Abstract
Nuclear microsatellite loci (2- to 5-bp tandem repeats) would seem to be ideal markers for population genetic monitoring because of their abundant polymorphism, wide dispersal in vertebrate genomes, near selective neutrality, and ease of assessment; however, questions about their mode of generation, mutation rates and ascertainment bias have limited interpretation considerably. We have assessed the patterns of genomic diversity for ninety feline microsatellite loci among previously characterized populations of cheetahs, lions and pumas in recapitulating demographic history. The results imply that the microsatellite diversity measures (heterozygosity, allele reconstitution and microsatellite allele variance) offer proportionate indicators, albeit with large variance, of historic population bottlenecks and founder effects. The observed rate of reconstruction of new alleles plus the growth in the breadth of microsatellite allele size (variance) was used here to develop genomic estimates of time intervals following historic founder events in cheetahs (12,000 yr ago), in North American pumas (10,000–17,000 yr ago), and in Asiatic lions of the Gir Forest (1000–4000 yr ago).
[Supplemental material available online at http://rex.nci.nih.gov/lgd/front_page.htm and at http://www.genome.org.]
Microsatellite loci are 2- to 5-bp tandem repeats that are abundant (estimated at 100,000–200,000) and dispersed nearly randomly in all eukaryotic genomes. Their high mutability, owing to DNA slippage during replication and estimated variously at 6 × 10−5 to 2.1 × 103 among mammals (Dallas 1992; de la Chapelle et al. 1992; Edwards et al. 1992a; Weber and Wong 1993; Ellegren 1995; Heyer et al. 1997; Kayser and Sajantila 2000; 2001), leads to the accumulation of new alleles in populations, providing invaluable markers for genetic individualization, parentage assessment, gene mapping, and population monitors of genetic diversity. Their genomic abundance, conservation of their distinctive flanking sequence across closely related species, apparent selective neutrality, and high heterozygosity contribute to their utility in detecting historic demographic events in natural populations (Goldstein and Schlotterer 1999).
We examined the extent and character of variation using 90 feline-specific microsatellite loci in well-described populations of three free-living species of Felidae: cheetahs (Acinonyx jubatus), lions (Panthera pardus), and pumas (Puma concolor) (O'Brien 1994). Populations were selected because previous studies with multiple molecular genetic markers had indicated that historic bottlenecks had reduced overall genetic variation in certain populations (Table 1). Our analysis had four aims: 1) to determine the ability of a large sampling of microsatellites to detect historic population reductions, 2) to use previously dated demographic contractions to calibrate empirically the approximate rate and patterns of microsatellite allele reconstitution following genetic homogenization, 3) to apply the calibration to estimate the time elapsed since imputed population bottlenecks, and 4) to assess the influence of known variation in locus-specific mutation rates in biasing a microsatellite-based molecular clock by comparing reconstitution patterns of homologous microsatellite loci after independent bottleneck events. The results show that microsatellite surveys provide a consistent and informative measure of genomic natural history in each of these aspects.
Table 1.
Percent Polymorphic Loci and Percent Average Heterozygosity for Different Nuclear Gene Families in Free-Living Populations of Cheetahs, Lions, and Pumas
| Cheetahsa | Lions | Pumas | Dom. Cat | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Ajr | Ajj | GIRa | NGC | SER | BCSa | IDO | DOM | ||
| % Polymorphism | Allozymeb | 4.1 | 2.0 | 0.0 | 4.0 | 11.0 | 4.9 | 4.9–10.0 | 21.3 |
| MHC-RFLPc | 5.5 | 4.2 | 0.0 | 5.8 | 17.0 | — | — | 24.9 | |
| Microsatellited | 84.1 | 80.5 | 19.3 | 83.0 | 84.1 | 42.9 | 75.0 | 98.9 | |
| % Heterozygosity | Allozymeb | 1.4 | 0.0 | 0.0 | 1.5 | 3.8 | 1.8 | 2.0–4.0 | 8.2 |
| MHC-RFLPc | 6.7 | 5.1 | 0.0 | 8.0 | 21.8 | — | — | 28.9 | |
| Minisatellitee | 43.3 | 43.6 | 2.9 | 43.5 | 48.1 | 10.3 | 46.9 | 44.9 | |
| Microsatellited | 46.7 | 47.5 | 7.9 | 40.4 | 47.4 | 14.7 | 34.8 | 68.1 | |
Abbreviations: GIR, Gir Forest lions; NGC, Ngorongoro Crater lions; SER, Serengeti Park lions; ETO, Etosha Park lions; KAL, Kalahari-Gemsbok Park lions; KRU, Kruger Park lions; BCS, Big Cypress Swamp pumas; IDO, Idaho pumas; SA, South American pumas; EA, East African cheetahs; CNM, captive Namibian cheetahs; WNM, wild Namibian cheetahs; Ajr, Acinonyx jubatus raineyi; Ajj, A. j. jubutus; DOM, domestic cats.
All cheetahs, Gir forest lions (GIR), and Florida panthers (Big Cypress Swamp BCS) have shown reduced molecular genetic diversity as a consequence of historic demographic reduction and inbreeding (O'Brien 1994).
This study.
RESULTS AND DISCUSSION
Population History in Three Felidae Species
Previously, African cheetahs sampled from eastern, southwestern, and southeastern Africa have been shown to retain 10- to 100-fold less genetic diversity as a species than other felids; diversity was determined using allozymes, two-dimensional PAGE, MHC-RFLP, and mtDNA RFLP screens (Table 1; O'Brien et al. 1983; O'Brien et al. 1985; Wayne et al. 1986; O'Brien et al. 1987c; Yuhki and O'Brien 1990; O'Brien 1994). Cheetahs also display increased fluctuating asymmetry in metric skull measurements (Wayne et al. 1986). In addition, reciprocal skin grafts between unrelated individuals were accepted immunologically, a sign of extreme genetic homogeneity (O'Brien et al. 1985). These cumulative results have been interpreted as evidence of a historical bottleneck or series of demographic reductions over time and space, dated at 10–12,000 yr ago based on mutational reconstitution of genetic variation in rapidly evolving minisatellites and mtDNA (Menotti-Raymond and O'Brien 1993; O'Brien 1994).
The large outbred Serengeti lion population (N = 3,000) displayed appreciable levels of molecular genetic variation with the same gene markers (Table 1; O'Brien et al. 1987b; Wildt et al. 1987; Yuhki and O'Brien 1990; O'Brien 1994). In contrast, an isolated relict population of 250 Asiatic lions living in the Gir Forest Sanctuary in eastern India were genetically uniform in allozymes, MHC- RFLP, minisatellites, and mtDNA diversity, signaling an extreme and more recent demographic reduction than for cheetahs (O'Brien et al. 1987a; Wildt et al. 1987; Gilbert et al. 1991). We have speculated that the Gir lion genetic depletion occurred as a consequence of human depredation by hunting in the late nineteenth century (O'Brien et al. 1987a; Wildt et al. 1987). The isolated Ngorongoro Crater lion population in East Africa is descended from 12 founders who survived a population crash caused by a Stomoxys (biting fly) epidemic in 1962 (Packer et al. 1991b).
The endangered Florida puma (P. concolor coryi, also called the Florida panther) was reduced to <50 individuals in the hardwood cypress swamps of south Florida by habitat encroachment over the previous century (Maehr 1990). This population displayed diminished genetic variation relative to western and South American puma populations (Table 1); Roelke et al. (1993) documented that it suffers episodes of consanguinous matings. The inbred populations from the three species (i.e., all cheetahs, Gir lions, and Florida pumas) experienced consanguinous matings and genetic reduction at different periods in their past, offering a rare opportunity to quantify genomic variation in an historical context.
Microsatellite Diversity Estimates
Estimates of microsatellite population genetic variation parameters (77–93 loci) from three cheetah, six lion, and three puma populations were compared to a group of unrelated domestic cats, the species from which the microsatellites were derived (Table 2; Menotti-Raymond and O'Brien 1995; Menotti-Raymond et al. 1999). Each full species (combining populations) show appreciable microsatellite variation based on percent polymorphic loci and average heterozygosity (P = 86.5, 92.9, and 85.4; He = 54.3, 56.2, and 52.3 for lion, puma, and cheetah, respectively). (See Supplementary Table 1.)
Table 2.
Measure Genetic Variation of Microsatellite Loci among Populations of Four Species of Felids
| Species | Population | N | No. loci amplified | Tot.no.alleles | % P (SE) | Av. he (SE) | Av. ho (SE) | Av. no.alleles per locus (SE) | Av.microsat.variance (MV) | Av.range repeats | Max.range bp |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lion | |||||||||||
| P. I. persica | GIR | 10 | 88 | 111 | 19.3 (4.2) | 7.9 (0.09) | 7.6 (0.09) | 1.26 (2.8) | 0.21 | 2.4 | 10 |
| P. I. leo | NGC | 10 | 88 | 252 | 83.0 (4.0) | 40.4 (0.46) | 42.5 (0.48) | 2.86 (5.3) | 3.26 | 4.8 | 36 |
| P. I. leo | SER | 10 | 88 | 305 | 84.1 (3.9) | 47.4 (0.54) | 47.3 (0.54) | 3.47 (5.3) | 4.45 | 5.8 | 38 |
| P. I. leo | ETO | 10 | 77 | 202 | 71.0 (5.2) | 37.3 (0.48) | 37.8 (0.49) | 2.62 (5.5) | 3.98 | 5.0 | 38 |
| P. I. leo | KAL | 10 | 77 | 229 | 80.5 (4.5) | 43.4 (0.56) | 41.4 (0.54) | 2.97 (5.6) | 3.92 | 4.7 | 36 |
| P. I. leo | KRU | 10 | 77 | 258 | 79.2 (4.6) | 44.4 (0.57) | 47.2 (0.61) | 3.35 (5.7) | 3.99 | 5.3 | 38 |
| P. leo | All | 60 | 89 | 456 | 86.5 (3.6) | 54.3 (0.58) | 37.3 (0.42) | 5.12 (5.1) | 5.34 | 7.3 | 46 |
| Puma | |||||||||||
| P. c. coryi | BCS | 10 | 84 | 127 | 42.9 (5.4) | 14.7 (0.17) | 16.1 (0.19) | 1.51 (4.0) | 0.82 | 2.7 | 26 |
| P. c. hippolestes | IDO | 10 | 84 | 211 | 75.0 (4.7) | 34.8 (0.41) | 32.9 (0.39) | 2.51 (5.1) | 2.75 | 4.0 | 26 |
| P. c. subsp. | SA | 10 | 84 | 503 | 91.0 (3.1) | 68.3 (0.81) | 58.2 (0.69) | 5.99 (5.4) | 7.99 | 8.7 | 38 |
| P. concolor | All | 30 | 84 | 558 | 92.9 (2.8) | 56.2 (0.67) | 35.7 (0.42) | 6.64 (0.42) | 5.58 | 9.2 | 38 |
| Cheetah | |||||||||||
| A. j. raineyi | EA | 10 | 82 | 291 | 84.1 (4.0) | 46.7 (0.57) | 43.3 (0.53) | 3.55 (5.5) | 2.38 | 4.5 | 22 |
| A. j. jubatus | CNM | 10 | 82 | 275 | 79.2 (4.5) | 47.7 (0.58) | 45.7 (0.56) | 3.35 (5.5) | 2.18 | 4.4 | 28 |
| A. j. jubatus | WNM | 10 | 82 | 276 | 81.7 (4.3) | 46.2 (0.56) | 43.8 (0.53) | 3.37 (5.5) | 1.93 | 4.2 | 20 |
| A. jubatus | All | 30 | 82 | 382 | 85.4 (3.9) | 52.3 (0.64) | 44.3 (0.54) | 4.66 (5.5) | 2.33 | 5.3 | 28 |
| Domestic Cat | |||||||||||
| F. catus | DOM | 10 | 93 | 498 | 98.9 (1.1) | 68.1 (0.73) | 62.8 (0.67) | 5.41 (5.0) | 6.4 | 7.4 | 42 |
Population/species abbreviations: GIR, Gir Forest lions; NGC, Ngorongoro Crater lions; SER, Serengeti Park lions; ETO, Etosha Park lions; KAL, Kalahari-Gemsbok Park lions; KRU, Kruger Park lions; BCS, Big Cypress Swamp pumas; IDO, Idaho pumas; SA, South American pumas; EA, East African cheetahs; CNM, captive Namibian cheetahs; WNM, wild Namibian cheetahs; Ajr, Acinonyx jubatus raineyi; Ajj, A. j. jubutus; DOM, domestic cats.
Other Abbreivations: N, individuals sampled; %P, percent polymorphic loci; Av. he, average Expected heterozygosity; Av. ho, Average observed heterozygosity; Av. variance, average variance of repeat number; Av. range, average range in repeats, of polymorphic loci only, in the sample; Max. range, greatest range of allele sizes of a locus, in bases. Standard errors (SE) are reported in parentheses.
The microsatellite variance (MV) in average allele repeat size offers a measure of microsatellite diversity that has an expectation of N*μ in a constant size population, but which would increase proportionally with time in an expanding population after a bottleneck (Goldstein and Pollack 1997). The average MV is 2.33 in cheetahs, a 60% reduction compared to lions, pumas, or domestic cats (MV = 5.34, 5.58, and 6.4, respectively; Table 2). The cheetah's maximal microsatellite heterozygosity, but reduced microsatellite variance, may reflect the postulated late Pleistocene homogenization of allele variation that was followed by maximal reconstitution of microsatellite heterozygosity but incomplete size expansion (allele size breadth or MV), which requires a longer period for saturation (Goldstein and Pollack 1997).
The Gir lion and Florida puma populations descend from documented, recent population bottlenecks and display highly reduced microsatellite heterozygosity (7-fold for Gir lion compared to all lions and 4-fold for Florida panther compared to other pumas), confirming the previous inference of genetic reductions in the more recent past. Microsatellite variance is also reduced appreciably in these demographically contracted populations (25-fold in Gir lion and 7-fold in Florida panther; Table 2), affirming their history of close inbreeding and major loss of endemic, genome-wide allelic diversity. The quantitative parameters of microsatellite variation presented in Table 2 not only provide strong support for the sensitivity of composite microsatellite locus monitors for revealing genomic reductions, both recent (Florida puma and Gir lion) and ancient (cheetah), but also may provide a means to describe more precisely the character and timing of such events.
Microsatellite Reconstitution in Cheetahs
The cheetah's natural history and molecular genetic data are particularly useful in assessing the reconstitution of microsatellite allele variation subsequent to a bottleneck and in providing a chronometer for dating the presumed bottleneck. Microsatellite heterozygosity in modern cheetahs is as high as in other outbred populations or species (Table 2). If we presume that the cheetah's ancestral bottleneck, which homogenized traditional molecular genetic variation (Table 1), also reduced most nuclear microsatellites to homozygosity, then the new polymorphic alleles observed today would have developed by new mutations in the elapsed interval.
The time required to generate maximal microsatellite heterozygosity for cheetahs (assuming that the cheetahs' heterozygosity equivalence with outbred lions and pumas reflect heterozygosity saturation for cheetahs) can be estimated in several ways. By an infinite allele model of evolution, the number of generations required for heterozygosity reconstitution would be on the order of the reciprocal of the mutation rate (Nei et al. 1975; Nei 1987). The microsatellite mutation rate has not been estimated in cheetahs or other cats, however, numerous estimates have been offered in humans and in other species (see Supplementary Table 2). Average mutation rates per microsatellite locus vary from 10−2 to 10−5 (Dallas 1992; Edwards et al. 1992; Hastabacka et al. 1992; Weber and Wong 1993; Ellegren 1995, 2000; Heyer et al. 1997; Kayser et al. 1997, 2000; Bianchi et al. 1998; Brinkmann et al. 1998, Henke and Henke 1999; see Methods). There can also be appreciable variation in mutation rate among different microsatellite loci that depends on the repeat structure (Wierdl et al. 1997; Brinkmann et al. 1998; Schlotterer et al. 1998; Ellegren 2000).
We used two estimates based on large counts of meioses in human microsatellites, 5.6 × 10−4 (Weber and Wong 1993) and the average of four more recent human estimates, 2.05 × 10−3 (Brinkmann et al. 1998; Henke and Henke 1999; Sajantila et al. 1999; Kayser et al. 2001). The two mutation rates yield a range of 488–1786 generations. Given a 6-yr generation time for cheetahs (Marker and O'Brien 1989), we estimate it would take minimally between 2928 and 10,716 yr to generate the present microsatellite variation in cheetahs.
Applying the more appropriate stepwise mutation model (Ohta and Kimura 1973; Valdes et al. 1993), the number of generations required equals (1/μ)/(1−1/e) or 772–2825 generations (4631–16,950 yr). In addition, the divergence date of the two cheetah subspecies (A. j. raineyi and A. j. jubatus) can be estimated based on the computation of (δμ)2 = 2μG (μ = mutation rate and G = generations), a measure of change in mean allele sizes at microsatellite loci over time (Zhivotovsky and Feldman 1995; Goldstein and Pollack 1997). This distance parameter has been shown to be fairly robust to potential violation of mutation-drift equilibrium encountered in this case (Takezaki and Nei 1996). For the two subspecies of cheetahs, this (δμ)2 computation to 708.8 generations or 4253 yr since the geographic divergence of the two subspecies (see below). These microsatellite-based estimates are not inconsistent with previous estimates of the original cheetah bottleneck date (10,000–12,000 yr ago) based on reconstitution of rapidly evolving minisatellites and mtDNA-RFLP variation (Menotti-Raymond and O'Brien 1993) and also coincide with the continental range reduction of cheetah species coincident with the late Pleistocene extinctions of large vertebrates (Marshall et al. 1982). Below, we set the estimated date of the original cheetah bottleneck at ∼12,000 yr ago.
As the accumulation of new microsatellite mutations in an expanding population is stochastic and proportional to elapsed time, the distribution of variation between modern cheetah subspecies can be used to estimate the time of separation of geographically isolated subspecies (A. jubatus jubatus in southern Africa and A. jubatus raineyi in eastern Africa; Fig. 1). Cheetah populations were likely expanding since the late Pleistocene bottleneck 10,000 yr ago to their rise to 100,000–200,000 individuals a century ago. This presumption was tested explicitly using the cheetahs' 82 microsatellites by computation of g, which evaluates the variance of the variance as relatively small in growing populations as compared to constant-sized populations (Reich and Goldstein, 1998). A total of 39.8% (33 of 83) microsatellite loci have k > 0. Because the expectation for a constant non-expanding population is 51.5% (Reich and Goldstein, 1998), cheetah microsatellite variation shows a pattern of a recently and ongoing expanding population expansion (p = 0.02).
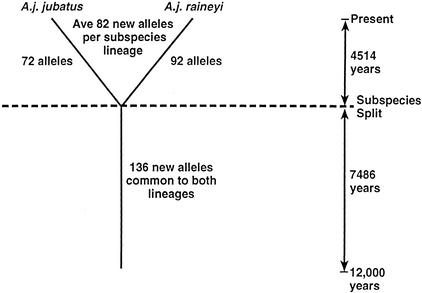
Figure 1.
Schematic representation of new allele reconstitution at 82 microsatellite loci in cheetah populations since the homogenizing bottleneck, estimated at 12,000 yr ago (see text). Common alleles are those shared between the two geographically isolated subspecies (A. j. jubatus and A. j. raineyi). Unique alleles, which were likely produced by mutation after the geographic isolation of the subspecies, are alleles represented in one subspecies but not the other. The calculation of allele reconstitution would include a back-mutation bias, particularly as time and/or distances grow larger; however, the close agreement of the (δμ)2 estimate with the allele reconstitution calculations (see text) offers some assurance that the estimates would approximate the time period considered here.
Consider any alleles found beyond the first at each locus (supernumerary alleles) to derive from a mutation event after the defining bottleneck 12,000 yr ago. Modern cheetahs have a total of 382 alleles across 82 loci (Table 2) representing 300 supernumerary alleles produced in 12,000 yr, or 1 new mutation every 40 yr or 6.7 generations. Of the 300 new microsatellite alleles, 164 are unique to one subspecies or the other and 136 are new and common to both (Fig. 1). Taking the 164 subspecies specific mutations as the sum of an average 82 new variants on each subspecies lineage, we estimate the time since their separation as 82/(136 + 82) × 12,000 yr = 4514 yr ago (in close agreement with the δμ2 estimate of 4253 yr; see above). Because molecular clocks will become saturated as variation is regained, time estimates based on that clock will behave asymptotically as saturation approaches; therefore, both the 12,000 and 4500 yr dates should be considered as minimum estimates.
Dating the Gir Lion Population Bottleneck
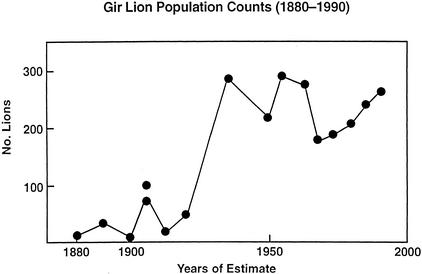
The patterns of microsatellite reconstitution can also be informative in interpreting more recent genetic homogenizations, such as occurred in the Gir forest lions. This population of ∼250 animals is descended from an Asiatic subspecies that at one time was continuously distributed from Anatolia and Palestine to eastern India (Caldwell 1938; Chellam and Johnsing 1983; Joslin 1984; Rashid and David 1992). Asiatic lions were reduced to <20 individuals by sport hunting and habitat encroachment until the early parts of the twentieth century (Fig. 2), leaving the population so genetically depauperate that even minisatellite DNA fingerprints of unrelated individuals were identical (Gilbert et al. 1991). Although population decreases in the late nineteenth century were held responsible, the extreme degree of genetic homogenization observed and simulation-based genetic/demographic models (Halley and Hoelzel 1996) indicated that the responsible bottleneck would have to be greater in severity or more extended than a brief reduction to ∼20 lions a century ago (Fig. 2).
Figure 2.
Demographic census for Asiatic lions in the Gir Forest Sanctuary from 1980–1990. Estimates were based on official estimates (in 1893, 1900, and 1905; Wynter-Blyth 1956; Buxton et al. 1992), unofficial informal estimates (in 1880, 1905, 1913, and 1920; Wynter-Blyth 1956; Buxton et al. 1992), pug marks (in 1936–1963; Wynter-Blyth 1956; Chellam and Johnsing 1983; Buxton et al. 1992), and official counts at bait points (Chellam and Johnsing 1983).
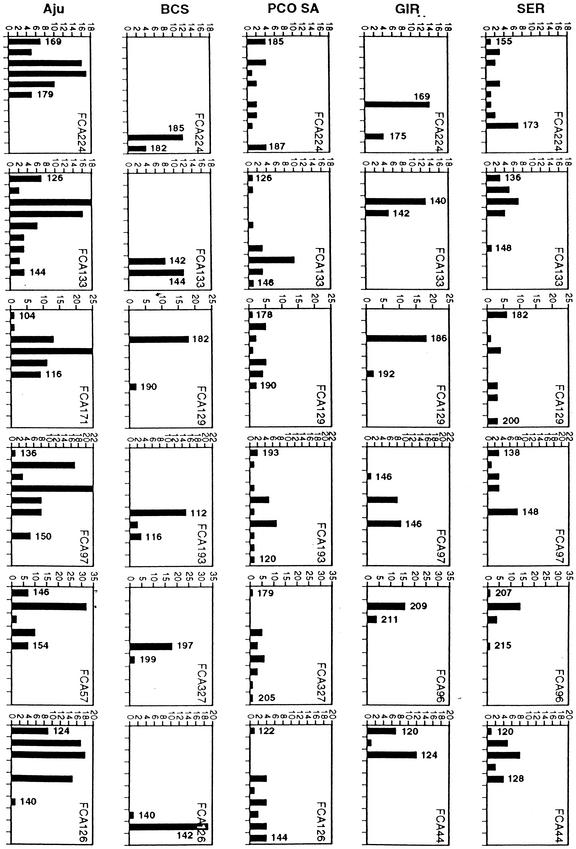
Microsatellite heterozygosity estimates (Table 2) affirmed the extreme loss in Gir lion genetic diversity, although the occurrence of 111 alleles (from 88 sampled loci, indicating 23 new or supernumerary alleles) seemed too many to be explained by mutational reconstitution since the turn of the century (e.g., at a rate of 1 new allele/40 yr as estimated for cheetahs we expect 3, not 23, new alleles). In addition, for the few loci that are still polymorphic in Gir lions, the size distribution of alleles is disjunct or non-continuous; that is, the remaining 18 polymorphic microsatellite loci have one or two alleles at high frequency, with missing intermediate allele size classes (Fig. 3). This disjunct pattern contrasts the continuous distribution of homologous loci in more outbred lion populations (SER, KAL, KRU; Fig. 3) as predicted by the SSM mode for microsatellite allele expansion (Ohta and Kimura 1973; Valdez et al. 1993) in a rapidly expanding population starting with limited (i.e., post-bottleneck homogenized) allele diversity. The GIR lion allele distribution pattern is more indicative of a stochastic allele escape through a recent, incomplete founder effect. Thus, it is likely that at least two bottlenecks have occurred in the Gir lion history, an early extreme event which homogenized diversity extensively and a more recent (c. 100 yr ago), relatively moderate founder event attributable to hunting.
Figure 3.
Frequency distribution of alleles at homologous microsatellite loci among lions (SER and GIR), pumas (PCO-SA and BCS), and cheetahs (Aju). Each graph shows 2-bp (CA repeat) increments on either side of the most common allele in the outbred population plotted (horizontal axis) against number of alleles (vertical axis). For homologous loci, allele sizes are comparable within species, but not between species. Allele size of the largest and smallest alleles for each population are indicated by number above bar. FCA### is the name of the actual microsatellite locus (Menotti-Raymond et al. 1999).
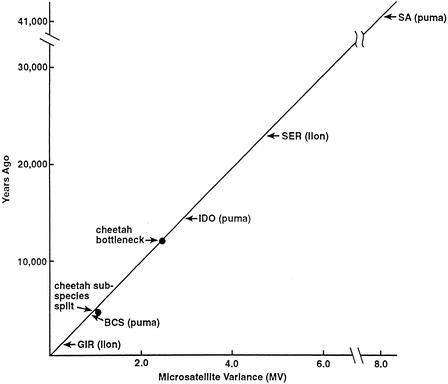
Stochastic allele survival through an incomplete recent bottleneck would reduce the number of reconstituted microsatellite alleles and invalidate using counts of Gir lion supernumerary alleles (as we did above for cheetahs in Fig. 1) to date the origins of allele variation. To examine the relationship for elapsed time following the earliest bottleneck for Gir lions and microsatellite allele variance growth, we used the regression of MV in the cheetah populations from the estimated time of the first bottleneck (12,000 yr ago) and the time of subspecies divergence (4500 yr ago, from Fig. 1). These two points define an approximately linear relationship (i.e., the line passes through the origin) of MV and elapsed time supporting this calibration of MV expansion with elapsed time (Fig. 4). Measured MV of different lion populations was then plotted to estimate the time to accumulate the observed MV (Fig. 4). When the Gir lion MV (0.21) is fitted to the curve, it corresponds to ∼1081 yr as the minimum time period required to generate the observed average Gir lion MV.
Figure 4.
Regression of average microsatellite variance (MV) for both cheetah subspecies and for all cheetahs since the original bottleneck, estimated at 12,000 yr ago (see text; Menotti-Raymond and O'Brien 1993). In addition, MV accumulation from the time of the original bottleneck to the time of subspecies divergence (12,000 − 4514 = 7486 yr ago) was estimated by computing MV considering only allele variants shared between both subspecies of cheetahs.
Because 80% of the Gir lion microsatellites are monomorphic (Table 2), this date estimate (1081 yr) includes a bias because of the large fraction of monomorphic loci considered. This bias would underestimate the actual time by 2–3 fold because our counting of genetically monomorphic in the denominator (for computing MV across all test loci) will diminish overall average MV owing to stochastic fixation. An alternative computation, using only the remaining polymorphic loci in Gir lions in the denominator, produces an upper estimate of 4279 yr for the Gir lion coalescence date. This would be a maximal estimate, providing a range of 1081–4279 yr ago (median = 2680 yr) for the timing of the genetic reduction of the ancestors of the Gir forest lions. This value is consistent with the following approximate estimate of the time required to produce observed Gir lion's supernumary alleles. Thus, if 26 supernumary alleles represent about 1/3 of reconstituted alleles in GIR lions; thus 3 × 26 = 76 new alleles at 1 new allele/40 yr would require 40 × 76 = 3040 yr to reconstitute to 60% polymorphic loci.
The estimated dates can now be interpreted in the context of the geographic and geological history of the Saurashtra Peninsula (in western India) where the Gir Forest is situated (Gupta 1972; Ghose et al. 1979). Saurashtra is bounded by water on three sides, by the Gulf of Kutch, the Arabian Sea, and the Gulf of Cambay. Directly north of the peninsula is the Rann of Kutch, currently the world's largest saline waste; further north is the 300-mile-wide Thar desert. Until relatively recently (4000–5000 yr ago), the Rann of Kutch was a bay or inlet rendering the Saurashtra Peninsula and Gir Forest an island inaccessible to mainland fauna. More recently, the peninsula has been cut off periodically from the mainland by the monsoon-driven expansion of Lake Nall from the Gulf of Kutch to the Gulf of Cambay. Consequently, lions isolated in the Gir peninsula may have restricted or interrupted gene flow with continental populations.
Taken together, the geologic and genomic results indicate that island separation of Gir lions until 4000–5000 yr ago reduced ancestral variation appreciably, leading to observed phenotypic correlates of inbreeding, such as low sperm count, low-testosterone, highly malformed spermatozoa, belly fold, reduced mane size, and paired infra-orbital foramen (O'Brien et al. 1987a; Wildt et al. 1987). Genetically homogenized microsatellite loci accumulated new alleles gradually as the population expanded, but were reduced to a discontinuous distribution by the more recent founder effect (Fig. 3). This explanation is consistent with the recorded history of Asiatic lions in the Gir, the SSM model of microsatellite mutation, and the overall genetic uniformity. Thus, the total genetic picture presented by the Gir lions may result from long-term geographic isolation, exacerbated recently by human expansion and depredation.
Dating a Founder Event in North American Pumas
The sampled puma populations show extreme differences in microsatellite diversity (Table 2). South American pumas have very high levels of variation (P = 91% and He = 68.3%), whereas the Florida puma, known to have experienced a recent range contraction and demographic and genetic reduction, showed the least variation (P = 42.9% and He = 14.7%). Pumas from a western North American population (IDO; Table 2) had an intermediate level of variation (P = 75% and He = 34.8%). As with the Gir lions, the distributions of polymorphic microsatellite alleles are disjunct among Florida panthers with the majority of polymorphic loci (21/36 variable loci, 58%) showing one or two predominant alleles for each locus and “missing” allele size classes (Fig. 3). This disjunction contrasts SA pumas and the SSA model for allelic production in an expanding population, in both of which a continuous near-normal distribution of multiple rare alleles representing continuous size classes are observed. As for the Gir forest lions, this disjunct allele pattern is a signal for stochastic allele sampling caused by a recent founder effect or bottleneck in which allele leakage is observed.
The MV for Florida panthers is 0.82, which corresponds to a minimum age of 4223 yr (Fig. 4). The North American, population in Idaho showed an MV of 2.75 (corresponding to 14,163 yr), considerably lower than the South American population (7.99; 41,000 yr), implying that the North American puma population may derive from a late Pleistocene (∼12,000–18,000 yr ago) genetic reduction or homogenization. This result affirms a more extensive survey of 310 puma specimens for mtDNA and microsatellite variation (Culver et al. 2000). That study indicated that the entire North American puma population was replaced by a founder event of pumas migrating out of South American after the late Pleistocene massive extinctions that eliminated 80% of large vertebrates, including pumas, from North America (Martin and Wright 1967; Marshall et al. 1982; Martin 1989; Pielou 1991). The calibration of MV with time in Figure 4 offers support to this scenario by providing evidence for two bottlenecks in the history of North American pumas, but not South American ones. The first occurred 10,000–15,000 yr ago when cheetahs, American lions, saber tooth cats, and probably pumas were extirpated from North America (Martin and Wright 1967; Martin 1989; Pielou 1991), followed by repopulation by puma migrants from South America (Culver et al. 2000). The second was a recent nineteenth century reduction of pumas east of the Mississippi, leading to extinction of the Eastern cougar (P. concolor cougar) and severe genetic reduction of the Florida panther (Maehr 1990; Roelke et al. 1993).
Is Microsatellite Allele Reconstitution Affected by Mutation Rate Variance?
The demographic history of these populations allows a straightforward test of the influence of differential mutation rates across loci, which would have important implications for evolutionary inference with microsatellite loci (Goldstein and Pollack 1997). Several studies have estimated the microsatellite mutation rate from 10−5 to 10−2 (Dallas 1992; de la Chapelle et al. 1992; Edwards et al. 1992; Weber and Wong 1993; Ellegren 1995), determined largely by averaging new mutations over several loci. A few studies, particularly those involving triplet repeat expansions in hereditary disease pedigrees, report differential mutation rates, some as high as 10−2 (Edwards et al. 1992b; Mahtani and Willard 1993; Brinkmann et al. 1998; Gonser et al. 2000; Kayser et al. 2000, 2001; Xu et al. 2000). To detect the influence of locus-specific mutation rate differential on development of new alleles we compared independent microsatellite locus reconstitutions in populations of different species using a Kendall rank test for two diversity parameters, He and MV (Table 3). This test measures the null hypothesis for independence of specific locus reconstitution following confirmed bottlenecks in the three separate species and locales. The compared populations were Gir lions, cheetahs from subspecies A. j. jubatus, and North American pumas (IDO and BSC). As a positive control (for non-independence of specific locus allele reconstitution), we also compared the non-independent populations P. concolor-BSC and -IDO separately. The results indicated only a single significant association in rank test (MV P. l. leo Gir vs. A j. jubutus, P = 0.05) out of six comparisons (Table 3); except with the positive controls (western versus Florida pumas), in which P = 10–5. We believe that although there are differences in locus-specific mutation rate, they do not play a large role in estimating the time of historic population bottlenecks described here.
Table 3.
Comparing Diversity Measures of Reconstituted Microsatellites in Three Species by Kendall Rank Test
| Population | No. loci | No. new alleles | Loci with new alleles | |
|---|---|---|---|---|
| No. | % | |||
| P. leo persica (Gir) | 78 | 44 | 14 | 17.9 |
| A. j. jubatus | 78 | 73 | 47 | 60.3 |
| P. c. cougar (NA)* | 84 | 166 | 70 | 83.3 |
| Compared population | He | MVa | |||
|---|---|---|---|---|---|
| N | τ | p-value | τ | p-value | |
| P. I. persica vs. A. j. jubatus | 75 | 0.139 | 0.078 | 0.158 | 0.044 |
| P. I. persica vs. NA.-P. concolor | 74 | 0.079 | 0.317 | 0.113 | 0.153 |
| NA-P. concolor vs. A. j. jubatus | 74 | 0.129 | 0.105 | 0.192 | 0.015 |
| IDO-P. concolor vs. BCS-P. concolor | 84 | 0.335 | 6 × 10−6 | 0.262 | 4.2 × 10−4 |
P.c. cougar (NA) is a mix of IDO and BSC combined; MV, microsatellite variance.
CONCLUSIONS
The results show empirically the utility of estimates of microsatellite variation, particularly microsatellite allele reconstruction and microsatellite allele variance, for interpreting historic population contractions. We used populations of wild cats that have been previously characterized as having demographic reductions to assess the panel of 93 microsatellites in 13 populations (11,250 genotypes in total). The results affirm previous inferences, but also provide new insights into the populations and the application of microsatellite markers.
Microsatellite average heterozygosity and percent polymorphism accumulated in a time-dependent manner following a bottleneck according to the SSM model. Heterozygosity parameters maximize in a shorter time (<10,000 yr) than does MV, which displays a window of proportionality empirically calibrated to nearly 40,000 yr (Fig. 4). Both allele reconstitutions and MV accumulate in a clock-like manner that can approximate the time of historic genetic reductions (Figs. 1,4).
The results support previous conclusions about the cheetah's demographic reduction in the late Pleistocene, minimally estimated here based on microsatellite mutation rate as 10,000–17,000 yr ago, but with a subspecies split around 4500 yr ago. Using the growth rate of microsatellite variance in cheetahs since that original bottleneck as a scale, we believe that the Gir lion's defining founder effect was actually earlier than predicted, on the order of 2600 yr ago, driven by geographic isolation from a larger metapopulation occupying the Indian subcontinent. Our analysis also would support the concept of North American puma genetic diversity descending from a founder effect precipitated by migration of South American pumas that colonized North America after the late Pleistocene extinction of North American vertebrates, including resident pumas (Martin and Wright 1967; Martin 1989; Pielou 1991; Culver et al. 2000). The affirmation and extension of previous molecular and natural historical conclusions for cheetahs, lions, and pumas offer empirical support for the potential of genomic microsatellite patterns for inferring evolutionary history of free-living species.
METHODS
Samples
Ten individuals from each of the six lion populations, three puma populations, and three cheetah populations were genotyped for 105 microsatellites (see Supplementary Table 3). We selected unrelated individuals, for which pedigrees were known, from each study population. Pedigrees of lion and cheetah groups were based on direct field observation (Marker and O'Brien 1989; Packer et al. 1991 a,b) and DNA fingerprints (Gilbert et al. 1991; Menotti-Raymond and O'Brien 1993; Roelke et al. 1993); studbook records (Smith 1985; Marker and O'Brien 1989) of wild-caught zoo animals were used in the case of the Asiatic lion representatives. Selection of Gir lions included founder lions from the Sakkarbaug Zoo Asiatic lion pedigree (O'Brien et al. 1987a) plus five half-sibs, reducing the genetic equivalent number of individuals to six. Except for the Big Cypress Swamp population, puma samples were assembled from presumably unrelated material collected over several years. Zoo-held animals were recent captures from the wild. Florida pumas were selected to include founders of the management pedigree (Roelke et al. 1983). Domestic cats are closed-colony, random-bred domestics (Liberty Labs, Waverly, NY) or fancy breed domestics from various U.S. Egyptian Mau breeders.
Lions from the relict P. I. persica population in the Gir Forest in India and the moderately bottlenecked Ngorongoro Crater (P. I. leo) were compared with outbred P. l. leo individuals from the Serengeti (Tanzania), from Etosha National Park (Namibia), and from Kalahari Gemsbok and Kruger National Parks (RSA). Two cheetah sub-species were examined in three population samples: wild A. j. raineyi from the Serengeti ecosystem, wild A. j. jubatus from central Namibia, and captive A. j. jubatus from Namibia. Pumas from the Big Cypress Swamp in Florida (i.e., Florida panthers; Roelke et al. 1993) were compared with more outbred representatives from Idaho and with ten individuals from different locales in South America (Culver et al. 2000). DNA from individual animals was amplified; amplification products were electrophoresed in four panels: 1) Gir Forest, Ngorongoro Crater, and Serengeti lions; 2) Namibian, Kruger Park, and Kalahari-Gemsbok Park lions; 3) cheetahs; and 4) pumas. Domestic cat samples were electrophoresed on every gel as cross-gel reference controls. Genomic DNA (extracted from leukocytes, tissue, or fibroblast cell lines; Sambrook et al. 1989) was obtained from the DNA inventory at the LGD.
One hundred and five microsatellite loci were selected for this study from a panel of over 300 polymorphic loci isolated from a female domestic cat (Menotti-Raymond and O'Brien 1995; Menotti-Raymond et al. 1999). Primers were designed in unique sequence regions flanking 2-bp repeats. Five loci failed to amplify clear single bands in any of the non-domestic species satisfactorily and were not considered further.
Conditions for Amplification
PCR amplification of individual microsatellite loci was performed in 10 μl reactions of 1× Perkin Elmer Getus (PEG) PGR buffer (10 mM Tris-hydrochloric acid at pH 8.3, 50 mM potassium chloride), 2 mM MgGI2, 250 mM each dATP, dGTP, dTTP, and dGTP (Pharmacia), 0.4 U/mL AmpliTaq DNA polymerase (Perkin-Elmer Cetus, Norwalk, CT), 50 ng DNA, and 4.0 picomoles each of forward and reverse primer (Life Technologies, Gaithersburg, MD and PE Applied Biosytems Inc., Foster City, GA). One primer of each pair was labeled with a fluorescent dye phosphoramidite. Amplification was performed in a PEC 9600 Thermocycler according to the following procedure. Samples were denatured at 94°C for 3 min, followed by 10 rounds of denaturation at 94°C for 15 s, annealing at 55°C for 15 s, and extension at 72°C for 30 s. Next were 20 rounds of denaturation at 89°C for 15 s, annealing at 55°C for 15 s, and extension at 72°C for 30 s. The final step was a 10 min extension at 72°C. Amplified products were diluted, as determined empirically for each locus, with sterile deionized water (Quality Biological) in individual tubes in a 9600 tray assembly. 2 μL diluted product were mixed with 4 mL cocktail (6:1:1 formamide, ABI Prism Genescan-350 TAMRA internal lane standard, and ABI Genescan loading buffer). Diluted mixed samples were denatured by heating 3 min at 93°C and then placed immediately on ice. 2 μL denatured sample cocktail was run per lane at 2000 V, 400 mA, and 25 W in 1× TBE buffer on a 6% denaturing polyacrylamide gel on an ABI 373A DNA sequencer running GeneScan, v. 1.2.2-1.
Genotype analysis was performed with GeneScan; final base-calling was performed with ABI Genotyper, v. 1.1. Allele sizes were estimated using the Local Southern method (Elder and Southern 1987) to generate a best-fit curve from the size standards in each lane. Domestic cat DNA amplification products were run on each gel as a reference in aligning alleles across panels. PCR product length was used as an indication of the actual repeat length (Elder and Southern 1987). We assumed that all one-base-pair differences between species were the result of insertion or deletion events in regions flanking the repeat. Thus, across species in all four panels, all such mismatches of allele sizes were rectified empirically based on allele sizes in domestic cat (run on each panel) and on the consensus of odd- or even-binning in exotic species. This correction was necessary because distance estimators based on allele sharing would interpret odd/even mismatches incorrectly as infinite distance because no alleles would be shared. Distance estimates from mutation-based estimators that use mean allele size per locus are not likely to be affected by single base size shifts, particularly when averaged over loci. Because all individuals of a single species (except lions) were electrophoresed on the same panels, intra-specific analyses are unaffected by this correction.
Measures of Population Genetic Diversity
Genetic variation of microsatellite loci for each population was measured as the percent polymorphic loci (P), the observed heterozygosity (Ho), the expected heterozygosity (He) by Hardy-Weinberg, and the average number of alleles per locus (A; Hartl and Clarke 1989; Gao and Thompson 1992; Slatkin 1995; Schneider et al. 1996). Allele polymorphism with frequency >0.05 were considered as the cut-off for the locus to be considered polymorphic. The Arlequin computer package (Schneider et al. 1996) was used to estimate diversity parameters. Microsatellite variance (MV) is the variance in allele repeat number averaged across all microsatellite loci:
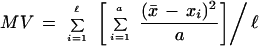 |
in which X is the repeat number of the ith allele at one of a alleles at the first of ℓ loci (Goldstein and Clark 1995; Goldstein and Pollock 1997).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL obrien@ncifcrf.gov; FAX (301) 846-1686.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.185702.
REFERENCES
- Bianchi NO, Catanesi CI, Bailliet G, Martinez-Marignac VL, Bravi CM, Vidal-Rioja LB, Herrera RJ, Lopez-Camelo JS. Characterization of ancestral and derived Y-chromosome haplotypes of New World native populations. Am J Hum Genet. 1998;63:1862–1871. doi: 10.1086/302141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann B, Klintschar M, Neuhuber F, Huhne J, Rolf B. Mutation rate in human microsatellites: Influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton J, Shelbourne P, Davies J, Jones C, Van T, Aslanidis C, de Jong P, Jansen G, Anret M, Riley B, et al. Detection of an unstable fragment of DNA specific to individuals with myotonic dystrophy. Nature. 1992;355:547–548. doi: 10.1038/355547a0. [DOI] [PubMed] [Google Scholar]
- Caldwell K. The Gir lions. 1938. J Soc Preserv Fauna Empire. 1938;34:62–65. [Google Scholar]
- Chakraborty R, Kimmel M, Stivers D, Davison LJ, Deka R. Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc Natl Acad Sci USA. 1997;94:1041–1046. doi: 10.1073/pnas.94.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellam R, Johnsing AJT. Management of Asiatic lions in the Gir forest. Zool Soc Lond. 1983;65:409–424. [Google Scholar]
- Culver M, Johnson WE, Pecon Slattery J, Menotti-Raymond M, O'Brien SJ. Genomic ancestry of the American puma. A phylogeographic study using mitochondrial DNA and microsatellites. J Hered. 2000;91:186–197. doi: 10.1093/jhered/91.3.186. [DOI] [PubMed] [Google Scholar]
- Dallas JF. Estimation of microsatellite mutation rates in recombinant inbred strains of mouse. Mamm Genome. 1992;5:32–38. doi: 10.1007/BF00356155. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E. Linkage disequilibrium mapping in isolated founder populations: Diastrophic dysplasia in Finland. Nat Genet. 1992;2:343. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- Edwards A, Hammond HA, Jin L, Caskey CT, Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992;12:241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- Elder J K, Southern EM. Computer-aided analysis of one-dimensional restriction fragment gels. In: Bishop MJ, Rawlings CJ, editors. Nucleic acid and protein sequence analysis—A practical approach. Oxford, UK: IRL Press; 1987. pp. 165–172. [Google Scholar]
- Ellegren H. Mutation rates at porcine microsatellite loci. Mamm Genome. 1995;6:376–377. doi: 10.1007/BF00364807. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nat Genet. 2000;24:400–402. doi: 10.1038/74249. [DOI] [PubMed] [Google Scholar]
- Ghose G, Kar A, Husain Z. The lost courses of the Saraswati River in the Great Indian Desert: New evidence from landsat imagery. Geographic J. 1979;145:446–451. [Google Scholar]
- Gilbert DA, Packer C, Pusey AE, Stephens JC, O'Brien SJ. Analytical DNA fingerprinting in lions: Parentage, genetic diversity and kinship. J Hered. 1991;82:378–386. doi: 10.1093/oxfordjournals.jhered.a111107. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Pollock DD. Launching microsatellites: A review of mutation processes and methods of phylogenetic inference. J Hered. 1997;88:335–342. doi: 10.1093/oxfordjournals.jhered.a023114. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Schlotterer C. Microsatellites: Evolution and applications. Oxford, UK: Oxford University Press; 1999. [Google Scholar]
- Gonser R, Donnelly P, Nicholson G, Di Rienzo A. Microsatellite mutations and inferences about human demography. Genetics. 2000;154:1793–1807. doi: 10.1093/genetics/154.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Gupta SK. Chronology of the raised beaches and inland coral reefs of the Saurashtra Coast. J Geol. 1972;80:357–361. [Google Scholar]
- Halley J, Hoelzel AR. Simulation models of bottleneck events in natural populations. In: Smith TB, Wayne RK, editors. Molecular genetic approaches in conservation. Oxford, UK: Oxford University Press; 1996. pp. 347–364. [Google Scholar]
- Hartl DL, Clark AG. Principles of population genetics. 2d ed. Sunderland, MA: Sinauer Associates, Inc.; 1989. [Google Scholar]
- Hastabacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E. Linkage disequilibrium mapping in isolated founder populations: Cliastrophic dysplasia in Finland. Nat Genet. 1992;2:204–211. doi: 10.1038/ng1192-204. [DOI] [PubMed] [Google Scholar]
- Henke L, Henke J. Mutation rate in human microsatellites. Am J Hum Genet. 1999;64:1473–1474. doi: 10.1086/302373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Puymirat J, Dieltjes P, Bakker E, de Knijff P. Estimating Y chromosome specific microsatellite mutation frequencies using deep rooting pedigrees. Hum Mol Genet. 1997;6:799–803. doi: 10.1093/hmg/6.5.799. [DOI] [PubMed] [Google Scholar]
- Joslin P. The environmental limitations and future of the Asiatic lions. J Bombay Nat Hist Soc. 1984;81:648–664. [Google Scholar]
- Joslin P, O'Brien SJ. Reproductive and genetic consequences of founding isolated lion populations. Nature. 1987;329:328–331. [Google Scholar]
- Kayser M, Sajantila A. Mutations at Y-STR loci: Implications for paternity testing and forensic analysis. Foren Sci Inter. 2001;118:116–121. doi: 10.1016/s0379-0738(00)00480-1. [DOI] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Kruger C, Krawczak M, Nagy M, Dobosz T, et al. Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet. 2000;66:1580–1588. doi: 10.1086/302905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehr DS. The Florida panther and private lands. Conservation Biol. 1990;4:167–170. [Google Scholar]
- Mahtani MM, Willard H. A polymorphic X-linked tetranucleotide repeat locus displaying a high rate of new mutation: Implications for mechanisms of mutation at short tandem repeat loci. Hum Mol Genet. 1993;2:431–437. doi: 10.1093/hmg/2.4.431. [DOI] [PubMed] [Google Scholar]
- Marker L, O'Brien SJ. Captive breeding of the cheetah (Acinonyx jubatus) in North American zoos. Zoo Biol. 1989;8:3–16. [Google Scholar]
- Marshall LG, Webb SD, Sepkoski JJ, Jr, Raup DM. Mammalian evolution and the great American interchange. Science. 1982;215:1351–1357. doi: 10.1126/science.215.4538.1351. [DOI] [PubMed] [Google Scholar]
- Martin LD. Fossil history of terrestrial carnivora. In: Gittleman JL, editor. Carnivore behavior, ecology, and evolution. Ithaca, NY: Cornell University Press; 1989. pp. 536–568. [Google Scholar]
- Martin PS, Wright HE. Pleistocene extinctions: The search for cause. New Haven, CT: Yale University Press; 1967. [Google Scholar]
- Menotti-Raymond M, David VA, Lyons LA, Schäffer AA, Tomlin JF, Hutton MK, O'Brien SJ. A genetic linkage map of microsatellites in the domestic cat (Felis catus) Genomics. 1999;57:9–23. doi: 10.1006/geno.1999.5743. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond MA, O'Brien SJ. Dating the genetic bottleneck of the African cheetah. Proc Natl Acad Sci USA. 1993;90:3172–3176. doi: 10.1073/pnas.90.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti-Raymond MA, O'Brien SJ. Evolutionary conservation of ten microsatellite loci in four species of Felidae. J Hered. 1995;86:319–322. doi: 10.1093/oxfordjournals.jhered.a111594. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY: Columbia University Press; 1987. [Google Scholar]
- Nei M, Maryuyama T, Chakroborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ. A role for molecular genetics in biological conservation. Proc Natl Acad Sci USA. 1994;91:5748–5755. doi: 10.1073/pnas.91.13.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Joslin P, Smith GL, III, Wolfe R, Schaffer N, Heath E, Ott-Joslin J, Rawal PP, Bhattacharjee KK, Martenson JS. Evidence for African origins of founders of the Asiatic lion species survival plan. Zoo Biol. 1987a;6:99–116. [Google Scholar]
- O'Brien SJ, Martenson JS, Packer C, Herbst L, de Vos V, Joslin P, Ott-Joslin J, Wildt DE, Bush M. Biochemical genetic variation in geographic isolates of African and Asiatic lions. Natl Geog Res. 1987b;3:114–124. [Google Scholar]
- O'Brien SJ, Roelke ME, Marker L, Newman A, Winkler CA, Meltzer D, Colly L, Evermann JF, Bush M, Wildt DE. Genetic basis for species vulnerability in the cheetah. Science. 1985;227:1428–1434. doi: 10.1126/science.2983425. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Wildt D, Bush M, Caro TM, Fitzgibbon C, Aggundey I, Leakey RE. East African cheetahs: Evidence for two population bottlenecks? Proc Natl Acad Sci USA. 1987c;84:508–511. doi: 10.1073/pnas.84.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien SJ, Wildt DE, Goldman D, Merril CR, Bush M. The cheetah is depauperate in genetic variation. Science. 1983;221:459–462. doi: 10.1126/science.221.4609.459. [DOI] [PubMed] [Google Scholar]
- Ohta T, Kimura M. The model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a genetic population. Genet Res. 1973;22:201–204. doi: 10.1017/s0016672300012994. [DOI] [PubMed] [Google Scholar]
- Packer C, Gilbert DA, Pusey AE, O'Brien SJ. Kinship, cooperation, and inbreeding in African lions: A molecular genetic analysis. Nature. 1991a;351:562–565. [Google Scholar]
- Packer C, Pusey AE, Rowley H, Gilbert DA, Martenson J, O'Brien SJ. Case study of a population bottleneck: Lions of the Ngorongoro Crater. Conservation Biol. 1991b;5:219–230. [Google Scholar]
- Pielou EC. After the Ice Age, the return of life to glaciated North America. Chicago, IL: University of Chicago Press; 1991. [Google Scholar]
- Rashid MA, David R. ‘The asiatic lion (Panthera leo persica).‘. Dept. of Environment, Gov’t. of India; 1992. [Google Scholar]
- Reich DE, Goldstein DB. Genetic evidence for a paleolithic human population expansion in Africa. Proc Natl Acad Sci USA. 1998;95:8119–8123. doi: 10.1073/pnas.95.14.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelke ME, Martenson JS, O'Brien SJ. The consequences of demographic reduction and genetic depletion in the endangered Florida panther. Curr Biol. 1993;3:340–350. doi: 10.1016/0960-9822(93)90197-v. [DOI] [PubMed] [Google Scholar]
- Sajantila A, Lukka M, Syvanen A-C. Experimentally observed germline mutations at human micro- and minisatellite loci. Eur J Hum Genet. 1999;7:263–266. doi: 10.1038/sj.ejhg.5200257. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schlotterer C, Ritter R, Harr B, Brem G. High mutation rate of a long microsatellite allele in Drosophila melanogaster provides evidence for allele-specific mutation rates. Mol Biol. 1998;15:1269–1274. doi: 10.1093/oxfordjournals.molbev.a025855. [DOI] [PubMed] [Google Scholar]
- Schneider S, Kueffer J-M, Roessli D, Excofier L. Arlequin: A software package for population genetics. Genetics and Biometry Lab, Dept. of Anthropology, University of Geneva; 1996. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GL., III . International studbook of the Indian lion, Panthera leo persica (Meyer, 1826). Knoxville, TN: Knoxville Zoological Garden; 1985. [Google Scholar]
- Takezaki N, Nei M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics. 1996;144:389–399. doi: 10.1093/genetics/144.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AM, Slatkin M, Freimer N. Allele frequencies at microsatellite loci: The stepwise mutation model revisited. Genetics. 1993;133:737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne RK, Modi WS, O'Brien SJ. Morphological variability in the cheetah (Acinonyx jubatus), a genetically uniform species. Evolution. 1986;40:78–85. doi: 10.1111/j.1558-5646.1986.tb05719.x. [DOI] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Hum Mol Genet. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Wierdl M, Dominska M, Petes TD. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics. 1997;146:769–779. doi: 10.1093/genetics/146.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt D E, Bush M, Goodrowe K L, Packer C, Pusey A E, Brown J L, Joslin P, O'Brien S J. Reproductive and genetic consequences of founding isolated lion populations. Nature. 1987;329:328–331. [Google Scholar]
- Wynter-Blyth M A. The lion census of 1955. J Bombay Nat Hist Soc. 1956;53:527–536. [Google Scholar]
- Wynter-Blyth M A, Dharmakumarsinhji K S. The Gir forest and its lions, pt. II. J Bombay Nat Hist Soc. 1950;49:456–470. [Google Scholar]
- Xu X, Peng M, Fang Z. The direction of microsatellite mutations is dependent on allele length. Nat Genet. 2000;24:396–399. doi: 10.1038/74238. [DOI] [PubMed] [Google Scholar]
- Yuhki N, O'Brien SJ. DNA variation of the mammalian major histocompatibility complex reflects genomic diversity and population history. Proc Natl Acad Sci USA. 1990;87:836–840. doi: 10.1073/pnas.87.2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Feldman MW. Microsatellite variability and genetic distances. Proc Natl Acad Sci USA. 1995;92:11549–11552. doi: 10.1073/pnas.92.25.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]