Abstract
A novel histone methyltransferase, termed Set9, was isolated from human cells. Set9 contains a SET domain, but lacks the pre- and post-SET domains. Set9 methylates specifically lysine 4 (K4) of histone H3 (H3-K4) and potentiates transcription activation. The histone H3 tail interacts specifically with the histone deacetylase NuRD complex. Methylation of histone H3-K4 by Set9 precludes the association of NuRD with the H3 tail. Moreover, methylation of H3-K4 impairs Suv39h1-mediated methylation at K9 of H3 (H3-K9). The interplay between the Set9 and Suv39h1 histone methyltransferases is specific, as the methylation of H3-K9 by the histone methyltransferase G9a was not affected by Set9 methylation of H3-K4. Our studies suggest that Set9-mediated methylation of H3-K4 functions in transcription activation by competing with histone deacetylases and by precluding H3-K9 methylation by Suv39h1. Our results suggest that the methylation of histone tails can have distinct effects on transcription, depending on its chromosomal location, the combination of posttranslational modifications, and the enzyme (or protein complex) involved in the particular modification.
Keywords: Histone H3-K4-specific methyltransferase, SET domain, transcription, NuRD, Suv39h1, G9a
In the nucleus of eukaryotic cells, DNA is stored in the form of chromatin; this structure is repressive to most, if not all processes that require access of protein machinery to DNA. The basic building block of chromatin is the nucleosome, which is composed of two copies of each of the four core histone proteins (H2A, H2B, H3, and H4) wrapped by 146 bp of DNA (Luger et al. 1997). Protruding from the nucleosome are the N-terminal tails of the core histone proteins. The histone tails appear to be unstructured and are believed to participate in the formation of higher-order chromatin structure by mediating internucleosomal interactions (Luger et al. 1997), as well as contacts with DNA that is positioned between the nucleosomes (linker DNA; Angelov et al. 2001).
Cytological studies have defined two types of chromatin: euchromatin, which appears as an extended structure and is transcriptionally active, and heterochromatin, a compacted, transcriptionally silent structure (Grunstein et al. 1995). The mechanisms by which euchromatin is converted to heterochromatin, and vice versa, are poorly defined. However, an extensive literature documents that histone tails undergo a variety of posttranslational modifications (including acetylation, phosphorylation, methylation, and ubiquitination; for reviews, see Strahl and Allis 2000; Zhang and Reinberg 2001), and recent work has provided compelling evidence that these alterations affect chromatin structure and its functional properties. Well accepted, for example, are the findings that acetylation of histone tails regulates gene expression by influencing the interconversion between heterochromatin and euchromatin. In general, acetylation of core histone tails correlates with the opening of the chromatin structure to allow transcription to occur (for review, see Roth et al. 2001).
Researchers have also begun to decipher the effects of other types of covalent modifications of chromatin components, including methylation of histones H3 and H4. First described in 1964, histones have long been known to be substrates for methylation (Murray 1964). Early studies using metabolic labeling followed by sequencing of bulk histones showed that several lysine residues, including lysines 4, 9, 27, and 36 of H3 (i.e., H3-K4, H3-K9, H3-K27, H3-K36) and lysine 20 of H4 (H4-K20), are preferred sites of methylation (for review, see van Holde 1988; Strahl et al. 1999). In addition, members of the protein arginine methyltransferase family of enzymes can methylate histones (Gary and Clarke 1998; Strahl et al. 2001; Wang et al. 2001) and other transcriptional regulatory proteins, such as the general coactivator CBP/p300 (Xu et al. 2001).
An important breakthrough in the identification of the enzymes that carry out histone-lysine methylation came from studies of suppressors of position effect variegation (PEV) in Drosophila (Reuter and Spierer 1992). Suppressers of PEV, such as Su(var)3-9, the polycomb-group protein Enhancer of zeste, and the trithorax-group protein Trithorax, all contain an evolutionarily conserved sequence motif termed the SET domain (Suppressor of Variegation, Enhancer of Zeste and Trithorax; Jenuwein et al. 1998). The SET domain of the human homolog of Drosophila Su(var)3-9 (Suv39h1) was later found to share sequence similarity with several previously identified SET-domain-containing methyltransferases from plants (Klein and Houtz 1995; Zheng et al. 1998). This observation permitted Jenuwein and colleagues to discover that Suv39h1 and its Schizosaccharomyces pombe homolog Clr4 each contains an intrinsic histone methyltransferase (HMT) activity (Rea et al. 2000) that specifically methylates histone H3 at K9 (H3-K9) and correlates with repression of transcription. Mutagenesis studies with Suv39h1 revealed that the SET domain and two adjacent cysteine-rich regions (the pre-SET and post-SET domains) are required for enzymatic activity. Subsequent studies resulted in the isolation of another enzyme, G9a, which displays substrate specificity similar to that of Suv39h1, but also methylates H3-K27 (Tachibana et al. 2001). In the case of G9a, the enzyme appears to recognize the consensus sequence N-TKXXARKS-C within the histone H3 tail (for review, see Zhang and Reinberg 2001).
In addition to the SET domain, the Su(var)3-9 protein also contains an evolutionarily conserved chromodomain, which is found in a group of chromatin-related proteins (Koonin et al. 1995). Studies with the human Suv39h1 and its mouse homologs showed that these polypeptides associate with the mammalian heterochromatic protein HP1, a homolog of the S. pombe protein Swi6 (Aagaard et al. 1999). The chromodomain of HP1/Swi6 recognizes H3-K9 after it has been methylated by Suv39h1/Clr4. The molecular events leading to the formation of heterochromatin at yeast centromeres and mating-type loci were described recently by Nakayama et al. (2001). After histone deacetylases remove the acetyl groups from K9 and K14 of histone H3, Suv39h1/Clr4 methylates H3-K9. Methylated H3-K9 then serves as a binding site for the recruitment of HP1/Swi6. HP1/Swi6, through the shadow chromodomain located at its C terminus, can then oligomerize to form heterochromatin (Brasher et al. 2000). Similar results were observed with the vertebrate-derived enzymes (Bannister et al. 2001; Jacobs et al. 2001; Lachner et al. 2001).
However, not all lysine-directed histone methylation appears to function as a signal for repression of transcription. Methylation at H3-K4 was found to be a preferred site of methylation in the transcriptionally active macronuclei of Tetrahymena, and importantly, methylation at H3-K4 correlates with histone acetylation, which itself correlates with the opening of the chromatin structure (Strahl et al. 1999). Chromatin immunoprecipitation (ChIP) experiments performed with the fission yeast mating-type locus and antibodies specific to either H3-K9 or H3-K4 have revealed that methylated H3-K9 is strictly localized to a 20-kb silent heterochromatic region, whereas methylated H3-K4 is found exclusively in surrounding euchromatic regions (Noma et al. 2001). The mechanism(s) by which methylation at H3-K4 results in transcription activation is presently unknown. Two possible models are that methylation at residues such as H3-K4 marks a gene for the recruitment of complexes involved in transcription activation and/or displaces complexes involved in transcription repression, such as histone deacetylases.
In the present study we describe the identification and characterization of a novel, mammalian HMT specific for K4 of histone H3. We found that this enzyme, termed Set9, potentiates the activation function of a transcriptional activator protein. We also show that the histone deacetylase NuRD complex specifically binds to the histone H3 tail and that methylation by Set9 at K4, but not at K9, displaces the NuRD complex.
Results
Analysis of histone methyltransferases (HMTs) in HeLa cells
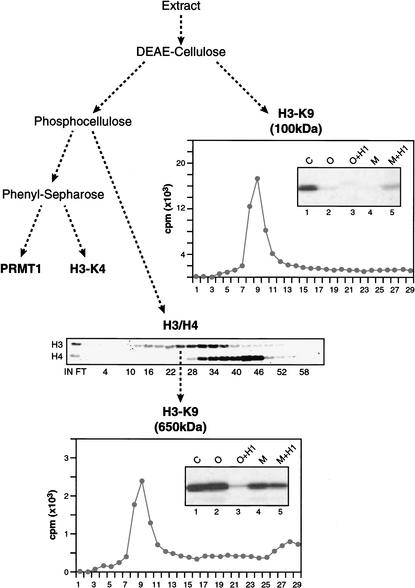
To identify and analyze the different HMTs present in human cells, HeLa cell-derived extracts were fractionated on several distinct chromatographic resins. Fractions derived from the column were then assayed for HMT activity using as substrates core-histone polypeptides and mono- and oligonucleosomes, both in the presence and absence of histone H1 (Fig. 1). Fractionation of the extract on a positively charged resin (DEAE-cellulose) results in the retention of one major activity with specificity for histone H3, and this activity was reduced significantly when nucleosomes were used as substrates. Further fractionation of this activity on a gel filtration column revealed it to have an apparent native mass of ∼100 kD (data not shown). Histone H3 was incubated with 3H-labeled S-adenosyl methionine (SAM) and the 100-kD HMT, and the labeled H3 was subjected to Edman degradation. This experiment revealed that this DEAE-bound HMT methylated H3-K9 specifically (Fig. 1). We have identified this activity as G9a (Tachibana et al. 2001).
Figure 1.
Isolation of different HMTs. Schematic representation of the HMT activities present in HeLa-derived extracts. The DEAE-cellulose column binds a 100-kD core histone H3-K9-specific HMT (G9a-like activity), and the phosphocellulose column binds two HMTs, reactive on oligonucleosomes: a 650-kD H3-K9 activity (Suv39h1-like activity) and an H4-K20 activity. (Middle panel) The activity profile of the latter two HMTs. The graphs show the results of radiolabeled histone H3 polypeptide sequencing, where the X-axis indicates the position of an amino acid residue relative to the N terminus. These experiments were performed with protein derived from the gel filtration step. Additionally, the specificity of the HMTs was analyzed by Western blots using antibodies specific for methylated H3-K4 or H3-K9. Insets in these graphs are HMT assays showing substrate specificity of the indicated HMTs. (C) Core histones; (O) oligonucleosomes; (M) mononucleosome; (H1) histone H1. The Set9 activity is found in the phenyl-Sepharose-bound fraction.
The separation of proteins in the DEAE-cellulose flow-through (unbound) fraction on a negatively charged column (phosphocellulose) resulted in the resolution of two other HMT activities, each with a different histone specificity. The H3-specific activity eluted earlier from the column and used core histones and oligonucleosomes as substrates. Further fractionation on a gel filtration column showed that the activity eluted with an apparent mass of ∼650 kD (data not shown). Edman degradation of histone H3 labeled with this HMT revealed specificity for K9. Further biochemical analyses showed this HMT to be Suv39h1 (data not shown). Thus, the analyses described above allowed us to separate two enzymes that target H3-K9. It is important to stress, however, that these enzymes are unique and show distinct substrate specificity. For example, the G9a activity is drastically reduced when H3 is incorporated into nucleosomes, whereas the Suv39h1 activity is active on free histones as well as on nucleosomes, and is impaired by the presence of histone H1 when oligonucleosomes, but not mononucleosomes, are used as substrates. Although the G9a activity is drastically impaired when nucleosomes are used as the substrate, histone H1 appears to stimulate this activity on mononucleosomes. The other major activity present in the phosphocellulose column fractions recognized exclusively nucleosomal histone H4 with specificity for K20 (data not shown; we have termed this activity Set7; Nishioka et al., in prep.).
Further separation of the proteins in the unbound fraction of the phosphocellulose fraction on a hydrophobic column (phenyl-Sepharose) allowed us to resolve two additional HMTs. One of the activities was specific for histone H4 and was identified as the arginine-specific HMT PRMT1 (data not shown). The other activity was specific for histone H3 and targeted exclusively K4 (see below). We further characterized this activity, which we termed Set9.
Isolation of Set9, an H3-K4-specific HMT
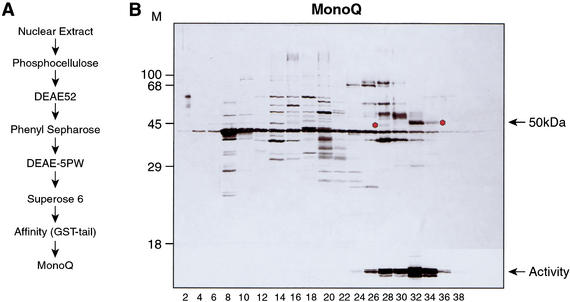
Initial characterization of Set9 indicated that nucleosomes are not optimal substrates for this activity. Therefore, we used GST-histone H3 tail carrying mutations at K9R and K27R to monitor HMT activity during purification. Set9 activity was isolated from HeLa cells as illustrated in Figure 2A and described in Materials and Methods. The last step of the purification, fractionation on a MonoQ column, allowed us to correlate the HMT activity with a single polypeptide (Fig. 2B). Two closely spaced peaks of H3 HMT activity were detected, and microsequencing of the putative polypeptides that coeluted with these activities showed that a species of ∼50 kD cofractionated with both HMT peaks (Fig. 2B, see asterisks). The 50-kD polypeptide migrates differently in each peak of activity, suggesting that the slower-migrating polypeptide is modified posttranslationally or that the faster-migrating polypeptide is a degraded form. This remains to be elucidated.
Figure 2.
Purification of Set9. (A) Schematic representation of purification steps of Set9. (B) Silver staining of MonoQ fractions separated by 10% SDS-PAGE. (Bottom panel) The corresponding HMT activity for each fraction using the GST–histone-H3 tail as substrate. Asterisks indicate Set9 polypeptides that were identified by protein microsequencing.
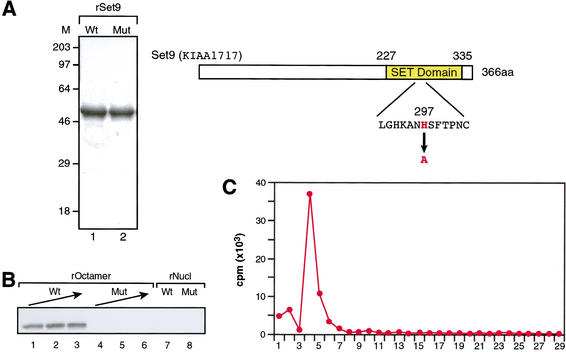
From the microsequencing information, a full-length cDNA clone encoding Set9 was isolated (see below). cDNA sequence analysis (Fig. 3) revealed that Set9 was encoded by a novel gene that is absent from lower eukaryotes, and is present only in vertebrates. Amino acid sequence analysis showed that Set9 was related to other lysine-directed HMTs through the presence of a SET domain (Fig. 3B). However, Set9 was devoid of the pre- and post-SET domains, which are present in most other characterized lysine-directed HMTs and are thought to be important for enzymatic activity (Rea et al. 2000). Interestingly, Set1 in yeast has recently been found to carry out H3-K4 methylation (Briggs et al. 2001), and this protein, although containing the post-SET domain, is lacking the pre-SET domain (see Discussion).
Figure 3.
Set9 protein contains a SET domain. (A) Amino acid sequence of Set9 protein. Underlined sequences were identified by mass spectrometry. SET-domain residues are in blue, and histidine 297 is highlighted in red. (B) Sequence alignment of SET domains among known lysine-HMTs. Sequences are aligned using CLUSTALW. Conserved amino acid residues are shown in blue, and histidine 297 is highlighted in red.
We synthesized in bacteria recombinant, full-length, wild-type Set9, as well as a mutant version of the protein (Set9-H297A) that contained a single substitution to alanine at amino acid 297, a position that corresponds to a key conserved histidine residue within the SET domains of SET family proteins (Fig. 4A). Recombinant proteins were then isolated and assayed for HMT activity using octamers and nucleosomes as substrates. The wild-type protein was found to be active, whereas Set9-H297A was inactive as expected (Fig. 4B). The recombinant wild-type protein was used to analyze the substrate specificity of Set9. Edman degradation of histone H3 that was labeled with 3H-SAM and recombinant Set9 showed that the enzyme was highly specific for K4 (Fig. 4C). Identical results were observed with native protein isolated from HeLa cells (data not shown; see Fig. 1). As expected, recombinant Set9 (Fig. 4B), like the native protein (data not shown), was found to be severely impaired when nucleosomes were used as substrates. Therefore, we concluded that Set9 is a lysine-specific HMT with specificity for H3-K4.
Figure 4.
Specificity of Set9. (A) Bacterially expressed recombinant Set9 protein (rSet9) was resolved on 10% SDS-PAGE and visualized by CBB staining (left panel). (Lane 1) Wild-type rSet9; (lane 2) the H297A mutant. The neighboring panel shows the schematic structure of Set9 protein with the highly conserved region in the SET domain indicated. The substitution in the mutant protein is indicated. (B) HMT assay showing Set9 substrate specificity. Either wild-type (Wt) or mutant (Mut) recombinant Set9 protein was titrated using recombinant octamer as substrate. The amounts of recombinant Set9 in a reaction with recombinant oligonucleosomes are the same as the maximum amount of enzyme used with the recombinant octamer. (C) The result of radiolabeled histone H3 polypeptide sequencing after treatment with Set9 and 3H-SAM. The X-axis indicates the position of amino acid residues relative to the N terminus.
Functional analysis of Set9
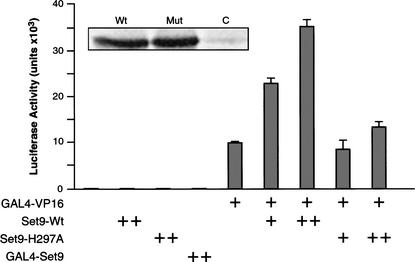
To gain insight into the function of Set9, we analyzed its effect on the expression of reporter constructs that were transfected into HeLa and 293 cells. In both cell types, when expression of the reporter gene (luciferase) was driven by the thymidine kinase (TK) promoter, cotransfection of a Set9 expression vector had no effect on luciferase expression (Fig. 5; data not shown). Similarly, cotransfection of a vector expressing the Gal4–Set9 fusion protein had no effect on expression of a Gal4–TK–luciferase reporter (Fig. 5; data not shown). As expected, the expression of Gal4–VP16 resulted in the stimulation of luciferase expression (Fig. 5). Interestingly, coexpression of Set9 with Gal4–VP16 resulted in the stimulation of luciferase expression from the Gal4–TK–luciferase reporter (Fig. 5). The extent of stimulation was directly related to the level of expression of Set9 (Fig. 5; data not shown). Importantly, cotransfection of a Set9 expression vector with a mutation in the SET domain that abolished Set9-HMT activity (Fig. 4B) impaired the ability of Set9 to stimulate Gal4–VP16-activated luciferase expression (Fig. 5). This was not the result of poor expression of the mutant polypeptide as both wild-type and mutant proteins were expressed to similar levels (Fig. 5). Taken together, these results suggest that methylation of H3-K4 by Set9 might function in the stimulation of activated transcription. This observation is in agreement with studies suggesting that methylation on H3-K4 correlates with transcriptionally active regions (Strahl et al. 1999; Litt et al. 2001; Noma et al. 2001).
Figure 5.
Set9 expression potentiates GAL4–VP16 activation in transfected cells. The effector plasmids indicated at the bottom of the graph were cotransfected with 100 ng of reporter plasmid carrying the GAL4–TK–luciferase gene. The amount of DNA used in each experiment is indicated by either a single plus (125 ng) or a double plus (250 ng). The data shown represent the average of two independent experiments. The inset on the graph denotes the expression of transfected wild-type and mutant Set9 protein as detected in Western blot using antibodies against the FLAG tag.
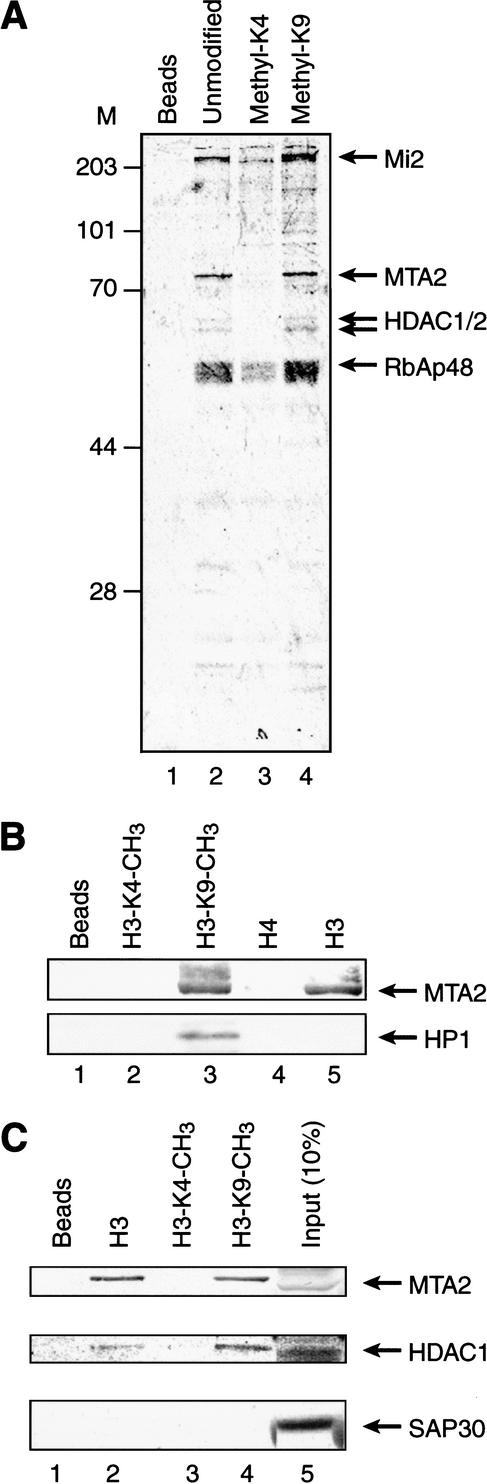
To gain further insights into the possible mechanism(s) by which Set9 enhanced the activation potential of a transcriptional activator, we sought to identify proteins that interact with the histone H3 tail methylated at K4. Toward this end, nuclear extracts were incubated with either an unmodified histone H3-tail peptide or similar peptides methylated at either K4 or K9, and the associated proteins were analyzed on SDS-PAGE and with CBB staining. The unmodified H3 tail bound polypeptides that were similar to those bound by the histone tail peptide methylated at K9 (Fig. 6A, cf. lanes 2 and 4). Importantly, binding of these polypeptides was greatly reduced when H3-K4 peptide was used under identical conditions.
Figure 6.
The histone H3 tail interacts with NuRD in a K4 methylation-dependent manner. (A) The result of pull-down experiments using the indicated synthetic histone H3 peptides is shown. The bound proteins were separated on 10% SDS-PAGE and visualized by CBB staining. Arrows indicate subunits of the NuRD complex identified by mass spectrometry. (B,C) Western blot analysis of pull-down experiment with the indicated synthetic histone N-terminal peptides. The antibodies used as probes are indicated on the right.
To identify the major H3-tail-binding polypeptides binding to the unmodified and the H3-K9 peptides, protein bands were excised from the gel and analyzed by mass spectrometry. This analysis revealed that the higher-molecular-weight polypeptide corresponded to Mi2, a subunit of the nucleosome remodeling and histone deacetylase complex NuRD (Zhang et al. 1998). The 70-kD band was identified as MTA2, and the other polypeptides were identified as HDAC1, HDAC2, and RbAp48, all subunits of the NuRD complex (Fig. 6A). These results suggest that the histone H3 tail interacts with the NuRD complex and that this interaction, although not disturbed by methylation at K9, is significantly disrupted by methylation at K4. To further characterize the association of NuRD with the histone H3 tail, the pull-down experiments were analyzed by Western blot. We first analyzed whether the interaction of the NuRD complex was specific for the histone H3 tail and found that the histone H4 tail failed to pull down NuRD components (Fig. 6B). The above interactions therefore appear to be specific for the histone H3 tail.
Of the histone deacetylases that associate with the histone H3 tail, HDAC1 and HDAC2 are present in two distinct protein complexes in HeLa cells referred to as the Sin3 and NuRD complexes (Zhang et al. 1999). The analysis described above (Fig. 6A) did not reveal the presence of the Sin3 polypeptide, suggesting that the histone H3 tail interacts specifically with the NuRD complex. To further investigate this issue, we analyzed the pull-down experiments for the presence of SAP30, a subunit that is specific to the Sin3-HDAC1/HDAC2-containing complexes. This analysis failed to detect SAP30 (Fig. 6C), and therefore we concluded that the histone H3 tail interacts specifically with the NuRD complex. Because methylation at K4, but not methylation at K9, inhibited the association of NuRD, we concluded that the interaction is regulated by methylation at K4.
Methylation of histone H3 at K9 marks the histone for the recruitment of HP1 (Bannister et al. 2001; Lachner et al. 2001). We therefore used antibody against HP1 to determine whether the K9-methylated histone H3-tail peptide interacts with HP1 as well as with the NuRD complex. This analysis showed that the histone H3-tail peptide methylated at K9 pulled down both HP1 and the NuRD subunits together (Fig. 6B).
Methylation of H3-K4 affects methylation at H3-K9
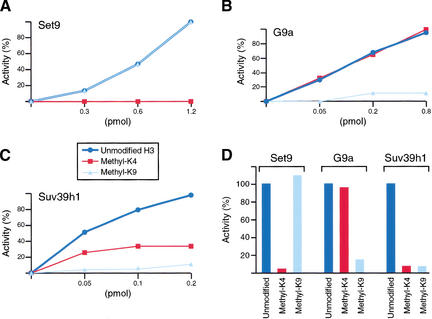
We next analyzed whether methylation at one residue within the histone H3 tail affected methylation at other sites. Peptides that contained the first 20 amino acids of the histone H3 tail were synthesized. Three different peptides were produced; one peptide was methylated at K4, the second peptide was methylated at K9, and the third peptide was unmethylated. These peptides were used as substrates in methylation assays mediated by Set9 (Fig. 7A), G9a (Fig. 7B), or Suv39h1 (Fig. 7C). In agreement with the specificity of each of the enzymes used, a peptide methylated at K4 inhibited the activity of Set9, and similarly, a peptide methylated at K9 inhibited the activities of G9a and Suv39h1 (Fig. 7). Methylation at K9 did not affect the activity of Set9; similarly, methylation at K4 was without effect on the activity of G9a. A different situation was observed when Suv39h1 was analyzed. Suv39h1 was active in methylating an unmodified peptide; however, methylation at K4 drastically inhibited the ability of Suv39h1 to methylate K9. Similar results were obtained using each native enzyme (Fig. 7D). The implications of this finding are discussed below.
Figure 7.
Interplay of histone H3 methylation. Recombinant HMTs (A) Set9 and (B) G9a, and (D) purified from human cells, as well as theSuv39h1 complex isolated from (C) baculovirus-infected SF9 cells or (D) human cells, were incubated as described in Materials and Methods. Reactions were incubated at 30°C for 10 min. The percentage of enzyme activity detected with each peptide is plotted, setting the reaction with unmodified peptide to 100% (D) or taking the highest value of the experiment as 100% activity (A–C). The amount of Suv39h1 added to the reactions was calculated assuming that the recombinant complex has a mass of ∼500 kD.
Discussion
In this study, we analyzed lysine-HMT activities in HeLa cells and isolated a novel enzyme, Set9, with specificity for lysine 4 of histone H3. Set9 activity is contained within a single polypeptide of ∼50 kD. In agreement with previous studies showing that the SET domain can be a signature for lysine-HMTs, Set9 contains a SET domain, and, more importantly, a single substitution of a conserved histidine to alanine within the SET domain impaired its enzymatic activity. Interestingly, Set9 is devoid of the pre- and post-SET domains, showing that these domains, although important for the functions of other HMTs, are not absolutely required for HMT enzymatic activity. Set9 was found to be inactive when nucleosomes were used as substrate. This does not appear to be caused by the lack of the pre- and post-SET domains, as we have isolated an HMT with strict specificity for lysine 20 of nucleosomal histone H4, and this enzyme is devoid of the pre- and post-SET domains (Nishioka et al., in prep.). We hypothesize that Set9 is tethered to nucleosomes by another, as yet unidentified, factor and that this factor is necessary for Set9 to recognize nucleosomes as substrates.
Although it has been known for many years that histones are methylated at specific residues in vivo (for review, see van Holde 1988), the enzymes that catalyze these modifications and the function of these modifications have only recently begun to be revealed (for review, see Zhang and Reinberg 2001). The theme that is beginning to emerge is that a particular modification appears to have a different consequence depending on its chromosomal location, combination of modifications, and the enzyme (or protein complex) involved in the particular modification. For example, methylation on H3-K9 by Suv39h1 in human or Clr4 in S. pombe establishes a signal for the recruitment of HP1 and the establishment of heterochromatin (Brasher et al. 2000; Bannister et al. 2001; Lachner et al. 2001).
As seen in this study, a similar mode of regulation appears to operate with respect to methylation of H3-K4, wherein the human HMT Set9 modifies H3-K4 and functions in transcription activation. Our findings are in agreement with several previous studies. In the transcriptionally active macronuclei of Tetrahymena, H3-K4 was found to be a preferred site of methylation, and methylation at H3-K4 correlates with acetylation (Strahl et al. 1999). Moreover, ChIP experiments using the fission yeast mating-type locus (Noma et al. 2001) or the β-globin locus during erythropoiesis (Litt et al. 2001) and antibodies specific to either H3-K9 or H3-K4 revealed that H3-K9 methylation is strictly localized to silent heterochromatic regions. In contrast, H3-K4 methylation was found to be specific to the surrounding euchromatic regions. Finally, immunofluorescence studies on human female metaphase chromosomes revealed that H3-K4 is preferentially methylated within transcriptionally active regions of autosomal chromosomes and methylated H3-K4 is largely absent from the inactive X chromosome, which is rich in H3-K9 methylation (Boggs et al. 2002).
However, recent work in the yeast Saccharomyces cerevisiae has shown that methylation of H3-K4 can have a very different consequence from transcriptional activation. Interestingly, Set1 appears to be a major, if not exclusive, HMT responsible for methylation of histone H3-K4 in yeast. SET-1 is required for silencing at telomeres and at the silent mating type (HML) locus (Laible et al. 1997; Nislow et al. 1997). In addition, SET-1 regulates rDNA silencing and, most importantly, this function requires the methylation of H3-K4 (Briggs et al. 2001; M. Bryk and F. Winston, pers. comm.). It remains unknown whether Set1 itself methylates H3-K4, as neither recombinant Set1 (Briggs et al. 2001) nor a Set1-containing complex isolated from yeast (Miller et al. 2001) was able to methylate this residue in vitro.
The mechanism by which methylation at H3-K4 results in modulation of transcription is currently unknown. However, in the present study we shed light on this important problem. Two independent mechanisms were found to operate in response to methylation of H3-K4. First, we observed that this modification precludes methylation by Suv39h1 at H3-K9. This finding is consistent with the ChIP studies described above, which were carried out in fission yeast (Noma et al. 2001) and higher eukaryotes and showed that the two modifications are mutually exclusive. Interestingly, we also observed that methylation at H3-K9 by G9a is not affected by methylation at H3-K4. This observation is important, as it implies that, although Suv39h1 and G9a methylate the same residue in the histone H3 tail, the recognition of this residue by these two enzymes can be independently regulated by other modifications within the same tail (and perhaps regulated by modifications present in the tails of other histone polypeptides within the same nucleosome). This finding is in agreement with the histone code hypothesis, which predicts that the pattern of histone tail modifications serves as a recognition code for the binding of proteins that modulate chromatin structure and function (for reviews, see Strahl and Allis 2000; Turner 2000). The second mechanism by which methylation at H3-K4 results in transcription modulation involves the displacement of the histone deacetylase NuRD complex from the histone H3 tail. We found that an unmodified histone H3 tail and an H3 tail methylated at K9 both represent binding sites for the NuRD complex. Methylation at H3-K4 interfered with the interaction between NuRD and the histone H3 tail. Similar findings were observed when the full-length histone H3 polypeptide was incubated with Set9; however, because methylation at H3-K4 by Set9 was not quantitative, we did not observe complete displacement of NuRD (data not shown).
On first consideration, the mechanism by which NuRD is displaced from the histone H3 tail might appear to be unrelated to the mechanism by which methylation of H3-K4 precludes methylation by Suv39h1 at H3-K9. However, we suggest that the two mechanisms are related and that the discrete events influence one another. Studies in fission yeast have shown that methylation at H3-K9 by Clr4, a homolog of Suv39h1, requires the deacetylation of two histone H3-lysine residues. Mutations in Clr3, a gene that encodes an H3-specific histone deacetylase, impaired methylation at H3-K9 (Nakayama et al. 2001). Because mutations in Clr3 are known to increase specifically the level of histone H3-K14 acetylation, we can infer that deacetylation of K14 is a prerequisite for H3-K9 methylation. Because the acetylation of H3-K9 prevents this site from being methylated (Rea et al. 2000), deacetylation of H3-K9 is also a prerequisite for methylation. The fact that methylation of histone H3-K4 results in the displacement of the histone deacetylase NuRD complex from the histone H3 tail and also inhibits methylation of H3-K9 by Suv39h1 strongly suggests that these two processes are coupled. In support of this hypothesis are our previous studies with NuRD showing that nucleosomal histone H3 appears to be a preferred substrate of NuRD (Zhang et al. 1998).
As stated above, it appears that methylation of a specific residue in the histone H3 tail can have a different effect on transcription depending on which enzyme carries out the modification. As shown in Figure 5, methylation of H3-K4 by Set9 correlates with transcriptional activation in human cells. Although the nature of the chromatin structure associated with transiently expressed plasmids is not well-characterized, this assay has been used successfully to study the effects of modifications of histone tails in human cells. The cumulative results presented in this study support the idea that methylation of H3-K4 by Set9 functions in transcription activation by precluding the association of NuRD with the histone H3 tail, as well as by inhibiting the methylation of H3-K9 by Suv39h1. These findings not only support the histone code hypothesis, but also begin to elucidate the mechanisms by which H3-K4 methylation translates into the modification of transcriptional activity.
Materials and methods
Cloning and plasmid construction for Set9
The coding sequence of Set9 (GenBank accession no. AF462150) was PCR-amplified with combination of either a 5′ NdeI site-introduced primer and a 3′ FLAG-tag and HindIII site-introduced primer, or a 5′ BamHI site-introduced primer and a 3′ XbaI site-introduced primer using Marathon cDNA library derived from HeLa cells (Clontech) and cloned into a pCR2.1 vector (Invitrogen). The FLAG-tagged Set9 cDNA was inserted into NdeI–HindIII sites of the bacterial expression vector pET-28b (Novagen). The recombinant proteins were expressed in the Escherichia coli strain BL21Star(DE3) (Invitrogen) at 37°C for 3 h and purified on Ni-NTA agarose according to the manufacturer's instructions (QIAGEN). For construction of mammalian expression vectors, the XbaI–HindIII fragment of the pET-28b vector was transferred to the mammalian expression vector pcDNA3.1 (Invitrogen), whereas the non-tagged fragment of the latter pCR2.1 vector was transferred into BamHI–XbaI sites of a pSG424 vector encoding the DNA-binding domain of GAL4 (Sadowski and Ptashne 1989). Site-directed mutagenesis of each plasmid was carried out using a Quick Change Site-Directed Mutagenesis Kit according to the manufacturer's instructions (Stratagene).
Isolation of different HMTs from HeLa-derived extracts
HeLa-derived extracts were prepared as previously described (LeRoy et al. 1998). All procedures were performed at 4°C. Approximately 10 g of proteins were loaded onto a 2000-mL DEAE-52 (Whatman) column equilibrated with buffer D (50 mM Tris-HCl at pH 7.9, 0.2 mM EDTA, 10% glycerol, 1 mM DTT, 0.2 mM PMSF) containing 0.1 M ammonium sulfate. The column was eluted with a 5-column volume linear gradient of ammonium sulfate from 0.1 M to 0.65 M in buffer D. G9a-like HMT activity eluted between 0.2 M and 0.25 M ammonium sulfate with the bulk of the proteins. The flow-through of the DEAE-52 column was loaded onto a 500-mL phosphocellulose column (Sigma). The column was eluted with a 10-column volume linear gradient of ammonium sulfate from 0.1 M to 0.6 M in buffer D. Suv39h1-like activity eluted between 0.1 M and 0.2 M ammonium sulfate with the bulk of the proteins, whereas H4-K20-specific activity eluted between 0.15 M and 0.25 M ammonium sulfate. Set9 activity, which is H3-K4-specific, was found in the flow-through. The sample was adjusted to the equivalent of 1 M ammonium sulfate and was loaded onto a 200-mL phenyl-Sepharose column (Amersham Pharmacia Biotech). The column was eluted with a 10-column volume linear gradient of ammonium sulfate from 1 M to 0 M in buffer D. The activity eluted between 0.1 M and 0.2 M ammonium sulfate with the bulk of the proteins. Active fractions for each of the activities described above were pooled, concentrated by 50% ammonium sulfate precipitation, and individually loaded onto a 480-mL Superose 6 column (Amersham Pharmacia Biotech) equilibrated in buffer D containing 0.5 M ammonium sulfate. The G9a-like activity eluted at ∼100 kD, and the Suv39h1-like activity eluted between 600 and 700 kD. Set9 activity eluted at ∼50 kD.
Purification of human Set9
Because the majority of Set9 activity was found in nuclear extracts, they were used for further purification. The nuclear extract was prepared as previously described (Dignam et al. 1983). Approximately 15 g of proteins was loaded onto a 1500-mL phosphocellulose column (Sigma) equilibrated with buffer C (20 mM Tris-HCl at pH 7.9, 1 mM EDTA, 10% glycerol, 1 mM DTT, 0.2 mM PMSF) containing 0.1 M KCl. The activity was detected in the flow-through, and was loaded onto a 1000-mL DEAE-52 column (Whatman). After extensive washing with buffer C containing 0.1 M KCl, the column was eluted with buffer C containing 0.35 M KCl. The eluate was adjusted to 1 M ammonium sulfate and was loaded onto a 200-mL phenyl-Sepharose column (Amersham Pharmacia Biotech). The column was washed with buffer D containing 0.5 M ammonium sulfate, then eluted with the buffer D alone. The eluate was dialyzed against buffer C containing 0.1 M KCl and loaded onto a 54.4-mL DEAE-5PW column (TosoHaas). The column was eluted with a 10-column volume linear gradient from 0.1 M to 0.5 M KCl in buffer C. The Set9 activity elutes between 0.15 M and 0.25 M KCl. The active fractions were pooled, concentrated by 40% ammonium sulfate precipitation, and loaded onto a 480-mL Superose 6 column (Amersham Pharmacia Biotech) equilibrated in buffer C containing 1 M KCl. The activity eluted at ∼50 kD. The active fractions were pooled and dialyzed against buffer C containing 0.1 M KCl and incubated overnight with 1 mL of glutathione-Sepharose beads carrying GST–histone-H3-tail fusion proteins. Then the beads were washed with binding buffer and the activity was eluted by buffer C containing 0.3 M KCl. The active fractions were pooled and dialyzed against buffer C containing 0.1 M KCl and loaded onto a 1-mL MonoQ column (Amersham Pharmacia Biotech). The column was eluted with a 10-column volume linear gradient of KCl from 0.1 M to 1 M in buffer C. The activity eluted between 0.35 M and 0.45 M KCl.
Protein identification
Gel-bound polypeptides were subjected to in-gel tryptic digestion and mass spectrometric analysis as previously described (Erdjument-Bromage et al. 1998). Identification of spectra corresponding to known peptide sequences in the NCBI nonredundant database was facilitated using the PeptideSearch algorithm (M. Mann, University of Southern Denmark). The data obtained from Set9 polypeptides revealed that the peptide masses matched a protein called KIAA1717 in the HUGE database (http://www.kazusa.or.jp/huge).
Histone methyltransferase assay and preparation of its substrates
The samples were incubated at 30°C for 10 to 60 min in a reaction buffer containing 50 mM Tris-HCl (pH 8.5), 5 mM MgCl2, 4 mM DTT, and 1 μM 3H-labeled SAM (Amersham Pharmacia Biotech). Two micrograms of octamer, oligonucleosomes, mononucleosome, or GST–histone-H3-tail fusion protein were used as substrates. The total volume of a reaction mixture was adjusted to 25 μL. The reaction was stopped by addition of SDS sample buffer and then fractionated on 15% SDS-PAGE. Separated histones or GST fusion protein were then transferred onto an Immobilon-P membrane (Millipore) and visualized by CBB staining. The membrane was sprayed with EN3HANCE (NEN), and exposed to Kodak XAR film overnight.
Native octamer, oligonucleosomes, and mononucleosomes were purified from HeLa cells as previously described (Orphanides et al. 1998). Recombinant Xenopus octamer and oligonucleosomes were prepared as described (Luger et al. 1999), where 5S ribosomal DNA repeats were used for assembly of oligonucleosomes. Purified calf thymus histone H1 was purchased from Roche Molecular Biochemicals. GST–histone-H3-tail fusion proteins (Tachibana et al. 2001) were overexpressed in the E. coli strain BL21(DE3)pLysS at 37°C for 3 h and purified on glutathione-Sepharose 4B according to the manufacturer's instructions (Amersham Pharmacia Biotech).
Radiolabeled histone sequencing
Recombinant Xenopus histone H3 was labeled with 3H-SAM using different enzymes and fractionated on 15% SDS-PAGE. The polypeptide was then transferred onto an Immobilon-P membrane and subjected to Edman degradation amino acid analysis coupled with scintillation counting.
Peptide pull-down using synthetic histone tails
Histone H3 N-terminal peptides (1–20) were synthesized and followed by GG-linker and biotinylated lysine. Modified peptides were synthesized using dimethylated lysine as indicated in some figures. Biotinylated histone H4 N-terminal peptide (1–21) was purchased from Upstate Biotechnology. Two hundred microliters of nuclear extract was incubated with 10 nmoles of each peptide at 4°C for 4 h in the presence of 20 μL of streptavidin-conjugated Sepharose (Amersham Pharmacia Biotech). After extensive washing with buffer C containing 0.5 M KCl and 1% Triton X-100, the bound proteins were fractionated on 10% SDS-PAGE and subjected to either CBB staining or Western blot.
Transfection and reporter gene assay
The reporter plasmid carrying the 5× GAL4-binding site, TK promoter, and firefly luciferase gene was previously described (Zhang et al. 1997). The plasmid encoding GAL4–VP16 was previously described (Ayer et al. 1996). Transfection to 293 cells was performed using Effectene Tranfection Reagent (QIAGEN) according to the manufacturer's instructions. The cells were collected 48 h after transfection and subjected to luciferase and β-gal assays using a Promega kit, Luciferase Assay Systems.
Interplay of methyltransferases
Methyltransferase assays were performed as described above. Reactions were incubated for 10 min, using 2.5 nmoles of each synthetic peptide as substrates. Reaction mixtures were then incubated with 20 μL of streptavidin-conjugated Sepharose (Amersham Pharmacia Biotech) in the presence of excess SAM at 4°C for 1 h. After extensive washing with buffer C containing 0.5 M KCl and 1% Triton X-100, the bound materials were subjected to scintillation counting. These assays were performed using recombinant polypeptides as well as with native proteins purified as described in Figure 1. Suv39h1 and G9a proteins were derived from the gel filtration step. Recombinant Suv39h1 and G9a were purified from SF9 cells infected with recombinant baculovirus carrying Suv39h1 or G9a cDNA. G9a was purified to a single polypeptide, whereas Suv39h1 behaved as a protein complex.
Acknowledgments
We thank Tony Kouzarides and Yi Zhang for communicating results prior to publication. We also thank Yoshihiro Nakatani for providing recombinant Suv39h1 and G9a proteins, Yoichi Shinkai for providing expression vectors for GST-H3 tails, members of the Reinberg laboratory for helpful discussions, and Sherrie Hu for technical assistance. We thank Michael Hampsey and Brian Lewis for comments on the manuscript. This work was supported by a grant from NIH (GM-37120) and the Howard Hughes Medical Institute to D.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note
While this manuscript was under review, a report describing an identical activity was published online by Molecular Cell (2001. 8: 1207–1217). The authors named the H3-K4-HMT activity Set7 based on the fact that yeast has six SET-domain-containing genes and the H3-K4-HMT is absent in yeast. However, a novel sequence encoding a SET-domain-containing protein, PR/SET7, was deposited in GenBank (accession no. AF287261) in July 2000. Therefore, to avoid confusion, we suggest that the H3-K4 activity described here and in the Molecular Cell paper should not be named Set7. We have therefore named this activity Set9.
Footnotes
E-MAIL reinbedf@umdnj.edu; FAX (732) 235-5294.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.967202.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D, Vitolo JM, Mutskov V, Dimitrov S, Hayes JJ. Preferential interaction of the core histone tail domains with linker DNA. Proc Natl Acad Sci. 2001;98:6599–6604. doi: 10.1073/pnas.121171498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer DE, Laherty CD, Lawrence QA, Armstrong AP, Eisenman RN. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, Laue ED. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SYR, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes & Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan RS, McNulty DE, Carr SA, Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- Grunstein M, Hecht A, Fisher-Adams G, Wan J, Mann RK, Strahl-Bolsinger S, Laroche T, Gasser S. The regulation of euchromatin and heterochromatin by histones in yeast. J Cell Sci Suppl. 1995;19:29–36. doi: 10.1242/jcs.1995.supplement_19.4. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G. SET-domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Houtz RL. Cloning and developmental expression of pea ribulose-1,5-biphosphate carboxylase/oxygenase large subunit N-methyltransferase. Plant Mol Biol. 1995;27:249–261. doi: 10.1007/BF00020181. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Zhou S, Lucchesi JC. The chromo superfamily: New members, duplication of the chromodomain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4232. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K. The occurrence of ɛ-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Reuter G, Spierer P. Position-effect variegation and chromatin proteins. BioEssays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ptashne M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Ohba R, Cook RG, Allis CD. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc Natl Acad Sci. 1999;96:14967–14972. doi: 10.1073/pnas.96.26.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. SET domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- van Holde KE. Chromatin. New York: Springer Verlag; 1988. [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Simel E, Klein P, Royer M, Houtz RL. Expression, purification, and characterization of recombinant ribulose-1,5-biphosphate carboxylase/oxygenase large subunit N-methyltransferase. Protein Exp Purif. 1998;14:104–112. doi: 10.1006/prep.1998.0936. [DOI] [PubMed] [Google Scholar]