Abstract
The origins of replication of many different bacteria have been shown to reside at specific subcellular locations, but the mechanisms underlying their positioning and segregation are still being elucidated. In particular, little is known about the replication of multipartite genomes in bacteria. We determined the cellular positions of the origins of the replicons in the alpha proteobacteria Agrobacterium tumefaciens and Sinorhizobium meliloti and found that they are located at the poles of the cells. Our work demonstrates the conserved extreme polar localization of circular chromosome origins in these alpha proteobacteria and is also the first to specify the cellular location of origin regions from the repABC family. The cellular location of a derivative of the RK2 plasmid is distinct from that of the alpha proteobacterium genomic replicon origins but is conserved across bacteria. Colocalization experiments with the genomic replicons of A. tumefaciens revealed that the repABC replicons, although preferentially positioned at the cell pole, colocalize only rarely. For the repABC replicons in this organism, occupying discrete spatial locations may contribute to their coexistence and stable inheritance.
The use of fluorescence in situ hybridization (FISH) and protein fusions to the green fluorescent protein and its variants has revealed that the bacterial cell is far more organized than previously thought and that both DNA and proteins are targeted to specific subcellular locations (30). Studies of Escherichia coli have shown that its chromosomal origin localizes to nucleoid borders near the cell poles (24), while low-copy-number plasmids localize either to the cell midpoint and near the poles (16) (R1) or to the mid- and quarter-cell positions (11, 25) (F and prophage P1). In sporulating Bacillus subtilis (35) and Caulobacter crescentus (18), chromosomal origins are found at the extreme cell poles. In synchronized populations of Caulobacter, as soon as the polar origin is replicated, it can be detected at the opposite pole of the cell (17). In vegetatively growing B. subtilis, chromosomal origins are found in subpolar positions (35). Little is known about origin positioning in bacteria with genomes of higher complexity, including several members of the alpha proteobacteria which contain multiple replicons, as well as Borrelia burgdorferi, Burkholderia cepacia, and Vibrio species, which contain two or more chromosomes (5, 12, 19, 21, 34, 37). The mechanisms utilized to accurately duplicate and segregate multiple DNA molecules during the bacterial cell cycle are not known.
The alpha proteobacterium Agrobacterium tumefaciens has four replicons: a circular chromosome, a linear chromosome, a cryptic plasmid (pAtC58), and the tumor-producing Ti plasmid. The circular chromosome origin of replication is similar to those of other alpha proteobacteria, whereas the other replicons carry plasmid-type replication systems of the repABC family (3, 10, 36). In previous work, we described the synchronization of DNA replication with the cell cycle in A. tumefaciens (20). Duplication of all replicons has been independently confirmed to occur during a defined period early in the cell cycle, thus requiring coordinated initiation of these two classes of replication origin (10). In this paper, we have determined the cellular locations of the predicted replication origins of all four replicons and found that they all localize to the poles of the cell. This represents the first such analysis of a multipartite bacterial genome and of the very-low-copy-number repABC family of plasmids. Their localization pattern is distinct from that of the low-copy-number plasmids F and P1, which have been previously characterized in E. coli. We show that Sinorhizobium meliloti, an alpha proteobacterium with a circular chromosome and two repABC megaplasmids (pSymA and pSymB), also positions its replication origins at the cell pole, suggesting that the mechanisms governing the cellular organizations of both classes of replicons are conserved among the alpha proteobacteria. In comparison, analysis of a broad-host-range RK2-based multicopy plasmid in three different alpha proteobacteria revealed that RK2 is localized to mid- and quarter-cell positions, as is its parent plasmid in E. coli and other gamma proteobacteria (28). Thus, the means of targeting this plasmid are conserved across an even wider range of bacteria than previously shown yet are clearly different from those governing the localization of the genomic replicons. We performed dual-labeling experiments to determine whether the genomic replicon origins of A. tumefaciens colocalized and found that despite their location at the cell poles, they generate coincident foci only rarely. Thus, there is a conserved localization pattern of multiple replicons among the alpha proteobacteria; however, as with compatible plasmids in E. coli, coexisting repABC replicon origins are found in the same general region of the cell but occupy discrete areas of their own.
MATERIALS AND METHODS
Bacterial growth conditions and media.
Strains and plasmids used are listed in Table 1. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium containing 10 g of NaCl/liter. A. tumefaciens and S. meliloti strains were grown at 28°C in LB medium containing 5 g of NaCl/liter. C. crescentus strains were grown at 28°C in PYE medium (8). Antibiotics were used at the following concentrations: nalidixic acid, 20 μg/ml; kanamycin, 25 μg/ml. Plasmids were introduced into A. tumefaciens and S. meliloti either by electroporation or by mating with E. coli S17-1 as a donor strain. Agrobacterium, Sinorhizobium, and Caulobacter strains grown in these rich media had doubling times between 70 and 90 min depending on the bacterium. All bacteria were harvested in exponential growth phase.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli TOP10 | Cloning strain | Invitrogen |
| E. coli S17-1 | Conjugal transfer of plasmids | 32 |
| C. crescentus CB15N | Synchronizable wild-type strain | Laboratory collection |
| A. tumefaciens C58 | Wild-type strain | Laboratory collection |
| S. meliloti 1021 | Wild-type strain | Laboratory collection |
| LS2305 | C. crescentus CB15N with pMR10 | Laboratory collection |
| LS3607 | A. tumefaciens C58 with pMR10 | This study |
| LS3592 | S. meliloti 1021 with pMR10 | This study |
| Plasmids | ||
| pMR10 | Kan derivative of RK2; broad-host-range low-copy-number vector | R. Roberts and C. Mohr |
| pCR-XL-TOPO | Kan vector for cloning PCR products | Invitrogen |
| Derivatives of pCR-XL-TOPO (Invitrogen) containing FISH probes | ||
| pLK317 | A. tumefaciens circular chromosome origin probe | This study |
| pLK322 | A. tumefaciens linear chromosome repABC probe 1 | This study |
| pLK323 | A. tumefaciens linear chromosome repABC probe 2 | This study |
| pLK339 | A. tumefaciens pAtC58 repABC probe 1 | This study |
| pLK340 | A. tumefaciens pAtC58 repABC probe 2 | This study |
| pLK341 | A. tumefaciens pTiC58 repABC probe 1 | This study |
| pLK342 | A. tumefaciens pTiC58 repABC probe 2 | This study |
| pLK333 | S. meliloti circular chromosome origin probe 1 | This study |
| pLK334 | S. meliloti circular chromosome origin probe 2 | This study |
| pLK335 | S. meliloti pSymA repABC probe 1 | This study |
| pLK336 | S. meliloti pSymA repABC probe 2 | This study |
| pLK337 | S. meliloti pSymB repABC probe 1 | This study |
| pLK338 | S. meliloti pSymB repABC probe 2 | This study |
FISH.
Cells were fixed directly in growth media with 2.5% paraformaldehyde and 30 mM KH2PO4 and processed for FISH as described previously (17). Lysozyme was used to permeabilize the cells at a concentrations of between 2 and 20 μg/ml for Caulobacter and Sinorhizobium and between 50 and 250 μg/ml for Agrobacterium. A range of lysozyme concentrations were tested, and the concentration was optimized for each hybridization. Chromosomal DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml) in SlowFade light (Molecular Probes). Cells analyzed were chosen from fields in which there was adequate permeabilization of cells and preservation of a compact nucleoid and cell morphology. In such areas, approximately 20% of cells were not labeled with a given probe.
Preparation of probes.
Regions of replicons near their putative replication origins were synthesized by PCR as 5- or 10-kb products. The replication origins used were putatively identified as such by GC skew analysis of the completed genome sequences of the organisms (4, 10, 23, 36). This identification is supported by the conserved genetic organization around the origins of the circular chromosomes in alpha proteobacteria (2). Furthermore, repABC loci from other plasmids have been isolated and shown to be capable of independent replication (22, 29). These were cloned with the TOPO-XL cloning kit (Invitrogen) and verified by direct sequencing and restriction analysis. Sequences were chosen to be within approximately 10 kb of a putative replication origin and avoid cross-hybridization with other regions of the genome. Ten micrograms of probe sequence DNA was digested with Sau3AI and MspI to give fragments ranging in size from 20 to 300 bp. These were labeled with either Cy3-dCTP (Molecular Probes) for FISH or digoxigenin-11-dUTP (Roche) for immunofluorescence microscopy (IFM) using terminal deoxynucleotidyltransferase (Promega). Sequences of oligonucleotides are available on request.
Immunofluorescence.
For detection of digoxigenin-labeled probes, FISH was performed as described above. Following this, in the manner of Yang and Losick (39), slides were prehybridized for 15 min with 2% bovine serum albumin in phosphate-buffered saline and then incubated for 2 h with fluorescein-conjugated antidigoxigenin Fab fragments (Roche).
Microscopy.
Cells were immobilized on poly-l-lysine-coated slides as described previously (17). Fluorescence and differential interference contrast (DIC) images were acquired with a Nikon E800 microscope with a 100× DIC objective and a 5-MHz Micromax 5600 cooled charge-coupled device camera controlled by Metamorph (Universal Imaging Corp., Scranton, Pa.). Images were processed with Metamorph, which was also used to measure cells. A minimum of 100 cells were analyzed for each probe. Results were confirmed with separately prepared slides, hybridizations, and cultures.
RESULTS
Localization of replication origins in A. tumefaciens by FISH.
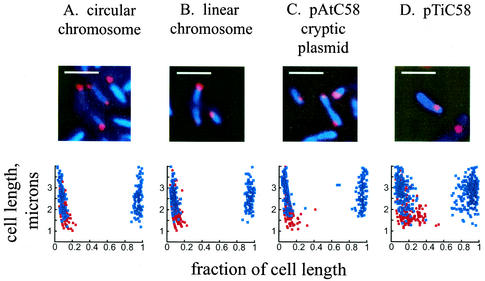
A. tumefaciens contains four replicons. It has a circular chromosome with a bacterial chromosome-like origin resembling the Caulobacter origin of replication (Cori). It also has a linear chromosome, a cryptic plasmid (pAt), and a Ti plasmid, which all contain plasmid replicators of the repABC class (10, 33, 36). These replicons have been estimated at unit copy number, and all replicate coincidently during the cell cycle (10, 20, 33). To determine if temporal coordination is accompanied by spatial coordination, we identified the locations in the cell of the origin region of the circular chromosome and the regions adjacent to the repABC loci of the linear chromosome, pAtC58, and pTiC58 (Fig. 1). We used FISH with Cy3-labeled probes and DAPI staining of the nucleoid, as described in Materials and Methods. As in Caulobacter (17), the nucleoid fills the cells in Agrobacterium. Only one or two foci were seen in each cell, consistent with our prior flow cytometry and Southern blot data suggesting that replication of all the replicons occurs just once per cell cycle (20). The presence of two foci is indicative of cells which have already initiated replication. No signal was observed when the empty plasmid used to clone the probes was itself used as a probe (data not shown). The positions of foci along the length of the cell were measured, revealing that the origins of all four replicons are located preferentially at or near the cell poles (Fig. 1). As observed in Caulobacter, the presence of two polar foci suggests the movement of the replicated origin to the opposite cell pole. The origin of pTiC58, while also predominantly polar, was frequently found in a subpolar location. While this finding was more pronounced in smaller cells with a single focus, it was also a prominent feature of longer cells with two origins.
FIG. 1.
Localization of A. tumefaciens origins of replication by FISH. Red, probe signals from the origin sequences; blue, DAPI staining of the nucleoid. Graphical representations of the cellular positions of the origin are shown below the FISH staining. A minimum of 100 cells was counted for each graph. Red dots, origin positions in cells with a single focus; blue dots, origin positions in cells with two foci. Zero and 1 on the x axis represent the poles of the cell. Bars, 2 μm.
Localization of origins in S. meliloti by FISH.
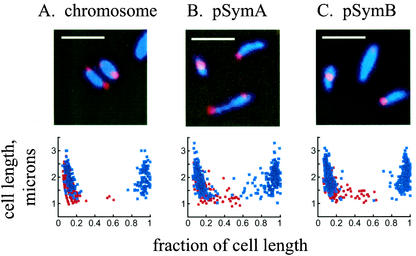
To determine if the polar location of replication origins of all genomic replicons in a bacterial cell is conserved among the alpha proteobacteria, we examined the cellular positions of the replication origins in S. meliloti. S. meliloti contains three replicons; its circular chromosome origin resembles those of Caulobacter and Agrobacterium (3), and its two megaplasmids, pSymA and pSymB, have repABC origins (9) (Table 2). As in Agrobacterium, we used FISH with Cy3-labeled probes (Fig. 2). The DAPI-stained nucleoid fills the cell in Sinorhizobium, as in Agrobacterium and Caulobacter. We consistently observed one or two foci per cell, and in all cases, the replication origins of the circular chromosome and two megaplasmids are located at the cell poles. As observed in the graphic representation of the origin locations in individual cells, the polar localization was most stringent for chromosome origin foci (Fig. 2A). For the pSymA and pSymB origins, we observed that, in cells with single foci (red dots) and in larger cells with two foci (blue dots), there was some drift away from the cell pole (Fig. 2B and C). Thus, in two different alpha proteobacteria with multipartite genomes, the origins of the replicons are found predominantly at the cell poles and the chromosomal origins are localized most strictly in this fashion.
TABLE 2.
Replication origins and their positioning
| Organism | Replicon | Origin type | Polar localizationa | Position(s) |
|---|---|---|---|---|
| A. tumefaciens | Circular chromosome | Cori | +++ | Polar |
| Linear chromosome | repABC | +++ | Polar | |
| pAtC58 | repABC | ++ | Polar | |
| pTiC58 | repABC | + | Polar | |
| S. meliloti | Circular chromosome | Cori | +++ | Polar |
| pSymA | repABC | ++ | Polar | |
| pSymB | repABC | ++ | Polar | |
| C. crescentus | Circular chromosome | Cori | +++ | Polar |
| A. tumefaciens | pMR10 | RK2 | − | 1/4 and 3/4 |
| S. meliloti | pMR10 | RK2 | − | 1/4 and 3/4 |
| C. crescentus | pMR10 | RK2 | − | 1/4 and 3/4 |
+++, <10% nonpolar; ++, 10 to 25% nonpolar; +, 35% nonpolar; −, never polar.
FIG. 2.
Fluorescence micrographs and graphical representation of origin localization by FISH in S. meliloti. Red, probe signals from the origin sequences; blue, DAPI staining of the nucleoid. Red dots, origin positions in cells containing a single focus; blue dots, origin positions in cells with two foci. Zero and 1 on the x axis represent the poles of the cell. Bar, 2 μm.
Dual labeling of the chromosomal and repABC origins in A. tumefaciens.
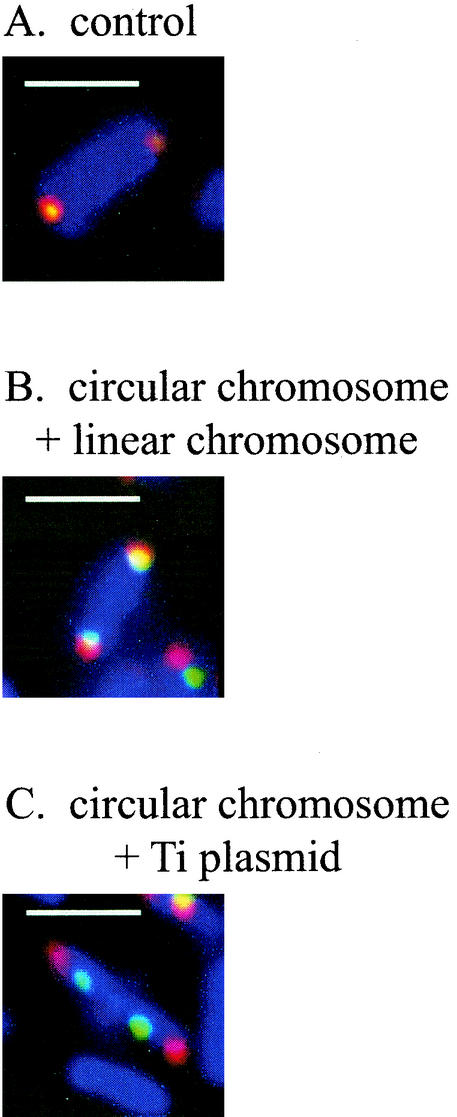
To determine if the multiple replication origins in A. tumefaciens are targeted to overlapping positions at the pole, we performed dual labeling using a combination of FISH and IFM to detect different pairs of foci (Fig. 3; Table 3). The probes were labeled with Cy3 for FISH and digoxigenin-11-dUTP for IFM, as described in Materials and Methods. For each individual probe, digoxigenin-labeled probes gave the same localization results as when Cy3 was used (data not shown). In hybridizations with pairs of origin probes, each labeled with a different fluor, the degree of signal overlap was assessed. Foci were considered coincident if at least one-half of each focus overlapped with the other. Examples of coincident and noncoincident foci are shown in Fig. 3, and the results are summarized in Table 3. When the circular chromosome origin labeled with Cy3 was hybridized with the same probe labeled with digoxigenin, the pairs of foci were entirely coincident 97% of the time. When the circular chromosome and the linear chromosome were examined, it was found that the foci were coincident 68% of the time. This percentage dropped to 58% when the circular chromosome and pAtC58 were examined and to only 33% when the chromosome and the Ti plasmid were examined. When different pairs of repABC replicons (linear chromosome with pAtC58, linear chromosome with pTiC58, and pAtC58 with pTiC58) were examined in separate hybridizations, it was found that their foci were coincident less than 50% of the time. Given the size of the cells and the size of the foci, we estimate that overlap could occur randomly approximately 30% of the time. Thus, given the limitations of this assay, we conclude that none of the three repABC replicons displayed a strong tendency to colocalize with each other, although they can be found in the same general region of the cell.
FIG. 3.
Dual labeling of origins in A. tumefaciens showing localization patterns of Cy3-labeled probes (red) and digoxigenin-labeled probes (green). Superimposed red and green appear yellow. (A) Control with both probes to the circular chromosome origin; (B) origins of the circular chromosome (red) and the linear chromosome (green); (C) origins of the circular chromosome (red) and the Ti plasmid (green). Blue, DAPI staining of the nucleoid. Bars, 2 μm.
TABLE 3.
Percentages of coincident foci in dual-labeling FISH-IFM experiments on A. tumefaciens
| Pair of origins | % Coincident | No. of pairs counted |
|---|---|---|
| Circular chromosome vs circular chromosome | 97 | 200 |
| Circular chromosome vs linear chromosome | 68 | 391 |
| Circular chromosome vs pAtC58 | 58 | 523 |
| Circular chromosome vs pTiC58 | 33 | 316 |
| Linear chromosome vs pAtC58 | 48 | 315 |
| Linear chromosome vs pTiC58 | 45 | 196 |
| pAtC58 vs pTiC58 | 42 | 193 |
Subcellular localization of an RK2-based multicopy plasmid in alpha proteobacteria.
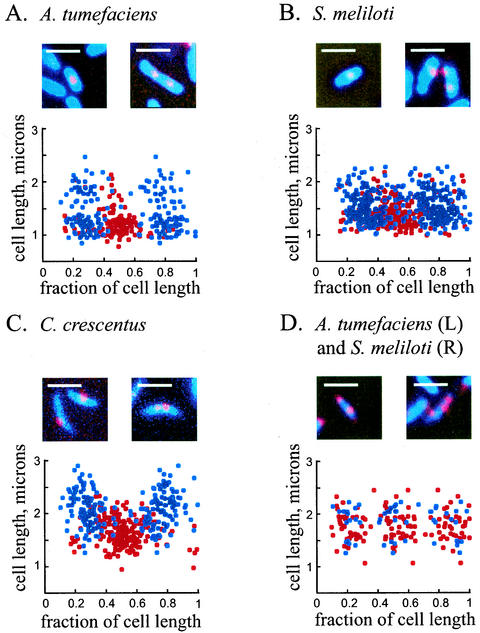
Because of the tendency of alpha proteobacterium genomic replicon origins to localize at or near the cell poles, we asked if the pole is a general site for origin localization in the alpha proteobacteria. Thus, we examined the localization pattern of a different replicon, the plasmid pMR10, which is an RK2-derived broad-host-range multicopy plasmid (R. Roberts and C. Mohr, unpublished results), as described in Materials and Methods. PMR10, which is 8.5 kb in size, was used in its entirety as a probe in C. crescentus, A. tumefaciens, and S. meliloti (Fig. 4). Hybridizations, with the plasmid as a labeled probe, to strains which did not contain any plasmid did not give a signal (data not shown). Cells bearing pMR10 contained primarily one or two foci; three foci were seen only infrequently (∼1% of cells) in Sinorhizobium and Agrobacterium and not at all in Caulobacter. The localization patterns in all three bacteria were similar: in cells with a single focus, this focus was found predominantly at midcell, whereas in cells with two foci, these were located on average at the one-quarter and three-quarter points (Fig. 4A to C). In the few cells with three foci, there was a trend for those foci to occupy the mid- and quarter-cell positions (Fig. 4D). Thus, an RK2-based multicopy plasmid occupies regions of the cell distinct from those of the other replicons, which are found at the cell poles.
FIG. 4.
(A to C) Localization of an RK2-based plasmid in A. tumefaciens, S. meliloti, and C. crescentus. Red, probe signal from the plasmid; blue, DAPI staining of the nucleoid. Each panel also shows a graphical representation of positions of one and two foci in all three bacteria (red, positions of spots in cells with one focus; blue, positions of spots in cells with two foci). (D) Positions of spots in cells with three foci. Red, A. tumefaciens; blue, S. meliloti. In all panels, zero and 1 on the x axis represent the poles of the cell. Bars, 2 μm.
DISCUSSION
The alpha proteobacteria A. tumefaciens and S. meliloti have multipartite genomes with replicons containing different types of origins. The replication origins of the circular chromosomes of both these bacteria are localized to the extreme poles of the cell, as observed in the alpha proteobacterium C. crescentus, which contains a single replication unit (17). This is in contrast to the subpolar origin position in B. subtilis and E. coli (11, 24, 35). Analysis of the origin regions of the additional replicons in A. tumefaciens and S. meliloti showed that their repABC class origin regions are also positioned at or near the extreme poles of the cell.
This is the first observation of the cellular positions of origins of the repABC class, which were found to preferentially localize to the cell poles. This origin class occurs primarily in large plasmids of the Rhizobiaceae, as well as in Paracoccus spp. (1, 22). Well-characterized plasmids that have been localized in E. coli are found at mid- and quarter-cell positions (F, prophage P1, and RK2) (11, 25, 28) or at midcell and close to the poles (R1) (16); these patterns are distinct from the predominantly polar location described here. The polar positioning is most strongly manifested for the linear chromosome of A. tumefaciens and least strongly manifested for its Ti plasmid. The linear chromosome of A. tumefaciens is 2 Mb in size, compared to the 2.8-Mb circular chromosome, while pAtC58 is 0.5 Mb and the Ti plasmid is only 0.2 Mb. Neither pAtC58 nor pTiC58 is essential for viability of Agrobacterium (14); thus, nonessentiality does not explain the greater variability in extreme polar localization seen for the Ti plasmid. In addition, the pSymA megaplasmid of S. meliloti is not predicted to carry essential genes (9), and its localization is very similar to that of pSymB, which does carry essential genes. Their sizes are similar (1.35 and 1.68 Mb, respectively), and both are smaller than the 3.65-Mb chromosome, whose origin has a more strictly polar localization. Thus, rather than correlating with the presence of essential genes, localization patterns of the repABC origins may be determined by replicon size or by differing degrees of interaction between their individual partitioning proteins and host cellular structures.
Dual-labeling experiments show that the replicon origins in A. tumefaciens do not colocalize. Hybridization with a single probe (to the circular chromosome origin) labeled two ways yields an extremely high frequency (97%) of coincident foci, demonstrating that the technique accurately represents colocalization. Thus, the relatively low colocalization frequencies among repABC replicons suggest that they are targeted to nonoverlapping sites. In repABC systems, the RepA and RepB proteins are responsible for partitioning and the RepC protein functions in the initiation of replication (29). While RepA proteins are phylogenetically related to plasmid ATPases such as SopA of plasmid F and ParA of P1, they comprise a separate group. Distinct incompatibility groups exist among the repABC plasmids, and this feature has been exploited in curing plasmids from certain strains (14, 26). Based on studies of compatible plasmids in E. coli, Ho et al. proposed a model wherein they occupy spatially discrete regions of the cell, as opposed to competing for a finite number of overlapping target sites (13). Our data, which suggest only limited spatial overlap of the repABC replication origins with each other inside the cell, support this model and extend it to a different class of replicons and bacteria. While the replicons must all exploit common cellular machinery to replicate, each also carries its own dnaE gene, encoding a homolog of the α subunit of DNA polymerase III (10). DnaE in Staphylococcus aureus and B. subtilis, organisms with a single chromosome which already contain PolC, has been found to be essential for viability and appears to be localized to the replication machinery along with PolC in B. subtilis (7, 15). It is thus possible that specific, separate polymerase complexes exist in the alpha proteobacteria, allowing synchronous replication of different molecules while remaining spatially separated. However, origin movement in live cells and localization of the replication apparatus itself in these bacteria have not yet been assessed.
In E. coli and other gamma proteobacteria, RK2 has been localized to mid- and quarter-cell positions (13), suggesting conservation of host cell-interacting components and subcellular architecture. We have found that, in three alpha proteobacteria, the same localization pattern occurs, making it likely that the bacteria which support replication of a broad-host-range plasmid do so because of conserved structural elements with which they interact. These results are consistent with the function of the IncC/KorB partition system of plasmid RK2 in a variety of gram-negative bacteria including A. tumefaciens (31). It is possible that there is an even wider conservation of DNA segregation machinery, as the Soj/SpoOJ partitioning system of B. subtilis is functional in E. coli (38).
Our findings demonstrate that the cell poles in the alpha proteobacteria are not a default location for replication origins; rather, the chromosome and repABC origins are specifically targeted there in two different bacteria, while the RK2 origin is found at mid- and quarter-cell positions. While the structures with which these DNA molecules interact remain to be identified, the multipartite genome structure (including coexistent chromosomal and repABC origins) is found in multiple members of the alpha subdivision, including Brucella spp. (6, 19, 27). These findings lay the groundwork for further study of replication and segregation in the alpha proteobacteria specifically and the maintenance of complex bacterial genomes in general.
Acknowledgments
This work was supported by grant K08-AI-01510 from the National Institute for Allergy and Infectious Diseases, National Institutes of Health (L.S.K.), NIH grant GM51426 (L.S.), and Defense Advanced Research Projects Agency grant MDA972-00-1-0032 (L.S.).
We are grateful to members of the Shapiro lab, particularly Kathleen Ryan and Patrick Viollier, for their critical reading of the manuscript and helpful discussions.
REFERENCES
- 1.Bartosik, D., J. Baj, E. Piechucka, E. Waker, and M. Wlodarczyk. 2002. Comparative characterization of repABC-type replicons of Paracoccus pantotrophus composite plasmids. Plasmid 48:130-141. [DOI] [PubMed] [Google Scholar]
- 2.Brassinga, A. K., R. Siam, and G. T. Marczynski. 2001. Conserved gene cluster at replication origins of the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii. J. Bacteriol. 183:1824-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brassinga, A. K., R. Siam, W. McSween, H. Winkler, D. Wood, and G. T. Marczynski. 2002. Conserved response regulator CtrA and IHF binding sites in the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J. Bacteriol. 184:5789-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 6.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dervyn, E., C. Suski, R. Daniel, C. Bruand, J. Chapuis, J. Errington, L. Janniere, and S. D. Ehrlich. 2001. Two essential DNA polymerases at the bacterial replication fork. Science 294:1716-1719. [DOI] [PubMed] [Google Scholar]
- 8.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 9.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 10.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, G. S., D. Sitnikov, C. D. Webb, A. Teleman, A. Straight, R. Losick, A. W. Murray, and A. Wright. 1997. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90:1113-1121. [DOI] [PubMed] [Google Scholar]
- 12.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, and O. White. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, T. Q., Z. Zhong, S. Aung, and J. Pogliano. 2002. Compatible bacterial plasmids are targeted to independent cellular locations in Escherichia coli. EMBO J. 21:1864-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hynes, M. F., R. Simon, and A. Puhler. 1985. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid 13:99-105. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, R., C. Kaito, M. Tanabe, K. Kamura, N. Akimitsu, and K. Sekimizu. 2001. Genetic identification of two distinct DNA polymerases, DnaE and PolC, that are essential for chromosomal DNA replication in Staphylococcus aureus. Mol. Genet Genomics 266:564-571. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, R. B., and K. Gerdes. 1999. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 18:4076-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, R. B., and L. Shapiro. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. USA 96:10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, R. B., S. C. Wang, and L. Shapiro. 2002. Dynamic localization of proteins and DNA during a bacterial cell cycle. Nat. Rev. Mol. Cell Biol. 3:167-176. [DOI] [PubMed] [Google Scholar]
- 19.Jumas-Bilak, E., S. Michaux-Charachon, G. Bourg, M. Ramuz, and A. Allardet-Servent. 1998. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J. Bacteriol. 180:2749-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahng, L. S., and L. Shapiro. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 183:3065-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 22.Li, P. L., and S. K. Farrand. 2000. The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J. Bacteriol. 182:179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, C. M. Fraser, and J. Eisen. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niki, H., and S. Hiraga. 1998. Polar localization of the replication origin and terminus in Escherichia coli nucleoids during chromosome partitioning. Genes Dev. 12:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90:951-957. [DOI] [PubMed] [Google Scholar]
- 26.Palmer, K. M., S. L. Turner, and J. P. Young. 2000. Sequence diversity of the plasmid replication gene repC in the Rhizobiaceae. Plasmid 44:209-219. [DOI] [PubMed] [Google Scholar]
- 27.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogliano, J., T. Q. Ho, Z. Zhong, and D. R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez-Romero, M. A., N. Soberon, A. Perez-Oseguera, J. Tellez-Sosa, and M. A. Cevallos. 2000. Structural elements required for replication and incompatibility of the Rhizobium etli symbiotic plasmid. J. Bacteriol. 182:3117-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro, L., H. H. McAdams, and R. Losick. 2002. Generating and exploiting polarity in bacteria. Science 298:1942-1946. [DOI] [PubMed] [Google Scholar]
- 31.Siddique, A., and D. H. Figurski. 2002. The active partition gene incC of IncP plasmids is required for stable maintenance in a broad range of hosts. J. Bacteriol. 184:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 33.Suzuki, K., K. Iwata, and K. Yoshida. 2001. Genome analysis of Agrobacterium tumefaciens: construction of physical maps for linear and circular chromosomal DNAs, determination of copy number ratio and mapping of chromosomal virulence genes. DNA Res. 8:141-152. [DOI] [PubMed] [Google Scholar]
- 34.Tagomori, K., T. Iida, and T. Honda. 2002. Comparison of genome structures of vibrios, bacteria possessing two chromosomes. J. Bacteriol. 184:4351-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 36.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 37.Yamaichi, Y., T. Iida, K. S. Park, K. Yamamoto, and T. Honda. 1999. Physical and genetic map of the genome of Vibrio parahaemolyticus: presence of two chromosomes in Vibrio species. Mol. Microbiol. 31:1513-1521. [DOI] [PubMed] [Google Scholar]
- 38.Yamaichi, Y., and H. Niki. 2000. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:14656-14661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, M. C., and R. Losick. 2001. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J. Bacteriol. 183:5180-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]