Abstract
Mechanisms that regulate signal propagation through the ERK/MAPK pathway are still poorly understood. Several proteins are suspected to play critical roles in this process. One of these is Kinase Suppressor of Ras (KSR), a component previously identified in RAS-dependent genetic screens in Drosophila and Caenorhabditis elegans. Here, we show that KSR functions upstream of MEK within the ERK/MAPK module. In agreement with this, we found that KSR facilitates the phosphorylation of MEK by RAF. We further show that KSR associates independently with RAF and MEK, and that these interactions lead to the formation of a RAF/MEK complex, thereby positioning RAF in close proximity to its substrate MEK. These findings suggest that KSR functions as a scaffold that assembles the RAF/MEK functional pair.
Keywords: KSR, MEK, RAF, RAS, MAPK, signal transduction
Cells use a range of signaling pathways to convey distinct information to appropriate intracellular targets. The evolutionarily conserved mitogen-activated protein kinase (MAPK) pathways are among the best described examples (for review, see Schaeffer and Weber 1999). The prototypical MAPK pathway is a three-kinase module that transmits signals through a phosphorylation cascade. At least five groups of MAPK pathways have been distinguished in mammals, which include the extensively studied extracellular-regulated kinase (ERK) pathway. This pathway is composed of specific combinations of RAF, MEK, and ERK/MAPK isoforms and is one of the principal mediator of signals emanating from the small GTPase RAS, thereby influencing multiple aspects of cell physiology such as cell growth, proliferation, differentiation, and survival (for review, see English et al. 1999; Kolch 2000).
As for the ERK pathway, most groups of MAPK pathways comprise multiple and closely related kinase isoforms that are often expressed in the same cells (Garrington and Johnson 1999). This raised questions early on about how signal transmission specificity could be achieved. The identification of proteins capable of binding at the same time to at least two of the three kinases of the module, thereby physically linking them together, provided a framework to explain signal transmission specificity and possibly efficiency (for review, see Whitmarsh and Davis 1998). The prototype for such scaffolding proteins is STE5, which simultaneously binds to STE11, STE7, and FUS3, the three-kinase module required for mating in Saccharomyces cerevisiae (Choi et al. 1994). Ever since the discovery of STE5, only a handful of potential scaffolding proteins with respect to specific MAPK pathways have been identified (for review, see Garrington and Johnson 1999; Schaeffer and Weber 1999). The clearest examples include the yeast protein PBS2 required in the osmosensing-response MAPK pathway in S. cerevisiae (Posas and Saito 1997); the JNK-interacting proteins (JIP-1/2; Whitmarsh et al. 1998; Yasuda et al. 1999), which function in the JNK pathway; and finally MEK partner 1 (MP1), which apparently bridges MEK1 and ERK1 (Schaeffer et al. 1998).
Other proteins have been suggested to function as scaffolds in specific MAPK pathways (for review, see Garrington and Johnson 1999); however, their precise molecular function remains ambiguous. One of these is Kinase Suppressor of RAS (KSR), whose activity appears to be required in the ERK/MAPK pathway (for review, see Morrison 2001). KSR was originally identified in RAS-dependent genetic screens in Drosophila and C. elegans (Kornfeld et al. 1995; Sundaram and Han 1995; Therrien et al. 1995). Interestingly, KSR proteins are mostly related to RAF serine/threonine kinase family members (Therrien et al. 1995), but differ in at least three main aspects: (1) they do not contain the so-called RAS-binding domain found in RAF proteins; (2) they contain a conserved region of ∼40 amino acids at their N terminus called Conserved Area 1 (CA1) that is unique to them; and (3) the mammalian homologs contain an arginine residue instead of an invariant lysine residue in kinase subdomain II that is thought to be critical for the phosphotransfer reaction. This peculiarity suggests that KSR proteins might be devoid of kinase activity.
The characterization of KSR with respect to the ERK/MAPK pathway has been undertaken by a number of groups using mKSR1, a murine isoform (for review, see Morrison 2001). Like RAF, mKSR1 associates with HSP90 and p55/CDC37 as well as with 14–3–3 proteins (Xing et al. 1997; Stewart et al. 1999; Cacace et al. 1999). Interestingly, mKSR1 has also been shown to interact constitutively with MEK (Denouel-Galy et al. 1997; Yu et al. 1997; Muller et al. 2000) and in a RAS-dependent manner with RAF and ERK/MAPK (Therrien et al. 1996; Xing et al. 1997; Cacace et al. 1999). These results led different groups to propose that mKSR1 might coordinate the assembly of the ERK/MAPK module (for review, see Morrison 2001). However, this hypothesis remains to be tested because the molecular relationship between mKSR1 and the three kinase components of the ERK/MAPK module is currently unknown. Furthermore, functional assays conducted by different groups produced contradictory results, which resulted in a fragmented and somewhat controversial view of the role of KSR. A notable case regards the catalytic function of mKSR1. Kolesnick and colleagues reported that mKSR1 can phosphorylate and activate RAF in a TNFα- or EGF-dependent manner (Zhang et al. 1997; Xing et al. 2000). Intriguingly, however, these results could not be reproduced by other laboratories (Denouel-Galy et al. 1997; Michaud et al. 1997; Yu et al. 1997; Sugimoto et al. 1998). Instead, mKSR1 activity was reported to be independent of its putative catalytic function (Michaud et al. 1997; Stewart et al. 1999). Another discrepancy is the observation made by a number of groups that forced expression of mKSR1 strongly and specifically blocked signaling through the ERK/MAPK pathway (Denouel-Galy et al. 1997; Yu et al. 1997; Joneson et al. 1998; Sugimoto et al. 1998), whereas others showed that mKSR1 strongly cooperated with activated RAS to induce meiotic maturation of Xenopus oocytes (Therrien et al. 1996). As suggested by Cacace et al. (1999), this contradiction might be attributable to differences in mKSR1 expression levels. Given that mKSR1 interacts with several components of the ERK/MAPK module, it is possible that these components are sequestered from each other when mKSR1 levels are in excess. In any event, it remains unclear whether the information obtained using mKSR1 truly reflects the function of KSR as it had been genetically defined. It is possible that the experimental systems used could simply not support normal mKSR1 function. For example, it is intriguing that in the Xenopus oocyte maturation assay, mKSR1 activity mainly depended on its cysteine-rich motif (CRM; Therrien et al. 1996; Michaud et al. 1997), whereas several loss-of-function mutations affecting other parts of the KSR protein have been identified in Drosophila and C. elegans (Kornfeld et al. 1995; Sundaram and Han 1995; Therrien et al. 1995).
A major problem hindering the elucidation of the role of KSR is the lack of an assay that faithfully recapitulates its function. Here, using a simple transfection protocol, we reconstituted a KSR-dependent functional assay in a homologous system, that is, in Schneider (S2) cells using only Drosophila-derived components. We found that KSR strongly promoted MEK phosphorylation by RAF in a RAS-dependent manner. Strikingly, KSR activity appeared to depend mainly on its ability to associate independently with RAF and MEK, thereby allowing it to link the two kinases. Together, our findings suggest that KSR promotes signal propagation through the ERK/MAPK module by coordinating the assembly of a RAF/MEK complex.
Results
Forced association between KSR and RAF kinase domains induces MEK phosphorylation
The KSR kinase domain has been reported to interact with MEK but not phosphorylate MEK (see above). KSR has also been found to interact with RAF, but the regions involved have not been identified. We wanted to verify whether forced association between KSR and RAF catalytic domains (KSRc and RAFc) would affect MEK activity. We took advantage of two previously described Drosophila transgenic lines that express, during eye development, either RAFc or KSRc fused to the N-terminal portion of Torso4021, which comprises the extracellular and transmembrane regions of the Torso receptor tyrosine kinase (N-Tor4021). Tor4021 is a gain-of-function allele of torso. The aberrant Torso4021 protein has a Y327C change in the extracellular portion that is thought to promote ligand-independent receptor oligomerization (Sprenger and Nusslein-Volhard 1992).
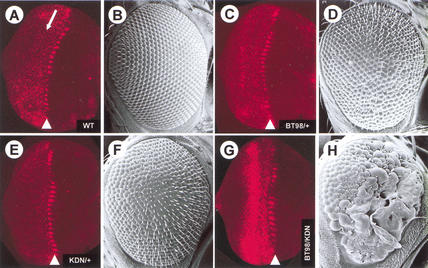
N-Tor4021RAFc behaves as an activated form of RAF and transforms cone cells into additional R7 photoreceptor cells when expressed in the developing Drosophila eye (Dickson et al. 1992). In contrast, N-Tor4021KSRc is a potent dominant-negative form of KSR that blocks signaling through the MAPK pathway, presumably by sequestering MEK. It strongly antagonizes photoreceptor cell differentiation (Therrien et al. 1996). We reasoned that crossing flies expressing N-Tor4021RAFc (BT98) to flies expressing N-Tor4021KSRc (KDN) should lead to the formation of hetero-oligomers, which might result in activation of endogenous MEK and MAPK. We crossed BT98 and KDN transgenic flies to a wild-type strain or to each other to assess the effect on MAPK compared with wild-type flies (Fig. 1). We stained third instar eye discs of the different genotypes with an anti-phospho-MAPK (anti-pMAPK) antibody and found that, compared with wild-type pMAPK levels, BT98/+ flies had slightly elevated pMAPK levels posterior to the morphogenetic furrow at a position consistent with the expected transgene expression pattern (Fig. 1C). In agreement with elevated pMAPK, BT98/+ adult eyes had additional R7 photoreceptor cells (data not shown) that resulted in a rough eye phenotype (Fig. 1D; Dickson et al. 1992). In contrast, the KDN/+ flies had reduced pMAPK levels (Fig. 1E), which is also consistent with the roughening of the external adult eye surface (Fig. 1F) due to a block in photoreceptor cell differentiation (data not shown; Therrien et al. 1996). Strikingly, the BT98/KDN flies showed a massive accumulation of pMAPK posterior to the morphogenetic furrow (Fig. 1G) that correlated with a robust enhancement of activated RAF rough eye phenotype (Fig. 1H). These findings strongly suggest that N-Tor4021KSRc brings endogenous MEK to N-Tor4021RAFc, thus strongly enhancing endogenous MEK and MAPK activation.
Figure 1.
TorKSRc, a dominant-negative form of KSR, strongly cooperates with TorRAFc, an activated form of RAF, to activate the MAPK pathway during Drosophila eye development. (A,C,E,G) Anti-pMAPK staining of Drosophila third instar eye imaginal discs and (B,D,F,H) scanning electron micrographs of adult Drosophila eyes of the following genotypes: (A,B) wild type (WT); (C,D) BT98/+; (E,F) KDN/+; (G,H) BT98/KDN. Anterior is to the right. During normal eye disc development (A), activation of MAPK first appears in groups of cells in the morphogenetic furrow (arrowhead). Posterior (left) to the morphogenetic furrow, pMAPK staining is restricted to individual cells undergoing cell fate specification. The RAFc and KSRc constructs have been expressed under the sevenless enhancer sequences, which drives transgene expression in a subset of cells posterior to the morphogenetic furrow: arrow in A marks approximately the rows of ommatidial clusters where transgene expression occurs.
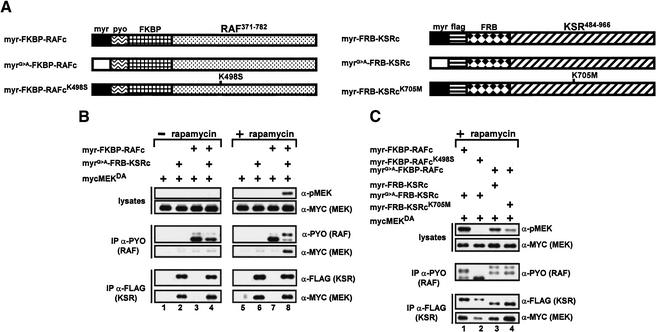
To verify that the association between RAFc and KSRc is responsible for the results shown above, we fused RAFc and KSRc to the FK506-binding protein (FKBP) and FKBP-rapamycin-binding domain (FRB), respectively, to allow their heterodimerization in a rapamycin-dependent manner (Fig. 2A; Muthuswamy et al. 1999). We also introduced a functional or inactivated myristoylation signal to examine the influence of membrane localization. RAFc and KSRc derivatives were tagged with the polyoma (pyo) and the Flag epitopes, respectively, to allow their detection.
Figure 2.
Induced association between KSR and RAF catalytic domains triggers MEK phosphorylation. (A) Schematic of the FKBP–RAF and FRB–KSR fusion proteins used in this study. (B) S2 cells were transfected with mycMEKDA (1 μg) alone or with the indicated combinations of KSRc (0.2 μg) and RAFc (0.3 μg) constructs. Twenty-four hours postinduction of expression, cells were treated with either vehicle (ethanol; −rapamycin) or with 1 μM rapamycin (+rapamycin) for 2 h prior to harvest. Cell lysates were prepared, and small fractions were used to evaluate pMEK and mycMEKDA levels by immunoblot analysis. The remaining portion of the lysates was split and immunoprecipitated with either anti-pyo or anti-Flag antibody to monitor RAFc or KSRc protein levels, associated mycMEKDA, and heterodimerization with the opposite FRB or FKBP fusion proteins. Heterodimerization data are not shown, for simplicity. The results shown here and in Figures 3–6 are representative of at least three similar experiments. (C) The experiments were conducted essentially as described in B to evaluate the ability of myr-FKBP–RAFcK498M and myr-FRB–KSRcK705M to induce MEK phosphorylation. Because of the posttranslational modification, the myristoylated fusion proteins show faster mobility. We noticed that the dimerization of different fusion proteins was less rapamycin-dependent when they were either both membrane-targeted or not. This was presumably caused by the tendency of these proteins to associate together. However, dimerization was strictly rapamycin-dependent when RAFc proteins were membrane-targeted, but not the KSRc proteins or vice versa. Therefore, we presented only the data for which either RAFc or KSRc is membrane-targeted.
The FKBP–RAFc and FRB–KSRc fusion proteins were expressed separately or together along with myc-epitope-tagged MEKDA in the absence or the presence of rapamycin (Fig. 2B). Expression of KSRc or RAFc variants alone or together did not result in MEK phosphorylation in the absence of rapamycin (Fig. 2B, lanes 2–4; top panel; α-pMEK). As expected, FRB–KSRc physically interacted with MEK as revealed by the associated mycMEKDA in anti-Flag immunoprecipitates (Fig. 2B, lanes 2,4; bottom panel; α-MYC), whereas the FKBP–RAFc did not interact with MEK (Fig. 2B, lanes 3,4; middle panel; α-MYC). Rapamycin treatment did not affect the behavior of the KSRc and RAFc constructs expressed alone (Fig. 2B, lanes 6,7), but promoted their heterodimerization when expressed together (data not shown). This resulted in complex formation between RAF and MEK and led to MEK phosphorylation (Fig. 2B, lane 8). Interestingly, this effect was accompanied by a clear and strong mobility shift of FKBP–RAFc. Permutation of the myristoylation signal or no myristoylation signal on the two types of fusion proteins gave identical results (data not shown), which suggested that no additional membrane components are required in this event. Together, these results showed that induced association between RAFc and KSRc results in MEK phosphorylation.
To address whether MEK phosphorylation depended on the catalytic function of RAFc or KSRc, we tested the activity of two kinase-inactivated mutants, RAFcK498S and KSRcK705M (Fig. 2A). As expected, kinase-inactivated RAFc did not support rapamycin-dependent MEK phosphorylation (Fig. 2C, lane 2). Interestingly, this mutant did not display a mobility shift. In contrast, KSRcK705M, which interacted normally with MEK, did promote MEK phosphorylation (Fig. 2C, lane 4), although not as efficiently as wild-type KSR. These results suggest that the ability of KSR to promote MEK phosphorylation does not absolutely require a catalytically intact function (see Discussion).
KSR activity is naturally required for RAS-dependent activation of MAPK in S2 cells
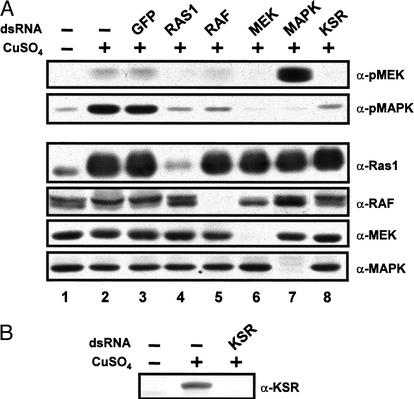
The data shown above provide compelling evidence that KSR facilitates signaling between RAF and MEK. Before characterizing further the mechanism by which KSR carries out this function, we wanted to make sure that KSR is genuinely required in S2 cells for activation of the MAPK pathway. Although previous genetic and biochemical data on KSR have suggested its involvement in the MAPK module, they have never demonstrated it. We used the RNAi technique (Clemens et al. 2000) to deplete endogenous KSR levels in S2 cells as well as the levels of the other major components and examine the effect on MAPK activation. A stable S2 cell line expressing RASV12 under the control of a heavy metal-inducible promoter was used to activate the MAPK pathway. Addition of copper to cell culture media augmented total RAS1 levels, which resulted in a modest increase in phospho-MEK (pMEK) and a robust elevation of pMAPK (Fig. 3, cf. lanes 1 and 2).
Figure 3.
KSR is a bona fide component of the MAPK pathway. (A) Untreated (−) or CuSO4-treated (+) RASV12 cells were either incubated alone (−) or with (+) 10 μg of the indicated dsRNAs. After incubation, cells were lysed and pMEK or pMAPK levels, or protein levels for RAS1, RAF, MEK, and MAPK, were analyzed by immunoblot analysis using the indicated antibodies. Endogenous KSR protein levels were under the detection limit of our KSR antibody. Immunoprecipitation from ∼25× more cells is required to detect endogenous KSR, which is incompatible with the RNAi protocol. (B) S2 cells were transfected with pMet-KSR and were either untreated (−) or CuSO4-treated (+) to induce KSR expression. Cotransfection of pMet-KSR with 0.5 μg of dsKSR RNA prevented KSR accumulation, thus showing the effectiveness of dsKSR RNA.
In contrast to the double-stranded green-fluorescent protein (dsGFP) control RNA, incubation of cells with dsRAS1 RNA prevented MEK and MAPK activation (Fig. 3, cf. lanes 3 and 4). dsRAS1 RNA specifically decreased the levels of RAS1, but did not affect the levels of endogenous RAF, MEK, and MAPK. Specific removal of the three kinases of the module also impaired signal transduction (Fig. 3, lanes 5, 6, and 7, respectively). However, as previously reported (Clemens et al. 2000), elimination of MAPK in S2 cells led to an increase in pMEK levels (Fig. 3, lane 7), which suggests the presence of a MAPK-dependent negative-feedback mechanism. Interestingly, ablation of endogenous KSR also precluded activation of MEK and MAPK by RASV12 (Fig. 3, lane 8), and similar results were obtained when the pathway was activated using an activated form of the Sevenless receptor tyrosine kinase (data not shown), which indicates that KSR plays a critical role as well in situation where RAS has not been overexpressed. Therefore, these results show that KSR is a bona fide component of the MAPK pathway. In addition, given that dsKSR RNA inhibited MEK activation, it implies that it is required at a step upstream of MEK, which is consistent with its ability to promote MEK phosphorylation.
KSR promotes MEK phosphorylation by RAF in a RAS-dependent manner
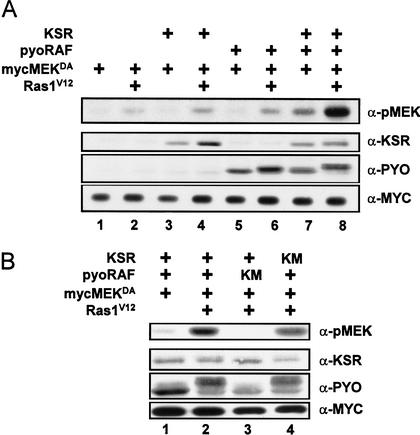
To further unravel the molecular function of KSR, we wanted to reconstitute a KSR-dependent MEK phosphorylation assay using native KSR and RAF proteins. However, mouse KSR1 has been previously reported to block signaling through the ERK/MAPK pathway when overexpressed in various cell lines (see above). This effect is likely owing to sequestration of specific components of the pathway that are in limiting amounts. RAF and MEK are prime candidates as they have been reported to interact with mKSR1. To circumvent this problem, we coexpressed KSR together with RAF and MEK. RASV12 was also used to activate the MAPK pathway. To prevent possible MAPK-dependent negative-feedback effects, as suggested by the RNAi result shown in Figure 3 (lane 7), we used a kinase-inactivated version of MEK (MEKDA) throughout this work. Compared with myc-epitope-tagged MEKDA expressed alone (Fig. 4A, lane 1), RASV12 coexpression slightly increased pMEK levels (Fig. 4A, lane 2). Coexpression of KSR or pyo-epitope-tagged RAF in those two conditions (absence or presence of RASV12) did not significantly alter the pMEK profiles (Fig. 4A, cf. lanes 3–6 to lanes 1 and 2). Strikingly, coexpression of MEK with KSR and RAF without RASV12 induced MEK phosphorylation (Fig. 4A, lane 7), which strongly increased upon addition of RASV12 (Fig. 4A, lane 8). These results suggest that native KSR promotes the ability of RAF to phosphorylate MEK. Although RAS activity strongly increased the effect of KSR, the fact that we detected an effect of KSR in the absence of RASV12 (Fig. 4A, lane 7) suggests among different possibilities that either endogenous RAS activity contributed to that effect or that the higher levels of the transiently expressed proteins somewhat bypassed the normal RAS-dependency of MEK phosphorylation by RAF.
Figure 4.
KSR enhances MEK phosphorylation by RAF in a RAS-dependent manner. (A) S2 cells were transfected with the mycMEKDA reporter construct (0.3 μg) either alone (lane 1) or together with the indicated combinations of RASV12 (0.6 μg), KSR (0.1 μg), and pyoRAF (0.03 μg) constructs. Cells were lysed 36 h postinduction of expression, and pMEK levels were examined by immunoblot analysis. Protein levels were evaluated for KSR, pyoRAF, and mycMEKDA as indicated. (B) S2 cells were transfected as in A with the indicated combinations of expression plasmids. KM denotes pyoRAFK498M or KSRK705M.
We next examined whether the kinase function of RAF and the putative kinase function of KSR are required for MEK phosphorylation in this assay. We generated kinase-inactivated mutants for pyoRAF (K498M) and KSR (K705M), and tested their respective effect as described above. As expected, RAF kinase function was absolutely required for MEK phosphorylation (Fig. 4B, lane 3). In contrast, KSRK705M still promoted MEK phosphorylation, although slightly less efficiently than wild-type KSR (Fig. 4B, lane 4). As for the FRB–KSRc construct, the ability of KSR to stimulate MEK phosphorylation does not absolutely depend on its putative kinase function.
KSR activity correlates with its ability to associate with MEK
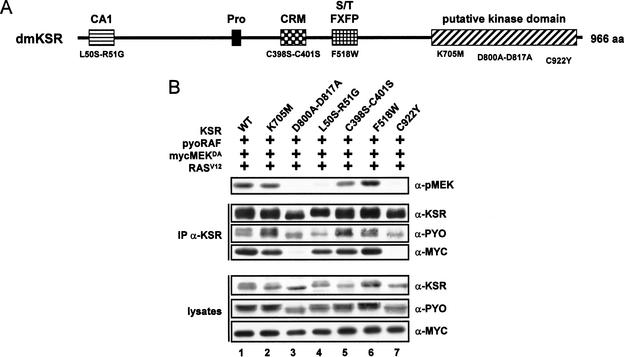
We took advantage of the simple cotransfection assay in S2 cells to examine the effect of five additional mutations in KSR (Fig. 5A). Kolesnick and colleagues presented evidence that mKSR1 can phosphorylate RAF (Zhang et al. 1997; Xing et al. 2000). As a kinase-inactive mutant, they changed two conserved aspartic residues in subdomains VI and VII to alanine residues. Because the KSRK705M mutant is still active (Fig. 3B), we generated a KSR kinase-defective mutant similar to theirs (D800A–D817A) to test its effect. The four other mutants affected independent regions of KSR. KSRL50S–R51G lies within the CA1 domain. This mutation has been recovered as a hypomorphic loss-of-function allele in a RAS-dependent genetic screen in Drosophila (Therrien et al. 1995). KSRC398S–C401S disrupts the integrity of the cysteine-rich motif. A similar mutation in mKSR1 has been shown to abrogate its ability to promote RAS-dependent Xenopus oocyte maturation (Michaud et al. 1997). KSR contains an FXFP motif that has been proposed to function as a MAPK docking site (Jacobs et al. 1999). The function of this motif in KSR is unknown. We changed the first phenylalanine residue of the motif, KSRF518W, to verify its functional relevance. Finally, a mutation in KSR isolated in a RAS-dependent genetic screen in C. elegans (Sundaram and Han 1995) has been reported to prevent the association between KSR and MEK (Stewart et al. 1999). A similar mutation was generated, KSRC922Y, to examine the effect of impairing the KSR/MEK interaction. The effect of these mutations is shown in Figure 5B. Interestingly, in contrast to KSRK705M, which still supported MEK phosphorylation, KSRD800A–D817A and KSRC922Y completely abrogated KSR activity (Fig. 5A, cf. lanes 2, 3, and 7). In addition, KSRL50S–R51G showed a severe reduction of activity (Fig. 5A, lane 4), whereas KSRC398S–C401S showed only a slight, but reproducible reduction in KSR activity (Fig. 5A, lane 5), and KSRF518W was as active as wild-type KSR (Fig. 5A, lane 6). Given that the two KSR mutations (L50S–R51G and C922Y) corresponding to loss-of-function mutations isolated in genetic screens also disrupted the ability of KSR to induce MEK phosphorylation by RAF, it strongly suggests that the assay recapitulates the normal function of KSR.
Figure 5.
KSR activity depends on multiple regions of the KSR protein. (A) Structure of KSR and amino acid positions of various mutants used in this study. In addition to a putative kinase domain, the conserved area 1 (CA1), the proline-rich stretch (Pro), the cysteine-rich motif (CRM), and the serine/threonine-rich stretch/FXFP motif (S/T FXFP) are other features of the KSR proteins (Therrien et al. 1995). (B) S2 cells were transfected with the indicated combinations of mycMEKDA (0.6 μg), RASV12 (0.6 μg), pyoRAF (0.2 μg), and wild-type (WT) KSR (0.7 μg) or one of the indicated KSR mutants (0.7 μg). Cells lysates were prepared 36 h postinduction of expression and either directly used to determine pMEK levels or immunoprecipitated with a monoclonal anti-KSR antibody to examine the amounts of mycMEKDA and pyoRAF associated with KSR. Protein levels for KSR, mycMEKDA, and pyoRAF were also evaluated.
We then investigated whether the effect of the mutations could be caused by defects in their ability to associate with MEK and/or RAF. To verify this, we immunoprecipitated KSR or the mutant variants from cell lysates used to analyze pMEK levels (Fig. 5B) and examined the amount of coimmunoprecipitated mycMEKDA and pyoRAF (Fig. 5B, middle part of the panel). As reported for mKSR1 (see above), Drosophila KSR also associated with MEK and RAF (Fig. 5B, lane 1). Two mutants, KSRD800A–D817A and KSRC922Y, did not interact with MEK (Fig. 5B, lanes 3,7). Interestingly, they also failed to stimulate MEK phosphorylation. In contrast, KSRK705M, which is almost as active as wild-type KSR, was not significantly affected in its association with MEK (Fig. 5B, lane 2). These results suggest that the ability of KSR to interact with MEK is critical to stimulate MEK phosphorylation. Intriguingly, KSRL50S–R51G associated normally with MEK, yet was severely diminished in its capacity to promote MEK phosphorylation (Fig. 5B, lane 4). Although we did not observe a drastic effect regarding the binding properties of this mutant, it very reproducibly showed an approximately twofold decrease in its capacity to associate with RAF. This observation suggests that the CA1 domain mediates an association with RAF. KSRD800A–D817A and KSRC922Y also showed a similar RAF-association defect (Fig. 5B, lanes 3 and 7, respectively). Their reduced interaction with RAF might be caused by their inability to bind MEK, which normally might stabilize the KSR/RAF interaction. Alternatively, these mutations might induce structural perturbations that reduce the KSR/RAF association independently of the inability of MEK to bind KSR. Taken together, these data indicate that KSR associates independently with MEK and RAF, and that these interactions appear to be critical for KSR activity.
KSR allows the formation of a RAF/MEK complex
Previous work reported that the association between mKSR1 and RAF is RAS-dependent (Therrien et al. 1996; Xing et al. 1997). Interestingly, we found that Drosophila KSR can associate with RAF without coexpressing an activated form of RAS (Fig. 4A; data not shown). We therefore wanted to determine whether endogenous RAS1 activity might contribute to this interaction. To verify this, we coexpressed pyoRAF and KSR in the presence of dsRNA for GFP, RAS1, MEK, or MAPK, immunoprecipitated RAF, and measured the levels of associated KSR. As for the negative controls, addition of dsRAS1 RNA did not perturb the KSR/RAF association (Fig. 6A, lane 3), which suggests that the Drosophila KSR/RAF interaction can occur in the absence of RAS-mediated signals. The inability of dsMEK RNA to alter the KSR/RAF association (Fig. 6A, lane 4) is another indication that KSR interacts with RAF independently of MEK.
Figure 6.
KSR interacts independently with RAF and MEK and induces the formation of a RAF/MEK complex. (A) S2 cells were transfected with pyoRAF alone or together with KSR in the presence of the indicated dsRNAs (0.5 μg; lanes 2–5). Cell lysates were prepared and immunoprecipitated 36 h postinduction as indicated. Levels of associated proteins and their relative amounts were evaluated by immunoblot analysis. (B) S2 cells were transfected with mycMEKDA (0.2 μg) and pyoRAF (0.2 μg) constructs either alone (lane 3) or with increasing quantities for wild-type (WT) KSR (0.02 μg, 0.08 μg, and 0.325 μg) or for KSRL50S-R51G or KSRC922Y (0.02 μg, 0.08 μg, 0.325 μg, and 1.3 μg), as indicated; control (lane 1) used 1.3 μg of wild-type KSR. Cell lysates were prepared 36 h postinduction and immunoprecipitated by the anti-pyo antibody to measure associated MEK and KSR levels by immunoblot analysis. Protein levels in lysates were also determined. (C) S2 cells were transfected with mycMEKDA (0.2 μg), pyoRAF (0.2 μg), or KSR (0.7 μg) without or with 0.6 μg of HA-epitope-tagged RASV12 or 0.5 μg of dsRAS1 RNA as indicated. Cell lysates were prepared as in C. ha-RASV12 levels were examined using anti-HA antibody. Note that addition of dsRAS1 RNA eliminates ha-RASV12 completely.
Given that KSR appears to associate independently with RAF and MEK, we tested whether KSR might physically link RAF and MEK together. We coexpressed fixed amounts of RAF and MEK alone or in the presence of increasing amounts of KSR, and evaluated the levels of associated MEK by probing the anti-pyo immunoprecipitates with anti-myc. As shown in Figure 6B, coexpression of RAF and MEK alone did not result in the formation of a stable RAF/MEK association (Fig. 6A, lane 3). In sharp contrast, expression of KSR allowed the formation of a RAF/MEK complex (Fig. 6A, lanes 4–6). The levels of MEK associated with RAF increased upon augmenting the expression levels of KSR as predicted if KSR physically connects RAF and MEK. If this model is correct, KSRC922Y should not induce the formation of a RAF/MEK complex because it no longer binds to MEK (Fig. 5B, lane 7). Interestingly, when coexpressed with RAF and MEK, wild-type KSR showed greater protein stability compared with KSRC922Y. We therefore transfected more DNA for the mutant construct to compare the effect of equal amounts of proteins. As predicted, KSRC922Y could not promote the formation of a RAF/MEK complex (Fig. 6B, lanes 7–10).
We were intrigued by the fact that KSR induced the formation of a RAF/MEK complex in the absence of cotransfected RASV12. We therefore verified whether endogenous RAS1 activity was involved in the RAF/MEK complex formation by eliminating its contribution using dsRAS1 RNA. Surprisingly, as shown in Figure 6C, dsRAS1 RNA did not affect the complex (Fig. 6C, cf. lanes 1 and 2). Similarly, coexpression of RASV12 did not significantly enhance its formation (Fig. 6C, lane 3). Together, these findings strongly suggest that KSR connects RAF and MEK and that the assembly does not require RAS activity.
Discussion
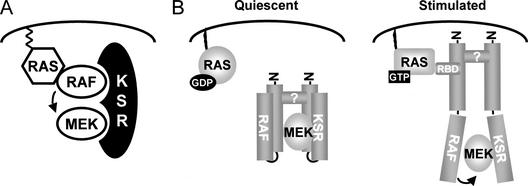
KSR was originally recognized as an essential component of RAS-mediated signaling pathways in Drosophila and C. elegans (Kornfeld et al. 1995; Sundaram and Han 1995; Therrien et al. 1995). Although the characterization of mouse KSR1 linked it to the ERK module (Morrison 2001), its precise molecular function has not heretofore been elucidated. Here, we show that Drosophila KSR is genuinely required for signal transmission through the MAPK pathway at a step upstream of MEK. In agreement with this, we found that KSR directly participates in the phosphorylation of MEK by RAF and that this event does not absolutely require the putative catalytic function of KSR. We further show that KSR associates independently with MEK and with RAF. These independent interactions lead to the formation of a stable RAF/MEK complex, thereby positioning the activator RAF in close proximity to its substrate MEK. Hence, KSR operationally behaves as a molecular scaffold that assembles the RAF/MEK functional pair (Fig. 7A). Given that MP1 apparently coordinates the formation of MEK/ERK complexes (Schaeffer et al. 1998), it will certainly be interesting to investigate whether KSR and MP1 function together to orchestrate the formation of a complete ERK/MAPK module.
Figure 7.
Model accounting for KSR function. See text for details.
Mouse KSR1 had been previously reported to associate constitutively with MEK and in a RAS-dependent manner with RAF (see above). However, the physical consequences of these associations had not been explored further. We found here that Drosophila KSR also interacts with MEK and RAF, but, in contrast to mKSR1, the Drosophila KSR/RAF association did not appear to depend on RAS activity. A reason for this difference might be that the association between mKSR1 and c-RAF did not resist the immunoprecipitation procedure. However, in the presence of activated RAS other contacts might have formed, which could stabilize the complex. RAS-induced oligomerization and/or other proteins might be involved in stabilizing the mKSR1/c-RAF complex. Regardless of the exact mechanism, our results clearly show that the associations between KSR and RAF and between KSR and MEK are mediated by independent parts of KSR and result in the association of RAF and MEK. Given that our data suggest that RAS is dispensable for these interactions, this implies that an inactive KSR/MEK/RAF ternary complex might exist in nonstimulated cells (Fig. 7B). Interestingly, STE5 has also been shown to assemble a three-kinase MAPK complex prior to signal transduction (Choi et al. 1994). This might be a general mechanism used by scaffolding proteins to allow highly efficient switch-like signal transmission. Consistent with this possibility, we found that endogenous KSR, which apparently represents <1% of endogenous RAF and MEK protein levels, is predominantly associated with RAF and MEK in S2 cells (data not shown).
The CA1 domain appears to be involved in connecting the KSR/MEK complex to RAF. It is still unclear whether the interaction is direct or not. Although the mutation affecting the CA1 domain reduced the KSR/RAF association, it did not abrogate it (Fig. 5B; data not shown). This suggests that either the mutation does not fully disrupt the interaction with RAF or that another region(s) of KSR makes contact with RAF. This is consistent with the fact that this mutation allowed to some extent the formation of a RAF/MEK association, albeit not as effectively as wild-type KSR (data not shown) and that it genetically behaved as a weak loss-of-function mutation (Therrien et al. 1995). Alternatively, it is possible that the mutation does not affect RAF binding per se, but localizes the mutant KSR protein aberrantly. Although MEK might stabilize the RAF/KSR interaction (Fig. 5B), it is not primarily mediating it. Indeed, dsMEK RNA did not perturb the KSR/RAF association (Fig. 6A), and the two mutants, KSRD800A–D817A and KSRC922Y, which no longer interacted with MEK, still associated with RAF (Figs. 5B and 6B). Systematic mutagenesis and deletion mutants of KSR should allow the identification of additional regions, if any, involved in the formation of the RAF/MEK complex. Given that mKSR1, like the RAF isozymes, interacts with the 14–3–3 proteins, HSP90, and p55/CDC37 (Xing et al. 1997; Cacace et al. 1999; Stewart et al. 1999), it will be interesting to discover the contribution of these proteins in the KSR/RAF association and in the regulation of the KSR-dependent RAF/MEK complex.
Previous work in Drosophila showed that ksr loss-of-function mutations suppressed activated RAS-mediated signaling, but did not alter activated RAF function (Therrien et al. 1995). These observations led the authors to propose that KSR is required at a step between RAS and RAF or in a pathway that acts in parallel. However, as they mentioned, the data were also consistent with the possibility that the activated RAF transgene was expressed to high levels, thereby bypassing the requirement for KSR function. Our results are consistent with the second interpretation. We found that KSR promoted MEK phosphorylation when low amounts of the RAF construct were transfected (Fig. 2A), but that this effect declined upon increasing the amounts of transfected RAF (data not shown). The genetic data also raised the possibility that KSR is involved in the RAS-dependent RAF activation mechanism. This latter process has been the subject of numerous studies, but remains largely enigmatic (Kolch 2000). A number of laboratories have tested the possibility that KSR directly phosphorylates RAF (see above). Most of these attempts have failed to show catalytic function for KSR, except for Kolesnick and colleagues, who did report that KSR can phosphorylate and activate RAF (Zhang et al. 1997). Our data do not support their findings, but may provide an explanation for the apparent contradiction. Interestingly, there is a correlation between the ability of KSR to bind MEK and the appearance of a mobility shift in transfected RAF (Fig. 5B), which appears to be caused by phosphorylation (data not shown). KSRD800A–D817A and KSRC922Y did not induce the mobility shift, whereas KSRK705M did. This suggests that this event does not depend on the putative catalytic function of KSR, but, rather, on its ability to interact with MEK. Moreover, the mobility shift observed for FKBP–RAFc appeared to depend on its autocatalytic function because FKBP–RAFcK498S did not display the mobility shift, even though it heterodimerized with the FRB–KSRc/MEK complex upon rapamycin treatment (Fig. 2C, lane 2; data not shown). Together, these data suggest that RAF autophosphorylation is strongly stimulated when MEK is brought to RAF by KSR, and thus KSR might be involved indirectly in RAF activation, not by virtue of its catalytic function, but rather by the effect of recruiting MEK to RAF. Given that the Kolesnick group used KSRD800A–D817A as a kinase-inactivated mutant, which does not bind to MEK, it raises the possibility that their results correspond, in fact, to a KSR-dependent RAF autophosphorylation. Consistent with this, the only KSR-dependent phosphorylated residues that they identified correspond to RAF autophosphorylation sites (Zhang et al. 1997; Xing and Kolesnick 2001).
Surprisingly, the simple coexpression of KSR with MEK, RAF, and activated RAS, is sufficient to reconstitute a KSR-dependent assay. The reliability of the assay is supported by the observation that wild-type KSR behaved as a positive component of the pathway, as genetics predicted, and KSR mutations corresponding to previously identified ksr loss-of-function alleles were inert. The critical aspect of the assay is the simultaneous expression of RAF and MEK with KSR. When KSR is overexpressed in a cell line, RAF and MEK also need to be coexpressed to high enough levels to prevent their uncoupling on separate KSR molecules. A similar situation has been observed for JIP-1, which was first thought to be an inhibitor of the JNK pathway (Dickens et al. 1997). Our results thus provide an explanation as to why many laboratories found that KSR inhibited signaling through the ERK pathway when overexpressed in various cell lines. When expressed at very low levels, mKSR1 has been shown to accelerate RAS-dependent Xenopus oocyte meiotic maturation (Therrien et al. 1996; Cacace et al. 1999), a process that depends on activation of the ERK module (Birchmeier et al. 1985). Intriguingly, mKSR1 activity in this system appeared to be mediated mainly by the cysteine-rich motif (CRM; Therrien et al. 1996; Michaud et al. 1997). Although this region is probably required for normal KSR activity, it does not account for its entire function. Therefore, this assay probably recapitulated only partially the normal function of KSR. Biochemical analysis of the CRM revealed that it is involved in targeting mKSR1 to the plasma membrane in a RAS-dependent manner (Michaud et al. 1997). Nonetheless, it is unclear why the CRM alone was capable of stimulating RAS-dependent Xenopus oocyte maturation. KSRC398S–C401S was reproducibly less active than wild-type KSR in the S2 cell assay, thus confirming the functional relevance of the CRM. However, the effect was weak (Fig. 5B). This could be caused by the fact that the main participants were overexpressed, thereby making the system less dependent on signals normally concentrating the various components to the plasma membrane.
Intriguingly, besides the lysine to arginine change in subdomain II of the mammalian homologs, KSR proteins have a highly conserved kinase domain (Therrien et al. 1995). It is formally possible that this domain is enzymatically active, but that we have not found the proper conditions and/or substrates to detect its activity. The observation that the two KSRK705M mutants used in this study are slightly less active than their wild-type counterparts, even though they associate with MEK to the same extent as wild-type KSR, suggests that KSR might possess a catalytic function that is required in concert with its scaffolding property for full activity. Alternatively, it is possible that their lesser activity is owing to structural changes in KSR that do not perturb the KSR–MEK interaction but affect the way KSR presents MEK to RAF. Stringent sequence conservation might thus be required to maintain a particular kinase domain conformation to allow highly specific and robust interaction with MEK for the sole purpose of presenting a particular portion of MEK to RAF. Given their structural relatedness and the good homology between their kinase domains, RAF and KSR probably evolved from a common ancestral kinase by gene duplication. One of the descendents of this hypothetical duplication event might have given rise to the three RAF kinase family members, which retained catalytic function, whereas the other descendent might have eventually led to the two KSR genes found in mammals (M.T., unpubl.) that evolved as scaffolds specialized in bridging RAF and MEK proteins together. Although it is currently unclear whether the functional shift observed for the KSR kinase domain will also be observed in other uncharacterized kinases for a similar purpose, this certainly highlights the importance of showing the catalytic activity of a kinase or any other enzyme before assuming it performs an enzymatic step in a given process.
It is becoming increasingly clear that components for several signaling pathways are specifically organized by scaffolding proteins (for review, see Pawson and Scott 1997). However, we still know very little about the way they operate. The genetic and molecular data gathered so far on Drosophila KSR are consistent with its involvement in signaling efficiency, that is, when KSR is nonfunctional, low amounts of signal reach MAPK. The chain of events is, however, not severed because activated RAF can rescue ksr loss-of-function phenotypes in Drosophila (Therrien et al. 1995). Our results show a role for KSR as a molecular scaffold coordinating the RAF/MEK interaction. This is, however, one part of the signal propagation mechanism through the ERK/MAPK module. Once MEK is activated, it must relay the signal to MAPK. It will be interesting to determine whether KSR also participates in this process or whether another molecule, such as MP1, executes that step independently.
Materials and methods
Plasmids
pMet-KSR was made by inserting a 2.9-kb NotI cDNA insert encoding full-length Drosophila KSR (Therrien et al. 1995) into pMet, a vector containing the metallothionein promoter inducible by heavy metals. Mutant KSR constructs and other mutants mentioned below were generated using the QuickChange mutagenesis kit (Stratagene). All constructs were verified by sequencing.
pMet-pyoRAF was generated in three steps. First, an EcoR1/XhoI PCR product corresponding to Drosophila RAF full-length cDNA was inserted into pBlueScript (Stratagene). Second, an oligonucleotide encoding two polyoma epitope tags (MEYMPME) was inserted into the EcoR1 site in place of the first methionine. The EcoR1/XhoI insert was then moved into the pMet EcoRI/SalI sites. pMet-pyoRAFK498M was generated by mutating the indicated codon in pMet-pyoRAF.
pMet-mycMEKDA was generated in two steps. First, a PCR product corresponding to Drosophila MEK full-length cDNA with EcoRI extremities and a myc-epitope tag (EQKLISEEDLN) at the 5′ end was inserted into a pBlueScript II vector. The resulting pBS-mycMEK was then mutagenized to change aspartic acid residue 224 in kinase subdomain VII to an alanine residue (DA). The mycMEKDA insert was then moved into the EcoRI site of pMet.
pMet-RASV12 was previously described (Therrien et al. 1999). pMet-haRASV12 was made by inserting an oligonucleotide encoding three HA epitopes (YPYDVPDYA) into the EcoRI site of pMet-RASV12 in place of the first methionine.
The FKBP and FRB fusion constructs were assembled as follows: oligonucleotides encoding either the Drosophila SRC myristoylation signal (first 16 amino acids) or a nonmyristoylatable version (Gly 2 changed to Ala), were inserted into the KpnI/EcoRI sites of pBlueScript; a BstEII site has been included immediately upstream of the EcoR1 site in the oligonucleotides. For the RAFc derivatives, a BstEII/EcoR1 fragment encompassing the RAF catalytic domain (wild type or kinase-inactivated; amino acids 371–782) was inserted into the BstEII/EcoRI sites of pBS-Myr or pBS-MyrGA. A PCR product corresponding to pyo-tagged mouse FK506-binding protein cDNA was then inserted in the BstEII site of the Myr or non-Myr RAFc constructs. KpnI/NotI complete inserts were then transferred into a pMet vector. For the KSRc derivatives, an EcoRI/XbaI fragment encompassing the KSR catalytic domain (wild type or mutated versions; amino acids 484–966) was inserted into the equivalent sites of pBS-Myr or pBS-MyrGA. A PCR product corresponding to Flag epitope (DYKDDDDK)-tagged rat FKBP-rapamycin-binding domain (FRB; amino acids 2025–2114 of rat FRAP) was inserted into the BstEII/EcoRI sites of the Myr or non-Myr KSRc constructs. KpnI/NotI complete inserts were then transferred into a pMet vector.
Cell transfection, cell lysates, and immunoprecipitations
For transfection experiments, 107 S2 cells were plated per 100-mm-diameter dish (Nunc) and incubated overnight. Cells were transfected with different combinations of plasmids (2 μg total) using the Effectene reagent (QIAGEN). Protein expression was induced 36 h after transfection by addition of 0.7 mM CuSO4.
Cells were harvested in Nonidet P-40 lysis buffer (Therrien et al. 1996) at 36 h postinduction. Immunoprecipitations were performed by incubating cell lysates with antibody supernatants at 4°C for 1 h. Protein A/G agarose beads (Santa Cruz Biotechnology) were then added, and gently rocked at 4°C for an additional 3 h. Immunoprecipitates were washed three times with cold lysis buffer before analysis.
Western blot analysis and antibodies
Cell lysates or immunoprecipitated proteins were resolved on 8% SDS-PAGE and transferred to nitrocellulose membranes. Proteins were probed using appropriate primary antibodies from the following sources: α-KSR monoclonal antibody (mAb) was previously described (Therrien et al. 1996); α-RAS1, α-PYO epitope, and α-HA epitope (12CA5) mAbs were kindly provided by Gerry Rubin (University of California, Berkeley); α-Drosophila RAF polyclonal antibody was a kind gift from Debbie Morrison (National Cancer Institute, Frederick, MD); α-MYC epitope mAb (9E10) was from Santa Cruz Biotechnology; α-MEK-1&2 and α-pMEK-1&2 polyclonal antibodies were from Cell Signaling; and α-ERK-1&2 and α-dpERK-1&2 mAbs were from Sigma.
RNAi experiments
DsRNAs were generated as previously described (Clemens et al. 2000) with slight modifications. DNA fragments (∼700 bp) containing coding sequences for the targeted proteins were amplified by PCR. Each PCR primer contained a 5′-T7 RNA polymerase binding site (GAATTAATACGACTCACTATAGGGAGA) followed by 21 nucleotides corresponding to the targeted sequence. The PCR products were purified using the QIAquick PCR purification kit (QIAGEN). One microgram of PCR product was used per in vitro transcription reaction. RNAs were phenol–chloroform-extracted, ethanol-precipitated, and resuspended in TE buffer. DsRNAs were generated by heating RNA samples to 95°C and annealed by slow cooling to room temperature. DsRNA quality was verified on 1% agarose gels.
For RNAi experiments, 2 × 106 RASV12 cells were plated per well of six-well tissue culture dishes (Nunc) and incubated overnight. Media were changed for 2 mL of complete medium with or without 10 μg of dsRNA. Cells were incubated for 5 d. RASV12 expression was induced by adding 0.7 mM CuSO4 to the cell culture 10 h prior to harvest. Given that much less dsRNA was used in transfection experiments (0.5 μg), interference was essentially restricted to the transfected cells.
Genetics, histology, and immunohistochemistry
Fly culture and crosses were performed according to standard procedures. Scanning electron microscopy was conducted as previously described (Kimmel et al. 1990). Staining of third instar eye imaginal disc was performed according to Gabay et al. (1997).
Acknowledgments
We are grateful to Morag Park, André Veillette, and David Wassarman for critical reading of the manuscript. We thank Gerry Rubin for fly stocks and anti-RAS1, anti-PYO, and anti-HA antibodies; and Debbie Morrison for anti-Drosophila RAF antibody. M.T. is the recipient of a Canadian Institute of Health Research Scholarship. This work was supported by the National Cancer Institute of Canada with funds from the Terry Fox Run to M.T.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL therrim@ircm.qc.ca; FAX (514) 987-5591.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.962902.
References
- Birchmeier C, Broek D, Wigler M. Ras proteins can induce meiosis in Xenopus oocytes. Cell. 1985;43:615–621. doi: 10.1016/0092-8674(85)90233-8. [DOI] [PubMed] [Google Scholar]
- Cacace AM, Michaud NR, Therrien M, Mathes K, Copeland T, Rubin GM, Morrison DK. Identification of constitutive and ras-inducible phosphorylation sites of KSR: Implications for 14–3–3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denouel-Galy A, Douville EM, Warne PH, Papin C, Laugier D, Calothy G, Downward J, Eychene A. Murine Ksr interacts with MEK and inhibits Ras-induced transformation. Curr Biol. 1997;8:46–55. doi: 10.1016/s0960-9822(98)70019-3. [DOI] [PubMed] [Google Scholar]
- Dickens M, Rogers JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- Dickson B, Sprenger F, Morrison D, Hafen E. Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature. 1992;360:600–603. doi: 10.1038/360600a0. [DOI] [PubMed] [Google Scholar]
- English J, Pearson G, Wilsbacher J, Swantek J, Karandikar M, Xu S, Cobb MH. New insights into the control of MAP kinase pathways. Exp Cell Res. 1999;253:255–270. doi: 10.1006/excr.1999.4687. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes & Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- Joneson T, Fulton JA, Volle DJ, Chaika OV, Bar-Sagi D, Lewis RE. Kinase suppressor of Ras inhibits the activation of extracellular ligand-regulated (ERK) mitogen-activated protein (MAP) kinase by growth factors, activated Ras, and Ras effectors. J Biol Chem. 1998;273:7743–7748. doi: 10.1074/jbc.273.13.7743. [DOI] [PubMed] [Google Scholar]
- Kimmel BE, Heberlein U, Rubin GM. The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes & Dev. 1990;4:712–727. doi: 10.1101/gad.4.5.712. [DOI] [PubMed] [Google Scholar]
- Kolch W. Meaningful relationships: The regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J. 2000;351:289–305. [PMC free article] [PubMed] [Google Scholar]
- Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- Michaud NR, Therrien M, Cacace A, Edsall LC, Spiegel S, Rubin GM, Morrison DK. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc Natl Acad Sci. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK. KSR: A MAPK scaffold of the Ras pathway? J Cell Sci. 2001;114:1609–1612. doi: 10.1242/jcs.114.9.1609. [DOI] [PubMed] [Google Scholar]
- Muller J, Cacace AM, Lyons WE, McGill CB, Morrison DK. Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol Cell Biol. 2000;20:5529–5539. doi: 10.1128/mcb.20.15.5529-5539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: Scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: Specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer HJ, Catling AD, Eblen ST, Collier LS, Krauss A, Weber MJ. MP1: A MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- Sprenger F, Nusslein-Volhard C. Torso receptor activity is regulated by a diffusible ligand produced at the extracellular terminal regions of the Drosophila egg. Cell. 1992;71:987–1001. doi: 10.1016/0092-8674(92)90394-r. [DOI] [PubMed] [Google Scholar]
- Stewart S, Sundaram M, Zhang Y, Lee J, Han M, Guan KL. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol Cell Biol. 1999;19:5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto T, Stewart S, Han M, Guan KL. The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling Elk-1 phosphorylation from MAP kinase activation. EMBO J. 1998;17:1717–1727. doi: 10.1093/emboj/17.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- Therrien M, Michaud NR, Rubin GM, Morrison DK. KSR modulates signal propagation within the MAPK cascade. Genes & Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- Therrien M, Wong AM, Kwan E, Rubin GM. Functional analysis of CNK in RAS signaling. Proc Natl Acad Sci. 1999;96:13259–13263. doi: 10.1073/pnas.96.23.13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ. Structural organization of MAP-kinase signaling modules by scaffold proteins in yeast and mammals. Trends Biochem Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- Xing HR, Kolesnick R. Kinase suppressor of Ras signals through Thr269 of c-Raf-1. J Biol Chem. 2001;276:9733–9741. doi: 10.1074/jbc.M008096200. [DOI] [PubMed] [Google Scholar]
- Xing H, Kornfeld K, Muslin AJ. The protein kinase KSR interacts with 14–3–3 protein and Raf. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- Xing HR, Lozano J, Kolesnick R. Epidermal growth factor treatment enhances the kinase activity of kinase suppressor of Ras. J Biol Chem. 2000;275:17276–17280. doi: 10.1074/jbc.C900989199. [DOI] [PubMed] [Google Scholar]
- Yasuda J, Whitmarsh AJ, Cavanagh J, Sharma M, Davis RJ. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Fantl WJ, Harrowe G, Williams LT. Regulation of the MAP kinase pathway by mammalian Ksr through direct interaction with MEK and ERK. Curr Biol. 1997;8:56–64. doi: 10.1016/s0960-9822(98)70020-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yao B, Delikat S, Bayoumy S, Lin XH, Basu S, McGinley M, Chan-Hui PY, Lichenstein H, Kolesnick R. Kinase suppressor of Ras is ceramide-activated protein kinase. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]