Abstract
Insertional mutagenesis and gene silencing are efficient tools for the determination of gene function. In contrast to gain- or loss-of-function approaches, RNA interference (RNAi)-induced gene silencing can possibly silence multigene families and homoeologous genes in polyploids. This is of great importance for functional studies in hexaploid wheat (Triticum aestivum), where most of the genes are present in at least three homoeologous copies and conventional insertional mutagenesis is not effective. We have introduced into bread wheat double-stranded RNA-expressing constructs containing fragments of genes encoding Phytoene Desaturase (PDS) or the signal transducer of ethylene, Ethylene Insensitive 2 (EIN2). Transformed plants showed phenotypic changes that were stably inherited over at least two generations. These changes were very similar to mutant phenotypes of the two genes in diploid model plants. Quantitative real-time polymerase chain reaction revealed a good correlation between decreasing mRNA levels and increasingly severe phenotypes. RNAi silencing had the same quantitative effect on all three homoeologous genes. The most severe phenotypes were observed in homozygous plants that showed the strongest mRNA reduction and, interestingly, produced around 2-fold the amount of small RNAs compared to heterozygous plants. This suggests that the effect of RNAi in hexaploid wheat is gene-dosage dependent. Wheat seedlings with low mRNA levels for EIN2 were ethylene insensitive. Thus, EIN2 is a positive regulator of the ethylene-signaling pathway in wheat, very similar to its homologs in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Our data show that RNAi results in stably inherited phenotypes and therefore represents an efficient tool for functional genomic studies in polyploid wheat.
A major challenge in the postgenome era of plant biology is to determine the functions of all the genes in a plant genome. A straightforward approach to this problem is to reduce or knock out expression of a gene to induce a mutant phenotype that is indicative of the gene function. Insertional mutagenesis is a useful tool for this type of study and is based on transposon/T-DNA insertions (Page and Grossniklaus, 2002). However, this approach is limited by the time required to saturate a genome, by lethal knockouts, and is restricted to a few plant species. In addition, it is complicated by the problem of genetic redundancy caused by multigene families and polyploidy. In contrast to insertional mutagenesis, RNA interference (RNAi) is based on sequence-specific RNA degradation that follows the formation of double-stranded RNA (dsRNA) homologous in sequence to the targeted gene (Marx, 2000; Carthew, 2001; Baulcombe, 2004). RNAi allows silencing one, several, or all members of a multigene family or homoeologous gene copies in polyploids by targeting sequences that are unique or shared by several genes (Lawrence and Pikaard, 2003; Miki et al., 2005). dsRNA is detected by the host plant genome as aberrant and is cleaved by the action of Dicer-like enzymes (Hamilton and Baulcombe, 1999; Zamore et al., 2000; Djikeng et al., 2001; Tang et al., 2003) into two distinct classes of small interfering RNA (siRNA): long and short siRNAs (Hamilton et al., 2002; Tang et al., 2003). These two classes of small RNAs were proposed to have distinct RNA silencing functions: approximately 21-mers to direct posttranscriptional signaling via mRNA degradation and the approximately 24-mers to trigger systemic silencing and the methylation of homologous DNA (Hamilton et al., 2002). Biochemical experiments in wheat (Triticum aestivum) germ and cauliflower (Brassica oleracea) extracts provided clear evidence that each class of siRNAs is generated by distinct Dicer-like enzymes (Tang et al., 2003). Through sequence complementarity, siRNA in association with an RNA-induced silencing complex directs the cleavage of endogenous RNA transcripts. siRNAs are also responsible for amplifying the silencing signal by priming endogenous RNA, which can be converted to dsRNA by the action of RNA-directed RNA polymerase (RdRP) encoded in the plant genome (Lipardi et al., 2001; Sijen et al., 2001). Tang et al. (2003) showed that wheat germ extracts contained an RdRP activity. Thus, a few trigger dsRNA molecules suffice to inactivate a continuously transcribed target mRNA for long periods of time. The inactivation persists through cell division, spreads to untreated cells and tissues of plants, and is inherited to subsequent generations.
RNAi has proven to be very efficient in interfering with gene expression in various plant systems such as Petunia hybrida, Arabidopsis (Arabidopsis thaliana), Papaver somniferum, Torenia hybrida, Coffea arabica, and rice (Oryza sativa; Stam et al., 1997; Chuang and Meyerowitz, 2000; Wesley et al., 2001; Stoutjesdijk et al., 2002; Allen et al., 2004; Fukusaki et al., 2004; Lee et al., 2004; Ogita et al., 2004; Miki et al., 2005). The wide use of this powerful technique reflects its ease of application and the possibilities for genome-wide reverse genetics. Gene constructs encoding intron-spliced RNA with a self-complementary hairpin (hp) structure have been shown to induce posttranscriptional gene silencing with almost 100% efficiency when directed against viruses or endogenous genes and transgenes (Smith et al., 2000).
In functional gene analysis and isolation of agronomically important genes, wheat is clearly lagging behind compared to other major food crops such as maize (Zea mays), rice, and also species such as tomato (Lycopersicon esculentum). This is mainly due to the lack of efficient tools to study gene function in polyploid species. Hexaploid wheat has a large genome (16,000 Mb) that consists of three closely related homoeologous genomes (A, B, and D) and has a high content of repetitive DNA (80%; Flavell et al., 1974). Genes are found to be organized in gene islands or as single genes separated by large regions of nested repetitive elements (Feuillet and Keller, 2002). Due to the hexaploid nature of its genome, bread wheat has three (or a multiple of three) copies of most genes. It was found that many of these homoeologous genes are expressed (Mochida et al., 2003) and that there is, therefore, a high degree of functional gene redundancy in hexaploid wheat.
In wheat and barley (Hordeum vulgare), Schweizer et al. (2000), Christensen et al. (2004), and Douchkov et al. (2005) showed that delivery of specific dsRNA into single epidermal cells transiently interfered with gene function. Single-cell RNAi, as well as virus-induced gene silencing (VIGS; Burch-Smith et al., 2004; Hein et al., 2005; Scofield et al., 2005), can be used as tools for reverse genetics, but there are several drawbacks to these transient approaches. The analysis of whole-organism gene function is not possible with these techniques, and they do not introduce stable genetic change in plants. Yan et al. (2004) and Loukoianov et al. (2005) used RNAi to stably transform wheat and demonstrate that reduction of VRN2 and VRN1 transcript levels, respectively, accelerated and delayed flowering initiation in winter wheat. However, in both studies, only one independent transgenic plant showed the expected silenced phenotype. Regina et al. (2005) also used RNAi to generate high-amylose wheat. These studies did not report any molecular analyses on the long-term phenotypic stability of RNAi-mediated gene silencing over several generations; neither did they report any molecular details on silencing of homoeologous genes. To develop RNAi technology for functional genomics in wheat, there is a need to characterize in molecular detail the silencing of homoeologous genes as well as the inheritance of RNAi-induced phenotype.
To investigate the potential of dsRNAi in wheat, we introduced into hexaploid wheat dsRNA-expressing constructs of two genes that have not yet been cloned in wheat, but with previously defined functions in other plant species and with unambiguous phenotypes in corresponding mutant plants. The first gene was Phytoene Desaturase (PDS), which is often used to evaluate VIGS efficiency because of its distinct phenotype (Holzberg et al., 2002). PDS is an enzyme of the carotenoid biosynthetic pathway, and reduction or loss of this enzyme results in inhibition of the carotenoid biosynthesis and subsequently in a photobleaching phenotype due to chlorophyll photooxidation (Bartley and Scolnik, 1995). To assess whether genes that do not encode enzymes can also be effectively silenced by RNAi in wheat, we decided to target Ethylene Insensitive 2 (EIN2), which encodes a transmembrane protein of the ethylene signaling pathway (Alonso et al., 1999). It has been identified in Arabidopsis, where all 25 ein2 mutant alleles showed a clear phenotype of complete insensitivity to ethylene at the morphological, physiological, and molecular levels (Alonso et al., 1999). In our study, RNAi constructs expressing these two genes were delivered into wheat by particle bombardment-mediated transformation and were stably integrated into the genome. In addition to high specificity and heritability, a phenotypic series (weak, intermediate, and strong) was obtained from dsRNAi. Furthermore, quantitative real-time experiments showed that the endogenous target mRNA levels of all three homoeologous genes are decreased in RNAi transgenic lines and that siRNA production is gene-dosage dependent. Thus, specific and inheritable dsRNAi offers a useful and efficient tool for functional genomics in hexaploid wheat and provides a powerful tool to manipulate gene expression experimentally.
RESULTS
Identification of Wheat Expressed Sequence Tags for PDS and EIN2
The two genes selected for testing RNAi in hexaploid wheat, PDS and EIN2, have not yet been described in the wheat genome. A BLASTN search using the barley PDS cDNA sequence (AY062039; Holzberg et al., 2002) identified several wheat expressed sequence tags (ESTs) showing high nucleotide (nt) identity (E < 10−100) to the barley sequence. Seven wheat ESTs that were homologous to the 5′ end of the barley sequence were used in a multiple sequence alignment to find out whether more than one PDS gene is expressed in wheat. Two groups of three and four ESTs, with more than 97% nt identity, could be clearly distinguished based on characteristic simple nt polymorphisms (SNPs). A pair of primers (Table I) was designed within the conserved region and based on the BG908924 hexaploid wheat sequence that shared the highest nt identity (95%) with the barley PDS cDNA (AY062039). This BG908924 wheat sequence (785 bp) belongs to the consensus sequence TC236658 of 2,464 bp (http://www.tigr.org/tigr-scripts/tgi/tc_report.pl?tc=TC236658&species=wheat) and aligns with the full-length cDNA of PDS of rice (AF049356; 2,027 bp) between nts 311 and 1,075 with an 89% identity (Fig. 1A). A 480-bp fragment of wheat PDS (wPDS) was amplified by reverse transcription (RT)-PCR from wheat leaf cDNA and was used for the RNAi construct. The RT-PCR product was also cloned, and several independent transformants were sequenced. Following a multiple sequence alignment, three distinct groups of sequences were identified based on SNPs (Supplemental Fig. 1A; the same SNPs were detected in three independent RT-PCR reactions), indicating that there are at least three active copies of the PDS gene in the wheat genome. The same RT-PCR fragment was used as a probe to estimate the number of copies of wPDS in the wheat genome by hybridization to hexaploid (cv Bobwhite and cv Chinese Spring) and diploid (Triticum monococcum cv DV92) wheat DNA digested with several restriction enzymes (Supplemental Fig. 2A). The hybridization pattern was simple for several enzyme combinations, showing between one and two bands in the diploid T. monococcum species and between two and four bands in hexaploid wheat. Southern hybridizations to nullitetrasomic lines of Chinese Spring (Sears, 1966) digested with HindIII allowed mapping wPDS on chromosomes 4A and 4D (Supplemental Fig. 2B). In a recent study, Cenci et al. (2004) localized durum wheat (Triticum durum) bacterial artificial chromosome clones containing the PDS genes on chromosome 4A as well as 4B. The nullitetrasomic 4B line used in our study was probably not genetically pure. Indeed, several SSR markers specific of chromosome 4B could be amplified (data not shown), suggesting the presence of chromosome 4B in this nullitetrasomic 4B line. Therefore, additional fragments of PDS could not be mapped on chromosome 4B.
Table I.
Primers designed on wheat ESTs for the specific amplification of sequences used for RNAi
EST sources and product sizes amplified by RT-PCR are shown. Restriction sites are shown in bold (GGATCC for BamHI and AGATCT for BglII). Primers designed to perform quantitative real-time PCR are designated by RT.
| Primer Name | Sequence (5′ → 3′) | Source | Product Size |
|---|---|---|---|
| bp | |||
| PDS-F | CCAAGGATCCGAATTTGTTTGCTGAGCTTGG | BG908924 | 480 |
| PDS-R | GGCAAGATCTGCCTTTCAGGAGGATTACCA | BG908924 | |
| PDS-RT-F | CAGCAGTGTCCAGGCACTA | CK163183 | 115 |
| PDS-RT-R | ACAACCTGCAGAGCACGAAG | CK163183 | |
| PDS-RT-F1 | ACCTTTAGTTCGACTTCCCC | Contig 1 | 68 |
| PDS-RT-R1 | AGAGCACGAAGTCCACGT | Contig 1 | |
| PDS-RT-F2 | CAGGCACTAAAAACCAGTCAC | Contig 2 | 96 |
| PDS-RT-R2 | AGAGCACGAAGTCCACCG | Contig 2 | |
| PDS-RT-F3 | CAGCAGTGTCCAGGCACTA | Contig 3 | 106 |
| PDS-RT-R3 | AGAGCACGAAGTCCATGA | Contig 3 | |
| EIN2-F | CTAAGGATCCACAAAGCCCAGCAATGAATC | AL816731 | 518 |
| EIN2-R | CCTAAGATCTTGAAGAAGCTCTGCCTCACA | AL816731 | |
| EIN2-RT-F | TGGGTTCATCCAACTGGTC | CD925940 | 80 |
| EIN2-RT-R | AAGATGGCATATTGAAATTTG | CD925940 | |
| EIN2-RT-F1 | GCAGCTTACTTGAGCAAAATC | Contig 1 | 92 |
| EIN2-RT-R1 | ATGAATAGTAGCAGGCTGATAGA | Contig 1 | |
| EIN2-RT-F2 | GCAGCTTACTTGAGCCAAATG | Contig 2 | 92 |
| EIN2-RT-R2 | ATGAATAGTAGCAGGCTGATAGG | Contig 2 | |
| EIN2-RT-F3 | GGATATCAGCTGGCATCTT | Contig 3 | 132 |
| EIN2-RT-R3 | CATAACAGGATCAGCATAGTTAGA | Contig 3 | |
| TAK14-F | TGGAAGATCTTGATATCCGTTCTGTTTCTA | AF325198 | 568 |
| TAK14-R | TTCAGGATCCTTGTGCCAGATATTTGCTCC | AF325198 | |
| SC255-RT-F | GTACAACGCTGGAACGAACA | TC264870 | 84 |
| SC255-RT-R | GAAAGGTTCTCGGTGTCGTC | TC264870 | |
| GAPDH-RT-F | TTAGACTTGCGAAGCCAGCA | AF251217 | 81 |
| GAPDH-RT-R | AAATGCCCTTGAGGTTTCCC | AF251217 | |
| TAK14-3′-F | CATGGTGCCATGGTTCAAAG | AF325198 | Variable |
| NOS-R | CAAGACCGGCAACAGGATTC | PBI101TD | |
| Mlo-F | CCTGACGCTATTCCAGAACG | AF361932 | 518 |
| Mlo-R | AGACCGACCTTCTCCTGTCA | AF361932 |
Figure 1.
Location of the wheat ESTs used to construct the RNAi vectors in their corresponding wheat consensus sequence and in the corresponding full-length cDNAs identified in rice. A, Wheat EST BG908924 aligned with its consensus sequence TC236658 and the full-length cDNA of PDS of rice (OsPDS; AF049356). B, Wheat EST AL816731 aligned with its consensus sequence TC257467 and the full-length cDNA of EIN2 of rice (OsEIN2; AY396568). Base pairs, percentage of identity, and overlapping areas (shaded boxes) are indicated (not on scale). C, Self-complementary hp construct derived from hp transgene used in the bombardment experiments. Gene-specific sequences (black arrows indicating the orientation) in the antisense and sense orientations were cloned with a 548-bp fragment of the TAK14 (AF325198) wheat intron (white box) and were controlled by the constitutive ubi promoter (hatched box) and the nopaline synthase terminator (dotted box). Restriction enzymes HindIII (H) and EcoRI (E) used for Southern-blot hybridization analysis are indicated.
A BLASTN search using the barley EIN2 cDNA sequence (BM816947) identified eight wheat EST sequences, but only one EST (AL816731) showed high nt identity (E < 10−100; 93%) to the barley sequence. This AL816731 wheat sequence (564 bp) belongs to the consensus sequence TC257467 of 1,213 bp (http://www.tigr.org/tigr-scripts/tgi/tc_report.pl?tc=TC257467) and aligns with the full-length cDNA of EIN2 of rice (AY396568; 4,646 bp) between nts 3,061 and 3,527 with an 80% identity (Fig. 1B). A pair of primers (Table I) was designed based on the AL816731 wheat sequence, and a 518-bp fragment of wEIN2 was amplified by RT-PCR from wheat leaf cDNA to make the RNAi construct. As described above for the wPDS genes, the RT-PCR product was also cloned, and several independent transformants were sequenced. A multiple sequence alignment identified three distinct groups of sequences based on SNPs (Supplemental Fig. 3A; the same SNPs were detected in three independent RT-PCR reactions), indicating that there are at least three active EIN2 genes in the wheat genome. The same RT-PCR wEIN2 fragment was used as a probe to estimate the number of copies of wEIN2 in the wheat genome (Supplemental Fig. 2C). The hybridization pattern was simple for several enzyme combinations, showing between one and three bands in the diploid T. monococcum species and between two and five bands in hexaploid wheat. Southern hybridizations to nullitetrasomic lines of Chinese Spring (Sears, 1966) digested with HindIII allowed mapping wEIN2 on chromosomes 5A, 5B, and 5D (Supplemental Fig. 2D). These results suggest that for both wPDS and wEIN2 genes, there are three expressed copies in hexaploid wheat, with one copy on each of the homoeologous genomes.
Specific Silencing of the PDS Genes Induces Photobleaching in Hexaploid Wheat
One hundred and fifty three putative transgenic plants were produced by particle bombardment with the intron-spliced self-complementary construct containing the sense and antisense repeats of the wPDS cDNA sequence. Genomic DNA from these plants was digested with HindIII, and the wPDS sequence was used as a probe for DNA Southern-blot hybridization. Eighty-two lines, which correspond to 53% of the analyzed plants, contained the expected three intact HindIII fragments of the construct (0.5, 1, and 2.2 kb; Fig. 1C; Supplemental Fig. 4A) and 64 of them (78%) exhibited photobleaching at the seedling stage, indicating specific silencing of endogenous PDS (Fig. 2). The PDS-RNAi transgenic lines were arranged into a phenotypic series based on the severity of the photobleached phenotype (strong, intermediate, and weak). Most of the lines (41 lines) developed a strong photobleached phenotype, resulting in a lethal albino phenotype (Fig. 2A). Fifteen lines showed an intermediate phenotype with continuous parallel streaks (Fig. 2, B and C) where photobleaching affected either one-half of the leaves (Fig. 2B) or only the middle part of the leaf, whereas the leaf margins maintained the wild-type phenotype (Fig. 2C). The remaining eight lines produced a weak phenotype where only a small part of the leaves was affected by photobleaching (Fig. 2D). Both single and multiple copy lines with extensive DNA rearrangements showed photobleaching (Supplemental Fig. 4A; Fig. 3A).
Figure 2.
An RNAi construct expressing a wPDS cDNA fragment induces photobleaching in hexaploid wheat. The PDS-RNAi transgenic T0 lines were arranged based on the severity of photobleaching. A, Strong photobleached phenotype resulting in albino plants and lethality. B, Intermediate phenotype with patterns of streaks where photobleaching is affecting one-half of the leaf surface. C, Intermediate phenotype with patterns of streaks where photobleaching is affecting the central part of the leaves with the margins still green. D, Weak phenotype where only a small sector of the leaves is affected by photobleaching.
Figure 3.
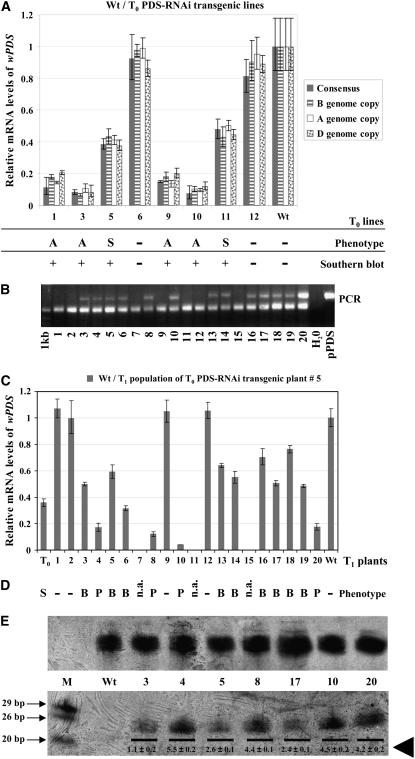
RNAi-mediated specific silencing of the wPDS genes in hexaploid wheat. A, Quantitative real-time PCR of eight wPDS-RNAi transgenic T0 primary transformants with primers designed within the conserved nt sequence region identified on wheat EST CK163183 and with primers specific to each of the homoeologous wPDS genes (see Supplemental Fig. 1). Relative mRNA levels of wPDS were normalized to the mRNA level of wild-type (Wt) plants (=1). The GAPDH gene was used as an internal standard. Data are the average of triplicate samples (±sd). The numbering of the transgenic lines analyzed by quantitative real-time PCR refers to the Southern-blot data shown in Supplemental Figure 4, and the Southern analysis is summarized at the bottom of A. Transgenic (+) and non transgenic (−) T0 lines. In A, the photobleached phenotype is indicated. A, Albino; S, streaks; and –, wild-type phenotype. B, C, D, and E, T1 generation analysis of the T0 PDS-RNAi transgenic line 5. B, Detection of the hp transgene by PCR. The top band corresponds to the transgene and the bottom band to the wheat homolog of the barley Mlo gene used here as an internal control. pPDS, hp construct used in the transformation experiments. C, Relative mRNA levels of wPDS, which were normalized to the mRNA level of wild-type (Wt) plants (=1). Line T0 from which this population is derived is shown. Plants 7, 11, and 15 were not analyzed, because they did not produce enough leaf material for RNA extraction. Data are the average of triplicate samples (±sd). D, Phenotypes. S, Streaks; P, photobleaching; B, bleaching only at the base of the leaf; n.a., not analyzed; and –, wild type. E, Detection of small RNAs in T1 heterozygous plants (plants 3, 5, and 17 from C) and T1 homozygous plants (plants 4, 8, 10, and 20 from C) of the T0 PDS-RNAi transgenic line 5. Low Mr RNA fractions were hybridized with a mixture of 11 DNA oligos complementary to the sequence of interest (Supplemental Fig. 6A). The 20-nt, 26-nt, and 29-nt DNA oligos were used as size controls (size indicated in nts); Wt, wild type. The same blot was hybridized with the housekeeping GAPDH gene as a control (top segment). The relative intensity of the hybridization signals in the transgenics versus wild-type plants was determined with a phosphoimager (Cyclone gene array system, Perkin-Elmer). The relative mean value of wPDS small RNAs per plant ± sd is indicated by the thick arrowhead.
Thus, PDS dsRNA-mediated genetic interference causes photobleaching in hexaploid wheat in a similar way to plants treated with a chemical inhibitor of PDS (Böger and Sandmann, 1998) or to virus-induced PDS gene silencing (VIGS) in barley (Holzberg et al., 2002). The consistent appearance of bleached tissue in the large number of regenerated plants suggests that the endogenous PDS genes are no longer functional in these tissues due to RNAi.
dsRNA Interferes with wPDS mRNA Accumulation of All Three Homoeologous Genes
The expression of PDS in the RNAi transgenic wheat lines was determined by real-time quantitative RT-PCR. Primers were designed to specifically measure effective endogenous PDS mRNA levels and not the transgene transcripts. Primers were located on the wheat EST sequence CK163183 (Table I; Supplemental Fig. 1A) upstream of the conserved nt sequence regions used to design the primers for the RNAi construct. Cloning and sequencing of the products produced by RT-PCR revealed that the activity of all three genes described above was measured. The primer binding sites were completely conserved in the three expressed genes, and three distinct sequences were identified based on SNPs (Supplemental Fig. 1A; the same SNPs were detected in three independent RT-PCR reactions). The SNPs allowed the design of additional primers for specifically amplifying each of the homoeologous copies of the wPDS gene (Supplemental Fig. 1B). The reduction of relative mRNA levels of wPDS was very similar in each of the three homoeologous genes (Fig. 3A). Therefore, the RNAi silencing mechanism affects all three copies of the gene in the same way.
The severity of the photobleached phenotype inversely correlated with PDS mRNA expression levels in the leaves of all the transgenic RNAi lines analyzed. As shown in Figure 3A, expression levels of lines that exhibited an albino phenotype (lines 1, 3, 9, and 10) were 11%, 8.4%, 15%, and 7.2% of the wild type, respectively. A transcript reduction relative to the wild type was also observed in lines showing streaks of photobleaching (5 and 11 in Fig. 3A), but they accumulated more mRNA than the albino lines (38% and 48% of the wild type, respectively). In all the other transgenic RNAi lines not showing any photobleaching, PDS mRNA expression levels were not significantly different from the wild type.
These results indicate that PDS mRNA levels decline with increasingly severe phenotypes and suggest that endogenous PDS mRNA is a target of dsRNA-mediated genetic interference that equally silences the three homoeologous copies.
Phytoene Accumulates in Bleached Tissue as a Result of RNAi-Based Silencing of Endogenous Wheat PDS
Blockage of the carotenoid pathway can be mimicked chemically with the herbicide norflurazon, a specific inhibitor of the PDS enzyme. This inhibition causes accumulation of phytoene in treated tissues (Böger and Sandmann, 1998). To confirm that the bleached phenotype in our experiments was due to PDS silencing, we used HPLC to determine phytoene levels in RNAi transgenic lines with or without bleached tissues and in norflurazon-treated and -untreated wild-type plants. In addition to a specific retention time on reverse-phase columns, carotenoids are characterized by their spectral properties, consisting of three peak maxima with a unique spectral shape (ratio of peaks II/III), which are influenced by the solvent used (Wurtzel et al., 2001). As shown in Supplemental Figure 5A, leaf tissue from wild-type norflurazon-treated plants contained two spectral peaks at 286 nm, with a retention time of 3.65 min (peak 1) and 5.61 min (peak 2; Supplemental Fig. 5G). These two peaks were absent in wild-type untreated plants (Supplemental Fig. 5B), which contained another type of spectral peak absorbing at a maximum of 446 nm, with a retention time of 4 min (Supplemental Fig. 5G). The spectra of numbered peaks 1 and 2 (Supplemental Fig. 5, D and E) show the characteristics of the compound phytoene (three peaks maxima and peaks shape), which is identical to previously published spectral profiles of phytoene (Li et al., 1996; Wurtzel et al., 2001). The spectrum detected in wild-type untreated plants (peak 3, Supplemental Fig. 5F) showed no phytoene peaks but the characteristics of the compound lutein (Suzuki and Shioi, 2003), which is derived from subsequent series of desaturations and cyclizations in the carotenoid biosynthesis pathway when PDS is active. PDS-RNAi transgenic lines, which showed an albino phenotype, also contained two peaks with virtually identical spectra to the spectrum of phytoene from norflurazon-treated wild-type tissue (Supplemental Fig. 5, A, D, and E). Leaves with photobleached streaks contained a mixture of peaks corresponding to the two compounds phytoene and lutein that were present in norflurazon-treated and -untreated wild-type tissue, respectively (Supplemental Fig. 5C). PDS-RNAi transgenic lines, which did not show any phenotype, produced a spectrum identical to the one detected in wild-type untreated plants (Supplemental Fig. 5B). Moreover, as shown in Supplemental Figure 5G, a comparison of peak retention times, peak maxima, and peak ratios II/III between the norflurazon-treated wild-type plants and the RNAi transgenic lines corroborated the presence or absence of phytoene in relation to the efficiency of silencing. These results demonstrate that phytoene accumulation and photobleaching are correlated and provide biochemical confirmation of PDS gene inactivation in hexaploid wheat.
Inheritance of the Genetic Interference of wPDS and Detection of siRNA in Silenced Plants
T0 PDS-RNAi lines that showed an albino phenotype did not set seeds and therefore could not be analyzed further. The progeny from each PDS-RNAi line that had intermediate phenotypes (streaks on leaves; 23 lines) all showed a 3:1 segregation for the presence and absence of the hp transgene, as shown in Figure 3B, with an example of PCR analysis in the progeny of line 5. This result suggests that the primary transformants integrated the hp transgene at a single locus, although separate integration at very closely linked loci cannot be excluded. All the T1 progeny showed Mendelian segregation for the photobleached phenotype on the leaves. Surprisingly, the intermediate phenotype with continuous parallel streaks (Fig. 2, B–D) of some of the T0 transgenic lines was never observed again in the following generations. Twenty-five percent of the T1 plants containing the hp transgene showed strong photobleaching of the leaves with large albino areas, which in a few cases eventually developed into completely albino plants (Fig. 3D). The other T1 plants that had integrated the transgene (approximately 50%) showed a weak phenotype, where photobleaching was affecting only the base of the leaves (Fig. 3D). Quantitative real-time PCR revealed again a correlation between the level of mRNA and the severity of the photobleached phenotype. As shown in Figure 3C, the T1 plants (derived from T0 line no. 5) having a reduction of endogenous wPDS transcripts to less than 20% of wild type (T1 plants 4, 8, 10, and 20) showed strong photobleaching. Among the T1 plants that showed a weak phenotype, plants 3, 5, 6, 13, 14, 16, 17, 18, and 19 accumulated mRNA levels between 30% and 70% of wild type (medium reduction). Plants 1, 2, 9, and 12, which did not contain the hp transgene, had the same mRNA levels as the wild type.
We subsequently analyzed each of the PDS-RNAi lines in the T2 generation by selfing different T1 plants showing strong/weak photobleaching and wild-type phenotypes. The T1 plants that showed strong photobleaching were homozygous, since all the T2 progenies contained the transgene, showed large albino areas, and had their mRNA levels of endogenous wPDS genes reduced to less than 40% of the wild type (data not shown). The T1 plants that showed weak photobleaching were heterozygous, since all the T2 progenies were still segregating for the phenotype and the transgene.
Small RNA-gel blots were used to test whether the amounts of siRNA were different in homozygous and heterozygous lines. Small RNAs were recovered from a subset of the T1 plants used as the source of total RNA for quantitative real-time PCR. We detected sequence-specific siRNAs of around 24 bp in both homozygous and heterozygous plants showing strong/weak photobleaching (Fig. 3E). Wild-type plants did not accumulate small PDS RNAs. Thus, small PDS RNAs accumulated only upon silencing of the PDS genes. The relative intensity of the hybridization signals in the transgenic versus wild-type plants indicated that the homozygous plants contain around double the amount of small RNAs compared to heterozygous plants. It is therefore likely that the strong photobleached phenotype observed in the homozygous plants is in part related to a higher accumulation of siRNAs.
This genetic analysis indicates that RNAi of wPDS is stably inherited over at least two generations in a Mendelian fashion of a single locus. Small RNAs specific for the silenced gene were detected and their accumulation was quantitatively different in homozygous and heterozygous lines. In homozygous plants, accumulation of siRNAs was significantly higher to give effective gene silencing and to develop the most severe phenotype. These results suggest that the effect of RNAi in hexaploid wheat is probably gene-dosage dependent. The reason for the discrepancy of hemizygous phenotypes at T0 versus later generations is not known, but it is intriguing that the streak phenotype of the T0 generation was never recovered in later generations.
EIN2 dsRNA-Mediated Genetic Interference in Hexaploid Wheat
To assess whether the EIN2 signal transducer of ethylene can also be effectively silenced by RNAi in wheat, 33 putative transgenic plants were produced by particle bombardment with the intron-spliced self-complementary construct containing the sense and antisense repeats of the wEIN2 cDNA sequence. Genomic DNA from these plants was digested with EcoRI, and blots were hybridized with a probe corresponding to the wEIN2 cDNA sequence of the hpRNA construct. Eighteen lines, which correspond to 54% of the analyzed plants, contained the expected intact EcoRI fragment of the construct (2 kb; Supplemental Fig. 4B; Fig. 1C).
We identified by quantitative real-time PCR six T0 primary transgenic lines (33% of the lines that contained the full-length RNAi fragment) with a significant reduction of mRNA expression of the endogenous wEIN2 genes (Fig. 4A). As for the wPDS genes, the RT-PCR primers were designed to specifically detect the endogenous EIN2 and not the transgene transcripts. Primers were located on the wheat EST sequence CD925940 (Table I; Supplemental Fig. 3A) upstream of the conserved nt sequence regions used to design the primers for the RNAi construct. Cloning and sequencing of the RT-PCR products revealed that the activity of all three homoeologous genes was measured (Supplemental Fig. 3A). Three distinct sequences were identified based on SNPs (the same SNPs were detected in three independent RT-PCR reactions). Quantitative real-time PCR with primers specific to each of the homoeologous copies of the wEIN2 gene (Supplemental Fig. 3B) revealed that the RNAi silencing mechanism is affecting all three homoeologous genes in the same way (Fig. 4A).
Figure 4.
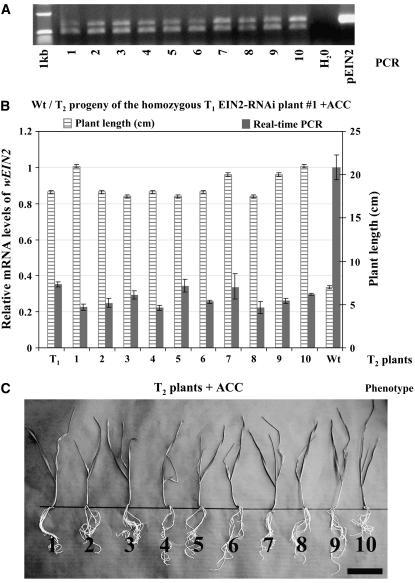
RNAi-mediated specific silencing of the wEIN2 genes in hexaploid wheat. A, Quantitative real-time PCR of eight wEIN2-RNAi transgenic T0 primary transformants with primers designed within the conserved nt sequence region identified on wheat EST CD925940 and with primers specific to each of the homoeologous wEIN2 genes (see Supplemental Fig. 3). Relative mRNA levels of wEIN2 were normalized to the mRNA level of wild-type (Wt) plants (=1). The GAPDH gene was used as an internal standard. Data are the average of triplicate samples (±sd). The numbering of the transgenic lines analyzed by quantitative real-time PCR refers to the Southern-blot data shown in Supplemental Figure 4, and the Southern analysis is summarized at the bottom of A. +, Transgenic; –, nontransgenic T0 lines. B, C, and D, Analysis of the ethylene response in the T1 generation of the T0 EIN2-RNAi transgenic line 10. B, Wild-type seeds germinated in presence and in absence of ACC. C, T1 seeds germinated in presence of ACC. Bars in B and C = 4 cm. D, Relative mRNA levels of wEIN2 that were normalized to the mRNA level of wild-type (Wt) plants (=1). Line T0 from which this population is derived is also shown. Length measurements of each T1 plant are shown on the same diagram. PCR analysis for the presence/absence of the transgene is shown in D.
For five lines, wEIN2 transcript level was reduced to between 30% and 50% compared to the wild type (intermediate reduction in lines 2, 7, 10, 13, and 19 of Fig. 4A). Line 18 had a strong reduction of the wEIN2 mRNA level, which was only 1% of the level detected in the wild-type plant (Fig. 4A). As for the wPDS genes, both single and multiple copy lines with extensive DNA rearrangements showed a significant reduction of wEIN2 mRNA levels (Supplemental Fig. 4B; Fig. 4A). The six T0 primary transgenic lines with lower EIN2 expression showed a normal phenotype, and their ethylene response signaling was studied in the next generations.
Ethylene Response Signaling in Wheat EIN2-RNAi Lines
EIN2 was identified as a central component of the ethylene signaling pathway both in Arabidopsis (Alonso et al., 1999) and in rice (Jun et al., 2004). All six T0 EIN2-RNAi transgenic wheat lines with lower EIN2 expression set seeds, and we could therefore examine whether their ethylene response was altered. Twenty T1 seeds of each of these six lines, wild-type plants, and five primary transgenic lines that did not show any reduction of the wEIN2 transcript were germinated at 25°C under light (16-h light, 8-h dark) on Murashige and Skoog medium in the presence of 20 μm 1-aminocyclopropane-1-carboxylic acid (ACC), the immediate precursor of ethylene. Plant length was determined 15 d after sowing. All T1 families showed a 3:1 segregation for the presence and absence of the hp transgene, suggesting that the primary transformants integrated the hp transgene at a single locus, although separate integration at very closely linked loci cannot be excluded. The six T1 families derived from the T0 lines with lower EIN2 expression showed Mendelian segregation for normal and stunted growth when grown in the presence of 20 μm ACC (e.g. in Fig. 4C, six T1 plants derived from T0 line 10 are shown). Wild-type plants and the other five transgenic lines tested (negative controls) showed a stunted morphology on ACC-containing medium (Fig. 4B). As observed with the wPDS genes, 25% of the T1 plants containing the hp transgene showed complete insensitivity to ethylene (Fig. 4C; T1 plant 1) and a normal growth in presence of ACC. Seedling lengths of these ethylene-insensitive plants were significantly higher (p = 1.18 × 10−5) than the rest of the population in the presence of ACC. The other T1 plants that had the transgene (approximately 50%) showed a growth only partially affected by the presence of ACC (Fig. 4C; T1 plants 3 and 4), and the length of the seedlings was also significantly higher (p = 2.78 × 10−8) than the T1 plants that did not contain the hp transgene (Fig. 4C; T1 plants 2, 5, and 6).
There was a very high negative linear correlation (r2 = −0.976) between the length of the seedlings in the presence of ACC and the amount of wEIN2 transcript measured in the T1 progenies. As observed with the wPDS genes, EIN2 mRNA levels declined with increasingly severe phenotypes (Fig. 4D). T1 plants completely insensitive to ethylene showed the strongest mRNA reduction (around 70%) compared to wild type. T1 plants with an intermediate phenotype accumulated mRNA levels around 50% of wild type, whereas all the other T1 plants that did not contain the hp transgene and were sensitive to ethylene with a stunted morphology had the same mRNA levels as the wild type.
By analyzing the T2 generation, we confirmed that those T1 plants that had the strongest mRNA reduction and were completely insensitive to ethylene were homozygous plants, because all their T2 progeny contained the transgene (Fig. 5A), had their mRNA levels reduced to less than 40% of the wild-type (Fig. 5B), and showed complete ethylene insensitivity in the presence of ACC (Fig. 5, B and C). T1 plants that were partially affected by ethylene were heterozygous, since their T2 progeny were still segregating for normal and stunted growth when grown in the presence of ACC.
Figure 5.
T2 generation analysis of the homozygous T1 EIN2-RNAi transgenic plant 1 generated from the T0 line 10. A, Detection of the hp transgene by PCR; the top band corresponds to the transgene and the bottom band to the wheat homolog of the barley Mlo gene used here as an internal control. pEIN2, hp construct used in the transformation experiments. B, Relative mRNA levels of wEIN2 that were normalized to the mRNA level of wild-type (Wt) plants (=1). The T1 plant from which this population is derived is also shown. Length measurements of each T2 plants are shown on the same diagram. C, Phenotype of the T2 plants which are all ethylene insensitive. Bar in C = 7 cm.
These results demonstrate that the EIN2-RNAi transgenic wheat lines produced in this study are ethylene insensitive. This is consistent with the hypothesis that EIN2 is a positive signal component in ethylene signaling and that inhibiting its expression reduces the ethylene response. We conclude that RNAi silencing in hexaploid wheat is also effective for the silencing of a gene involved in a signaling process.
DISCUSSION
We have demonstrated that RNAi-mediated gene silencing is effective in hexaploid wheat and can efficiently induce reduction of mRNA levels of three homoeologous genes. Expression of the three homoeologous genes was reduced to the same extent, suggesting that RNAi can resolve the issue of genetic redundancy in hexaploid wheat in an efficient way, as it was also suggested by studies in the two allotetraploid species Arabidopsis suenica (Lawrence and Pikaard, 2003) and cotton (Gossypium hirsutum; Liu et al., 2000).
Knowledge of the complete and exact sequence of the target genes was not essential to induce specific gene silencing, as sequence information from ESTs was sufficient. This is important for the development of high-throughput methods for functional genomics. Trigger-dsRNAs as short as 23 to 26 bp have been shown to induce degradation of target mRNAs (Hamilton and Baulcombe, 1999; Thomas et al., 2001), and several studies reported that sequences that are 88% to 100% identical to the endogenous gene target caused silencing (Jones et al., 1998; Ingelbrecht et al., 1999; Schweizer et al., 2000; Xu et al., 2001; Holzberg et al., 2002). In our study, we used wheat EST sequences of around 500 bp to construct the RNAi vectors. A specific EST is derived from a specific gene, but it is known that homoeologous genes in wheat share up to 99% identity at the nt sequence level in the coding regions (Kimbara et al., 2004). Therefore, there is a high chance that homoeologous genes will retain regions of identity, resulting in silencing of all the genes. The suppression of the homoeologous genes was probably also facilitated by the use of a sequence within the conserved regions identified among homologous wheat ESTs. Concerns have arisen that siRNA could cause other effects than those related to the knockdown of the target gene due to cross hybridization or binding in a sequence-dependent manner to various cellular proteins (off-target effects; Jackson et al., 2003; Scacheri et al., 2004). Standard software can now be used for improving detection of sequence identity to accurately and systematically evaluate and minimize RNAi off-target effects between siRNA sequences and target genes (Qiu et al., 2005).
We have used two different genes to assess the effectiveness of RNAi in hexaploid wheat, the enzyme PDS and the ethylene signaling component EIN2. These genes were chosen because mutant alleles have been reported to give distinct phenotypes in diploid plant species. It was also important to assess whether a gene that encodes a regulatory factor can be effectively silenced in wheat. Indeed, a reduction of enzyme amount might allow the detection of a mutant phenotype, whereas silencing of a signaling component might not be sufficient to cause a mutant phenotype, because even a strongly reduced amount of protein might still be sufficient for proper function. We have shown that RNAi-mediated silencing of both genes results in the reduction of transcripts by up to 93% for PDS and 99% for EIN2. Therefore, dsRNAs corresponding to these two genes caused strong and specific genetic interference, suggesting that dsRNA-mediated gene silencing under the control of the ubiquitin promoter can occur in the tissues where these genes normally function.
We found a strong correlation between decreased levels of mRNA and increased severity of phenotypes. A phenotypic series was obtained from the RNAi lines with a full spectrum of the effect of RNAi (weak, intermediate, and strong) on gene expression, which is in agreement with the results in Arabidopsis (Chuang and Meyerowitz, 2000; Wang et al., 2005) and tomato (Xiong et al., 2005). Part of the variation of the RNAi effect can be explained by transgene copy number and/or positional effects of particular DNA insertion events. However, our results suggest that the severity of phenotypes is not related to the transgene copy number, because both single and multiple copy lines with extensive DNA rearrangements showed a mutant phenotype. Chuang and Meyerowitz (2000) and Wang et al. (2005) also found no relationship between severity of the phenotype and transgene copy number in Arabidopsis. In contrast, Kerschen et al. (2004) described that multicopy RNAi Arabidopsis lines reduced target mRNA levels to a lesser extent and with more variability between lines than did single-copy lines. A number of studies in rice and wheat have reported that there is no evidence for any direct relationship between transgene copy number and transgene expression or stability (Kohli et al., 1999; Stoger et al., 1999). Thus, we can assume that the phenotypic series observed in the T0 transgenic lines is due to positional effects of the DNA insertion event. The variation in the degree of silencing observed in the transformants showing both reduction and loss of function may be a useful feature for gene discovery and functional genomics. Complete silencing of genes encoding a key element in basic cell functions or at particular developmental stages may result in lethality, whereas the reduced gene expression may give viable plants with phenotypes indicative of the role of the target gene. Systems to deliver inducible RNAi offer the advantage of silencing gene expression at specific developmental stages or in specific tissues, because they provide flexibility for the timing and the degree of gene inactivation and have the potential for reversal of silencing by withdrawal of the inducer (Guo et al., 2003; Wielopolska et al., 2005).
It has been shown by Smith et al. (2000) and Wesley et al. (2001) that 66% to 100% of the plants (tobacco [Nicotiana tabacum], Arabidopsis, cotton, and rice) transformed with an intron-containing hp construct can be silenced. In our investigation, 78% of the wheat PDS-RNAi and 33% of the wheat EIN2-RNAi transgenic lines showed silencing. One explanation for the difference in efficiency of silencing between the two genes may be that some phenotypes (such as silencing of EIN2) are less sensitive to the level of gene expression and that a reduced amount of protein is still sufficient to confer the proper function. The effectiveness of silencing appears to be gene dependent and could reflect accessibility of target mRNA or the relative abundance of the target mRNA and the hpRNA in cells in which the gene is active (Helliwell and Waterhouse, 2003). Kerschen et al. (2004) and Wang et al. (2005) suggested that RNAi efficiency and the endogenous transcription level of the target gene are not necessarily related.
We confirmed that the expression of the RNAi phenotype is stably inherited over at least two generations for both wPDS and wEIN2 genes, possibly making this approach a reliable tool not only for functional genomics but also for the genetic modification of agronomically interesting traits. In our study, dsRNA-expressing constructs were inherited in a Mendelian fashion as a single locus, and the most severe phenotype was observed in homozygous plants that showed the strongest mRNA reduction. Among the phenotypic series that we obtained from the transgenic PDS-RNAi lines, we have also observed T0 lines showing a strong phenotype in a hemizygous state. This strong phenotype was lethal for the wPDS genes but had no effect in the case of the wEIN2 genes (Fig. 5A, plant 18). The intermediate phenotype of wPDS with continuous parallel streaks (Fig. 2, B–D) of some of the T0 transgenic lines was never observed again in the following generations. This suggests that in the early development of newly transformed T0 seedlings, RNAi is not fully established, and therefore, the mutant phenotypes differ from later generations. Thus, it might be advisable to not only study the T0 generation in wheat RNAi projects but also in later generations. Our results indicate that the most efficient silenced phenotypes are stably recovered in homozygous lines, suggesting that the effect of RNAi in hexaploid wheat is gene-dosage dependent. This is possibly due to the progressive repression of the target gene with increasing allelic concentration of the transgene. This hypothesis is supported by our observation of gene-dosage dependence of specific siRNAs that were approximately double the amount in homozygous plants compared to heterozygous plants. These results are in agreement with García-Pérez et al. (2004), who reported that tobacco leaves of homozygous rootstocks have accumulated approximately double the amount of secondary siRNAs than the heterozygous rootstocks, suggesting that the induction of systemic silencing was strikingly dosage dependent. Similarly, several studies have suggested that transgene dosage can affect resistance of a transgenic line carrying transgenes homologous to viral sequences, because homozygous, but not hemizygous, R2 progenies were able to confer high level of resistance to different strains (Dinant et al., 1997; McDonald et al., 1997; Tenllado and Díaz-Ruíz, 1999; Tennant et al., 2001).
New studies provide convincing biochemical and genetic evidence that RdRP plays a critical role in amplifying the RNAi effect, explaining the extreme efficiency and the self-sustaining nature of RNAi (Lipardi et al., 2001; Sijen et al., 2001). An RdRP can use the siRNAs as primers and the target mRNAs as templates to produce newly formed dsRNAs that are subsequently cleaved to produce secondary siRNAs. Recently, it has been shown (Himber et al., 2003; García-Pérez et al., 2004) that secondary siRNAs belong exclusively to the 21-nt siRNA class. This observation argues against the previously proposed role for the 25-nt siRNAs as systemic silencing signal (Hamilton et al., 2002) and contrasts with the observation that de novo dsRNA synthesis in wheat germ extracts is linked to production of siRNAs that are almost exclusively 25-nt long (Tang et al., 2003). In addition, experiments with dsRNA suggested that the 21-mers are produced in wheat at about one-quarter the rate of the 24 to 25-nt small RNAs (Tang et al., 2003). Our results are in agreement with the observations of Hamilton et al. (2002) and Tang et al. (2003) because the RNAi wheat transgenic lines produced in our study have accumulated only one class of siRNAs, which is the 25-nt siRNAs. Identification of the RdRP responsible for systemic silencing signal in vitro will help to address the issue with the contrasting findings of Himber et al. (2003) and García-Pérez et al. (2004).
Down-regulation of EIN2 by RNAi resulted in ethylene-insensitive wheat plants that showed normal wild-type growth in presence of ACC, the immediate precursor of ethylene. This is consistent with the hypothesis that EIN2 is a positive regulator of the ethylene-signaling in wheat, very similar to its homologs in Arabidopsis (Alonso et al., 1999) and rice (Jun et al., 2004). Whether the EIN2 gene product acts indirectly in ethylene signaling by affecting metal homeostasis, as other Nramp family members are thought to do in some animal systems, or whether the EIN2 protein is a family member that has been recruited to function directly in ethylene signaling, e.g. by regulating a second messenger, remains unclear (Bleecker and Kende, 2000). Our results indicate that the RNAi sequence of EIN2 efficiently suppressed the ethylene signaling pathway, affecting the target EIN2. The phytohormone ethylene plays an important role in many aspects of plant growth, development, and environmental responses (Johnson and Ecker, 1998). In particular, ethylene is important in germination and seedling growth of cereals (Locke et al., 2000). Stem-shortening plant growth regulators are often used to control lodging in modern high input cereal management, and they include GA biosynthesis inhibitors and ethylene-releasing compounds. While promotion of seedling shoot growth by ethylene has been reported for barley, oat (Avena sativa), and rice (Locke et al., 2000; Jun et al., 2004), ethylene inhibits hypocotyl elongation in wheat (Huang et al., 1997; our results) as well as in dark-grown seedlings of Arabidopsis (Alonso et al., 1999; Wesley et al., 2001). Pathogen infection is also an ethylene-modulated process that results in distinct morphological and biochemical changes (Wan et al., 2002). Pasquer et al. (2005) recently showed differences in expression patterns of wheat defense-related genes after treatment with a systemic acquired resistance enhancer and two commonly used fungicides. They raised the hypothesis that BTH [benzo(1,2,3)thiadiazole-7-carbothioic acid S-methylester, a systemic acquired resistance enhancer] is a strong trigger of signaling mediated by ethylene and salicylic acid. The ethylene-insensitive RNAi wheat plants produced in this study can be used in future studies on the role of ethylene in development, defense, and fungicide responses in wheat.
RNAi silencing has an enormous potential as a tool in functional genomics of hexaploid wheat, a species for which other methods such as insertional mutagenesis are not available. dsRNA-expressing constructs, when delivered into wheat by particle bombardment-mediated transformation, created a heritable phenotypic series in the transformants that may be a useful feature for gene discovery and functional genomics. Technical barriers for high-throughput functional genomics have recently been lowered considerably by the development of pHELLSGATE vectors that utilize the Gateway recombination system and give the possibility of making hpRNA constructs for large sets of genes (Helliwell and Waterhouse, 2003). Tang and Galili (2004) raised the hypothesis that next generation RNAi vectors should contain characteristics of microRNA structures, because microRNAs do not trigger the PKR pathway, the RNA-dependent protein kinase pathway that causes nonspecific cell death in mammalian cells and could function as part of the plant stress response (Langland et al., 1995). Fast and efficient systems of transformation and regeneration of transgenic plants are necessary to successfully use RNAi constructs for plant functional genomics. Gene transfer technology is still limited in wheat by the low frequency of generation of transgenic plants. With further development and increase of the efficiency of the wheat transformation methods, an exciting perspective will be opened up to improve tools for functional analysis of wheat genes, with a strong impact on wheat breeding.
MATERIALS AND METHODS
Plant Material
Wheat (Triticum aestivum) cv Bobwhite, accession SH98 26 (provided by Dr. A. Pellegrineschi, International Center for Development of Maize and Wheat, Mexico) was used for all experiments. Donor plants for embryo transformation and transgenic plants were grown in a greenhouse with 16-h light at 21°C and 8-h night at 16°C. Every 10 to 15 d, routine fertilizer treatments were applied to the donor plants.
Generation of RNAi Lines
PDS and EIN2 nt sequences were obtained from The Institute for Genome Research database (www.tigr.org). Using barley (Hordeum vulgare) sequences, a BLASTN search was carried out to find hexaploid wheat ESTs. Multiple sequence alignments were done with ClustalW (Thompson et al., 1994). PCR primers were designed on the basis of wheat ESTs similar to the target genes using the Primer3 WWW primer tool (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; Whitehead Institute for Biomedical Research). Vector pAHC17 containing the maize (Zea mays) ubi-1 promoter and the nopaline synthase terminator (Christensen et al., 1992) were used to make the self-complementary intron-containing hp RNA constructs. Fragments of 480 and 518 bp corresponding to the wheat cDNAs of wPDS and wEIN2, respectively, were isolated by RT-PCR using specific primers with incorporated BamHI and BglII restriction sites (Table I), which produced ends compatible with each other. This allowed the gene fragments to be directionally cloned within the unique BamHI site of vector pAHC17. PCR amplification was carried out for 35 cycles of 45 s, denaturation at 94°C, 45 s annealing at 62°C, and 90 min extension at 72°C. The amplified fragments were then subcloned into pGEM-T vector (Promega) and sequenced. The cDNA fragments recovered by BamHI and BglII digestion were cloned in the BamHI restriction site of plasmid pAHC17. Each RNAi construct contained a cDNA fragment derived from the respective target gene and oriented in the antisense and sense directions at the 5′ and 3′ ends of the construct, respectively (Fig. 1C), separated by an intron derived from the wheat TAK14 gene (AF325198). The intron TAK14 fragment was produced with the same cloning strategy as described for the two RNAi target genes. The resulting plasmids were named pPDS-RNAi and pEIN2-RNAi for the wPDS and wEIN2 genes, respectively.
RNAi lines were produced using particle bombardment-mediated transformation of immature embryos as described (Pellegrineschi et al., 2002). Plasmid pPDS-RNAi or pEIN2-RNAi was cotransformed with a plasmid containing phospho-Man isomerase as selectable marker (Wright et al., 2001). Regeneration and selection of the transformed plants were performed essentially as described (Wright et al., 2001; Pellegrineschi et al., 2002). Transformants were identified by DNA Southern hybridization (Stein et al., 2000) using plant genomic DNA digested with HindIII for wPDS and EcoRI for wEIN2 and probes corresponding to the selected cDNA sequences of the hpRNA construct (Fig. 1C). Hybridizations were performed overnight at 65°C in 5× sodium chloride/sodium phosphate/EDTA, 0.5% SDS, and 5× Denhardt's solution. Membranes were washed at 65°C three times for 20 min in 0.5× SSC and 0.1% SDS, and then they were autoradiographed.
To determine the inheritance of the RNAi construct and the phenotype, transgenic lines were allowed to self pollinate, and segregation analyses were performed in the T1 and T2 generations by means of PCR, phenotype observation, and quantitative real-time PCR. Rapid DNA extractions at the two-leaf stage (modified for small volumes from Stein et al., 2000) and PCR techniques accelerated the screening of T1 and T2 plants. The presence of the RNAi construct was determined using a forward primer at the 3′ end of the TAK14 intron and reverse primer at the 5′ end of the nopaline synthase terminator (Table I). A PCR product corresponding to several wheat homologs of the barley Mlo gene was used as an internal standard (Table I).
Quantitative Real-Time PCR
Total RNA was isolated from leaves using TRIzol reagent (Invitrogen Life Technologies). Leaves were collected 3 weeks after transferring the transgenic and control plants from culture tubes to soil.
For RT, 10 μg of total RNA was denatured at 70°C for 5 min in the presence of 0.07 μg of oligo(dT)21 primers. The tubes were immediately chilled on ice and reverse transcribed with 7 units of reverse transcriptase (Invitrogen Life Technologies), 1× buffer, 0.7 mm of each dNTPs, 10 mm dithiothreitol, and 1.5 units of RNase OUT (Invitrogen Life Technologies) in a total volume of 30 μL at 42°C for 90 min.
Real-time PCR assays were performed with the ABI PRISM 7700 Sequence Detection System (Applied Biosystems) using SYBR Green PCR Master mix (Applied Biosystems) in a final volume of 26 μL including cDNA template and appropriate primer pairs (Table I). The amplification conditions were 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. To normalize results, the glyceraldehyde-3-P dehydrogenase (GAPDH; AF251217) gene was used as an internal standard (Table I). Three replicates were performed for each sample. The specificity of the unique amplification product was determined by melting curve analysis according to the manufacturer's instructions.
Short Interfering RNA Detection
To detect small RNAs, the procedures described by Hamilton and Baulcombe (1999) were followed. The lower Mr RNAs were recovered after removal of high Mr RNAs by precipitation with 10% polyethylene glycol 8000 and 0.5 m NaCl. From the different samples analyzed, a similar amount of RNA (7 μg) of the lower Mr RNA fraction was separated on gel (15% polyacrylamide, 7 m urea, 1× Tris-borate/EDTA) and transferred to Hybond N+ membranes (Amersham Biosciences) by electroblotting. As size marker, three gene-specific DNA oligos were loaded on the same gels (Supplemental Fig. 6). For hybridization, the ULTRAhyb-Oligo buffer from Ambion (1× buffer) was used, and 1 μm of around 10 DNA oligos complementary to the sequence of interest (Supplemental Fig. 6) were labeled using 0.5 units of T4 polynucleotide kinase (Roche) and 6 μL of [γ-32P]ATP (5,000 Ci/mmol). RNA-blot hybridizations were carried out at 35°C as described by Hamilton and Baulcombe (1999). The hybridization blots were then also hybridized with the housekeeping GAPDH gene (AF251217) as a control. The relative intensity of the hybridization signals in the transgenics versus wild-type plants was determined with a phosphoimager (Cyclone gene array system, Perkin-Elmer).
Carotenoid Extraction and Analysis of Phytoene by HPLC from wPDS-RNAi Transgenic Lines
Leaf samples were collected when the transgenic and control plants were transferred from culture tubes to soil. Norflurazon-treated wild-type plants were produced by watering 15-d-old seedlings with 3 μm norflurazon (Syngenta) to induce bleaching. To compensate local variations within a plant, three samples were taken from each transgenic and control plant, corresponding to three different leaves of the same plant.
Carotenoid extraction was performed essentially as described (Wurtzel et al., 2001). Around 100 mg of leaf tissue was ground in liquid N2, suspended in 1 mL methanol, and centrifuged at 9,000 rpm for 10 min at 4°C. After addition of 100 μL 60% KOH, the supernatant was heated at 65°C for 20 min. The mixture was then extracted three times in 10% (v/v) diethyl ether in hexane, the organic phase evaporated under N2 and the precipitate dissolved in 70 μL methanol. An HPLC system, model 480 (Gynkotek HPLC) with photodiode array detection, model UVD340S (Gynkotek HPLC), was used to separate samples by reverse phase chromatography on a EC 125/4.6 Nucleosil 100-5 C18 column (Macherey-Nagel) using acetonitrile:methanol:2-propanol (80:14:7, v/v/v) as a developing solvent at a flow rate of 1 mL min−1 under isocratic conditions. Substances were detected using UV absorption at 286, 440, and 550 nm, and spectra were recorded between 250 and 580 nm UV/vis. Peaks were identified by spectrophotometric profiles (as characterized by three absorption maxima and peak II/III ratios) and retention times, by comparing them to that of the norflurazon-treated plants and previously published spectral profiles of phytoene (Li et al., 1996; Wurtzel et al., 2001).
Supplementary Material
Acknowledgments
We thank Dr. A. Pellegrineschi (Centro Internacional de Mejoramiento de Maiz y Trigo, Mexico) for providing us with seeds of Bobwhite SH 98 26 and Dr. P. Quail (University of California, Berkeley and U.S. Department of Agriculture Plant Gene Expression Center, Albany, California) for the plasmid pAHC17. Syngenta (Basel, Switzerland) is acknowledged for the PMI gene. We thank Stephi Narain and Geri Herren (Institute of Plant Biology, Zürich) for excellent technical assistance. We also thank Dr. Markus Klein and Dr. Valeria Gagliardini (Institute of Plant Biology, Zürich) for helping with the HPLC and quantitative real-time experiments, respectively. Finally, special thanks to Dr. Christoph Ringli (Institute of Plant Biology, Zürich) for useful technical advice and critical reading of the manuscript.
This work was supported by the Indo-Swiss Collaboration in Biotechnology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Beat Keller (bkeller@botinst.unizh.ch).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JAC, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the non-narcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22: 1559–1566 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Bartley GE, Scolnik PA (1995) Plant carotenoids: pigments for photo-protection, visual attraction, and human health. Plant Cell 7: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe DC (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Böger P, Sandmann G (1998) Carotenoid biosynthesis inhibitor herbicides: mode of action and resistance mechanisms. Pestic Outlook 9: 29–35 [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP (2004) Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J 39: 734–746 [DOI] [PubMed] [Google Scholar]
- Carthew RW (2001) Gene silencing by double-stranded RNA. Curr Opin Cell Biol 13: 244–248 [DOI] [PubMed] [Google Scholar]
- Cenci A, Somma S, Chantret N, Dubcovsky J, Blanco A (2004) PCR identification of durum wheat BAC clones containing genes coding for carotenoid biosynthesis enzymes and their chromosome localization. Genome 47: 911–917 [DOI] [PubMed] [Google Scholar]
- Christensen AB, Thordal-Christensen H, Zimmermann G, Gjetting T, Lyngkjær MF, Dudler R, Schweizer P (2004) The Germinlike protein GLP4 exhibits superoxide dismutase activity and is an important component of quantitative resistance in wheat and barley. Mol Plant Microbe Interact 17: 109–117 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18: 675–689 [DOI] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant S, Maisonneuve B, Albouy J, Chupeau Y, Chupeau M, Bellec Y, Gaudefroy F, Kusiak C, Souche S, Robaglia C, et al (1997) Coat protein gene mediated protection in Lactuca sativa against lettuce mosaic potyvirus strains. Mol Breed 3: 75–86 [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Ullu E (2001) RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA 7: 1522–1530 [PMC free article] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P (2005) A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol Plant Microbe Interact 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Feuillet C, Keller B (2002) Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann Bot (Lond) 89: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell RB, Bennett MD, Smith JB, Smith DB (1974) Genome size and proportion of repeated nucleotide sequence DNA in plants. Biochem Genet 12: 257–269 [DOI] [PubMed] [Google Scholar]
- Fukusaki EI, Kawasaki K, Kajiyama S, An CI, Suzuki K, Tanaka Y, Kobayashi A (2004) Flower color modulations of Torenia hybrida by downregulation of chalcone synthase genes with RNA interference. J Biotechnol 111: 229–240 [DOI] [PubMed] [Google Scholar]
- García-Pérez RD, Van Houdt H, Depicker A (2004) Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. Plant J 38: 594–602 [DOI] [PubMed] [Google Scholar]
- Guo HS, Fei JF, Xie Q, Chua NH (2003) A chemical-regulated inducible RNAi system in plants. Plant J 34: 383–392 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Voinnet O, Chappell L, Baulcombe DC (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I, Barciszewska-Pacak M, Hrubikova K, Williamson S, Dinesen M, Soenderby IE, Sundar S, Jarmolowski A, Shirasu K, Lacomme C (2005) Virus-induced gene silencing-based functional characterization of genes associated with powdery mildew resistance in barley. Plant Physiol 138: 2155–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C, Waterhouse P (2003) Constructs and methods for high-throughput gene silencing in plants. Methods 30: 289–295 [DOI] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30: 315–327 [DOI] [PubMed] [Google Scholar]
- Huang B, Johnson JW, Box JE, Nesmith DS (1997) Root characteristics and hormone activity of wheat in response to hypoxia and ethylene. Crop Sci 37: 812–818 [Google Scholar]
- Ingelbrecht IL, Irvine JE, Mirkov TE (1999) Post-transcriptional gene silencing in transgenic sugarcane: dissection of homology-dependent virus resistance in a monocot that has a complex polyploid genome. Plant Physiol 119: 1187–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS (2003) Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Johnson PR, Ecker JR (1998) The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet 32: 227–254 [DOI] [PubMed] [Google Scholar]
- Jones AL, Johansen IE, Bean SJ, Bach I, Maule AJ (1998) Specificity of resistance to pea seed-borne mosaic potyvirus in transgenic peas expressing the viral replicase (Nlb) gene. J Gen Virol 79: 3129–3137 [DOI] [PubMed] [Google Scholar]
- Jun SH, Han MJ, Lee S, Seo YS, Kim WT, An G (2004) OsEIN2 is a positive component in ethylene signaling in rice. Plant Cell Physiol 45: 281–289 [DOI] [PubMed] [Google Scholar]
- Kerschen A, Napoli CA, Jorgensen RA, Müller AE (2004) Effectiveness of RNA interference in transgenic plants. FEBS Lett 566: 223–228 [DOI] [PubMed] [Google Scholar]
- Kimbara J, Takashi R, Nasuda S (2004) Characterization of the genes encoding for MAD2 homologues in wheat. Chromosome Res 12: 703–714 [DOI] [PubMed] [Google Scholar]
- Kohli A, Gahakwa D, Vain P, Laurie D, Christou P (1999) Transgene expression in rice engineered through particle bombardment: molecular factors controlling stable expression and transgene silencing. Planta 208: 88–97 [Google Scholar]
- Langland JO, Jin S, Jacobs BL, Roth DA (1995) Identification of a plant-encoded analog of PKR, the mammalian double-stranded RNA-dependent protein kinase. Plant Physiol 108: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RJ, Pikaard CS (2003) Transgene-induced RNA interference: a strategy for overcoming gene redundancy in polyploids to generate loss-of-function mutations. Plant J 36: 114–121 [DOI] [PubMed] [Google Scholar]
- Lee S, Kim J, Han JJ, Han MJ, An G (2004) Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1 / AGAMOUS-LIKE 20 (SOC1 / AGL20) ortholog in rice. Plant J 38: 754–764 [DOI] [PubMed] [Google Scholar]
- Li ZH, Matthews PD, Burr B, Wurtzel ET (1996) Cloning and characterization of a maize cDNA encoding phytoene desaturase, an enzyme of the carotenoid biosynthetic pathway. Plant Mol Biol 30: 269–279 [DOI] [PubMed] [Google Scholar]
- Lipardi C, Wei Q, Paterson BM (2001) RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107: 297–307 [DOI] [PubMed] [Google Scholar]
- Liu Q, Singh S, Green A (2000) Genetic modification of cotton seed oil using inverted-repeat gene-silencing techniques. Biochem Soc Trans 28: 927–929 [PubMed] [Google Scholar]
- Locke JM, Bryce JH, Morris PC (2000) Contrasting effects of ethylene perception and biosynthesis inhibitors on germination and seedling growth of barley (Hordeum vulgare L.). J Exp Bot 51: 1843–1849 [DOI] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J (2005) Regulation of VRN-1 vernalization genes in normal and transgenic wheat. Plant Physiol 138: 2364–2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx J (2000) Interfering with gene expression. Science 288: 1370–1372 [DOI] [PubMed] [Google Scholar]
- McDonald JG, Brandle F, Gleddie S, Hermans J, Kermali I (1997) Resistance to homologous and heterologous strains of potato virus Y in transgenic tobacco carrying the PVY (N) coat protein gene. Can J Plant Sci 77: 167–171 [Google Scholar]
- Miki D, Itoh R, Shimamoto K (2005) RNA silencing of single and multiple members in a gene family of rice. Plant Physiol 138: 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yamazaki Y, Ogihara Y (2003) Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol Genet Genomics 270: 371–377 [DOI] [PubMed] [Google Scholar]
- Ogita S, Uefuji H, Morimoto M, Sano H (2004) Application of RNAi to confirm theobromine as the major intermediate for caffeine biosynthesis in coffee plants with potential for construction of decaffeinated varieties. Plant Mol Biol 54: 931–941 [DOI] [PubMed] [Google Scholar]
- Page DR, Grossniklaus U (2002) The art and design of genetic screens: Arabidopsis thaliana. Nat Rev Genet 3: 124–136 [DOI] [PubMed] [Google Scholar]
- Pasquer F, Isidore E, Zarn J, Keller B (2005) Specific patterns of changes in wheat gene expression after treatment with three antifungal compounds. Plant Mol Biol 57: 693–707 [DOI] [PubMed] [Google Scholar]
- Pellegrineschi A, Noguera LM, Skovmand B, Brito RM, Velazquez L, Salgano MM, Hernandez R, Warburton M, Hoisington D (2002) Identification of highly transformable wheat genotypes for mass production of fertile transgenic plants. Genome 45: 421–430 [DOI] [PubMed] [Google Scholar]
- Qiu S, Adema CM, Lane T (2005) A computational study of off-target effects of RNA interference. Nucleic Acids Res 33: 1834–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M (2005) High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Natl Acad Sci USA 103: 3546–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, et al (2004) Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci USA 101: 1892–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P, Pokorny J, Schulze-Lefert P, Dudler R (2000) Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J 24: 895–903 [DOI] [PubMed] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS (2005) Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol 138: 2165–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears ER (1966) Nullisomic-tetrasomic combinations in hexaploid wheat. In R Riley, KR Lewis, eds, Chromosome Manipulations and Plant Genetics. Oliver and Boyd, Edinburgh, pp 29–45
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476 [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407: 319–320 [DOI] [PubMed] [Google Scholar]
- Stam M, deBruin R, Kenter S, van der Hoorn RAL, van Blokland R, Mol JNM, Kooter JM (1997) Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J 12: 63–82 [Google Scholar]
- Stein N, Feuillet C, Wicker T, Schlagenhauf E, Keller B (2000) Subgenome chromosome walking in wheat: a 450-kb physical contig in Triticum monococcum L. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.). Proc Natl Acad Sci USA 97: 13436–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoger E, Williams S, Keen D, Christou P (1999) Molecular characteristics of transgenic wheat and the effect on transgene expression. Transgenic Res 7: 463–471 [Google Scholar]
- Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG (2002) hp-RNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol 129: 1723–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Shioi Y (2003) Identification of chlorophylls and carotenoids in major teas by high-performance liquid with photodiode array detection. J Agric Food Chem 51: 5307–5314 [DOI] [PubMed] [Google Scholar]
- Tang G, Galili G (2004) Using RNAi to improve plant nutritional value: from mechanism to application. Trends Biotechnol 22: 463–469 [DOI] [PubMed] [Google Scholar]
- Tang G, Reinhart BJ, Bartel DP, Zamore PD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17: 49–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenllado F, Díaz-Ruíz JR (1999) Complete resistance to pepper mild mottle tobamovirus mediated by viral replicase sequences partially depends on transgene homozygosity and is based on a gene silencing mechanism. Transgenic Res 8: 83–93 [Google Scholar]
- Tennant P, Fermin G, Fitch MM, Manshardt RM, Slightom JL, Gonsalves D (2001) Papaya ringspot virus resistance of transgenic Rainbow and SunUp is affected by gene dosage, plant development and coat protein homology. Eur J Plant Pathol 107: 645–653 [Google Scholar]
- Thomas CL, Jones L, Baulcombe DC, Maule AJ (2001) Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J 25: 417–425 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Dunning FM, Bent AF (2002) Probing plant-pathogen interactions and downstream defense signaling using DNA microarrays. Funct Integr Genomics 2: 259–273 [DOI] [PubMed] [Google Scholar]
- Wang T, Iyer LM, Pancholy R, Shi X, Hall TC (2005) Assessment of penetrance and expressivity of RNAi-mediated silencing of the Arabidopsis phytoene desaturase gene. New Phytol 167: 751–760 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C (2005) A high-throughput inducible RNAi vector for plants. Plant Biotechnol J 3: 583–590 [DOI] [PubMed] [Google Scholar]
- Wright M, Dawson J, Dunder E, Suttie J, Reed J, Kramer C, Chang Y, Novitzky R, Wang H, Artim-Moore L (2001) Efficient biolistic transformation of maize (Zea mays L.) and wheat (Triticum aestivum L.) using the phosphomannose isomerase gene, pmi, as the selectable marker. Plant Cell Rep 20: 429–436 [DOI] [PubMed] [Google Scholar]
- Wurtzel ET, Luo R, Yatou O (2001) A simple approach to identify the first rice mutants blocked in carotenoid biosynthesis. J Exp Bot 52: 161–166 [PubMed] [Google Scholar]
- Xiong AS, Yao QH, Peng RH, Li X, Han PL, Fan HQ (2005) Different effects on ACC oxidase gene silencing triggered by RNA interference in transgenic tomato. Plant Cell Rep 23: 639–646 [DOI] [PubMed] [Google Scholar]
- Xu J, Schubert J, Altpeter F (2001) Dissection of RNA-mediated ryegrass mosaic virus resistance in fertile transgenic perennial ryegrass (Lolium perenne L.). Plant J 26: 265–274 [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.