Abstract
C1, a lytic bacteriophage infecting group C streptococci, is one of the earliest-isolated phages, and the method of bacterial classification known as phage typing was defined by using this bacteriophage. We present for the first time a detailed analysis of this phage by use of electron microscopy, protein profiling, and complete nucleotide sequencing. This virus belongs to the Podoviridae family of phages, all of which are characterized by short, noncontractile tails. The C1 genome consists of a linear double-stranded DNA molecule of 16,687 nucleotides with 143-bp inverted terminal repeats. We have assigned functions to 9 of 20 putative open reading frames based on experimental substantiation or bioinformatic analysis. Their products include DNA polymerase, holin, lysin, major capsid, head-tail connector, neck appendage, and major tail proteins. Additionally, we found one intron belonging to the HNH endonuclease family interrupting the apparent lysin gene, suggesting a potential splicing event yielding a functional lytic enzyme. Examination of the C1 DNA polymerase suggests that this phage utilizes a protein-primed mechanism of replication, which is prominent in the φ29-like members of Podoviridae. Consistent with this evidence, we experimentally determined that terminal proteins are covalently attached to both 5′ termini, despite the fact that no homology to known terminal proteins could be elucidated in any of our open reading frames. Likewise, comparative genomics revealed no close evolutionary matches, suggesting that the C1 bacteriophage is a unique member of the Podoviridae.
Tailed bacteriophages are the most populous organism on Earth, with roughly 1030 inhabitants in the biosphere (9). However, we are just beginning to appreciate the role they play in bacterial diversity (24) and, as reported more recently, bacterial pathogenesis (8, 46). Indeed, whole-genome sequencing of two different strains of group A streptococci (Streptococcus pyogenes) revealed that polylysogenic phage represents the only diversity between the two strains (3, 19). Recent advances have allowed whole bacteriophage genomes of evolutionary or biological interest to be rapidly sequenced for comparison with known genomes.
The streptococcal C1 bacteriophage has roots at the forefront of bacteriophage research. In 1925, shortly after the discovery of bacteriophages by Twort and d'Herelle (12, 42), the C1 bacteriophage was isolated from a sewage plant in Milwaukee, Wis., by Clark and Clark and represents the first documented bacteriophage found to be active on any type of streptococci (10). Initially known as the “sludge” phage or “Clark” phage, it infected streptococci isolated from animals (which we now know to be group C streptococci) but not streptococci found in humans (now known to be group A streptococci) (26, 39). In a hallmark 1934 paper, Alice Evans, using the Clark phage which she renamed B563, was the first person to utilize a phage to classify bacterial strains, thus founding the analytical field of phage typing (15). Additionally, Evans noticed that phage lysates had lytic activity on streptococci that were not sensitive to the phage itself. She called this phenomenon “nascent lysis” and attributed it to a lysin activity originally defined by Twort (43). In 1957, Krause renamed the Evans B563 phage, calling it “C1” to imply an exquisite specificity for group C streptococci (25). Krause also noted that the C1 cell wall hydrolase, or lysin, had a less restrictive range given that group A, C, and E streptococci were rapidly lysed by this enzyme.
On the genetic level, the C1 phage has not been studied in detail. Two published restriction maps of this genome exist (34, 41), but to date, no sequence data are available. The majority of interest in this phage involves its lysin, which has been used extensively as a tool to dissolve the streptococcal cell wall in order to make protoplasts (48), extract genomic DNA, or study surface proteins (44). More recently, Nelson et al. have shown that the lytic properties of the C1 lysin have therapeutic potential to eliminate streptococcal colonization (31).
The C1 phage is currently classified as a member of the Podoviridae family of bacteriophage based on its physical characteristic of short, noncontractile tails (30). This designation makes the C1 phage a tempting choice for the study of sequence for several reasons. First, the Podoviridae represent a diverse set of phages for which only a few sequenced genomes exist and even fewer have been studied in detail. Additionally, both the historical interest of the C1 phage and the current medical implications of its lysin warrant further investigation.
MATERIALS AND METHODS
Unless otherwise stated, all reagents were obtained from Sigma and were of the highest purity available.
Preparation and purification of phage.
The lytic bacteriophage C1 and its host bacterium, group C streptococcus 26RP66, are both part of The Rockefeller University collection. For preparation of the C1 bacteriophage, 26RP66 was grown at 37°C in chemically defined medium for streptococci (JRH Biosciences) (27.13 g/liter) supplemented with 2.5 g of sodium bicarbonate/liter and 0.5 g of cysteine/liter. During early log phase (optical density at 650 nm, ∼0.25), 1/10 to 1/2 (vol/vol) of the prewarmed C1 phage was added and allowed to incubate until complete lysis occurred (approximately 40 min). The lysate was clarified by centrifugation (10,000 × g, 10 min) and passed through a 0.45-μm-pore-size filter (Amicon), and final phage purification was achieved by ultracentrifugation (100,000 × g, 2 h), with the phage pellet resuspended in phosphate-buffered saline and stored at 4°C.
Purification of phage DNA.
To purify phage particles, RNase and DNase (10 μg each) were added and allowed to incubate for 30 min at 37°C, after which 50 μl of 0.5 M EDTA was supplemented to inhibit the endonucleases. Protease K (200 μg) and sodium dodecyl sulfate (SDS; 0.5% final concentration) were added, and the mixture was incubated for 1 h at 65°C. Final DNA purification was achieved through common phenol-chloroform extraction protocols as previously described (37).
Terminal protein studies.
To purify the DNA-protein complex, the same procedure used to purify the phage DNA (see above) was followed, except that a phenol extraction step with gentle shaking was used instead of multiple phenol-chloroform extractions. The DNA-protein complex, found at the interface between the aqueous and phenol layers, was extracted and precipitated by ethanol. Half of the DNA-protein complex was digested with protease K (10 μg) for 30 min at 37°C and repurified by ethanol precipitation. Aliquots (10 μg each) of the DNA-protein complex or protease K-digested DNA (PK-DNA) were treated with either 2 μl (130 U) of exonuclease III or 2 μl (11 U) of lambda exonuclease (both from Gibco/BRL) at 37°C according to the manufacturer's instructions. Reactions were stopped with the addition of 10 mM EDTA at the indicated times prior to electrophoresis. Alternatively, PK-DNA (10 μg) was pretreated with 0.5 M piperidine for 2 h at 37°C and then subjected to lambda exonuclease treatment.
DNA sequencing.
For the library construction, genomic DNA was hydrodynamically sheared using high-pressure liquid chromatography and separated on a 1% agarose gel. Fragments of 3,000 to 3,500 bp were excised, purified by the GeneClean procedure (Bio 101, Inc.), blunt ended using T4 DNA polymerase, and ligated to unique BstXI linker adapters. The linker-adapted inserts were separated from the unincorporated linkers by a second gel purification using GeneClean and ligated to BstXI-cut pGTC vector to form subclone libraries, which were transformed into DH10β competent cells (DH5α transformation protocol [Gibco/BRL]). DNA was purified from positive transformants by using the PerfectPrep384 system (Brinkmann Instruments) and then sequenced using ABI dye terminator chemistry on automated MegaBace 1000 (Amersham) machines (Genome Therapeutics Corporation, Waltham, Mass.). Base calls and quality scores were determined using the PHRED program (17, 18). Reads were assembled by using PHRAP with default program parameters and quality scores. Closure of gaps was accomplished by using primer-directed sequencing directly from purified phage DNA.
Bioinformatics.
The Lasergene suite of programs from DNASTAR was used for analysis, annotation, and assembly of the nucleotide and amino acid sequences. Putative open reading frames (ORFs) were identified either by ORF Finder, available through the National Center for Biotechnology Information (NCBI; www.ncbi/nlm.nih.gov), or the heuristic approach of gene prediction (GeneMark) (4). The BLAST algorithms (2), also available through the NCBI, were used for similarity searches of putative ORFs. Sequence alignments were performed with CLUSTAL W and visualized with BOXSHADE.
Electron microscopy.
Purified phage was applied to a carbon film and fixed to a copper grid before being negatively stained with 1% uranyl acetate. Electron micrographs of the phage were taken using a JEOL 100 CXII transmission electron microscope with a final magnification as noted in the figure legends. For phage infection images, C1 bacteriophage was incubated with early log phase group C streptococcus 26RP66 for 20 min, pelleted by centrifugation, and suspended in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4). The samples were then postfixed in 1% osmium tetroxide, block stained with uranyl acetate, and processed according to standard procedures. All microscopy was performed by The Rockefeller University Bio-Imaging Resource Center or in collaboration with John Swanson.
Receptor studies.
For C1 infection studies, phage was added to an exponential growth of group C streptococci or group A-variant streptococci and monitored for either the clearing of a liquid culture as described above or plaque formation in a soft agar overlay. Alternatively, group C streptococci were pretreated with pronase (100 μg/ml) or trypsin and chymotrypsin (100 μg/ml each) for 30 min prior to exposure to the C1 phage. For adsorption studies, group C streptococcal cell walls were isolated as described previously (21) and the group C streptococcal carbohydrate was isolated by the nitrous acid extraction method as described previously (40). Briefly, 100 μl of C1 phage (at 108 PFU/ml) was mixed with isolated group C streptococcal cell walls (5 mg/ml), group C streptococcal carbohydrate (5 mg/ml), or 20 mM N-acetylgalactosamine (GalNAc) in a final volume of 0.5 ml. After 10 min of incubation at 37°C, log phase group C streptococci were added to a final volume of 1.0 ml, incubated for 5 min, and centrifuged to pellet the streptococci, and the cell pellet was plated according to the soft agar layer technique to enumerate any phage in the supernatant. Adsorption was quantified as a corresponding decrease in the number of residual PFU per milliliter compared to buffer control.
Nucleotide sequence accession number.
The DNA sequences of the genome reported here appear in GenBank under accession no. AY212251.
RESULTS
C1 bacteriophage characterization.
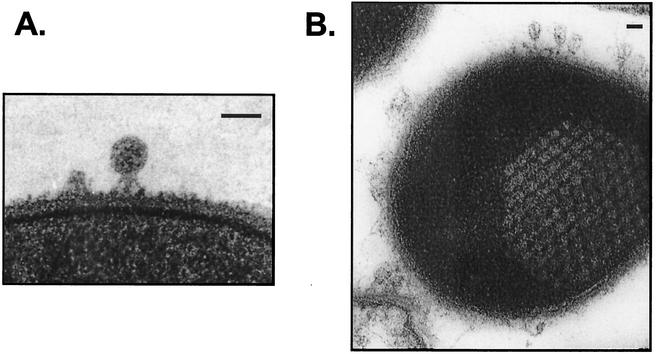
In agreement with the findings of a previously published report (30), the C1 phage was found to have a small polyhedral head (∼50 by 50 nm) and a very short tail and tail fibers (Fig. 1A and 2). A small collar or base plate with three protruding appendages was noted. The wider and longer central appendage is presumed to be the tail, but it is not clear whether the side appendages are minor tail fibers similar to those seen in phage T7 or P22 or are collar spikes as observed in the φ29 phage (1). Upon infection, complete lysis of susceptible group C streptococci was achieved by 40 min; however, mature phage particles could be observed by electron microscopy to be emerging from infected streptococci as soon as 10 min postinfection (data not shown). We were able to manually count >100 progeny particles in one thin-section micrograph of an infected streptococcus (Fig. 1B), which is consistent with the relatively high burst size noted previously (20).
FIG. 1.
Thin-section electron microscopy of the C1 bacteriophage. (A) C1 bacteriophage binding to the cell wall of a group C streptococcus. The electron-opaque capsid indicates that the phage DNA has not been injected. Bars, 50 nm. (B) Group C streptococcus at 15 min postinfection with C1 bacteriophage. The honeycomb structure shows the progeny virions during phage assembly.
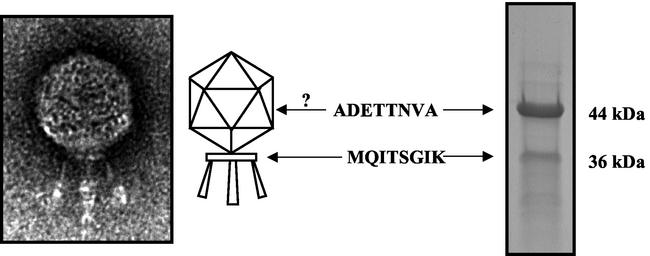
FIG. 2.
Characterization of C1 structural proteins. Shown at left is an electron micrograph of C1, showing its polyhedral head, base plate, and three tail fibers (magnification, ×100,000). Shown at right is the protein profile of the C1 phage obtained by SDS-PAGE. N-terminal sequencing of the two most prominent bands revealed that the 44- and 36-kDa bands correspond to ORF16 and -15, respectively. Whereas ORF15 is consistent with head-tail connector base plate proteins from several phage systems, ORF16 does not share homology with any known proteins. Because ORF16 is the most prevalent structural protein in the C1 proteome, we speculate that it may represent the major capsid protein.
Organization of the C1 genome and identification of ORFs.
A double-stranded, linear sequence of 16,687 bp was established with a mean redundancy of six, with each region being sequenced at least once on each strand. The G+C content of 34.6% is similar to that of the group C streptococcal genome as well as other low-GC streptococci. Also present were 143-bp inverted terminal repeats, which are characteristic of the φ29-like Podoviridae (1).
The criteria for the characterization of a potential ORF were the existence of a start codon (ATG, GTG, or TTG) and a minimum coding size of 50 amino acids. Based on this, 20 predicted ORFs were identified by both ORF Finder and GeneMark and were labeled 1 to 20 from the left end of transcription (Table 1). The first 11 ORFs are on the positive strand, and the remaining 9 ORFs are on the negative strand. Unexpectedly, the majority of the ORF-encoded proteins were not only dissimilar to known phage proteins but had no homology to any proteins contained in GenBank. Therefore, we assigned putative functions only to ORF proteins with significant homology or experimental proof. Included in this group are the following: (i) ORF6, (ii) ORF7, (iii) ORF8, (iv) ORF9, -10, and -11, (v) ORF12, (vi) ORF15, and (vii) ORF16.
TABLE 1.
Features of C1 ORFs and the putative functions of their products
| ORFa | Strand | Start position | End position | Length (nt) | Length of product (amino acids) | Putative function(s) | E valueb |
|---|---|---|---|---|---|---|---|
| 1 | +2 | 572 | 1092 | 522 | 173 | Unknown | >1 |
| 2 | +3 | 1746 | 1934 | 189 | 62 | Unknown | >1 |
| 3 | +2 | 1934 | 2470 | 537 | 178 | Unknown | >1 |
| 4 | +3 | 2601 | 2918 | 318 | 105 | Unknown | >1 |
| 5 | +2 | 2927 | 3550 | 624 | 207 | Unknown | >1 |
| 6 | +1 | 3550 | 4776 | 1,227 | 408 | Neck appendage | 1.2 |
| 7 | +3 | 4806 | 7160 | 2,355 | 784 | DNA polymerasec | 3.00E-05 |
| 8 | +1 | 7204 | 7530 | 327 | 108 | Holinc | 9.00E-06 |
| 9 | +2 | 7517 | 7735 | 219 | 72 | Lysinc | See text |
| 10 | +1 | 7735 | 8052 | 318 | 105 | HNH endonuclease | 1.00E-14 |
| 11 | +2 | 8024 | 9442 | 1,419 | 472 | Lysinc, amidase (group B streptocooci), tail (S. pyogenes 315.5) | See text |
| 8.00E-18 | |||||||
| 3.00E-12 | |||||||
| 12 | −3 | 11195 | 9471 | 1,725 | 574 | Major tail | 3.00E-14 |
| 13 | −1 | 12673 | 11381 | 1,293 | 430 | Unknown | >1 |
| 14 | −3 | 13370 | 12660 | 711 | 236 | Unknown | >1 |
| 15 | −1 | 14320 | 13367 | 954 | 317 | Head-tail connectorc | 2.00E-06 |
| 16 | −3 | 15545 | 14367 | 1,179 | 392 | Major capsidc | See text |
| 17 | −2 | 15738 | 15583 | 156 | 51 | Unknown | >1 |
| 18 | −1 | 15913 | 15743 | 171 | 56 | Unknown | >1 |
| 19 | −2 | 16296 | 16102 | 195 | 64 | Unknown | >1 |
| 20 | −3 | 16547 | 16412 | 168 | 55 | Unknown | >1 |
An ORF must contain a start codon (ATG, GTG, or TTG) and a minimum coding size of 50 amino acids.
E value is the expected value as defined by the NCBI (www.ncbi.nlm.nih.gov).
Experimentally determined or detailed further in the text.
(i) ORF6.
Although ORF6 did not have a high E value (1.2), it did have ∼20% identity to neck appendage proteins (late protein GP12) from Bacillus phages φ29 and PZA.
(ii) ORF7.
ORF7 had high homology to DNA polymerases from Bacillus phages φ29 and GA-1. Significantly, these phage polymerases utilize a protein-primed mechanism of replication (see below for evidence of a terminal protein).
(iii) ORF8.
ORF8 is a putative holin with similarity to a Listeria prophage holin and the Bacillus φ105 phage holin. Additionally, with 108 amino acids and three predicted transmembrane domains, this sequence fits the classic type I holin, as do holins from the φ29, φ105, and Cp-1 Podoviridae (47).
(iv) ORF9, -10, and -11.
The 72-amino-acid ORF9 has no homology to any known protein, yet sequencing of the purified C1 lysin yielded an N-terminal sequence that corresponded to ORF9 (data not shown). However, the native C1 lysin has a predicted molecular mass of ∼60 kDa, which is significantly larger than that of ORF9 (31). This may be explained by investigation of ORF10 and -11. ORF10 has noteworthy homology to the HNH family of homing endonucleases found in many phages (specifically, LambdaSa2 from Streptococcus agalactiae and bIL170 from Lactococcus spp.). These endonucleases are often part of bacteriophage intron systems that give rise to modular enzymes. ORF11 has the highest identity with a putative amidase (lysin) from the LambdaSa1 phage infecting group B streptococci. However, the LambdaSa1 protein consists of over 1,200 amino acids and the amidase region comprises less than 100 amino acids, none of which share identity with the orf11 gene product. The remaining 1,100 amino acids of the LambdaSa1 protein resemble a phage tail protein. As such, C1 ORF11 also has high homology to a putative tail protein from phage 315.5 infecting group A streptococci. Although no typical lysin or amidase regions are present on either ORF9 or -11, it is inviting to speculate that if orf10 is an intron, then gene products of orf9 and -11 could be spliced together to form an active lysin. Significantly, introns have been found in the middle of modular lysin genes for multiple streptococcal phages (22). Work is in progress to elucidate the exact interactions between ORF9, -10, and -11 that yield a functional lysin.
(v) ORF12.
ORF12 is the major tail protein based on homology to the GP9 protein from Bacillus phages in the Podoviridae family (B103, φ29, and GA-1).
(vi) ORF15.
ORF15 is a head-tail connector (collar) protein based on homology to the GP10 protein from Bacillus phages (GA-1 and φ29).
(vii) ORF16.
ORF16 does not have homology to known proteins, yet we experimentally determined this 44-kDa protein to be the major structural protein (see below) and believe that it is the head or capsid protein.
C1 phage structural proteins.
To examine C1 structural proteins, purified phage particles were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 2). Although several bands could be distinguished, two notable bands comprised >90% of the visualized protein. N-terminal sequencing of the smaller, 36-kDa band gave the sequence MQITSGIK, which corresponds to ORF15 (Table 1), a putative 35.9-kDa protein with significant homology to the upper collar proteins (GP-10) from Bacillus phages GA-1, B103, and φ29. The larger and more abundant 44-kDa protein had an N-terminal sequence of ADETTNVA. This sequence corresponds to ORF16, a putative 43.7-kDa protein that does not share similarity with any known protein in GenBank. Because this band accounts for ∼75% of the total phage structural proteins by scanning densitometry, we believe that it represents the major capsid or head protein despite any homology with similar proteins. This is partially supported by the lack of an identified capsid protein (Table 1) and the presence of other expected structural proteins, such as neck appendage (ORF6), major tail (ORF12), and collar (ORF15) proteins.
Evidence of a TP.
Bacteriophages that utilize a protein-primed mechanism of replication have a terminal protein (TP) covalently linked to the 5′ terminus of the DNA. One characteristic of this DNA-protein complex is a noted lack of migration in a standard agarose gel (23). We observed this trait for the C1 bacteriophage DNA (Fig. 3A, lane 1). The migrating band presumably represents DNA that had the TP sheared off during the phenol step in purification. Protease K treatment of the complex results in complete conversion of the nonmigrating complex to migrating DNA with a size of ∼17 kb (Fig. 3A, lane 4). Exonuclease III, which is specific for unblocked 3′ termini, degrades both the DNA-protein complex and PK-DNA (Fig. 3A, lanes 2 and 3 and lanes 5 and 6, respectively). Although we demonstrate that the 3′ termini are free in both the DNA-protein complex and PK-DNA, exonuclease III has slower activity on the complex, most likely due to steric hindrance of the TP near the 3′ termini.
FIG. 3.
Evidence of a covalently linked 5′ TP. (A) DNA-protein complex (lane 1), complex treated with exonuclease III for 15 min (lane 2), complex treated with exonuclease III for 30 min (lane 3), PK-DNA (lane 4), PK-DNA treated with exonuclease III for 15 min (lane 5), and PK-DNA treated with exonuclease III for 30 min (lane 6). (B) DNA-protein complex (lane 1), complex treated with λ exonuclease for 15 min (lane 2), complex treated with λ exonuclease for 30 min (lane 3), PK-DNA (lane 4), PK-DNA treated with λ exonuclease for 30 min (lane 5), PK-DNA treated with 0.5 M piperidine (lane 6), PK-DNA treated with piperidine and λ exonuclease for 30 min (lane 7). The amounts used are indicated in Materials and Methods.
In contrast to exonuclease III, both the DNA-protein complex and PK-DNA are insensitive to the effects of the 5′-specific lambda exonuclease (Fig. 3B, lanes 1 to 5). However, pretreatment of PK-DNA with 0.5 M piperidine, which has been shown to specifically hydrolyze the bond between the DNA and TP in the φ29 bacteriophage (33), renders the 5′ termini unprotected (Fig. 3B, lane 7), thus proving a covalent linkage at the 5′ termini.
Analysis of the DNA polymerase.
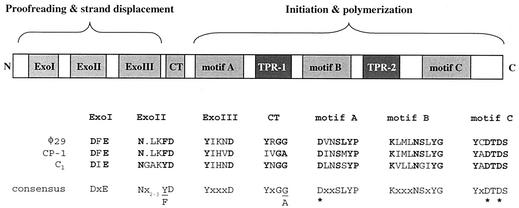
The data presented above confirm that C1 bacteriophage DNA contains a TP. Further supporting evidence can be found by examination of the putative C1 DNA polymerase (ORF7). Bioinformatic analysis suggests that the C1 polymerase belongs to family B of the DNA polymerases (also referred to as eukaryotic or α-like), which comprises eukaryotic, viral, and protein-primed polymerases (Fig. 4). This family has consensus sequences known to be vital for proofreading and strand displacement functions (ExoI, ExoII, and ExoIII), a cross-talk region important for coordination between 3′-to-5′ exonuclease and 5′-to-3′ polymerization events, and several consensus motifs (A, B, and C) involved in initiation and polymerization (for a review, see reference 5). Significantly, polymerases that utilize a protein-primed mechanism of replication have two additional motifs, called terminal protein regions (TPRs) (6). These structures are known to interact with TPs covalently linked to DNA (14). While the TPRs do not contain conserved sequence motifs, they are apparent as sequence insertions between motifs A and B (TPR-1) or motifs B and C (TPR-2) when aligned against family B polymerases that do not utilize the protein-primed mechanism (7). The C1 polymerase contains all necessary conserved elements, as well as two TPR insertion regions. For TPR-1, the C1 insertion is 64 amino acids, compared to 59 and 58 amino acids for the φ29 and Cp-1 DNA polymerases, respectively.
FIG. 4.
Structural and functional map of type B (α-like) phage DNA polymerases that utilize a protein-primed mechanism of replication. Alignments of important residues from the Bacillus φ29 (NP_040719), pneumococcal Cp-1 (NP_044817), and streptococcal C1 phage DNA polymerases are shown. The N-terminal domain of type B DNA polymerases contain conserved exonuclease I, exonuclease II, and exonuclease III sequences separated from the C-terminal domain by a cross-talk (CT) region. The C-terminal domain also has several conserved consensus motifs, A, B, and C. The three Asp residues, noted by asterisks, constitute the polymerization active site. See the text for further details on these regions. Consensus sequences were taken from references 5 and 29. All three of these polymerases have TPRs of 30 to 60 amino acids inserted between motifs A and B (TPR-1) or motifs B and C (TPR-2). These regions are thought to interact with the TP covalently attached to the phage DNA, which initiates replication.
Investigation of the putative C1 phage receptor.
Consistent with the findings of a previous report (25), we found that group C, but not group A, streptococci were susceptible to infection by the C1 bacteriophage. However, we also found that group A-variant streptococci were resistant to infection (data not shown). Furthermore, group C streptococci treated with pronase or trypsin and chymotrypsin remained susceptible to infection. This was expected, as most phages are believed to bind to carbohydrate, not protein, epitopes. Significantly, nuclear magnetic resonance structures of the carbohydrate from these three streptococcal groups have been elucidated (11). A polyrhamnose backbone is common to the surface carbohydrate of group C streptococci and the nonhost strains of group A and group A-variant streptococci; distinguishing these structures are side chains of two GalNAc residues in group C streptococci, one N-acetylglucosamine (GlcNAc) residue in group A streptococci, and a lack of side chains in group A-variant streptococci. Since the C1 phage does not infect group A or A-variant streptococci, the GalNAc moiety is likely an important element of the phage receptor.
In additional adsorption studies, we found that purified group C streptococcal cell walls were very efficient in adsorbing the C1 phage, reducing a phage titer from 107 PFU/ml to less than 102 PFU/ml. This same effect was not seen with purified group A streptococcal cell walls. Moreover, we found that the chemically extracted group C streptococcal carbohydrate also retained the ability to adsorb C1 phage. This is in contrast to Fischetti and Zabriskie's earlier findings (21); however, we used a method involving nitrous acid extraction of the carbohydrate layer, which is more efficient in retaining the integrity of the extracted carbohydrate than is the hot formamide method utilized by the aforementioned authors. While this further supports a potential role for GalNAc as a component of the receptor, we do not believe that the GalNAc monosaccharide itself is sufficient for binding. When the C1 phage was pretreated with 20 mM GalNAc, it retained its full capacity to infect group C streptococci (data not shown). Whether the receptor is ultimately determined to be the disaccharide form of GalNAc, a combination of the GalNAc and rhamnose, or some other combination will require further experimentation.
DISCUSSION
In the first example of phage typing, Evans used the C1 bacteriophage to distinguish streptococci isolated from animals from those isolated from humans (15, 16). It was speculated that this “race” of phage recognized something common among the animal streptococci and absent from the human streptococci. We know today that the majority of human infections are caused by group A streptococci and those of animals are caused by group C streptococci. Here we report that the receptor for the C1 phage is contained within the chemically extracted carbohydrate unique to group C streptococci.
An additional finding of Clark was that the C1 phage had the ability to lyse cultures that were not susceptible to infection with the phage itself. Seventy years later, this potent enzyme, the C1 lysin, may represent a new method to control streptococcal infections in humans because of its activity on both group A and group C streptococci (31). It is noteworthy that in phage infecting gram-positive bacteria, the binding domains of lysins and tail fibers from the same virion appear to recognize identical epitopes on their hosts due to the shared specificities of these proteins. We, along with others, have shown this for pneumococcal and Bacillus anthracis systems (13, 28, 38). However, based on the adsorption and infection data, the C1 phage has a more limited host range than the C1 lysin, indicating that the binding domains for the tail and the lysin recognize two different epitopes. Considering that the C1 phage has one of the smallest Podoviridae genomes, the evolutionary pressure to create separate binding domains when one suffices in other systems is not readily evident. Moreover, our finding that the C1 lysin is interrupted by an apparent intron adds further complexity to the already compact genome. Although introns are known to exist in phage, and to be specifically embedded in the lysin gene (22), this is usually seen in the larger-genome phages of Myoviridae and Siphoviridae. To our knowledge, this is the first potential intron or intron-like sequence in the genome of a Podoviridae family member.
The Podoviridae family of bacteriophages contains phages with short, noncontractile tails and represents one of the most diverse groups of phages known. With only 16 genomes sequenced and deposited in GenBank, these family members vary in size from under 12 to over 47 kb, and the host range includes gram-positive bacteria, gram-negative bacteria, and mycoplasma. However, a genus of the Podoviridae, known as the φ29-like phages, shows several common characteristics. All phages in this class are lytic, have double-stranded linear DNA genomes of ∼20 kb, and contain a TP that is used as the primer for initiation of translation (for a full review, see reference 29). As such, they also possess a unique type B DNA polymerase that contains TPRs, which bind the TP (36). According to the taxonomic placement chart developed by the International Committee on Taxonomy of Viruses (ICTV) (45), the C1 bacteriophage clearly belongs to the φ29-like family based on physical morphology, genome size, and the presence of a TP. Likewise, the ICTV also classifies the pneumococcal Cp-1 phage in this genus. Nonetheless, some groups contend that only Bacillus-infecting phages (φ29, GA-1, and B103) belong to the φ29-like genus (29), due in part to a shared genome organization and an evolution that can be traced to common ancestors (32).
Attempts to model the C1 phage against φ29-like phage, as well as other members of the Podoviridae, by using comparative genomic methods yielded no successful alignments, which was confirmed independently (Harald Brüssow, personal communication). This is not too surprising considering only 2 of the 20 putative ORFs had an E value greater than 1.00E-10 when a BLAST search was performed. Notably, to assign function to a majority of our ORFs, we had to rely on techniques other than significant BLAST scores. For the DNA polymerase (ORF7), we aligned conserved motifs from known DNA polymerases that utilized a protein-primed mechanism of replication; for the holin (ORF8), we had to perform transmembrane modeling; for the lysin (ORF9 and -11), we had to purify the native protein and perform N-terminal sequencing; and for the structural proteins, head-tail connector (ORF15), and capsid (ORF16), we had to N-terminally sequence the dominant proteins from purified phage particles. Nonetheless, the C1 lysin and capsid proteins do not share homology to known proteins with similar functions. Furthermore, no gene coding for a TP has been identified despite overwhelming evidence that such a gene must exist. This could be anticipated, since TPs are not only utilized by the φ29-like Podoviridae but also by the PDR1 phage of the Tectiviridae family and by adenoviruses, thus representing an early evolutionary divergence. Taken together, these data suggest that the C1 bacteriophage is a unique member of the φ29-like genus in the Podoviridae family.
It should be pointed out that controversy about the taxonomic classification of bacteriophages currently exists. Because there are no ribosomal sequences in phages and a recent analysis showed that there are no single protein markers or DNA sequence motifs shared between phages (35), any classification scheme is dubious at best. Nonetheless, the ICTV classification does attempt a hierarchical approach based initially on the physical appearance of tail fibers, which does not consider any potential genetic relatedness between species. Some authors have suggested that it may be impossible to apply any type of hierarchal approach to taxonomy due to genomic mosaicism exhibited by bacteriophages (27). However, others have had success with forming a phage proteomic tree, a hierarchal taxonomic approach based on BLAST scores of near protein neighbors (35). Indeed, this approach does resolve several apparent limitations with the ICTV method. For example, the PDR1 phage of the Tectiviridae family is now joined with other protein-primed phages such as the Bacillus GA-1 phage and the pneumococcal Cp-1 phage in a new group known as PZA-like Podophage. While the taxonomic debate continues, the C1 bacteriophage shares morphological, biological, and genetic relatedness to protein-primed members of the Podoviridae or Podophage family. As more phage genomes are made available, it is hoped that the evolutionary relationships between the C1 bacteriophage and its relatives can be further appreciated.
Acknowledgments
We are grateful to Harald Brüssow for his analysis and observations regarding the comparative genomics of the C1 bacteriophage sequence. We are indebted to Maclyn McCarty and Richard Krause for their knowledge of and commentary on the history of the C1 bacteriophage and early experiments to elucidate the streptococcal carbohydrate structure. Finally, we thank Eleana Sphicas at the Bio-Imaging Resource Center at The Rockefeller University for help with electron microscopy and members of V.A.F.'s laboratory for reviewing the manuscript.
This work was supported by a grant from the Defense Advanced Research Projects Agency to V.A.F. and a fellowship from the Charles H. Revson Biomedical Research Foundation to D.N.
REFERENCES
- 1.Ackermann, H.-W. 1999. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M.-Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. M. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanco, L., and M. Salas. 1996. Relating structure to function in φ29 DNA polymerase. J. Biol. Chem. 271:8509-8512. [DOI] [PubMed] [Google Scholar]
- 6.Blasco, M. A., L. Blanco, E. Pares, M. Salas, and A. Bernad. 1990. Structural and functional analysis of temperature-sensitive mutants of the phage φ29 DNA polymerase. Nucleic Acids Res. 18:4763-4770. [PMC free article] [PubMed] [Google Scholar]
- 7.Braithwaite, D. K., and J. Ito. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broudy, T. B., and V. A. Fischetti. In vivo lysogenic conversion of Tox− S. pyogenes to Tox+ with lysogenic streptococci or free phage. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 9.Brüssow, H., and R. W. Hendrix. 2002. Phage genomics: small is beautiful. Cell 108:13-16. [DOI] [PubMed] [Google Scholar]
- 10.Clark, P. F., and A. S. Clark. 1926. A “bacteriophage” active against a hemolytic streptococcus. J. Bacteriol. 11:89. [Google Scholar]
- 11.Coligan, J. E., T. J. Kindt, and R. M. Krause. 1978. Structure of the streptococcal groups A, A-variant and C carbohydrates. Immunochemistry 15:755-760. [DOI] [PubMed] [Google Scholar]
- 12.d'Herelle, F. H. 1917. Sur un microbe invisible antagoniste des bacilles dysenteriques. C. R. Acad. Sci. (Paris) 165:< /OTHER-REF>373-375.
- 13.Diaz, E., R. Lopez, and J. L. Garcia. 1990. Chimeric phage-bacterial enzymes: a clue to the modular evolution of genes. Proc. Natl. Acad. Sci. USA 87:8125-8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufour, E., J. Mendez, J. M. Lazaro, M. de Vega, L. Blanco, and M. Salas. 2000. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol. 304:289-300. [DOI] [PubMed] [Google Scholar]
- 15.Evans, A. C. 1934. Streptococcus bacteriophage: a study of four serological types. Public Health Rep. 49:1386-1401. [Google Scholar]
- 16.Evans, A. C. 1936. Studies on hemolytic streptococci. I. Methods of classification. J. Bacteriol. 31:423-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 18.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 19.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischetti, V. A., B. Barron, and J. B. Zabriskie. 1968. Studies on streptococcal bacteriophages. I. Burst size and intracellular growth of group A and group C streptococcal bacteriophages. J. Exp. Med. 127:475-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischetti, V. A., and J. B. Zabriskie. 1968. Studies on streptococcal bacteriophages. II. Adsorption studies on group A and group C streptococcal bacteriophages. J. Exp. Med. 127:489-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foley, S., A. Bruttin, and H. Brüssow. 2000. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J. Virol. 74:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia, E., A. Gomez, C. Ronda, C. Escarmis, and R. Lopez. 1983. Pneumococcal bacteriophage Cp-1 contains a protein bound to the 5′ termini of its DNA. Virology 128:92-104. [DOI] [PubMed] [Google Scholar]
- 24.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krause, R. M. 1957. Studies on bacteriophages of hemolytic streptococci. I. Factors influencing the interaction of phage and susceptible host cells. J. Exp. Med. 106:365-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lancefield, R. C. 1932. Note on the susceptibility of certain strains of hemolytic streptococcus to a streptococcus bacteriophage. Proc. Soc. Exp. Biol. Med. 30:169-171. [Google Scholar]
- 27.Lawrence, J. G., G. F. Hatfull, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170-2172. [DOI] [PubMed] [Google Scholar]
- 29.Meijer, W. J. J., J. A. Horcajadas, and M. Salas. 2001. φ29 family of phages. Microbiol. Mol. Biol. Rev. 65:261-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moynet, D. J., A. E. Colon-Whitt, G. B. Calandra, and R. M. Cole. 1985. Structure of eight streptococcal bacteriophages. Virology 142:263-269. [DOI] [PubMed] [Google Scholar]
- 31.Nelson, D., L. Loomis, and V. A. Fischetti. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 98:4107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pecenkova, T., and V. Paces. 1999. Molecular phylogeny of φ29-like phages and their evolutionary relatedness to other protein-primed replicating phages and other phages hosted by gram-positive bacteria. J. Mol. Evol. 48:197-208. [DOI] [PubMed] [Google Scholar]
- 33.Penalva, M. A., and M. Salas. 1982. Initiation of phage φ29 DNA replication in vitro: formation of a covalent complex between the terminal protein, p3, and 5′-dAMP. Proc. Natl. Acad. Sci. USA 79:5522-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomrenke, M. E., and J. J. Ferretti. 1989. Physical maps of the streptococcal bacteriophage A25 and C1 genomes. J. Basic Microbiol. 6:395-398. [DOI] [PubMed] [Google Scholar]
- 35.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39-71. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884-889. [DOI] [PubMed] [Google Scholar]
- 39.Shwartzman, G. 1927. Studies on streptococcus bacteriophage. I. A powerful lytic principle against hemolytic streptococci of erysipelas origin. J. Exp. Med. 46:497-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson, J., K. Hsu, and E. C. Gotschlich. 1969. Electron microscopic studies on streptococci. I. M antigen. J. Exp. Med. 130:1063-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Totolian, A. A., T. Gupalova, J. Coll, A. Suvorov, A. Boitsov, and V. I. Golubkov. 1981. Introduction to the molecular biology of virulent streptococcal bacteriophages, p. 231-233. In S. Holm and P. Christensen (ed.), Basic concepts of streptococci and streptococcal diseases. Reedbooks Ltd., Surrey, England.
- 42.Twort, F. W. 1915. An investigation on the nature of ultra-microscopic viruses. Lancet ii:1241-1243. [DOI] [PMC free article] [PubMed]
- 43.Twort, F. W. 1925. The transmissible bacterial lysin and its action on dead bacteria. Lancet ii:642-644. [Google Scholar]
- 44.van de Rijn, I., and V. A. Fischetti. 1981. Immunochemical analysis of intact M protein secreted from cell wall-less streptococci. Infect. Immun. 32:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.). 2000. Virus taxonomy: seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 46.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, I.-N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 48.Wheeler, J., J. Holland, J. M. Terry, and J. D. Blainey. 1980. Production of group C streptococcus phage-associated lysin and the preparation of Streptococcus pyogenes protoplast membranes. J. Gen. Microbiol. 120:27-33. [DOI] [PubMed] [Google Scholar]