Abstract
The targeting of the tumor suppressor PTEN protein to distinct subcellular compartments is a major regulatory mechanism of PTEN function, by controlling its access to substrates and effector proteins. Here, we investigated the molecular basis and functional consequences of PTEN nuclear/cytoplasmic distribution. PTEN accumulated in the nucleus of cells treated with apoptotic stimuli. Nuclear accumulation of PTEN was enhanced by mutations targeting motifs in distinct PTEN domains, and it was dependent on an N-terminal nuclear localization domain. Coexpression of a dominant negative Ran GTPase protein blocked PTEN accumulation in the nucleus, which was also affected by coexpression of importin α proteins. The lipid- and protein-phosphatase activity of PTEN differentially modulated PTEN nuclear accumulation. Furthermore, catalytically active nuclear PTEN enhanced cell apoptotic responses. Our findings indicate that multiple nuclear exclusion motifs and a nuclear localization domain control PTEN nuclear localization by a Ran-dependent mechanism and suggest a proapoptotic role for PTEN in the cell nucleus.
INTRODUCTION
PTEN is a tumor suppressor phosphatase involved in the control of cell growth and cell cycle traverse, apoptosis, cell size, and cell migration (Maehama et al., 2001; Mayo and Donner, 2002; Waite and Eng, 2002; Leslie and Downes, 2004; Parsons, 2004; Sansal and Sellers, 2004). The PTEN gene is mutated or lost in a wide variety of human tumors, including malignant glioblastomas, an aggressive tumor of the CNS in humans (Bonneau and Longy, 2000; Eng, 2003). The major tumor suppressor function of PTEN is mediated by the dephosphorylation of phosphatidylinositol-3,4,5-triphosphate (PIP3; Maehama and Dixon, 1998). Through this lipid phosphatase activity, PTEN counteracts the action of the prosurvival proto-oncogenes, the phosphatidylinositol 3-kinases, and protein kinase B/Akt, which control the function of key downstream effectors of this pathway, including cell cycle regulators (such as p27Kip1, cyclin D1, and CHK1) and transcription factors (such as NF-κB and FKHR; Li and Sun, 1998; Nakamura et al., 2000; Gustin et al., 2001; Weng et al., 2001; Radu et al., 2003; Puc et al., 2005). To control the PIP3 levels at the plasma membrane, PTEN possesses, in addition to its N-terminal phosphatase catalytic domain (residues 14–185), a C-terminal phospholipid-binding C2 domain (residues 186–350), which is critical for optimal binding to membranes and PIP3 dephosphorylation (Lee et al., 1999). In addition, the N-terminus of PTEN contains a phosphatidylinositol-4,5-diphosphate (PIP2) binding motif (residues 6–15), which is also essential for PTEN membrane binding and activity (Maehama et al., 2001; Funamoto et al., 2002; Iijima and Devreotes, 2002; Campbell et al., 2003; McConnachie et al., 2003; Iijima et al., 2004; Walker et al., 2004; Vazquez et al., 2006). On the other hand, PTEN possesses a C-terminal tail (residues 350–403) that plays a major role in the stabilization of the molecule and that is the target of posttranslational modifications, including phosphorylation by the protein kinase CK2 and cleavage by caspase-3 (Georgescu et al., 1999; Vazquez et al., 2000; Torres and Pulido, 2001; Torres et al., 2003). The PTEN C-terminal tail also contains a PDZ-domain binding motif that mediates binding to anchoring and/or regulatory proteins and that also controls PTEN binding to membranes (Wu, X. et al., 2000; Wu, Y. et al., 2000; Das et al., 2003; Valiente et al., 2005). Thus, the recruitment of PTEN to sites of the plasma membrane where signaling takes place is dictated by the combination of electrostatic interactions with plasma membrane lipids and protein–protein interactions with membrane-associated proteins.
PTEN has also been found in the nucleus of some cell lines and tissue cells (Gimm et al., 2000; Lachyankar et al., 2000; Perren et al., 2000; Torres et al., 2001; Ginn-Pease and Eng, 2003). Remarkably, PTEN exerts part of its function in coordination with the nuclear tumor suppressor transcription factor p53. Thus, p53 induces the transcription of the PTEN gene, whereas PTEN stimulates the transcriptional activity of p53 by both catalytically dependent and independent mechanisms that include direct binding of the two tumor suppressor proteins (Stambolic et al., 2001; Mayo et al., 2002; Freeman et al., 2003; Zhou et al., 2003; Tang and Eng, 2006). These findings underscore the complexity of PTEN functions at distinct subcellular compartments and imply that the tumor suppressor and/or proapoptotic functions of PTEN rely, at least in part, in actions executed by PTEN in the cell nucleus. Very recently, Chung and coworkers have reported the existence of several nuclear localization signal (NLS)-like sequences within the PTP and the C2 domains of PTEN, which could be important in regulating the cell cycle and apoptosis (Chung et al., 2005; Chung and Eng, 2005). On the other hand, Liu, F. et al. (2005) have reported that PTEN enters the nucleus by diffusion. Thus, the molecular basis of PTEN nuclear localization remains elusive. In this study, we investigated the molecular basis of the accumulation of PTEN in the cell nucleus, as well as the nuclear PTEN function upon apoptotic stimulation. We provide evidence for a Ran-dependent mechanism that controls PTEN nuclear/ cytoplasmic accumulation based on the existence of multiple nuclear exclusion motifs at distinct PTEN regions and a nuclear localization domain at the PTEN N-terminus. Furthermore, we demonstrate a proapoptotic role for catalytically active nuclear PTEN.
MATERIALS AND METHODS
Cell Culture, Transfections, Plasmids, Antibodies, and Reagents
Human glioblastoma U87MG cells were grown in DMEM containing high glucose supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1 mM l-glutamine, 1 mM sodium pyruvate, 1% nonessential amino acids, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. Human embryonic kidney HEK293, human cervix carcinoma HeLa S3, simian kidney COS-7 cells, mouse fibroblasts 3T3, and rat fibroblast Rat1 cell lines were grown in DMEM containing high glucose supplemented with 10% (5% for COS-7 cells) heat-inactivated FBS, 1 mM l-glutamine, 100 U/ml penicillin, and 0.1 mg/ml streptomycin. The cells were grown at 37°C and 5% CO2. All reagents for cell cultures were from Invitrogen. U87MG cells were transfected with FuGENE 6 (Roche Diagnostics, Indianapolis, IN), following the manufacturer’s instructions, and cells were processed 48 h after transfection. To generate stable HEK293 clones expressing ectopic PTEN, HEK293 cells were transfected by the calcium phosphate method with pIRES PTEN plasmids, and individual clones were isolated by selection in the presence of 2 μg/ml puromycin. The HEK293 clones expressing ectopic PTEN were maintained in the presence of 1 μg/ml puromycin. To induce apoptosis in U87MG cells, cell cultures were incubated in the presence of 200 ng/ml recombinant mouse TNF-α (Sigma, St. Louis, MO) plus 10 μg/ml cycloheximide (Sigma) during the indicated times, or in the presence of 1 μM of doxorubicin (Alexis Biochemicals, San Diego, CA) for 24 h. To induce apoptosis in 3T3, Rat1, and HeLa cell lines, cells were incubated in the presence of 0.5 M sorbitol for 30 min. To induce apoptosis in HEK293 cells, cell cultures were incubated in the presence of 50 μM etoposide (Sigma) for 24 h. PTEN was detected using the anti-PTEN 425A monoclonal antibody (mAb), which recognizes an epitope within the PTEN C2 domain (Andrés-Pons et al., 2005). In Figures 2C, 8C, and 10B, the anti-PTEN 421B mAb (which recognizes an epitope at the C-terminus of PTEN; Andrés-Pons et al., 2005) or a rabbit polyclonal anti-PTEN N-terminal antiserum (obtained by immunization with a peptide containing the PTEN residues 1–15) were used. In Figures 9C and 10B, a rabbit polyclonal anti-PTEN antibody (Upstate Biotechnology, Lake Placid, NY) was used that recognizes the PTEN C-terminal region (residues 239–403). The anti-GAPDH mAb has been previously described (Aniento et al., 1993). The anti-RhoA and anti-PCNA mAbs were from Santa Cruz Biotechnology (Santa Cruz, CA), the polyclonal anti-cleaved-PARP was from Cell Signaling (Beverly, MA), and the anti-Flag mouse mAb was from Sigma. Horseradish peroxidase–conjugated goat anti-rabbit and anti-mouse secondary antibodies were from Oncogene Research Products (Boston, MA) and Promega (Madison, WI), respectively. The fluorescein-conjugated goat anti-mouse and the rhodamine-conjugated goat anti-rabbit antibodies were from Sigma. pRK5 PTEN constructs have been previously described (Torres and Pulido, 2001; Torres et al., 2003). pIRES PTEN and pIRES PTEN QMA were made by subcloning of PCR-obtained PTEN cDNAs from the corresponding pRK5 constructs into pIRES. The PTEN truncation and deletion mutants and the PTEN N-terminal amino acid substitution mutants, were made by PCR oligonucleotide site-directed mutagenesis. pRK5 GST-GFP was made by fusing in frame the coding sequences of GST (from pGEX-5X2) and GFP (from pEGFP-N2) after PCR amplification and subcloning into pRK5. The 1–16 and 1–32 N-terminal residues of PTEN were fused to GST-GFP by PCR and subcloned into pRK5 (pRK5 PTEN 1–16/GST-GFP and pRK5 PTEN 1–32/GST-GFP). pRK5 GST-importin α1 and pRK5 GST-importin α5 (wild-type and Δ1–70 mutations) were made by subcloning from the appropriate pGEX constructs, which were made by subcloning into pGEX of PCR-obtained cDNA fragments from pDNR-LIB importin α1 and pOTB7 importin α5 (Mammalian Gene Collection, IMAGE ID 4105058 and 2822859, respectively). pCDNA3-Flag-Ran and pCDNA3-Flag-RanQ69L were provided by M.-C. Hung (Giri et al., 2005). All mutations and chimeras were confirmed by DNA sequencing.
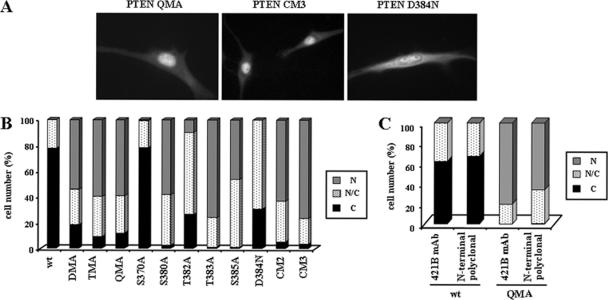
Figure 2.
PTEN nuclear accumulation is facilitated by mutations targeting the 370–385 region of the PTEN C-terminal tail. U87MG cells were transfected with wild-type (wt) PTEN, PTEN mutations targeting the CK2 phosphorylation sites (DMA [S370A/S385A], TMA [S380A/T382A/T383A], QMA [S370A/S380A/T382A/T383A/S385A], S370A, S380A, T382A, T383A, and S385A mutations), or PTEN mutations targeting the caspase-3 cleavage sites (D384N, CM2 [D371N/D375N], and CM3 [D371N/D375N/D384N] mutations), and cells were processed for immunofluorescence as in Figure 1. (A–C) Representative images of the localization of some recombinant PTEN proteins are shown. Quantitation of the subcellular localization of the indicated PTEN proteins, using the anti-PTEN 425A mAb (B), or the anti-PTEN 421B mAb or an anti-PTEN N-terminal polyclonal antibody (C). Data are presented as in Figure 1B.
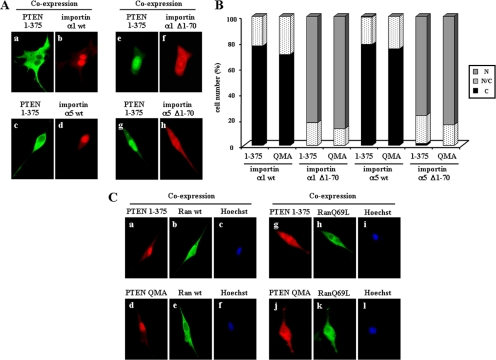
Figure 8.
PTEN nuclear accumulation is affected by importin α proteins and depends on Ran-GTPase activity. (A and B) U87MG cells were transfected with GST-importin α1 or α5 (wild-type [wt] or Δ1–70 mutations) together with the PTEN 1–375 mutant, and cells were processed for double immunofluorescence. PTEN was detected with the anti-PTEN 425A mAb and goat anti-mouse fluorescein-conjugated secondary antibody (green; a, c, e, and g). GST-importin proteins were detected with a rabbit polyclonal anti-GST antibody and goat anti-rabbit rhodamine-conjugated secondary antibody (red; b, d, f, and h). In A, representative images of the localization of importin and PTEN 1–375 proteins are shown. In B, quantitation of the subcellular localization of PTEN 1–375 and PTEN QMA coexpressed with the importin proteins is shown. Data are presented as in Figure 1B. (C) U87MG cells were transfected with Flag-Ran (wild-type [wt] or RanQ69L mutation) together with PTEN 1–375 or QMA mutants, and cells were processed for double immunofluorescence. PTEN was detected with rabbit anti-PTEN N-terminal polyclonal antibody and goat anti-rabbit rhodamine-conjugated secondary antibody (red; a, d, g, and j). Flag-Ran proteins were detected with a mouse anti-Flag mAb and goat anti-mouse fluorescein-conjugated secondary antibody (green; b, e, h, and k). Nuclei were costained with Hoechst 33258 (c, f, i, and l). Representative images of the localization of Ran and PTEN proteins are shown.
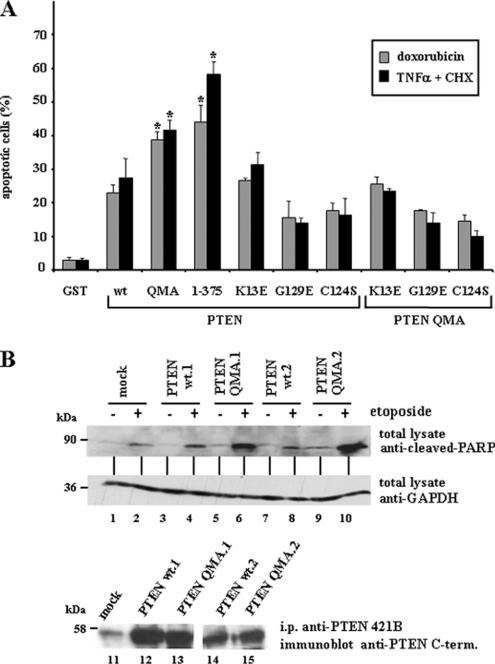
Figure 10.
Nuclear accumulation of PTEN enhances apoptosis. (A) U87MG cells, transfected with GST, wild-type (wt) PTEN, QMA (S370A, S380A, T382A, T383A, and S385A) or 1–375 PTEN mutants, or with K13E or the catalytically inactive PTEN mutants C124S or G129E (alone or in combination with the QMA mutation), were cultured in medium without serum for 24 h, followed by treatment with recombinant TNF-α plus cycloheximide (CHX) for 2 h or with doxorubicin for 24 h, as indicated. Cells were processed for immunofluorescence and stained with the anti-PTEN 425A mAb or with an anti-GST polyclonal antibody, and goat anti-mouse or anti-rabbit fluorescein conjugated secondary antibodies. Nuclei were stained with Hoechst 33258, and the percentage of apoptotic-stained cells (cells with condensed nuclei) is shown. Values represent the mean ± SD of at least two separate experiments. Significant increases with respect to wild-type PTEN are denoted with asterisks (p < 0.05). (B) HEK293 cell clones containing empty pIRES (mock) or expressing wild-type PTEN (clones wt.1 and wt.2) or PTEN QMA (clones QMA.1 and QMA.2) were left untreated (−) or were treated with 50 μM etoposide for 24 h (+). Cell were lysed, and PARP was detected from the total lysates (100 μg) using an anti-cleaved PARP-specific antibody. The amount of GAPDH, detected with an anti-GAPDH mAb, is shown as a control for protein loading. In the bottom panel (lanes 11–15), the expression of recombinant PTEN in the clones is shown. Proteins were immunoprecipitated (i.p.) with the anti-PTEN 421B mAb, followed by immunoblot with a polyclonal anti-PTEN C-terminal antibody.
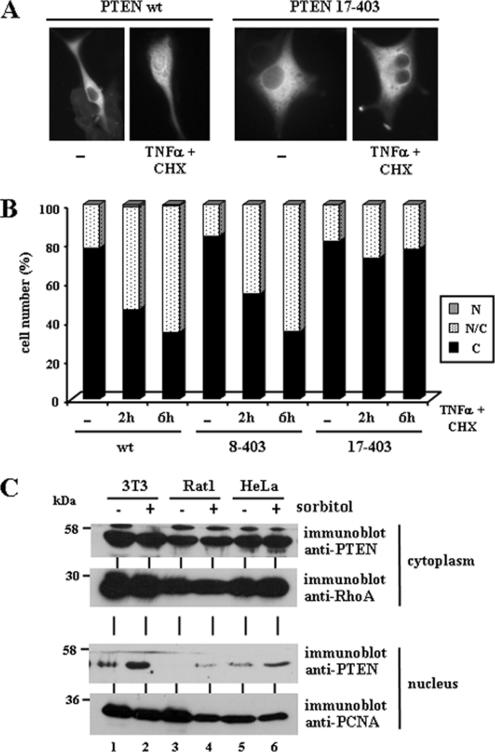
Figure 9.
Nuclear accumulation of PTEN is augmented after treatment with agents that induce apoptosis. (A and B) U87MG cells, transfected with wild-type (wt) PTEN, 8–403 or 17–403 PTEN mutants were left untreated (−) or were treated with recombinant TNF-α plus cycloheximide (CHX) for 2 or 6 h, and then cells were processed for immunofluorescence as in Figure 1. In A, representative images of the localization of wild-type or PTEN 17–403 proteins, in untreated (−) or TNF-α plus CHX (6 h) treated cells, are shown. In B, quantitation of the subcellular localization of the indicated PTEN proteins, is shown. Data are presented as in Figure 1B. (C) 3T3, Rat1, and HeLa cells were left untreated (−) or were treated with 0.5 M sorbitol for 30 min (+). After lysis, cells were fractionated in cytoplasmic (top panels) and nuclear (bottom panels) fractions, and equal amounts of proteins were analyzed by immunoblot for endogenous PTEN, RhoA or PCNA (used as cytoplasmic and nuclear markers, respectively), using specific antibodies.
Subcellular Fractionation and Immunoblot
For nucleus/cytoplasm fractionation, cells were harvested with EDTA-trypsin, centrifuged (1500 rpm, 6 min at 4°C), and washed twice with ice-cold phosphate-buffered saline (PBS). The cell pellet was lysed with buffer A (10 mM Tris-Cl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40) and vortexed for 10 s. The cells lysates were incubated 5 min on ice and centrifuged 6 min, 1500 rpm, at 4°C. The supernatant was considered as the cytoplasmic fraction. The cellular pellets were resuspended with buffer A and treated again as above to eliminate the cytoplasmic remainders. Finally, the cellular pellet was extracted with SDS-PAGE sample buffer during 5 min at 95°C, followed by centrifugation (14,000 rpm, 5 min at 20°C), and the supernatant was considered as the nuclear fraction. For detection of cleaved-PARP, HEK293 cells were lysed in buffer B (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 1 mg/ml aprotinin, 2 mM Na3VO4, 20 mM Na4P2O7). The cell lysates were centrifuged (14,000 rpm, 10 min at 4°C), and the supernatants were processed for immunoblot. The proteins were resolved by SDS-PAGE and analyzed by immunoblot as described (Torres et al., 2003).
Immunofluorescence
Cells were cultured on poly-d-lysine–coated glass coverslips, transfected, and subjected to treatments as indicated. The cells were washed twice with PBS and then fixed and permeabilized in ice-cold methanol for 5 min at −20°C. The coverslips were washed three times with PBS containing 3% bovine serum albumin (PBS-BSA) and incubated 1 h at 37°C with ascites fluid of mouse monoclonal anti-PTEN 425A, or with mouse monoclonal anti-PTEN 421B (for Figure 2C) or rabbit polyclonal anti-PTEN N-terminal (for Figures 2C and 8C; all diluted 1:200 in PBS-BSA). The coverslips were washed three times with PBS and incubated for 1 h at 20°C with fluorescein-conjugated anti-mouse antibody and/or with rhodamine-conjugated anti-rabbit antibody, diluted 1:200 in PBS-BSA, followed by three washes with PBS. The coverslips were mounted onto glass slides using Fa Mounting-fluid (Becton Dickinson, Lincoln Park, NJ) and examined using a Zeiss fluorescence microscope (Thornwood, NY). For quantitation of PTEN subcellular distribution, at least 100 positive cells were scored for each experiment. Cells were rated as showing nuclear staining (N), cytoplasmic staining (C), or staining within both the nucleus and the cytoplasm (N/C). Nuclei were identified by Hoescht 33258 (Molecular Probes, Eugene, OR) staining. All pictures were taken under a 400× magnification.
Apoptosis Assays
Quantitation of apoptosis in transiently transfected U87MG cells was made by nuclear condensation visualization. Cells were processed for immunofluorescence and PTEN- or GST-expressing cells (at least 100 positive cells) were monitored for nuclear condensation by Hoechst 33258 staining. Apoptosis assays in HEK293 cells stably expressing ectopic PTEN were made by immunoblot detection of cleaved-PARP, a proteolytic product of the effector caspase-3, using anti-cleaved-PARP antibodies.
RESULTS
PTEN Nuclear Accumulation Is Facilitated by Mutations at Multiple Nuclear Exclusion Motifs and Is Conditioned by PTEN Catalytic Activity
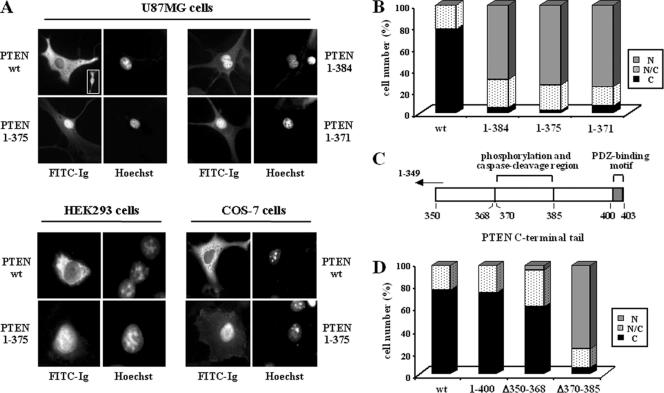
To study in detail the nuclear/cytoplasmic distribution of PTEN, immunofluorescence studies were performed using U87MG cells (PTEN-null) ectopically expressing wild-type or mutant PTEN proteins. First, mutations at the putative regulatory PTEN C-terminal tail were tested. Wild type, full-length PTEN was mostly located in the cytoplasm of the cells, although a faint nuclear staining was also consistently observed in some cells (Figure 1, A and B). However, a prominent nuclear accumulation of PTEN C-terminal truncations, including PTEN 1–384, PTEN 1–375, and PTEN 1–371 (that mimic the PTEN caspase-3 cleavage products; Torres et al., 2003) was observed (Figure 1, A and B). Similar results were obtained using HEK293 or COS-7 cells (Figure 1A; and unpublished data). On the other hand, the C-terminal truncation PTEN 1–400, which lacks the PDZ-domain binding motif, mostly displayed cytoplasmic localization (Figure 1, C and D). Remarkably, the C-terminal deletion PTEN Δ370–385 (see scheme in Figure 1C) accumulated in the nucleus, whereas the deletion PTEN Δ350–368 was mostly found in the cytoplasm (Figure 1D). These results indicate the existence of regulatory determinants of PTEN nuclear distribution in the region spanning the residues 370–385. Interestingly, this region contains both the sites of phosphorylation by CK2 (Ser370, Ser380, Thr382, Thr383, and Ser385) and of cleavage by caspase-3 (Asp371, Asp375, and Asp384; Torres and Pulido, 2001; Torres et al., 2003), suggesting the possibility that phosphorylation and/or caspase-3 cleavage of PTEN might determine PTEN nuclear accumulation. To test this hypothesis, immunofluorescence experiments were done using PTEN point mutations defective in phosphorylation and/or caspase-3 cleavage. PTEN mutations that affected combinations of the PTEN CK2-phosphorylated residues, including PTEN DMA (S370A/S385A), TMA (S380A/T382A/T383A), and QMA (S370A/S385A/S380A/T382A/T383A) mutations, equally favored PTEN nuclear accumulation (Figure 2, A and B). On the other hand, single amino acid substitutions targeting the individual phosphorylation sites, differentially affected PTEN nuclear localization: mutations S380A, T383A, and S385A, showed a strong nuclear accumulation; distribution of mutation T382A was shifted toward the nucleus, although few cells showed intense PTEN nuclear accumulation and mutation S370A was localized mostly in the cytoplasm (Figure 2B). Thus, mutations that affect PTEN phosphorylation also affect the nuclear/cytoplasmic distribution of PTEN. Next, the PTEN nuclear localization of mutations that abrogate the caspase-3 cleavage at the PTEN C-terminal tail were investigated. As shown in Figure 2, A and B, the localization of the PTEN mutations D384N, CM2 (D371N/D375N), and CM3 (D371N/D375N/D384N) was also nuclear; therefore, cleavage by caspase-3 is not necessary for PTEN nuclear accumulation. The observed PTEN nuclear accumulation was not due to a particular staining obtained with the anti-PTEN mAb used, because identical results were obtained using two other monoclonal and polyclonal anti-PTEN antibodies (Figure 2C). Therefore, distinct mutations in the 370–385 C-terminal region favor the nuclear accumulation of PTEN.
Figure 1.
Deletions of the C-terminal tail of PTEN favor its nuclear localization. Cells were transfected with wild-type (wt) PTEN or the C-terminal PTEN truncation (1–400, 1–384, 1–375, and 1–371) or deletion (Δ350–368 and Δ370–385) mutants, as indicated. Cells were processed for immunofluorescence using the anti-PTEN 425A mAb and goat anti-mouse fluorescein-conjugated antibody (FITC-immunoglobulin). Nuclei were costained with Hoechst 33258. (A) In the top panels, representative images of the localization of each PTEN protein in U87MG cells are shown: PTEN wt, cytoplasmic (C); PTEN 1–384, 1–375, and 1–371, nuclear (N). The inset in the PTEN wt panel illustrates a cell expressing PTEN wt that localizes in both the nucleus and the cytoplasm (N/C). As expected, untransfected U87MG cells did not give positive signal for PTEN staining (unpublished data). In the bottom panels, the distribution of wt or 1–375 PTEN in HEK293 or COS-7 cells is shown. The staining corresponds to cells overexpressing recombinant PTEN. (B) Quantitation of the subcellular localization of the PTEN proteins shown in the top panels in A. Localization was classified in three different categories: N, nuclear; C, cytoplasmic; and N/C, nuclear/cytoplasmic, as illustrated in A. Data are presented as the percentage of cells in each category, and values represent the mean of at least three different experiments. SD was always <10%. (C) Schematic of the PTEN C-terminal tail (residues 350–403) illustrating the regions that contain the phosphorylation and caspase cleavage sites (residues 370–385), and the PDZ-domain binding motif (residues 401–403). (D) Quantitation of the subcellular localization of wild-type PTEN versus the 1–400, Δ350–368, and Δ370–385 PTEN mutants. Data are presented as in B.
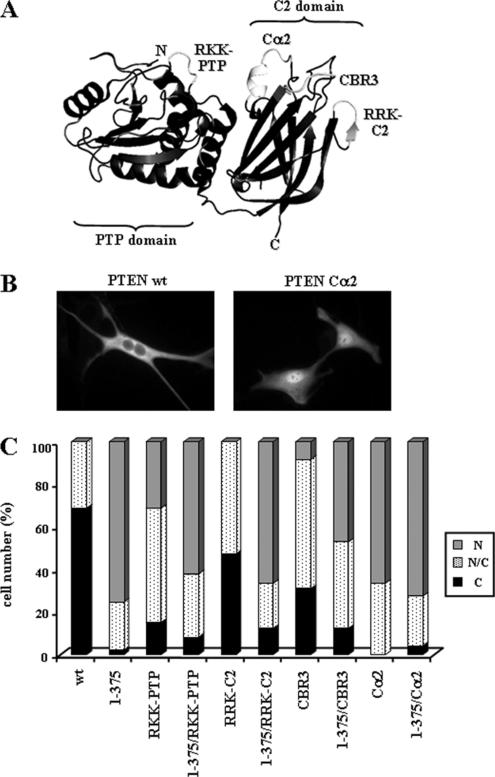
Several positively charged motifs located at distinct surface-exposed PTEN regions (residues 159–164 of the PTP domain [RKK-PTP region]; and residues 233–237 [RRK-C2 region], 263–269 [CBR3 region], and 327–335 [Cα2 region] at the C2 domain; see Figure 3A) regulate PTEN activities, including PTEN subcellular localization and function (Lee et al., 1999; Das et al., 2003; Orchiston et al., 2004; Chung et al., 2005; Chung and Eng, 2005). To test the role of these motifs in nuclear localization of PTEN, compound mutations were generated (R161A/K163A/K164A [RKK-PTP]; R233A/R234A/K237A [RRK-C2]; K263A/M264A/L265G/K266A/K267A/K269A [CBR3]; and K327A/N329G/K330A/K332A/ R335A [Cα2]), and assessed for effects on subcellular localization in U87MG cells (Figure 3, B and C). None of the mutations prevented the nuclear accumulation of PTEN 1–375, excluding their roles as putative nuclear localization signals. Furthermore, when present alone, these mutations had distinct effect on the nuclear accumulation of PTEN. The highest degree of nuclear localization was achieved by the mutation Cα2, followed by the mutations RKK-PTP and CBR3, whereas the mutation RRK-C2 displayed a moderate level of nuclear accumulation (Figure 3, B and C). Together, these results indicate the existence of multiple nuclear exclusion motifs on PTEN, including regions of the C-terminal tail, the PTP and the C2 domains.
Figure 3.
PTEN possesses multiple nuclear exclusion motifs. (A) Tertiary structure of PTEN (Lee et al., 1999), showing the surface-exposed motifs (light gray color) at the PTP and C2 domains; mutating in these motifs trigger PTEN nuclear localization. The mutations generated in these motifs that were tested for effects on PTEN localization are indicated; RKK-PTP (R161A/K163A/K164A); RRK-C2 (R233A/R234A/K237A); CBR3 (K263A/M264A/L265G/K266A/K267A/K269A); and Cα2 (K327A/N329G/K330A/K332A/R335A). (B and C) U87MG cells were transfected with wild-type (wt) PTEN or with PTEN mutations targeting the PTP and the C2 domains regions, as indicated, and cells were processed for immunofluorescence as in Figure 1. In B, representative images of the localization of the indicated PTEN proteins are shown. In C, quantitation of the subcellular localization of the indicated PTEN proteins is shown. Data are presented as in Figure 1B.
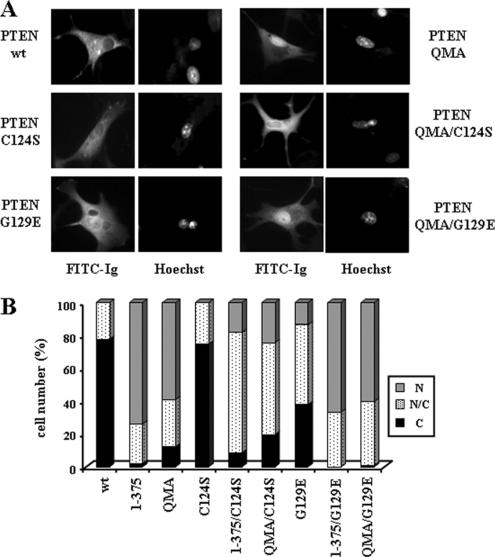
Next, the nuclear accumulation of the catalytically inactive PTEN C124S (protein- and lipid-phosphatase inactive) or G129E (lipid-phosphatase inactive) mutations (Myers et al., 1998) was assessed in U87MG cells. The nuclear/cytoplasmic distribution of the PTEN C124S mutation was similar to wild-type PTEN; however, on a C-terminal mutation background that caused the accumulation of PTEN in the nucleus, the nuclear accumulation of the C124S mutation was less pronounced (Figure 4, A and B). Interestingly, a different distribution pattern was observed with the G129E mutation. The nuclear localization of PTEN G129E was more pronounced than wild-type PTEN, and there where no effects of this mutation on a C-terminal mutation background (Figure 4, A and B). Therefore, the C124S mutation inhibits, whereas the G129E mutation augments, nuclear accumulation. Together, these results indicate that subcellular distribution of PTEN on U87MG cells is differentially modulated by its protein- and lipid-phosphatase activities.
Figure 4.
The nuclear accumulation of PTEN is affected in mutations that compromise catalytic activities. U87MG cells were transfected with wild-type (wt) PTEN, QMA (S370A/S380A/T382A/T383A/S385A), 1–375, or the catalytically inactive mutants (C124S and G129E) alone or in combination with the QMA (QMA/C124S and QMA/G129E) or the 1–375 (1–375/C124S and 1–375/G129E) mutations, and cells were processed for immunofluorescence as in Figure 1. (A) Representative images of the localization of the indicated PTEN proteins are shown (FITC-immunoglobulin). Nuclei were costained with Hoechst 33258. (B) Quantitation of the subcellular localization of the indicated PTEN proteins. Data are presented as in Figure 1B.
Nuclear Localization of PTEN Requires a Nuclear Localization Domain at the N-Terminus and the Activity of Ran GTPase
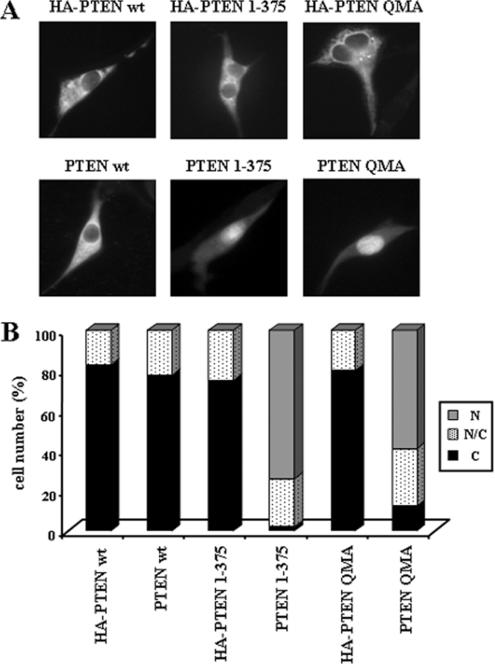
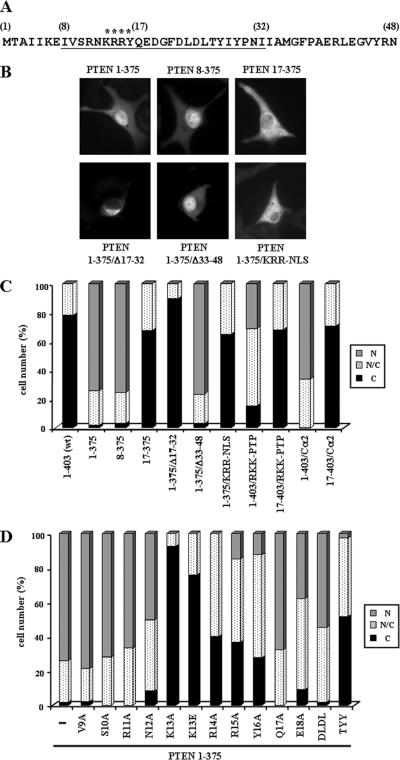
During the course of our studies, we noticed that C-terminal mutated PTEN recombinant proteins, tagged at their N-terminus with the hemagglutinin (HA) epitope, did not accumulate in the nucleus of U87MG cells, whereas the nontagged PTEN mutations showed nuclear localization (Figure 5). Intriguingly, N-terminal tagging with GST protein also hampered nuclear accumulation of PTEN mutations, whereas N-terminal tagging with the Flag epitope did not affect such accumulation (unpublished data). These findings suggested that the N-terminal region of PTEN was important for its entry in the nucleus. To evaluate this possibility, N-terminal truncations or deletions of PTEN were created covering this region (see Figure 6A), in PTEN wild-type or mutant backgrounds, and tested for subcellular localization by immunofluorescence analysis. None of the N-terminal mutations tested, with the exception of the Δ17–32 mutation (see below), affected the subcellular localization of PTEN (unpublished data). The removal of the first seven residues on a PTEN 1–375 background (PTEN 8–375 mutation) did not affect PTEN nuclear accumulation (Figure 6, B and C). On the other hand, removal of the first 16 residues of PTEN prevented the nuclear accumulation of PTEN 1–375, PTEN RKK-PTP, and PTEN Cα2 (PTEN 17–375, PTEN 17–403/RKK-PTP, and PTEN 17–403/Cα2 mutations, respectively; Figure 6, B and C). The deletion of residues 17–32 (PTEN Δ17–32) also impaired the nuclear accumulation of PTEN 1–375, and both PTEN Δ17–32 and PTEN 1–375/Δ17–32 showed, in most of the cells, a perinuclear localization with punctate staining in the cytoplasm (Figure 6B and unpublished data). Finally, the deletion of residues 33–48 did not affect the nuclear accumulation of PTEN 1–375 (Figure 6, B and C). These results pointed to residues 8–32 as being important for PTEN nuclear accumulation. In fact, mutation of positively charged residues within this amino acid region (K13A/R14A/R15A mutation; [KRR-NLS]; see Figure 6A), on the appropriate mutant background (1–375/KRR-NLS mutation), prevented PTEN nuclear accumulation (Figure 6, B and C). Alanine mutational scanning of the region 9–17 revealed that residues Lys13, Arg14, Arg15, and Tyr16 (mutations K13A, R14A, R15A, and Y16A) were essential for nuclear accumulation of PTEN 1–375 (Figure 6D). Interestingly, the tumor-associated mutation K13E (Duerr et al., 1998), on the background 1–375, was also excluded from the nucleus (Figure 6D). Additional mutational analysis was performed in the region 17–32. As shown, mutation of residues Thr26, Tyr27, and Tyr29 (mutation T26A/Y27A/Y29A; [TYY]) impaired the nuclear accumulation of PTEN 1–375, whereas mutation of residues Asp22 to Leu25 (D22A/L23A/D24A/L25A; [DLDL]) did not (Figure 6D). Thus, the N-terminus of PTEN is required for PTEN nuclear localization.
Figure 5.
PTEN tagged at its N-terminus with the HA epitope does not accumulate in the nucleus. U87MG cells were transfected with wild-type (wt) PTEN, QMA, or 1–375 PTEN mutants, untagged or tagged at their N-termini with the HA epitope (HA-PTEN), and the cells were assessed for PTEN localization as in Figure 1. (A) Representative images of wild-type and mutant PTEN proteins are shown. (B) Quantitation of the subcellular localization of the PTEN proteins shown in A. Data are presented as in Figure 1B.
Figure 6.
Nuclear accumulation of PTEN depends on its N-terminal domain. (A) Amino acid sequence of the N-terminal region of PTEN. Numbers in brackets correspond to PTEN amino acid numbering. The region required for PTEN nuclear accumulation is underlined. Asterisks indicate residues essential for PTEN nuclear accumulation. (B–D) U87MG cells were transfected with wild-type (wt) PTEN or with PTEN mutations, as indicated, and cells were processed for immunofluorescence as in Figure 1. The compound mutations analyzed include KRR-NLS (K13A/R14A/R15A), DLDL (D22A/L23A/D24A/L25A), and TYY (T26A/Y27A/Y29A). RKK-PTP and Cα2 mutations are as in Figure 3. In B, representative images of the localization of some PTEN proteins are shown. (C and D) Quantitation of the subcellular localization of the indicated PTEN proteins. Data are presented as in Figure 1B.
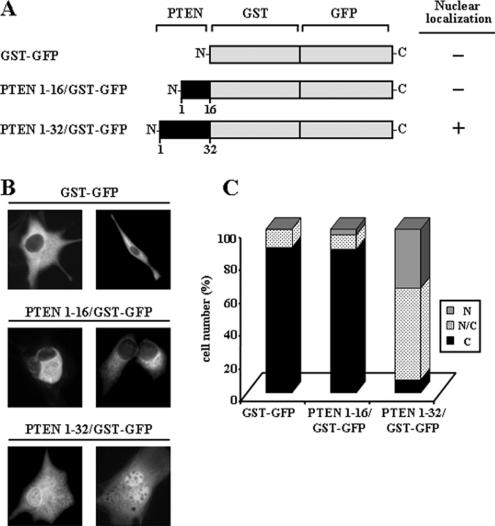
The above results suggested that the N-terminal region of PTEN could contain a functional nuclear localization domain. To test this, distinct portions of PTEN N-terminus were fused to the N-terminus of a GST-GFP fusion protein (see scheme in Figure 7A), and the subcellular localization of the resulting chimeric proteins was analyzed as above. The GST-GFP fusion protein displayed cytoplasmic localization and was excluded from the nucleus. The chimera containing the first 16 residues of PTEN (PTEN 1–16/GST-GFP) was also excluded from the nucleus, although abundant perinuclear localization was observed. Noticeably, the chimera containing the first 32 residues of PTEN (PTEN 1–32/GST-GFP) showed intense nuclear accumulation, and most of the cells expressing this protein displayed staining in both the cytoplasm and the nucleus (Figure 7, B and C). Together, these results indicate that the PTEN N-terminus harbors a functional nuclear localization domain, which is required for nuclear accumulation of PTEN.
Figure 7.
The N-terminus of PTEN contains a functional nuclear localization domain. (A) Schematic representation of the GST-GFP and PTEN/GST-GFP chimeric fusion proteins used. The black boxes denote the N-terminal region of PTEN. As a summary of the results shown in B and C, the nuclear localization of the chimeras is indicated at the right. (B and C) U87MG cells were transfected with GST-GFP or with GST-GFP containing the first 16 or 32 residues of PTEN (PTEN 1–16/GST-GFP, or PTEN 1–32/GST-GFP), as indicated, and protein localization was determined by immunofluorescence as in Figure 1. In B, representative images of the distribution of the chimeric fusion proteins are shown. In C, the quantitation of the subcellular localization of the chimeras is shown. Data are presented as in Figure 1B.
Next, we investigated whether PTEN nuclear accumulation could be affected by nuclear importin α proteins, which mediate classical nuclear transport of NLS-containing proteins in association with importin β transporters (Görlich and Kutay, 1999; Xu and Massagué, 2004). Wild-type or mutant forms of importin α1 and importin α5 were fused at their N-terminus to GST to facilitate their detection, and their subcellular localization in U87MG cells was tested. Both wild-type importin α1 and importin α5 were accumulated in the cell nucleus, whereas importin α1 Δ1–70 and importin α5 Δ1–70, which lack the binding site for the importin β transporters, were present both in the cytoplasm and in the nucleus (Figure 8A, b, d, f, and h). This distribution pattern was independent of the presence of recombinant PTEN (unpublished data). Remarkably, when PTEN nuclear mutants were coexpressed with nuclear, wild-type importin α1 or importin α5, PTEN did not accumulate in the nucleus and remained cytoplasmic (Figure 8A, a and c, and B). On the other hand, the accumulation of PTEN nuclear mutants was not affected when they were coexpressed with cytoplasmic importin α1 Δ1–70 or importin α5 Δ1–70 (Figure 8A, e and g, and B). Finally, the cytoplasmic localization of wild-type PTEN was unaffected by the coexpression with wild-type or the mutated importin α1 or importin α5 (unpublished data). These results indicate that PTEN entry into the nucleus is not mediated by direct binding to importin α1 or importin α5, but rather by an importin-related nuclear transport mechanism.
The binding and dissociation of the small GTPase Ran to importin β proteins is required for importin-mediated nuclear transport (Görlich and Kutay, 1999; Kuersten et al., 2001; Ström and Weis, 2001). To test the involvement of Ran in the nuclear accumulation of PTEN mutations, coexpression experiments were performed using the GTPase-deficient, dominant negative, RanQ69L mutation (Nachury and Weis, 1999). Wild-type Ran was distributed in both the nucleus and the cytoplasm, whereas RanQ69L displayed a diffuse cytoplasmic localization and was also at the nuclear membrane (Figure 8C, b, e, h, and k). Notably, in the presence of RanQ69L, PTEN 1–375 or PTEN QMA did not accumulate in the nucleus, whereas wild-type Ran did not affect the nuclear accumulation of PTEN (Figure 8C, a, d, g, and j). These results demonstrate that PTEN nuclear accumulation is dependent on the activity of Ran GTPase.
Nuclear PTEN Enhances Apoptosis
Treatment of U87MG cells with the proapoptotic cytokine TNF-α increases PTEN cleavage by caspase-3, that renders PTEN products lacking the PTEN C-terminal region (Torres et al., 2003). Thus, we tested the effects of TNF-α on localization of PTEN in U87MG cells. As predicted, PTEN accumulated in the nucleus following TNF-α treatment (Figure 9, A and B). The nuclear accumulation of PTEN upon TNF-α stimulation was not affected on the PTEN 8–403 N-terminal truncation, but it was prevented on the PTEN 17–403 truncation (Figure 9B), suggesting that the PTEN N-terminus controls PTEN nuclear/cytoplasmic transport during apoptosis. We also tested the nuclear accumulation of endogenous PTEN from 3T3, Rat1, and HeLa cell lines treated with the apoptotic stimulus sorbitol. As shown, the levels of nuclear PTEN augmented in all these cell lines after sorbitol treatment (Figure 9C). Next, we assessed the effect of PTEN nuclear accumulation on the response of cells to apoptotic stimuli. U87MG cells were transiently transfected with cytoplasmic or nuclear PTEN and treated with TNF-α or doxorubicin, followed by immunofluorescence and quantitation of nuclear condensation (as a measurement of apoptosis; Figure 10A). Remarkably, the percentage of cells with condensed nuclei under these conditions was higher when nuclear PTEN (PTEN QMA and PTEN 1–375) was overexpressed, compared with cytoplasmic PTEN (PTEN wt). Furthermore, these effects were dependent on the PTEN catalytic activity, because they were not observed after overexpression of the PTEN C124S or G129E mutants (Figure 10A). Also, overexpression of the PTEN mutant K13E, which prevents nuclear accumulation of PTEN 1–375 or QMA (Figure 6D, and unpublished data), did not augment apoptosis (Figure 10A). The apoptotic response of HEK293 cells stably expressing cytoplasmic or nuclear PTEN was also analyzed by immunoblot analyses of cleaved-PARP, a hydrolysis product of caspase-3. Stable expression of cytoplasmic wild-type PTEN slightly increased the amount of cleaved-PARP in HEK293 cells after etoposide treatment (Figure 10B). Noticeably, cleaved-PARP was more prominent in etoposide-treated cell clones expressing the nuclear PTEN QMA mutant than those expressing cytoplasmic wild-type PTEN (Figure 10B, lanes 5–6 and 9–10 vs. lanes 3–4 and 7–8). Together, our results indicate that PTEN nuclear accumulation is favored upon apoptotic stimulation and that catalytically active nuclear PTEN enhances apoptosis.
DISCUSSION
The subcellular distribution of PTEN between the cytoplasm and the nucleus has been documented in several cell types and tissues; however, the molecular basis for the cytoplasmic or nuclear localization of PTEN has remained elusive. In the present report, we identify specific amino acid regions in the PTEN PTP, C2, and C-terminal domains that behave as nuclear exclusion motifs, because they are required to keep PTEN outside of the nucleus. In addition, we demonstrate that the PTEN N-terminus contains a nuclear localization domain that is required for PTEN nuclear localization by a Ran-dependent mechanism. Remarkably, the N-terminus of PTEN contains a functional PIP2-binding motif (residues 6–15) that is involved in targeting of PTEN to the plasma membrane and/or in regulation of PTEN catalytic activity (Funamoto et al., 2002; Iijima and Devreotes, 2002; Campbell et al., 2003; McConnachie et al., 2003; Iijima et al., 2004; Walker et al., 2004). Our findings demonstrate that this region is also necessary for PTEN nuclear localization. First, N-terminal tagging of PTEN with an HA epitope, or with the GST-protein, blocked its nuclear accumulation in U87MG cells, suggesting they mask a functional N-terminal sequence required for PTEN nuclear localization. Second, a stretch of positively charged residues, which could be reminiscent of a NLS-like sequence, is present at the N-terminus of PTEN (see Figure 6A). Third, PTEN N-terminal truncations and deletions, as well as point mutations at specific N-terminal residues, abrogate PTEN nuclear accumulation in nuclear mutants of PTEN, or after TNF-α treatment. Fourth, the first N-terminal 32 residues of PTEN are sufficient to target a GST-GFP cytoplasmic protein to the nucleus. We hypothesize that both a PIP2-binding motif and a nuclear localization motif overlap within the N-terminal region of PTEN. An exciting possibility is that the precise functionality of one or the other of these targeting motifs may target the protein to a particular subcellular compartment and exclude it from the other. Using chicken PK fused to the N-terminal portion of PTEN, Liu, F. et al. (2005) have found that the N-terminus of PTEN lacks intrinsic NLS activity in HeLa cells. However, our findings demonstrate that PTEN residues 1–32 target GST-GFP to the nucleus in U87MG cells. These observations suggest that nuclear localization activity of PTEN N-terminus differs from classical NLS motifs and may be dependent on the molecular and cellular context. In fact, residues 1–16, which contain key residues for PTEN nuclear accumulation, were not sufficient to target GST-GFP to the nucleus. On the other hand, Chung et al. (2005) have reported the existence of several NLS-like sequences within the PTP and C2 domains of PTEN that overlap with our RKK-PTP, RRK-C2, and CBR3 mutations (Figure 3). The results obtained with our mutants show an increase in PTEN nuclear accumulation, indicating that these sequences may behave as nuclear exclusion motifs in the wild-type protein. The discrepancy between these studies could be explained by the mutations analyzed or by the distinct cell systems and conditions of PTEN nuclear accumulation tested, yet they all underscore the importance of these motifs in the control of PTEN nuclear localization.
The PTEN C-terminal tail (residues 350–403) is required for PTEN binding to regulatory proteins, including plasma membrane proteins and PDZ-domain scaffolding proteins (Wu, X. et al., 2000; Wu, Y. et al., 2000; Tolkacheva et al., 2001; Vazquez et al., 2001; Mahimainathan and Choudhury, 2004; Sumitomo et al., 2004; Valiente et al., 2005), and this region has also been shown to interfere with membrane binding and with PTEN-mediated regulation of cell migration (Das et al., 2003; Raftopoulou et al., 2004). We and others have reported the phosphorylation of the region 370–385 of the PTEN C-terminal tail (Torres and Pulido, 2001; Birle et al., 2002; Miller et al., 2002). The results here described, showing PTEN nuclear accumulation in mutants targeting the PTEN 370–385 region, outline the critical regulatory importance of this region on distinct PTEN functions, including those exerted in the nucleus (see below). Moreover, PTEN is cleaved by caspase-3 at residues in this region, rendering PTEN products that lack some phosphorylation sites and the PDZ-domain binding motif (Torres et al., 2003). Our findings indicate that the cleavage of PTEN by caspase-3 is sufficient, but not necessary, for PTEN nuclear accumulation, because PTEN mutations that either mimic or impair caspase-3 cleavage displayed nuclear localization. Our results also suggest that alterations in PTEN phosphorylation may be sufficient to trigger PTEN nuclear accumulation. However, we have not found a clear correlation between PTEN phosphorylation by CK2 and PTEN nuclear accumulation (this report; and unpublished data), suggesting that other factors may also regulate PTEN nuclear localization in U87MG cells. Phosphorylation of the C-terminal tail of PTEN by other protein kinases, such as MAST kinases, LKB1, or GSK3β (Al-Khouri et al., 2005; Mehenni et al., 2005; Valiente et al., 2005), could modulate PTEN subcellular localization. The possibility also exists that cytoplasmic or nuclear anchoring or transport proteins may regulate PTEN localization (Yu et al., 2002; Freeman et al., 2003; Zhou et al., 2003; Chung et al., 2005; Okumura et al., 2005; see also below). Treatment of cells with the nuclear export inhibitor leptomycin B did not affect the subcellular localization of PTEN in our experiments, suggesting that PTEN is not subjected to active CRM1-dependent nuclear export (unpublished data). Our overexpression experiments using wild-type and mutant importin α proteins suggest that PTEN entry into the nucleus could be mediated by proteins that can be sequestered upon binding to the N-terminal region of importin α1 and importin α5. Also, the Ran dependence of the PTEN nuclear accumulation suggests an active role for importin β transporters in this process. Whether an importin β and/or a PTEN-specific importin α are directly involved in PTEN nuclear transport deserves further study.
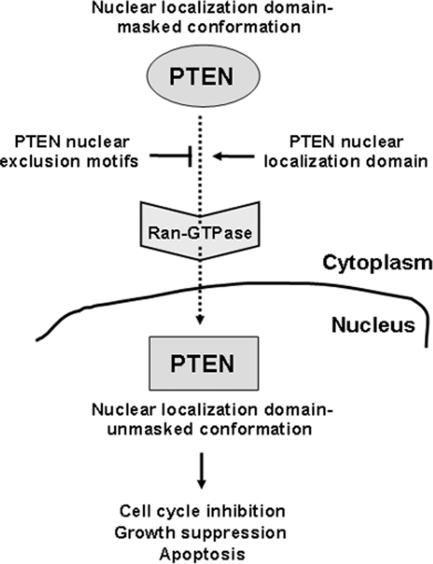
Remarkably, both the PTEN N-terminus and the PTEN C-terminal tail are unstructured regions within the PTEN molecule (Lee et al., 1999). Such regions might be solvent exposed and are likely to play a dynamic role in the PTEN intramolecular or intermolecular interactions that control PTEN subcellular distribution and access to substrates. In this regard, it has been found that phosphoisoforms of PTEN exist in different conformations (Vazquez et al., 2001). In addition, our findings that mutations of charged residues at surface-exposed regions within the PTP and C2 domains favor nuclear accumulation of PTEN, suggest that a large surface on the PTEN protein dictates its subcellular localization. Our results demonstrate a negative contribution of surface-exposed PTEN nuclear exclusion motifs and a positive contribution of the PTEN N-terminus, for PTEN nuclear localization. We thus hypothesize a nuclear localization domain-masked conformation (located in the cytoplasm) and a nuclear localization domain-unmasked conformation (located in the nucleus) for PTEN. The balance between the action of the PTEN nuclear exclusion motifs and the action of the PTEN nuclear localization domain (see scheme in Figure 11), could determine, in a Ran-dependent manner, the nuclear/cytoplasmic distribution of PTEN.
Figure 11.
The regulation of PTEN localization. The model shown is based on results reported here and on the findings of others (Vazquez et al., 2001; Ginn-Pease and Eng, 2003; Liu, F. et al., 2005; Chung and Eng, 2005). In this model, changes in the conformation of PTEN following receipt of apoptotic or cell growth inhibitory signals would unmask the N-terminal nuclear localization domain and trigger PTEN nuclear entry by a Ran-dependent mechanism.
The precise functional role of PTEN in the cell nucleus remains unclear, although a body of evidence favors the hypothesis that PTEN exerts antiproliferative actions within the nucleus, via phosphatase-dependent and -independent mechanisms. For example, in tumors an inverse correlation has been observed between PTEN nuclear staining and the transformed phenotype, suggesting a tumor-suppressor function for PTEN in the nucleus (Gimm et al., 2000; Perren et al., 2000; Whiteman et al., 2002). Also, in some cell lines expressing endogenous PTEN, its nuclear expression has been associated with G0-G1 cell cycle phase arrest (Ginn-Pease and Eng, 2003) or with cell differentiation (Lachyankar et al., 2000), and it has been found that nuclear PTEN is required for cell cycle arrest in MCF-7 cells (Chung and Eng, 2005). Also, it has been reported that nuclear targeted-PTEN leads to growth suppression of U251MG cells (Liu, J. L. et al., 2005). Our results show an increase in PTEN nuclear localization upon apoptotic stimulation, suggesting a relation between nuclear PTEN and apoptosis. Furthermore, we have found that catalytically active nuclear PTEN enhances the apoptotic responses of U87MG and HEK293 cells to distinct stimuli. The scheme shown in Figure 11 summarizes the proposed cellular functions of PTEN in the cell nucleus.
A nuclear PTEN/PI3K/Akt pathway that controls apoptosis has been described in some cell lines, and the existence of PIP3 phosphatase activity on nuclear PTEN has been reported (Deleris et al., 2003; Ahn et al., 2004; Ye and Snyder, 2004; Deleris et al., 2006). In addition, putative nuclear protein substrates of PTEN could account for the proapoptotic or growth suppressive effect of PTEN in the nucleus, and the possibility exists of a differential regulation of PTEN catalysis in the nucleus and in the cytoplasm. Interestingly, we have found that the protein- and lipid-phosphatase activities of PTEN affect its localization, suggesting a putative regulatory mechanism of PTEN nuclear/cytoplasmic distribution dictated by its dual enzymatic activity. On the other hand, recent reports have demonstrated a tumor-suppressor function for PTEN in the nucleus that is independent of its catalytic activity, but is dependent on the physical interaction of PTEN with nuclear effectors, including the tumor suppressor p53 and the oncogene MSP58/MCRS1; such binding cooperates with the transcriptional activity of p53 and inhibits the oncogenic potential of MSP58/MCRS1 (Freeman et al., 2003; Zhou et al., 2003; Okumura et al., 2005; Tang and Eng, 2006). Thus, it is conceivable that PTEN behaves as a multifunctional and versatile molecule that executes its biological functions via multiple mechanisms and at distinct subcellular localizations, including the cell nucleus. The identification of the molecular mechanisms that are important for PTEN nuclear accumulation will help to unravel the specific functions of PTEN in the nucleus.
ACKNOWLEDGMENTS
We thank M.-C. Hung, K. Weis, and the UK Medical Research Council gene service for providing plasmids, E. Knecht for critical reading of the manuscript, and I. Roglá for expert technical assistance. This work was supported in part by Grants SAF2002-00085 and BMC2003-02696 from the Ministerio de Ciencia y Tecnología (Spain and European Union) by a grant from Fundación Mutua Madrileña Automovilista (Spain; R.P.), and by Grant CP04/00318 from Instituto de Salud Carlos III (Spain; A.G.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0380) on July 28, 2006.
REFERENCES
- Al-Khouri A. M., Ma Y., Togo S. H., Williams S., Mustelin T. Cooperative phosphorylation of the tumor suppressor phosphatase PTEN by casein kinases and glycogen synthase kinase 3β. J. Biol. Chem. 2005;280:35195–35202. doi: 10.1074/jbc.M503045200. [DOI] [PubMed] [Google Scholar]
- Andrés-Pons A., Valiente M., Torres J., Gil A., Roglá I., Ripoll F., Cervera J., Pulido R. Functional definition of relevant epitopes on the tumor suppressor PTEN protein. Cancer Lett. 2005;223:303–312. doi: 10.1016/j.canlet.2004.09.047. [DOI] [PubMed] [Google Scholar]
- Aniento F., Roche E., Cuervo A. M., Knecht E. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 1993;268:10463–10470. [PubMed] [Google Scholar]
- Ahn J.-Y., Rong R., Liu X., Ye K. PIKE/nuclear PI 3-kinase signaling mediates the antiapoptotic actions of NGF in the nucleus. EMBO J. 2004;23:3995–4006. doi: 10.1038/sj.emboj.7600392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birle D., Bottini N., Williams S., Huynh H., deBelle I., Adamson E., Mustelin T. Negative feedback regulation of the tumor suppressor PTEN by phosphoinositide–induced serine phosphorylation. J. Immunol. 2002;169:286–291. doi: 10.4049/jimmunol.169.1.286. [DOI] [PubMed] [Google Scholar]
- Bonneau D., Longy M. Mutations of the human PTEN gene. Hum. Mutat. 2000;16:109–122. doi: 10.1002/1098-1004(200008)16:2<109::AID-HUMU3>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Campbell R. B., Liu F., Ross A. H. Allosteric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2003;278:33617–33620. doi: 10.1074/jbc.C300296200. [DOI] [PubMed] [Google Scholar]
- Chung J. H., Ginn-Pease M. E., Eng C. Phosphatase and tensin homologue deleted on chromosome 10 (PTEN) has nuclear localization signal-like sequences for nuclear import mediated by major vault protein. Cancer Res. 2005;65:4108–4116. doi: 10.1158/0008-5472.CAN-05-0124. [DOI] [PubMed] [Google Scholar]
- Chung J. H., Eng C. Nuclear-cytoplasmic partitioning of phosphatase and tensin homologue deleted on chromosome 10 (PTEN) differentially regulates the cell cycle and apoptosis. Cancer Res. 2005;65:8096–8100. doi: 10.1158/0008-5472.CAN-05-1888. [DOI] [PubMed] [Google Scholar]
- Das S., Dixon J. E., Cho W. Membrane-binding and activation mechanism of PTEN. Proc. Natl. Acad. Sci. USA. 2003;100:7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris P., Bacqueville D., Gayral S., Carrez L., Salles J. P., Perret B., Breton-Douillon M. SHIP-2 and PTEN are expressed and active in vascular smooth muscle cell nuclei, but only SHIP–2 is associated with nuclear speckles. J. Biol. Chem. 2003;278:38884–38891. doi: 10.1074/jbc.M300816200. [DOI] [PubMed] [Google Scholar]
- Deleris P., Gayral S., Breton-Douillon M. Nuclear PtdIns(3,4,5)P3 signaling: an ongoing story. J. Cell. Biochem. 2006;98:469–485. doi: 10.1002/jcb.20695. [DOI] [PubMed] [Google Scholar]
- Duerr E. M., et al. PTEN mutations in gliomas and glioneuronal tumors. Oncogene. 1998;16:2259–2264. doi: 10.1038/sj.onc.1201756. [DOI] [PubMed] [Google Scholar]
- Eng C. PTEN: one gene, many syndromes. Hum. Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- Freeman D. J., et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003;3:117–130. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
- Funamoto S., Meili R., Lee S., Parry L., Firtel R. A. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- Georgescu M. M., Kirsch K. H., Akagi T., Shishido T., Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc. Natl. Acad. Sci. USA. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimm O., et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am. J. Pathol. 2000;156:1693–1700. doi: 10.1016/s0002-9440(10)65040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn-Pease M. E., Eng C. Increased nuclear phosphatase and tensin homologue deleted on chromosome 10 is associated with G0-G1 in MCF-7 cells. Cancer Res. 2003;63:282–286. [PubMed] [Google Scholar]
- Giri D. K., Ali-Seyed M., Li L.-Y., Lee D.-F., Ling P., Bartholomeusz G., Wang S.-C., Hung M.-C. Endosomal transport of ErbB-2, mechanism for nuclear entry of the cell surface receptor. Mol. Cell. Biol. 2005;25:11005–11018. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Gustin J. A., Maehama T., Dixon J. E., Donner D. B. The PTEN tumor suppressor protein inhibits tumor necrosis factor-induced nuclear factor kappa B activity. J. Biol. Chem. 2001;276:27740–27744. doi: 10.1074/jbc.M102559200. [DOI] [PubMed] [Google Scholar]
- Iijima M., Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- Iijima M., Huang Y. E., Luo H. R., Vazquez F., Devreotes P. N. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J. Biol. Chem. 2004;279:16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- Kuersten S., Ohno M., Mattaj I. W. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 2001;11:497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- Lachyankar M. B., et al. A role for nuclear PTEN in neuronal differentiation. J. Neurosci. 2000;20:1404–1413. doi: 10.1523/JNEUROSCI.20-04-01404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. O., Yang H., Georgescu M. M., Di Cristofano A., Maehama T., Shi Y., Dixon J. E., Pandolfi P., Pavletich N. P. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Leslie N. R., Downes C. P. PTEN function: how normal cells control it and tumour cells lose it. Biochem. J. 2004;382:1–11. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. M., Sun H. PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induces G1 cell cycle arrest in human glioblastoma cells. Proc. Natl. Acad. Sci. USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Wagner S., Campbell R. B., Nickerson J. A., Schiffer C. A., Ross A. H. PTEN enters the nucleus by diffusion. J. Cell. Biochem. 2005;96:221–234. doi: 10.1002/jcb.20525. [DOI] [PubMed] [Google Scholar]
- Liu J. L., Sheng X., Hortobagyi Z. K., Mao Z., Gallick G. E., Yung W. K. Nuclear PTEN-mediated growth suppression is independent of Akt down-regulation. Mol. Cell. Biol. 2005;25:6211–6224. doi: 10.1128/MCB.25.14.6211-6224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T., Dixon J. E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Maehama T., Taylor G. S., Dixon J. E. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu. Rev. Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- Mahimainathan L., Choudhury G. G. Inactivation of platelet-derived growth factor receptor by the tumor suppressor PTEN provides a novel mechanism of action of the phosphatase. J. Biol. Chem. 2004;279:15258–15268. doi: 10.1074/jbc.M314328200. [DOI] [PubMed] [Google Scholar]
- Mayo L. D., Donner D. B. The PTEN, Mdm2, p53 tumor suppressor-oncoprotein network. Trends Biochem. Sci. 2002;27:462–467. doi: 10.1016/s0968-0004(02)02166-7. [DOI] [PubMed] [Google Scholar]
- Mayo L. D., Dixon J. E., Durden D. L., Tonks N. K., Donner D. B. PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J. Biol. Chem. 2002;277:5484–5489. doi: 10.1074/jbc.M108302200. [DOI] [PubMed] [Google Scholar]
- McConnachie G., Pass I., Walker S. M., Downes C. P. Interfacial kinetic analysis of the tumour suppressor phosphatase, PTEN: evidence for activation by anionic phospholipids. Biochem. J. 2003;371:947–955. doi: 10.1042/BJ20021848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehenni H., Lin-Marq N., Buchet-Poyau K., Reymond A., Collart M. A., Picard D., Antonarakis S. E. LKB1 interacts with and phosphorylates PTEN: a functional link between two proteins involved in cancer predisposing syndromes. Hum. Mol. Genet. 2005;14:2209–2219. doi: 10.1093/hmg/ddi225. [DOI] [PubMed] [Google Scholar]
- Miller S. J., Lou D. Y., Seldin D. C., Lane W. S., Neel B. G. Direct identification of PTEN phosphorylation sites. FEBS Lett. 2002;528:145–153. doi: 10.1016/s0014-5793(02)03274-x. [DOI] [PubMed] [Google Scholar]
- Myers M. P., Pass I., Batty I. H., Van der Kaay J., Stolarov J. P., Hemmings B. A., Wigler M. H., Downes C. P., Tonks N. K. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury M. V., Weis K. The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Ramaswamy S., Vazquez F., Signoretti S., Loda M., Sellers W. R. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 2000;20:8969–8982. doi: 10.1128/mcb.20.23.8969-8982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K., Zhao M., Depinho R. A., Furnari F. B., Cavenee W. K. Cellular transformation by the MSP58 oncogene is inhibited by its physical interaction with the PTEN tumor suppressor. Proc. Natl. Acad. Sci. USA. 2005;102:2703–2706. doi: 10.1073/pnas.0409370102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchiston E. A., Bennett D., Leslie N. R., Clarke R. G., Winward L., Downes C. P., Safrany S. T. PTEN M-CBR3, a versatile and selective regulator of inositol 1,3,4,5,6-pentakisphosphate (Ins(1,3,4,5,6)P5). Evidence for Ins(1,3,4,5,6)P5 as a proliferative signal. J. Biol. Chem. 2004;279:1116–1122. doi: 10.1074/jbc.M310933200. [DOI] [PubMed] [Google Scholar]
- Parsons R. Human cancer, PTEN and the PI-3 kinase pathway. Semin. Cell. Dev. Biol. 2004;15:171–176. doi: 10.1016/j.semcdb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Perren A., Komminoth P., Saremaslani P., Matter C., Feurer S., Lees J. A., Heitz P. U., Eng C. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am. J. Pathol. 2000;157:1097–1103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puc J., et al. Lack of PTEN sequesters CHK1 and initiates genetic stability. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Radu A., Neubauer V., Akagi T., Hanafusa H., Georgescu M. M. PTEN induces cell cycle arrest by decreasing the level and nuclear localization of cyclin D1. Mol. Cell. Biol. 2003;23:6139–6149. doi: 10.1128/MCB.23.17.6139-6149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- Sansal I., Sellers W. R. The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- Stambolic V., MacPherson D., Sas D., Lin Y., Snow B., Jang Y., Benchimol S., Mak T. W. Regulation of PTEN transcription by p53. Mol. Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- Ström A. C., Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2:3008.1–3008.9. doi: 10.1186/gb-2001-2-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitomo M., Iwase A., Zheng R., Navarro D., Kaminetzky D., Shen R., Georgescu M. M., Nanus D. M. Synergy in tumor suppression by direct interaction of neutral endopeptidase with PTEN. Cancer Cell. 2004;5:67–78. doi: 10.1016/s1535-6108(03)00331-3. [DOI] [PubMed] [Google Scholar]
- Tang Y., Eng. C. PTEN autoregulates its expression by stabilization of p53 in a phosphatase-independent manner. Cancer Res. 2006;66:736–742. doi: 10.1158/0008-5472.CAN-05-1557. [DOI] [PubMed] [Google Scholar]
- Tolkacheva T., Boddapati M., Sanfiz A., Tsuchida K., Kimmelman A. C., Chan A. M. Regulation of PTEN binding to MAGI-2 by two putative phosphorylation sites at threonine 382 and 383. Cancer Res. 2001;61:4985–4989. [PubMed] [Google Scholar]
- Torres J., Navarro S., Roglá I., Ripoll F., Lluch A., García-Conde J., Llombart-Bosch A., Cervera J., Pulido R. Heterogeneous lack of expression of the tumour suppressor PTEN protein in human neoplastic tissues. Eur. J. Cancer. 2001;37:114–121. doi: 10.1016/s0959-8049(00)00366-x. [DOI] [PubMed] [Google Scholar]
- Torres J., Pulido R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 2001;276:993–998. doi: 10.1074/jbc.M009134200. [DOI] [PubMed] [Google Scholar]
- Torres J., Rodriguez J., Myers M. P., Valiente M., Graves J. D., Tonks N. K., Pulido R. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3, implications for the control of protein stability and PTEN-protein interactions. J. Biol. Chem. 2003;278:30652–30660. doi: 10.1074/jbc.M212610200. [DOI] [PubMed] [Google Scholar]
- Valiente M., Andrés-Pons A., Gomar B., Torres J., Gil A., Tapparel C., Antonarakis S. E., Pulido R. Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinases. J. Biol. Chem. 2005;280:28936–28943. doi: 10.1074/jbc.M504761200. [DOI] [PubMed] [Google Scholar]
- Vazquez F., Ramaswamy S., Nakamura N., Sellers W. R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 2000;20:5010–5018. doi: 10.1128/mcb.20.14.5010-5018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F., Grossman S. R., Takahashi Y., Rokas M. V., Nakamura N., Sellers W. R. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 2001;276:48627–48630. doi: 10.1074/jbc.C100556200. [DOI] [PubMed] [Google Scholar]
- Vazquez F., Matsuoka S., Sellers W. R., Yanagida T., Ueda M., Devreotes P. N. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc. Natl. Acad. Sci. USA. 2006;103:3633–3638. doi: 10.1073/pnas.0510570103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite K. A., Eng C. Protean PTEN: form and function. Am. J. Hum. Genet. 2002;70:829–844. doi: 10.1086/340026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S. M., Leslie N. R., Perera N. M., Batty I. H., Downes C. P. The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem. J. 2004;379:301–307. doi: 10.1042/BJ20031839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L. P., Brown J. L., Eng C. PTEN coordinates G(1) arrest by down-regulating cyclin D1 via its protein phosphatase activity and up-regulating p27 via its lipid phosphatase activity in a breast cancer model. Hum. Mol. Genet. 2001;10:599–604. doi: 10.1093/hmg/10.6.599. [DOI] [PubMed] [Google Scholar]
- Whiteman D. C., Zhou X. P., Cummings M. C., Pavey S., Hayward N. K., Eng C. Nuclear PTEN expression and clinicopathologic features in a population-based series of primary cutaneous melanoma. Int. J. Cancer. 2002;99:63–67. doi: 10.1002/ijc.10294. [DOI] [PubMed] [Google Scholar]
- Wu X., Hepner K., Castelino-Prabhu S., Do D., Kaye M. B., Yuan X. J., Wood J., Ross C., Sawyers C. L., Whang Y. E. Evidence for regulation of the PTEN tumor suppressor by a membrane-localized multi-PDZ domain containing scaffold protein MAGI-2. Proc. Natl. Acad. Sci. USA. 2000;97:4233–4238. doi: 10.1073/pnas.97.8.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Dowbenko D., Spencer S., Laura R., Lee J., Gu Q., Lasky L. A. Interaction of the tumor suppressor PTEN/MMAC with a PDZ domain of MAGI3, a novel membrane-associated guanylate kinase. J. Biol. Chem. 2000;275:21477–21485. doi: 10.1074/jbc.M909741199. [DOI] [PubMed] [Google Scholar]
- Xu L., Massagué J. Nucleocytoplasmic shuttling of signal transducers. Nat. Rev. Mol. Cell Biol. 2004;5:209–219. doi: 10.1038/nrm1331. [DOI] [PubMed] [Google Scholar]
- Ye K., Snyder S. H. PIKE GTPase: a novel mediator of phosphoinositide signaling. J. Cell Sci. 2004;117:155–161. doi: 10.1242/jcs.00924. [DOI] [PubMed] [Google Scholar]
- Yu Z., Fotouhi-Ardakani N., Wu L., Maoui M., Wang S., Banville D., Shen S. H. PTEN associates with the vault particles in HeLa cells. J. Biol. Chem. 2002;277:40247–40252. doi: 10.1074/jbc.M207608200. [DOI] [PubMed] [Google Scholar]
- Zhou M., Gu L., Findley H. W., Jiang R., Woods W. G. PTEN reverses MDM2-mediated chemotherapy resistance by interacting with p53 in acute lymphoblastic leukemia cells. Cancer Res. 2003;63:6357–6362. [PubMed] [Google Scholar]