.1038/sj.onc.1203353. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, Willson JK, Markowitz S, Hamilton SR, Kern SE, Kinzler KW, Vogelstein B. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- van der Houven van Oordt CW, Smits R, Schouten TG, Houwing-Duistermaat JJ, Williamson SL, Luz A, Meera Khan P, van der Eb AJ, Breuer ML, Fodde R. The genetic background modifies the spontaneous and X-ray-induced tumor spectrum in the Apc1638N mouse model. Genes Chromosomes Cancer. 1999;24:191–198. doi: 10.1002/(sici)1098-2264(199903)24:3<191::aid-gcc3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Xu X, Brodie SG, Yang X, Im YH, Parks WT, Chen L, Zhou YX, Weinstein M, Kim SJ, Deng CX. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene. 2000;19:1868–1874. doi: 10.1038/sj.onc.1203504. [DOI] [PubMed] [Google Scholar]