Abstract
Low temperature is a primary determinant of plant growth and survival. Using accessions of Arabidopsis (Arabidopsis thaliana) originating from Scandinavia to the Cape Verde Islands, we show that freezing tolerance of natural accessions correlates with habitat winter temperatures, identifying low temperature as an important selective pressure for Arabidopsis. Combined metabolite and transcript profiling show that during cold exposure, global changes of transcripts, but not of metabolites, correlate with the ability of Arabidopsis to cold acclimate. There are, however, metabolites and transcripts, including several transcription factors, that correlate with freezing tolerance, indicating regulatory pathways that may be of primary importance for this trait. These data identify that enhanced freezing tolerance is associated with the down-regulation of photosynthesis and hormonal responses and the induction of flavonoid metabolism, provide evidence for naturally increased nonacclimated freezing tolerance due to the constitutive activation of the C-repeat binding factors pathway, and identify candidate transcriptional regulators that correlate with freezing tolerance.
Arabidopsis (Arabidopsis thaliana) is a widely spread small annual weed of the mustard family (Cruciferae) that is native to Europe and central Asia (for review, see Koornneef et al., 2004). Its exposure to a wide range of climatic conditions across its latitudinal range of 68°N to 0°, and its adoption as the major model plant, make it ideal for studying natural variation of adaptive traits (Koornneef et al., 2004). Low temperature defines a climatic range boundary for Arabidopsis (Hoffmann, 2002), making temperature an interesting candidate for a habitat parameter that influences the genetic structure of local populations through natural selection. Such selection should be manifest by a relationship between freezing tolerance and the minimum habitat temperature during the growing season.
Freezing tolerance in plants can be afforded at different levels depending on the anticipation of the freezing event. Nonacclimated freezing tolerance measures the plant's capacity to survive a rapid and severe temperature drop to below zero. Acclimated tolerance is higher and reflects the ability of many plants, including Arabidopsis, to increase their freezing tolerance in response to low but nonfreezing temperatures. The mechanism of nonacclimated tolerance is poorly understood, although it has been reported that it may be genetically determined by loci independent of acclimated tolerance in potato (Solanum tuberosum; Stone et al., 1993) and oilseed rape (Brassica napus; Teutonico et al., 1995). In contrast, cold acclimation is well studied and involves a wide array of metabolic changes governed by extensive reprogramming at the level of gene expression (Thomashow, 1999).
Forward and reverse genetics have defined some of the key regulatory components of cold acclimation. A prominent role has been demonstrated for the C-repeat binding factors (CBF) 1, 2, and 3 (Gilmour et al., 1998), which are also known as dehydration responsive element binding1 b, c, and a, respectively (Liu et al., 1998). The overexpression of CBF genes induces many genes and metabolites that also accumulate in response to cold (Fowler and Thomashow, 2002; Cook et al., 2004; Gilmour et al., 2004). There is evidence for negative regulation among the CBF genes; analysis of a CBF2 null mutant indicated it may negatively regulate CBF1 and CBF3 expression (Novillo et al., 2004), and reduced CBF3 expression in the inducer of CBF expression 1 (ice1) mutant was associated with enhanced CBF2 expression 6 to 12 h after cold treatment (Chinnusamy et al., 2003). ICE1 encodes a MYC-like bHLH transcriptional activator that acts upstream of the CBF genes, enhancing CBF3 expression (Chinnusamy et al., 2003). In the ice1 mutant, several genes in the CBF regulon are no longer cold induced (Lee et al., 2005). HOS1, encoding a RING-finger motif protein, also negatively regulates the CBF genes (Lee et al., 2001). CBF-dependent roles in freezing tolerance have also been demonstrated for LOS1, a translational elongation factor 2 gene, which is involved in protein synthesis in the cold (Guo et al., 2002), and for LOS4, a DEAD box RNA helicase that is essential for mRNA export (Gong et al., 2005).
Transcript profiling indicated the induction of multiple transcriptional pathways in addition to CBF during cold acclimation (Fowler and Thomashow, 2002) and more recently candidates have been characterized. Constitutive overexpression of the transcription factor ZAT12 was shown to cause a slight but significant increase in freezing tolerance and to affect the expression of cold-responsive genes. However, it also down-regulates the expression of the CBF genes, and its regulon partially overlaps with that of CBF2 (Vogel et al., 2005). Independence from the CBF pathway has been demonstrated for the transcription factors HOS9 and HOS10 that are both necessary for cold acclimation but do not affect CBF expression (Zhu et al., 2004, 2005).
Recently, interest in profiling the system response to low temperature has revealed thousands of transcript (Fowler and Thomashow, 2002; Kreps et al., 2002; Seki et al., 2002; Hannah et al., 2005; Lee et al., 2005) and hundreds of metabolite (Cook et al., 2004; Kaplan et al., 2004) changes. These profiling technologies have also been used to characterize regulatory pathways (Chinnusamy et al., 2003; Cook et al., 2004; Gilmour et al., 2004; Maruyama et al., 2004; Vogel et al., 2005). In common with the majority of profiling studies, individual accessions were used in these investigations, and results could therefore not be correlated quantitatively with different tolerance levels that occur in natural ecotypes. Here, we use nine accessions of Arabidopsis for a complete analysis of phenotypic variation for freezing tolerance, changes in gene expression, and metabolite status, and we perform a correlation analysis of changes at the molecular and physiological level. This provides more information than the use of single accessions for defining metabolites and transcripts that may be causally related to improved freezing tolerance and allows the separation of possible treatment-specific effects from those that may be functionally relevant for cold acclimation. We find considerable variation in nonacclimated as well as acclimated freezing tolerance of the various accessions and identify pathways that are associated with high freezing tolerance. This approach provides new insights into the processes that may be most important for improved freezing tolerance.
RESULTS
Natural Variation of Freezing Tolerance
To cover a wide array of phenotypic variation, we selected three laboratory and six natural accessions of Arabidopsis representing habitats from 16° to 66° northern latitude. These were C24, Canary Islands (Can), Coimbra (Co), Columbia-0 (Col-0), Cape Verde Islands (Cvi), Landsberg erecta (Ler), Niederzenz (Nd), Rschew (Rsch), and Tenela (Te). For the correlation of phenotypic variance with habitat temperatures, we assigned the origin of C24, Col-0, and Ler to their closest natural relatives Co, Gückingen (Gü), and Landsberg (La), respectively (Schmid et al., 2003; Nordborg et al., 2005; www.TAIR.org). These nine accessions represent different life history strategies; Te, in common with other accessions from Finland (Kuittinen et al., 1997), is likely to be a winter annual, as too are accessions that are late flowering (Napp-Zinn, 1985) such as Can. In contrast, the synchronous and early flowering of the other accessions suggests a summer annual habit (Table I).
Table I.
Harvest details for the experiments used in this study
| Experiment 1
|
Experiment 2
|
Experiment 3
|
Mean Age | |
|---|---|---|---|---|
| 16.10.03
|
06.01.04
|
05.04.04
|
||
| Agea (DAS) | Age (DAS) | Age (DAS) | ||
| Te | 61 | 57 | 56 | 58 |
| Rsch | 46 | 45 | 44 | 45 |
| Col-0 | 46 | 45 | 43 | 45 |
| Nd | 43 | 41 | 42 | 42 |
| Ler | 46 | 43 | 43 | 44 |
| Co | 46 | 44 | 43 | 44 |
| C24 | 43 | 42 | 42 | 42 |
| Can | 81 | 79 | 70 | 77 |
| Cvi | 44 | 42 | 44 | 43 |
Age of nonacclimated plants.
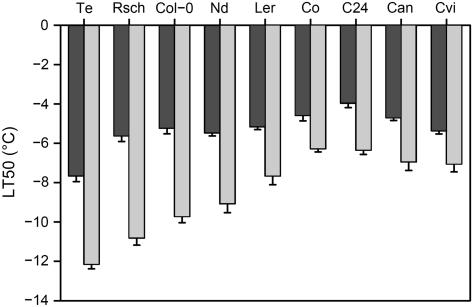
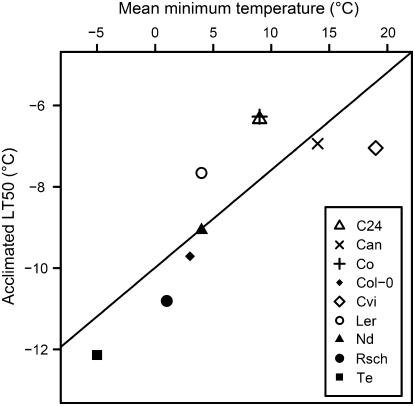
To obtain a quantitative measure of freezing tolerance in these accessions, we determined the temperature at which 50% electrolyte leakage occurred from detached leaves (Rohde et al., 2004). Freezing tolerance varied by 5.9°C in acclimated and 3.7°C in nonacclimated leaves (Fig. 1). The effect of genotype, treatment, and the genotype × treatment interaction on freezing tolerance, as determined by an ANOVA analysis, were all highly significant (P = 4 × 10−14, 3 × 10−19, and 3 × 10−5, respectively). Acclimated tolerance showed continuous variation and correlated significantly (P = 0.007; R2 = 0.67) with the average minimum habitat temperature recorded during the coldest month of the growing season (Fig. 2). This correlation was not dependent on our classification of accessions as summer or winter annuals, as it was significant even when all accessions were considered either as summer or winter annuals (P = 0.02 and 0.03; R2 = 0.54 and 0.51, respectively). Correlations were also significant (P < 0.03; R2 > 0.6) when Ler, which differs from its natural relative due to the erecta mutation, was excluded from these correlations. These data indicate the robust nature of this correlation. Nonacclimated tolerance was generally similar (−4.75°C ± 0.85°C) except for Te, which was >2°C more tolerant than all other accessions (Fig. 1). The ability to increase freezing tolerance during cold acclimation (acclimation capacity) also varied between accessions. Col-0, Rsch, and Te had high acclimation capacities of 4.5°C to 5.2°C, and C24, Can, Co, Cvi, and Ler had low capacities of 1.7°C to 2.5°C, with Nd intermediate (3.6°C).
Figure 1.
Freezing tolerance of Arabidopsis accessions. Histogram of the temperature at which 50% electrolyte leakage occurred from detached leaves (LT50) of nonacclimated plants (dark gray bars) and after 14 d of cold acclimation at 4°C (light gray bars). Experiments were performed as described (Rohde et al., 2004). Accessions used were Te, Rsch, Col-0, Nd, Ler, Co, C24, Can, and Cvi. Data are means ± se from four to six independent experiments.
Figure 2.
Correlation between habitat temperature and acclimated freezing tolerance. Acclimated freezing tolerance is expressed as the temperature at which 50% electrolyte leakage occurred from detached leaves (LT50) of plants after 14 d of cold acclimation at 4°C (compare Fig. 1). Temperature data are the average minimum temperature during January for winter annuals and April for summer annuals from the nearest recording station (www.weatherbase.com). Data are means from four to six independent experiments.
Plants were cold acclimated for 14 d, several days past the initial increase in freezing tolerance (Wanner and Junttila, 1999) yet before any new leaves had been formed. This time point should represent steady-state levels of transcripts and metabolites required to maintain the cold acclimated state. Plants were either harvested for nonacclimated measurements or cold acclimated when the inflorescence was 0.5 to 1 cm long (bolting stage). Using fully developed leaves from mature rosettes provided a defined system for which freezing tolerance, transcripts, and metabolites could be directly compared. As there are leaf order-dependent effects on freezing tolerance (Takagi et al., 2003) using fully developed leaves should reduce both within plant variation and between accession differences that may be associated with developmental stages. Except for Can and Te, the mean age at bolting was 42 to 45 d after sowing (DAS; Table I). Te had a mean age of 58 DAS and Can had a mean age of 77 DAS. Only Can showed signs of senescence, i.e. anthocyanin accumulation and yellowing of the oldest leaves. Because of the large differences Can showed in comparison to the other accessions, it was excluded from analyses that required direct comparisons of expression or metabolite levels; its presence as an outlier would influence results. However, its consideration in the overall analyses does provide important insights. For clarity, the remaining eight accessions are referred to as the core accessions.
Acclimation Capacity
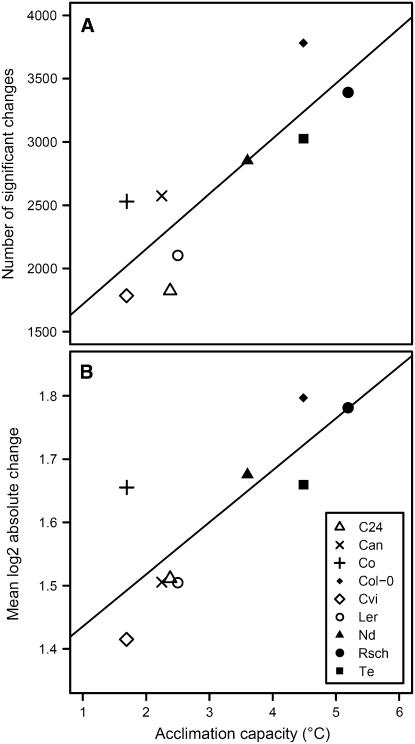
Using nine accessions, we were able to investigate how the massive transcriptional changes that occur during cold acclimation (Fowler and Thomashow, 2002; Kreps et al., 2002; Seki et al., 2002; Hannah et al., 2005; Lee et al., 2005) vary between accessions and if there was differential regulation that may explain their wide range of tolerance levels. Transcripts that were significantly changed in each accession were identified and compared with the ability of each accession to cold acclimate. This indicated that the number of significant changes in gene expression during cold acclimation was significantly (P = 0.004; R2 = 0.71) correlated with the ability to cold acclimate (Fig. 3A), as was the amplitude of change (P < 0.008; R2 = 0.66) among the 423 transcripts that changed significantly in all accessions during acclimation (Supplemental Table I; Fig. 3B). With the exception of Co, there was a 10% to 25% increase of mean expression change for accessions that acclimate well in comparison to those that acclimate poorly.
Figure 3.
Correlation between acclimation capacity and the number and magnitude of expression changes. Acclimation capacity was measured as the difference between the nonacclimated and acclimated LT50 of detached leaves (compare Fig. 1). This is plotted against the number of significant expression changes (A) and the average magnitude of change for the 423 transcripts that change significantly in all accessions during cold acclimation (B). Expression changes were evaluated by fitting a linear model to compare nonacclimated and acclimated expression using the Limma software. Changes were classified as significant if the gene-wise FDR corrected F test and t test P values were both less than 0.05. For A nonacclimated replicate 1 was excluded so sample sizes were equal for all accessions.
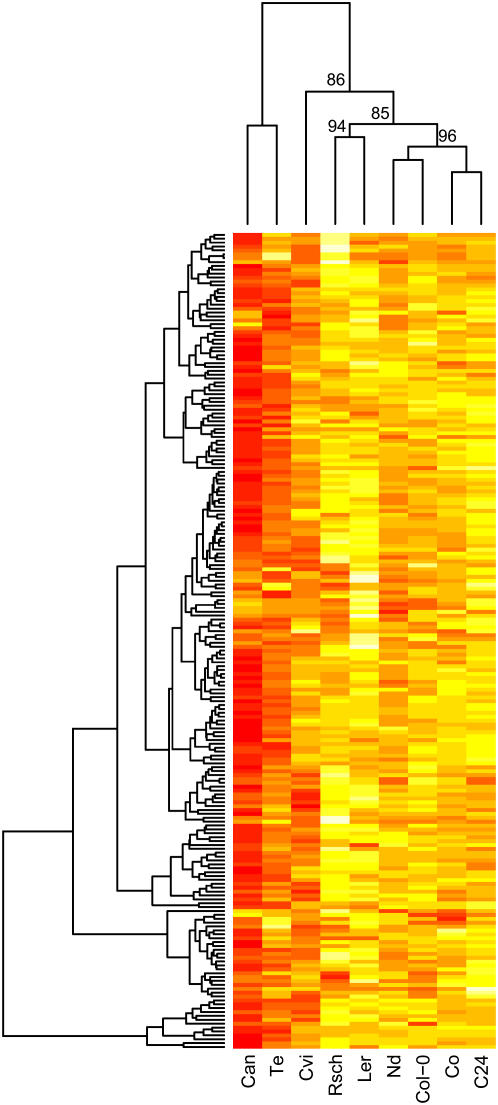
In contrast to transcript data, there are fewer reports on cold-induced metabolic changes in Arabidopsis. The lower acclimation capacity of Cvi in comparison to Wassilewskija-2 was associated with an attenuated accumulation of many cold-responsive metabolites (Cook et al., 2004). Hundreds of metabolite changes also occur in Col-0, but it has not been compared to other accessions (Kaplan et al., 2004). We compared metabolite changes during cold acclimation for 210 metabolites detected in all nine accessions. All accessions showed large increases (1.7- to 5.6-fold) in their total metabolite content during cold acclimation, with the exception of Can, whose content decreased. Te, similar to Can, behaved differently from all other accessions, with metabolite increases reduced in magnitude and a much higher proportion of metabolites decreasing during cold acclimation (Fig. 4; Supplemental Table II). Our data confirm the reduced magnitude of changes in Cvi (Cook et al., 2004). However, the overall magnitude of changes in Ler and C24 in comparison to Te, Nd, and Col-0 indicated there was no simple correlation between global metabolic changes and acclimation capacity (Fig. 4). To prevent bias due to missing values, this analysis did not include all metabolites. Detected metabolites varied from 302 to 373 for the nine accessions. The proportion of these showing a significant (false discovery rate [FDR]; P < 0.05) change during cold acclimation was compared (Supplemental Table II). The proportion of metabolites that accumulated ranged from 0.76 to 0.87 for the accessions C24, Co, Col-0, Ler, Nd, and Rsch, while Cvi had 0.66, Te 0.45, and Can only 0.25. The proportion of metabolites decreasing was 0.11 for Cvi, 0.15 for Te, and 0.37 for Can, but less than 0.07 for all other accessions. There are probably more metabolites decreasing, as many were only reliably detected in nonacclimated samples (Supplemental Table II). Because acclimated samples were 1/5 split injected to prevent peak saturation, missing peaks could also represent metabolites that fell below their detection limit. These are included in Supplemental Table II but are not considered further.
Figure 4.
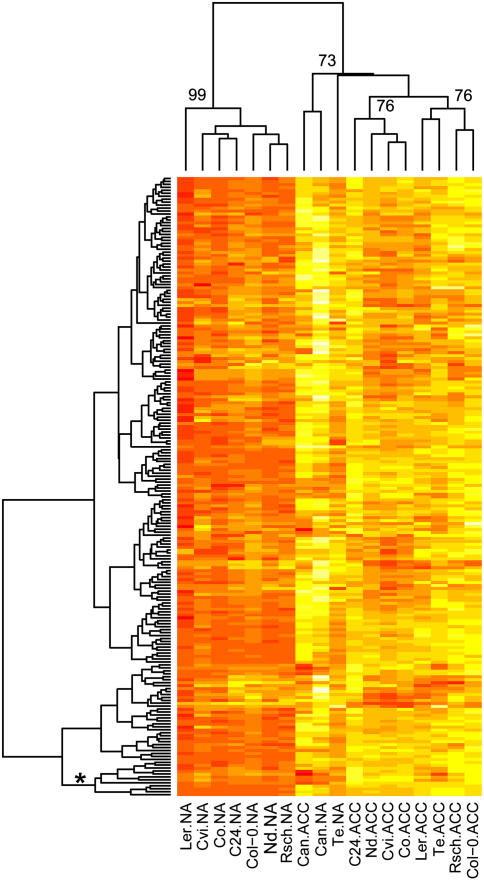
Hierarchical clustering of metabolite changes during cold acclimation. Data are the differences between nonacclimated and acclimated median log2 normalized peak area scaled to unit variance. Euclidean distance was used for clustering. Data for 210 metabolites detected in at least 10 replicates for all sample types are shown. Red indicates the lowest values and white the highest. Approximately unbiased bootstrap probabilities (%), calculated using the pvclust R package (Suzuki and Shimodaira, 2006), are given for selected nodes. A version including the metabolite names is available as Supplemental Figure 2.
To investigate functionally relevant changes (Hannah et al., 2005), we tested for overrepresentation of Mapman functional groups (Thimm et al., 2004; Usadel et al., 2005) among the 1,000 most significantly up- and down-regulated transcripts for each accession during cold acclimation. In most accessions, there was significant overrepresentation of secondary metabolism, transport, and much of primary metabolism among up-regulated transcripts and of the photosynthesis, lipid metabolism, nucleotide metabolism, hormone, and redox groups among down-regulated transcripts (Table II; Supplemental Fig. 1). In Cvi, a smaller number of transcripts from the primary metabolism functional groups were up-regulated, but more were down-regulated as compared to the other accessions.
Table II.
Analysis showing overrepresented functional groups for selected gene lists
The proportion of the selected gene list within a Mapman (Thimm et al., 2004; Usadel et al., 2005) functional group was compared to the proportion of all genes on the ATH1 array that were in the selected gene list. The P values from Fisher exact tests are given, with lower (more significant) P values indicating that genes within that functional group respond more than expected by chance; P values < 0.05, 0.01, and 0.001 are shown in underlined, italic, and bold text, respectively. A conservative correction for multiple testing over all performed tests indicate the highlighted P values correspond, respectively, to FDR P values of < 0.116, 0.038, and 0.006.
| Mapman Group | Acclimation Capacitya
|
Acclimatedc Correlation | Nonacclimatedd
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up-Regulated
|
Down-Regulated
|
|
|
Up-Regulated
|
Down-Regulated
|
|||||||||||||||||||
| Te | Rsch | Col-0 | Nd | Ler | Co | C24 | Cvi | Canb | Te | Rsch | Col-0 | Nd | Ler | Co | C24 | Cvi | Can | +ve | −ve | Te | Can | Te | Can | |
| Photosynthesis | – | – | – | – | – | – | – | – | –e | 1E-35 | 6E-06 | 1E-17 | 1E-12 | 0.001 | 0.015 | 3E-04 | 0.007 | 7E-50 | – | 1E-11 | – | – | 3E-09 | 9E-56 |
| Major carbohydrate | 6E-06 | 6E-06 | 0.001 | 0.009 | 1E-06 | 0.003 | 9E-05 | 0.157 | 0.003 | 0.156 | 0.815 | 0.061 | 0.061 | 0.484 | – | 0.157 | 0.031 | 4E-04 | 0.012 | – | 0.057 | 0.592 | – | 0.229 |
| Minor carbohydrate | 0.002 | 2E-04 | 0.014 | 0.029 | 0.004 | 0.004 | 0.001 | 0.031 | 0.511 | 0.189 | 0.513 | 0.515 | 0.280 | 0.826 | 0.828 | 0.829 | 0.030 | 0.386 | 0.076 | 0.330 | 0.089 | 0.019 | 0.524 | 0.205 |
| Glycolysis | 3E-04 | 2E-07 | 3E-04 | 0.049 | 1E-05 | 6E-05 | 3E-07 | 0.051 | 0.004 | 0.344 | – | 0.192 | 0.749 | – | 0.343 | – | 0.005 | 0.197 | 0.172 | – | 0.188 | – | 0.316 | 0.017 |
| Fermentation | 2E-04 | 0.003 | 0.123 | 0.021 | 0.003 | 0.003 | 0.125 | 0.022 | 0.021 | – | 0.468 | 0.125 | 0.470 | 0.122 | 0.471 | – | 0.003 | – | 0.161 | – | – | 0.091 | – | 0.344 |
| Gluconeogenesis/glyoxylate cycle | 0.386 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 0.393 | 0.126 | – | – | 0.205 | – | 1.000 |
| OPP | 0.170 | 0.013 | 0.051 | 0.652 | 0.053 | 0.653 | 0.653 | 0.654 | – | 0.172 | 0.652 | 0.013 | 0.052 | – | 0.653 | 0.054 | 0.053 | 0.177 | – | 0.136 | – | 0.317 | 0.124 | 0.018 |
| TCA/org. transformation | 0.100 | 0.004 | 1E-05 | 0.001 | 8E-05 | 0.001 | 3E-06 | 0.789 | – | 0.101 | 0.186 | 0.032 | 0.100 | 0.184 | 0.789 | 0.789 | 0.424 | 0.105 | – | 0.237 | – | 0.156 | 0.075 | 0.166 |
| Electron transport/ATP synthesis | 0.610 | 0.008 | 0.130 | 0.309 | – | – | 0.310 | 0.009 | 0.021 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Cell wall | – | 0.151 | 0.825 | – | 0.008 | 0.188 | – | – | – | 1E-07 | – | – | – | – | – | 0.662 | – | – | 0.414 | 0.002 | 0.842 | – | 0.044 | 1E-10 |
| Lipid metabolism | 0.327 | 0.715 | – | 0.714 | 0.037 | 0.087 | 0.036 | – | 0.902 | 2E-04 | 0.019 | 1E-05 | 2E-04 | 0.009 | 0.002 | 2E-06 | 2E-04 | 0.001 | – | 0.046 | 0.117 | – | 9E-07 | 4E-08 |
| N-metabolism | 0.300 | – | – | – | – | – | – | – | – | – | 0.022 | 0.300 | 0.094 | – | 0.301 | – | 0.095 | 0.309 | – | – | – | 0.439 | 0.377 | 0.160 |
| Amino acid metabolism | 0.424 | 0.286 | 0.789 | 0.593 | – | – | 0.424 | – | 0.591 | 0.186 | 0.182 | 0.002 | 4E-06 | 0.062 | 2E-04 | 0.084 | 7E-08 | 1E-08 | 1E-04 | 0.115 | 0.224 | 0.019 | 0.015 | 0.011 |
| S-assimilation | 0.125 | 0.125 | 0.466 | – | 0.022 | 0.470 | 0.470 | 0.472 | 0.466 | 0.471 | 0.021 | 0.021 | 0.003 | 0.122 | 0.022 | 0.472 | 0.126 | 0.003 | – | 0.031 | – | 0.303 | – | – |
| Metal handling | – | – | 1.000 | 0.296 | 0.063 | 0.296 | 0.601 | 0.299 | – | – | 1.000 | 1.000 | – | 0.293 | 0.602 | – | 0.064 | 0.129 | – | 0.108 | – | 0.073 | 0.245 | 0.448 |
| Secondary metabolism | 2E-05 | 4E-04 | 0.001 | 0.001 | 9E-05 | 0.007 | 0.019 | 0.014 | 2E-05 | 0.159 | 0.059 | 0.197 | 0.003 | 0.193 | 0.558 | 0.160 | 0.019 | 0.047 | 0.004 | 0.299 | – | 0.049 | 0.109 | 0.048 |
| Hormone metabolism | – | – | – | – | 0.184 | – | – | – | – | 5E-06 | 8E-08 | 3E-05 | 1E-04 | 2E-06 | 3E-05 | 4E-05 | 0.065 | 0.033 | 0.702 | 0.292 | – | 0.389 | – | – |
| Co-factor and vitamin metabolism | – | 0.036 | 0.110 | 0.112 | 0.010 | 0.709 | 0.432 | 0.711 | – | 0.434 | 0.431 | 0.432 | 0.433 | 0.430 | 0.037 | 0.116 | 0.433 | 0.120 | – | 0.573 | – | – | – | 0.405 |
| Tetrapyrrole synthesis | – | – | 0.656 | 0.181 | 0.659 | 0.182 | 0.182 | 0.059 | – | 1E-06 | 0.003 | 0.015 | 0.015 | 0.056 | 0.183 | 0.660 | 0.659 | 0.001 | – | 3E-06 | – | – | 3E-10 | 2E-11 |
| Stress | – | 0.023 | 0.151 | 0.275 | 0.065 | 0.029 | 0.093 | 0.133 | – | – | – | – | – | – | – | – | 0.867 | – | 0.041 | – | 0.879 | 0.034 | – | – |
| Redox.regulation | 0.593 | 0.007 | 0.030 | 0.073 | 0.032 | 0.031 | 0.013 | 0.113 | 0.209 | 4E-05 | 1E-05 | 1E-06 | 1E-07 | 3E-04 | 0.001 | 5E-05 | 1E-06 | 6E-10 | – | 0.440 | – | 0.279 | 0.001 | 3E-11 |
| Polyamine metabolism | 0.017 | 0.443 | 0.017 | 0.017 | 0.444 | 0.017 | 0.017 | 0.017 | 0.017 | – | 0.108 | 0.109 | 0.443 | 0.439 | 0.110 | 0.110 | 0.110 | 0.450 | – | – | – | – | – | 0.093 |
| Nucleotide metabolism | – | 0.107 | 0.105 | 0.107 | 0.322 | 0.227 | 0.550 | 0.229 | 0.164 | 0.009 | 0.025 | 0.045 | 0.025 | 0.317 | 0.009 | 0.325 | 0.026 | 0.002 | – | 0.012 | 1.000 | 1.000 | 0.008 | 0.002 |
| Biodegradation of Xenobiotics | 0.300 | 0.299 | 0.295 | 0.299 | – | 0.300 | – | 0.303 | – | – | – | 0.300 | 0.300 | 0.021 | 0.095 | 0.004 | 0.022 | 0.309 | 0.037 | – | 0.042 | 0.125 | – | – |
| C1-metabolism | – | – | – | – | 0.648 | – | – | – | – | 0.011 | – | 0.158 | 0.047 | – | 0.159 | 0.649 | 0.159 | 0.012 | – | – | – | 0.733 | – | 0.340 |
| Miscellaneous | – | 0.127 | 0.143 | 0.007 | 1.000 | 0.729 | 0.533 | 0.334 | – | 0.004 | 0.366 | 0.127 | 0.052 | 0.012 | 0.678 | 0.005 | 0.032 | 0.033 | 0.038 | 0.003 | 1E-05 | 0.001 | 0.059 | 0.611 |
| RNA | 0.295 | – | – | – | – | – | – | – | 0.055 | – | 0.774 | – | – | – | – | – | – | – | – | – | 0.048 | 0.002 | – | – |
| DNA | – | – | – | – | – | – | – | – | – | – | – | – | 0.322 | – | 0.227 | – | – | – | 0.683 | – | – | – | – | – |
| Protein | 0.342 | 0.689 | 0.160 | – | – | – | – | 0.178 | 0.269 | – | – | – | – | – | – | – | – | – | – | 0.164 | – | – | 0.003 | – |
| Signaling | – | – | – | – | – | – | 0.825 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 4E-09 | – | – |
| Cell | – | – | – | – | 0.697 | – | – | – | 1.000 | – | – | – | – | – | 0.559 | – | – | – | – | – | – | – | – | – |
| Development | 0.092 | – | – | 0.344 | 0.916 | 0.008 | 0.035 | 0.251 | 0.027 | – | – | – | – | – | – | – | – | – | 0.432 | – | 0.180 | 0.077 | – | – |
| Transport | 2E-04 | 5E-04 | 2E-05 | 1E-07 | 0.002 | 1E-09 | 2E-04 | 7E-08 | 3E-05 | 1.000 | 0.168 | 0.571 | 0.936 | 0.034 | 0.518 | 0.029 | 0.258 | – | 0.130 | 0.475 | 0.660 | 0.004 | 0.087 | 0.593 |
| Not assigned.no ontology | – | – | 0.964 | – | – | – | – | – | – | – | – | – | – | 0.789 | – | – | 0.825 | – | – | – | – | – | – | – |
| Not assigned.unknown | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
The 1,000 most significant changes for each accession were used.
Accessions are ordered by latitude, except Can, which is an outlier (see text).
Genes significantly (FDR P < 0.05) correlating with acclimated LT50 in the eight core accessions were used.
Genes that were significantly (FDR P < 0.05) differentially expressed in Te and Can compared to the other seven accessions were used.
Missing values represent underrepresented groups.
The CBF transcription factors CBF1 to 3 have a central role in cold acclimation (Gilmour et al., 2004), and the ZAT12 regulon contains cold-responsive genes and partially overlaps with that of CBF2 (Vogel et al., 2005). Our data revealed that the CBF and ZAT12 regulons were significantly overrepresented among genes changing during cold acclimation in all accessions (Table III). Consistent with the report that the most highly up-regulated genes belong to the CBF and ZAT12 regulons (Vogel et al., 2005), overrepresentation was highest among the 1,000 most significant changes (data not shown). Col-0 was the only accession with significantly increased ZAT12 expression (Supplemental Table I) and also had the largest overrepresentation of genes up-regulated by ZAT12 overexpression (Table III). In contrast, CBF genes were significantly induced in all accessions except Nd and Te, with varying contributions of individual members of the CBF family. CBF1 to 3 were induced in Can and Cvi, CBF1 and CBF2 in Col-0, Ler, and Rsch, and CBF2 in C24 and Co (Supplemental Table I).
Table III.
Overrepresentation of the CBF2 and ZAT12 regulons among selected gene lists
The number of genes in the selected lists changed in a common direction by CBF2 or ZAT12 overexpression were compared to the number expected, by chance, to be changed. Lower (more significant) P values and higher fold changes indicate gene lists with more overlap with the transcription factor regulon. Gene lists were as described in Table II, except for acclimation capacity lists, which contain all significant changes (FDR P < 0.05) instead of the 1,000 most significant changes.
| Analysis | Totala | Up-Regulated
|
Total | Down-Regulated
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CBF2
|
ZAT12
|
CBF2
|
ZAT12
|
|||||||||||
| Overlapb | ±c | P Valued | Overlap | ± | P Value | Overlap | ± | P Value | Overlap | ± | P Value | |||
| Acclimation capacity | ||||||||||||||
| Te | 1,622 | 65 | 4.6 | 1.1E-26 | 16 | 2.6 | 2.8E-04 | 1,969 | 18 | 3.7 | 6.8E-07 | 65 | 3.2 | 4.9E-18 |
| Rsch | 1,841 | 96 | 5.9 | 3.6E-52 | 18 | 2.6 | 1.2E-04 | 1,893 | 15 | 3.2 | 4.2E-05 | 50 | 2.6 | 3.2E-10 |
| Col-0 | 2,052 | 78 | 4.3 | 3.3E-31 | 26 | 3.4 | 1.2E-08 | 2,034 | 15 | 2.9 | 9.6E-05 | 53 | 2.5 | 1.3E-10 |
| Nd | 1,331 | 78 | 6.7 | 1.6E-44 | 15 | 3.0 | 1.0E-04 | 1,358 | 11 | 3.2 | 4.6E-04 | 43 | 3.1 | 2.8E-11 |
| Ler | 1,269 | 92 | 8.2 | 4.0E-62 | 15 | 3.2 | 6.1E-05 | 1,024 | 9 | 3.5 | 9.1E-04 | 34 | 3.2 | 1.4E-09 |
| Co | 1,530 | 63 | 4.7 | 2.0E-26 | 16 | 2.8 | 1.4E-04 | 1,609 | 13 | 3.2 | 1.3E-04 | 56 | 3.4 | 2.0E-16 |
| C24 | 719 | 49 | 7.8 | 6.9E-30 | 11 | 4.1 | 7.1E-05 | 995 | 5 | 2.0 | 1.0E-01 | 29 | 2.8 | 4.0E-07 |
| Can | 1,533 | 52 | 3.9 | 7.7E-18 | 9 | 1.6 | 1.8E-01 | 1,469 | 8 | 2.2 | 2.9E-02 | 36 | 2.4 | 1.1E-06 |
| Cvi | 1,019 | 45 | 5.0 | 1.0E-19 | 10 | 2.6 | 4.5E-03 | 1,055 | 7 | 2.6 | 1.6E-02 | 26 | 2.4 | 4.7E-05 |
| Acclimatede | 274 | 39 | 16 | 6.2E-36 | 8 | 7.8 | 8.4E-06 | 455 | 1 | 1 | 0.878 | 25 | 5.4 | 9.4E-12 |
| Nonacclimated | ||||||||||||||
| Te | 298 | 26 | 9.9 | 1.3E-18 | 8 | 7.2 | 1.5E-05 | 434 | 1 | 0.9 | 1.000 | 8 | 1.8 | 0.090 |
| Can | 1,767 | 27 | 1.7 | 0.005 | 18 | 2.7 | 7.1E-05 | 1,974 | 6 | 1.2 | 0.633 | 57 | 2.8 | 3.2E-13 |
Total number of significantly regulated genes.
Number of regulated genes that change in response to CBF2 or ZAT12 overexpression.
cFold change between the actual number of CBF2 or ZAT12 regulated genes present in each list versus the number expected by chance.
P value from Fisher exact test; P values < 0.001 are in bold.
Up- and down-regulated correspond to positive and negative correlating transcripts, respectively.
Acclimated Freezing Tolerance
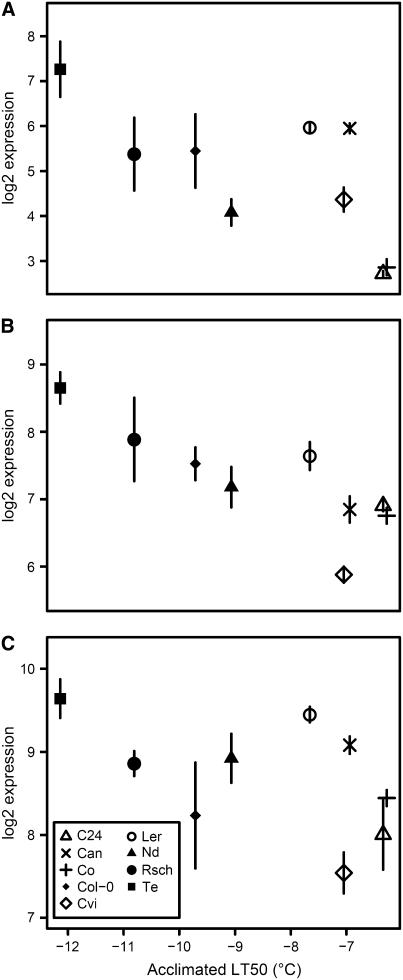
There were 731 transcripts that correlated significantly (FDR P < 0.05) with acclimated LT50 for the eight core accessions. Of these, 275 were positively correlated and 456 were negatively correlated with the freezing tolerance level (Supplemental Table I). Most of these transcripts also changed in abundance during cold acclimation. Photosynthesis, tetrapyrrole synthesis, cell wall, lipid, and nucleotide metabolism were significantly overrepresented among transcripts that negatively correlated with acclimated freezing tolerance (Table II; Supplemental Fig. 1). Carbohydrate, amino acid, and secondary metabolism as well as stress functional groups were overrepresented among positively correlating transcripts. CBF1 and CBF2 had significant positive correlations (FDR P = 0.045 and 0.053, respectively) with acclimated freezing tolerance (Fig. 5) but CBF3 and ZAT12 did not. Among transcripts positively correlating with acclimated freezing tolerance, those up-regulated by ZAT12 or CBF2 (Vogel et al., 2005) were highly overrepresented (Table III). Transcripts down-regulated by overexpression of ZAT12 were enriched among negatively correlating transcripts but those down-regulated by CBF2 were not.
Figure 5.
Expression of the CBF transcription factors in cold acclimated plants. Scatterplots of GCRMA (log2 scale) expression estimates for CBF1 (A), CBF2 (B), and CBF3 (C) against the LT50 of detached leaves from the nine accessions after 14 d cold acclimation. Can was not included in correlation analysis of acclimated freezing tolerance but is shown for information. Data are means ± se from three independent experiments.
At the metabolic level, there were large differences between nonacclimated and acclimated samples, but the differences between acclimated samples were smaller (Fig. 6). In general, there was not a strong cluster separation among acclimated samples, indicating no obvious global metabolic determination of differences in acclimated freezing tolerance. There was moderate support for the clustering of C24, Nd, Co, and Cvi versus Col-0, Rsch, Ler, and Te. Although this would support association between freezing tolerance and the global metabolite profile, the position of Ler and Nd are not in agreement with a clear separation of low and high acclimated tolerance (Fig. 6). To identify individual metabolites that may contribute to freezing tolerance, we correlated metabolite levels with acclimated LT50 for the core accessions. Fifteen metabolites, including Glc, Fru, Gal, raffinose, and Xyl, correlated significantly (FDR P < 0.1) with acclimated freezing tolerance (Supplemental Table II). Suc had the next most significant correlation (FDR P = 0.107). Glc, Fru, and raffinose are among the top four metabolites increased in response to CBF3 overexpression (Cook et al., 2004). CBF3 overexpression increased many other metabolites, and 35 increased more than 7.5-fold (Cook et al., 2004). The majority of these were part of a small cluster of metabolites in our dataset (Fig. 6).
Figure 6.
Hierarchical clustering of metabolites from nonacclimated and acclimated plants. Data are median log2 normalized peak areas scaled to unit variance with Euclidean distance used for clustering. Data for 210 metabolites detected in at least 10 replicates for all sample types are shown. Red indicates the lowest values and white the highest. Approximately unbiased bootstrap probabilities (%), calculated using the pvclust R package (Suzuki and Shimodaira, 2006), are given for selected nodes. The cluster referred to in the text is located at the bottom of the figure and is marked by an asterisk. A version including the metabolite names is available as Supplemental Figure 3.
Nonacclimated Freezing Tolerance
In contrast to acclimated freezing tolerance, nonacclimated freezing tolerance has received little attention. There have been some genetic analyses (Stone et al., 1993; Teutonico et al., 1995), but mechanisms underlying nonacclimated tolerance have so far not been uncovered. The winter-annual accession Te had significantly higher nonacclimated freezing tolerance than all other accessions (Fig. 1), thus providing a basis for an analysis of transcripts and metabolites related to nonacclimated tolerance. In all pairwise comparisons between Te and the other core accessions, 298 and 434 transcripts were significantly increased and decreased, respectively (Supplemental Table I; Supplemental Fig. 1). The 114 consensus genes we previously reported (Hannah et al., 2005) as cold regulated throughout cold acclimation were significantly (Fisher exact test, P = 6 × 10−8) overrepresented among the 298 transcripts increased in Te. With respect to metabolites, nonacclimated Te clustered with acclimated samples of the other accessions, indicating a metabolic state similar to that of cold-acclimated plants (Fig. 6). This is further supported by the increased levels of many previously reported cold-induced metabolites (Cook et al., 2004; Kaplan et al., 2004). In total, there were 167 metabolites increased in Te in comparison to all core accessions (Supplemental Table II).
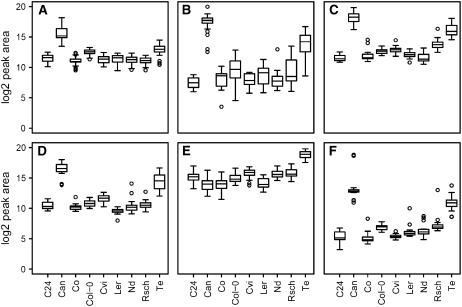
Among transcripts significantly different in Te were CBF1 and 2, which were elevated about 14.5- and 4.7-fold, respectively (Supplemental Table I). CBF3 was increased 6.6-fold but this was only significant in four of seven comparisons. This was paralleled by the significant enrichment of the CBF regulon (Fisher exact test, P = 6 × 10−18) among transcripts elevated in Te, but there was no enrichment of CBF2 down-regulated transcripts among those reduced in Te (Table III). There was also a large overlap with metabolites increased by CBF3 overexpression, of which galactinol, raffinose, Fru, Glc, Pro, and Man were the most prominent (Cook et al., 2004). With the exception of galactinol, these were identical to the top five metabolites discriminating Te from the other core accessions (Fig. 7).
Figure 7.
The levels of metabolites induced by CBF3 overexpression in nonacclimated plants. Boxplots of galactinol (A), raffinose (B), Fru (C), Glc (D), Pro (E), and Man log2 (F) normalized peak area in nonacclimated plants. These metabolites are the top six increased in response to CBF3 overexpression (Cook et al., 2004).
The second late flowering accession, Can, was very different from the other accessions before and after cold acclimation and was the only accession in which the total metabolite content decreased during acclimation. Despite these differences, nonacclimated Can showed similarities to Te and to acclimated rather than nonacclimated samples of other accessions at the metabolite level (Fig. 6). However, because its freezing tolerance was low, these similarities may represent transcripts and metabolites that correlate with life history traits or plant age but are not sufficient to increase freezing tolerance. We identified 1,767 transcripts that were higher in nonacclimated Can than in all accessions except Te (Supplemental Table I). In nonacclimated Can, total metabolite content was similar to acclimated samples, and our previously reported 114 consensus cold-regulated genes (Hannah et al., 2005) were weakly overrepresented (Fisher exact test, P = 0.057) among transcripts increased in nonacclimated Can. Although the expression of the CBF transcription factors was not different from other accessions, the CBF regulon was also significantly overrepresented among these transcripts (Fisher exact test, P = 0.005; Table III). With the exception of Pro, all metabolites discussed in relation to the CBF pathway were similarly or more elevated in nonacclimated Can in comparison to Te (Fig. 7). In addition, the functional groups of transcripts differentially expressed in Can compared to the other accessions, particularly those that were reduced, were similar to those identified in Te and in the analyses of acclimation capacity and acclimated freezing tolerance of the core accessions (Table II).
DISCUSSION
Our survey of nine geographically diverse accessions of Arabidopsis identified considerable natural variation for acclimated and nonacclimated freezing tolerance as well as for acclimation capacity. This is the widest survey of Arabidopsis freezing tolerance reported to date, and the observed variation was greater than reported in previous pairwise comparisons between Cvi and Wassilewskija-2 (Cook et al., 2004), Cvi and Col-0 (Zuther et al., 2004) or Ler (Alonso-Blanco et al., 2005), or between Col-0 and C24 (Klotke et al., 2004; Rohde et al., 2004). In Arabidopsis, natural variation has been reported for various traits (Koornneef et al., 2004), and latitudinal clines have been demonstrated for hypocotyl length (Maloof et al., 2001), circadian period length (Michael et al., 2003), and flowering time (Stinchcombe et al., 2004). However, the selective environmental factors, their ecological importance, i.e. the influence on the genetic structure of populations, and their relationship to underlying molecular mechanisms are not fully understood. Considering the general lack of evidence for environmental selection on genomic polymorphisms in Arabidopsis (Nordborg et al., 2005), our North-South transect revealing the significant correlation of freezing tolerance with habitat temperature (Fig. 2) assigns temperature a high probability of being an important selective force, with freezing tolerance levels being closely adjusted to habitat requirements. Such environmental selection is long known from studies of tree ecotypes being adapted to local climatic conditions. Genotype × environment interactions are well documented for low temperature as well as photoperiod, which are important signaling cues that trigger the onset of winter dormancy and bud break in the spring. More recently, a relationship between latitude of origin and the development of freezing tolerance has been demonstrated for both evergreen and deciduous trees (Repo et al., 2000; Li et al., 2002).
Our data demonstrate a significant association of the complexity of the transcriptional response with acclimation capacity. Accessions with strong cold acclimation had about 1.5- to 2-fold more significant changes in gene expression than those that acclimated poorly (Fig. 3A). The amplitude of gene expression changes common to all accessions was also greater in accessions with higher acclimation capacity (Fig. 3B). As larger magnitudes of change may favor statistical detection, the number and magnitude of expression changes may be related. However, in contrast to the changes that were significant for all accessions, there was no relationship between magnitude and acclimation capacity when all significant changes in each accession were considered. Although the association of number and/or magnitude of expression changes could be a cause or consequence of increased cold acclimation, metabolic changes during acclimation (Supplemental Table II), particularly starch accumulation, argue against generally reduced metabolic activity as the cause of smaller transcriptome changes in poorly acclimating accessions. This suggests that a large number of changes may be functionally relevant for acclimation capacity and is good genomic evidence for the complexity of the trait observed in physiological investigations. For genetic engineering of freezing tolerance, such complexity likely needs to be addressed; overexpressing transcription factors to up-regulate entire regulons may be more promising than trying to identify single, nonregulatory genes with large positive effects on freezing tolerance.
Because secondary treatment effects like reduced light intensity or altered water availability are inevitably coupled to temperature shifts, pairwise comparisons of acclimated and nonacclimated plants cannot unambiguously identify genes directly involved in determining acclimated freezing tolerance. The correlation analysis shown here circumvents that problem by quantitatively relating transcriptome changes with acclimated freezing tolerance of identically treated accessions. This allows for improved functional assignment of transcript changes based on phenotypic variation among accessions. The phenotypic variation of acclimated freezing tolerance between accessions was high (5.9°C) and was 3-fold greater than the minimum acclimation capacity of an individual accession (1.7°C; Fig. 1). Obviously, genes that do not vary between accessions but are still necessary for freezing tolerance cannot be identified by this approach. Likewise, as all accessions were able to cold acclimate to some degree, genes identified may not be essential for the minimal acclimation response. However, the accordance of functional groups of genes changing during cold acclimation and those correlating with acclimated freezing tolerance indicates that most changes in response to low temperature are likely related to cold acclimation (Table II). All analyses indicate the down-regulation of photosynthesis and the up-regulation of photoprotective flavonoids are associated with improved acclimated freezing tolerance. These pathways were not only cold regulated but were also more overrepresented in accessions with higher acclimation capacity (Table II). However, as the largest photosynthetic changes occurred in Can, which acclimated poorly, down-regulation of photosynthesis appears insufficient for increasing freezing tolerance on its own. We identified the induction of genes encoding enzymes of flavonoid biosynthesis as quantitatively related to freezing tolerance in Arabidopsis. Genes encoding flavonoid biosynthetic enzymes were prominent among transcripts up-regulated by cold (4°C) exposure (this study) as well as chilling (13°C), despite an overall low correlation of changes between these treatments (Provart et al., 2003). In addition, genes encoding flavonoid biosynthetic enzymes were among the largest changes in response to chilling in wild-type plants and were also less abundant in several chilling-sensitive mutants (Provart et al., 2003). Evidence for a role of flavonoids in plant stress responses has been accumulated in diverse species based on mutant analysis (for review, see Winkel-Shirley, 2002); however, a selective advantage under field conditions remains to be demonstrated. Our functional correlation with freezing tolerance of natural accessions is a first step in this direction.
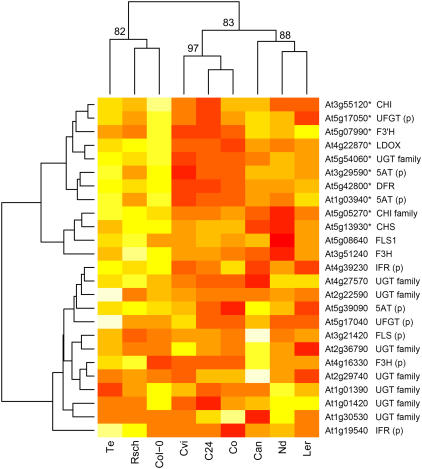
Flavonoid metabolism is regulated by the closely related MYB transcription factors production of anthocyanin pigment 1 and 2 (PAP1 and 2; Borevitz et al., 2000). PAP1 and 2 were both among transcription factors positively correlating with acclimated freezing tolerance, and PAP2 was significantly up-regulated in six accessions during cold acclimation (Supplemental Table I). The 37 genes induced by PAP1 overexpression (Tohge et al., 2005) were also significantly overrepresented (Fisher exact test; P = 0.001) among the genes we identified as positively correlating with acclimated freezing tolerance. More detailed analysis of flavonoid metabolism indicates separation based on a cluster of 12 genes, including CHALCONE ISOMERASE, CHALCONE SYNTHASE, DIHYDROFLAVONOL 4-REDUCTASE, and FLAVONOL SYNTHASE 1, which were up-regulated more in Col-0, Rsch, and Te than in accessions that cold acclimated less (Fig. 8). Strikingly, 10 of these 12 genes were up-regulated by PAP1 overexpression. There was also correspondingly higher induction of PAP2 during cold acclimation in the accessions that acclimated well when compared to those that did so poorly (Supplemental Table I). PAP1 was recently reported as the major quantitative trait locus (QTL) controlling variation of Suc-induced anthocyanin accumulation between Ler and Cvi (Teng et al., 2005). Because Suc was among metabolites positively correlating with acclimated freezing tolerance, its effect on PAP1 expression may partially explain the variation we observe. However, as only the expression of PAP2, not PAP1, was significantly increased during cold acclimation, it is likely that PAP2 may have a more specific, Suc-independent, role in the accumulation of flavonoids in response to cold. In agreement with a Suc-independent role, it was recently reported that PAP2 was not Suc induced (Solfanelli et al., 2005).
Figure 8.
Hierarchical clustering of up-regulated genes from flavonoid metabolism. The GCRMA (log2 scale) expression estimates for genes from the Mapman (Thimm et al., 2004; Usadel et al., 2005) flavonoid metabolism functional group that were significantly up-regulated in at least one accession were used. Data were scaled to unit variance with Euclidean distance used for clustering. Red indicates the lowest values and white the highest. Approximately unbiased bootstrap probabilities (%), calculated using the pvclust R package (Suzuki and Shimodaira, 2006), are given for selected nodes. Genes up-regulated by PAP1 overexpression (Tohge et al., 2005) are marked with asterisks. Gene names are as follows: 5AT, Anthocyanin 5-aromatic acyltransferase; CHI, chalcone isomerase; CHS, chalcone synthase; DFR, dihydroflavonol 4-reductase; F3′H, flavonoid 3′-hydroxylase; FLS, flavonol synthase; F3H, flavonone-3-hydroxylase; IFR, isoflavone reductase; UFGT, UDP-Glc:flavonoid 3-O-glucosyltransferase; UGT, UDP-glucoronosyl/UDP-glucosyl transferase; and LDOX, leucoanthocyanidin dioxygenase. Genes with putative function assigned by sequence similarity are designated (p).
Increased freezing tolerance during cold acclimation is associated with many metabolic changes (Cook et al., 2004; Kaplan et al., 2004). Using multiple accessions, we reach the conclusion that cold acclimation is always associated with numerous and complex metabolic changes, but there is no clear relationship between global metabolite changes and differences in acclimation capacity or differences between the accessions in acclimated freezing tolerance (Figs. 4 and 6). Our data show that the reduced scale of metabolic changes in Cvi compared to other accessions (Cook et al., 2004; Fig. 4) is not characteristic of low acclimation capacity but offers a possible explanation why it occurs. It could be traced to the transcript level, where carbohydrate metabolism was less up-regulated and glycolysis more down-regulated than in the other accessions (Table II). Differences in the TCA cycle may also contribute to differences in the magnitude of metabolic changes, as Can, Cvi, and Te, which showed reduced metabolic changes during cold acclimation (Fig. 4), were the only accessions without significant up-regulation of this group (Table II). With respect to the involvement of metabolites in freezing tolerance, the probable importance of central carbohydrate metabolism is indicated by the identification of Glc, Fru, and Suc among positively correlating metabolites (Supplemental Table II). This is further supported by its overrepresentation also among transcripts positively correlating with acclimated freezing tolerance. The function in plant stress tolerance of soluble sugars, particularly of raffinose, which was identified by our analyses at the metabolite as well as the transcript level, has been highlighted by many investigators (for review, see Smallwood and Bowles, 2002). However, raffinose accumulation alone is neither necessary nor sufficient for cold acclimation in Arabidopsis (Zuther et al., 2004).
The two winter annuals, Can and Te, differed markedly from all other accessions, because in the nonacclimated state, they were metabolically similar to cold acclimated plants of the summer annuals due to their elevated metabolite levels (Fig. 6). During cold acclimation, metabolite increases were reduced in magnitude and more metabolites decreased in Can and Te (Fig. 4; Supplemental Table II). Can was the only accession that showed a net decrease in metabolite content during cold acclimation. As plants were harvested at the bolting stage, the late flowering of Te and Can meant they were about 13 and 32 d older, respectively, than the other accessions that likely contributed to the transcriptional and metabolic differences we observed. However, our analyses provided strong evidence that a substantial proportion of the metabolic and transcriptional differences between Te and the summer annuals were likely related to its increased freezing tolerance. Te was collected in Finland at the northernmost edge of the Arabidopsis range. It showed exceptionally high nonacclimated freezing tolerance, being greater than 2°C more tolerant than all other nonacclimated accessions and more tolerant than C24, Can, Cvi, or Co even when they were cold acclimated (Fig. 1). In nonacclimated plants, 167 metabolites were more abundant in Te in comparison to the summer annual accessions (Supplemental Table II). Of these, four of the top five of known identity (raffinose, Man, Gal, Glc, and Fru) correlated with acclimated freezing tolerance. Likewise, among the 298 transcripts significantly more abundant in Te than in the summer annuals, there was massive enrichment of both cold induced genes (Fisher exact test; P = 2.1 × 10−32) and those positively correlating with acclimated freezing tolerance (Fisher exact test; P = 5.7 × 10−57). Nonacclimated Can showed similarities, both at the metabolite and the transcript level, to nonacclimated Te and all acclimated accessions (Fig. 6; Table II) but had only weak freezing tolerance (Fig. 1). Many of the similarities of nonacclimated Can and Te may be related to the common late flowering habit, causing increased age of these plants at bolting. Importantly, these data indicate that high metabolite content is not sufficient for improved freezing tolerance. It is therefore tempting to speculate that only a few mutations could be responsible for the loss of tolerance in Can that never experiences temperatures below 10°C in its natural habitat. The absence of significant amounts of the stress-related metabolite Pro in Can (Fig. 7) may be a candidate for such an effect. Also, sensitive to freezing mutants have been isolated in Arabidopsis (Warren et al., 1996) that have lost their ability to fully cold acclimate due to single-gene mutations (e.g. Thorlby et al., 2004). This indicates that despite the evident complexity, several genes are essential for freezing tolerance in Arabidopsis and that the inactivation of any of these genes may severely affect acclimated freezing tolerance.
Prime candidates for large effects on freezing tolerance are the major regulators, i.e. transcription factors that control large sets of temperature regulated genes. The best characterized of these are the CBF transcription factors. These have been shown to be strongly induced in response to short-term cold exposure but continue to be up-regulated after 7 d (Fowler and Thomashow, 2002). The up-regulation of CBF1 and 2 in most accessions was paralleled by the overrepresentation of the CBF regulon, indicating a high probability of their importance for maintaining the cold acclimated state. However, variation in different accessions of the occurrence and magnitude of CBF expression increases after 14 d of cold acclimation, and the corresponding status of the expression of their regulon indicates that control is not wholly dependent on CBF expression level in the long term (Supplemental Table I; Table III). These differences may be mediated by either CBF-independent up-regulation of CBF-targeted transcription factors or by direct overlap between the CBF regulon and that of so-far-unidentified regulatory factors. Overlap between the CBF and ZAT12 regulons has recently been reported (Vogel et al., 2005). Interestingly, variation was also seen for ZAT12 that was significantly induced in Col-0 but no other accession (Supplemental Table I). Strong evidence for the importance of the CBF genes comes from our analyses of acclimated and nonacclimated freezing tolerance. Transcripts of CBF1 and CBF2 had significant positive correlations (FDR P = 0.045 and 0.053, respectively) with acclimated freezing tolerance, and in nonacclimated Te, they were elevated about 14.5- and 4.7-fold, respectively. There was a coordinate up-regulation of the CBF regulon; transcripts induced by CBF2 overexpression (Vogel et al., 2005) are significantly more abundant among transcripts positively correlating with acclimated freezing tolerance (Fisher exact test; P = 6 × 10−36) and among those increased in nonacclimated Te (Fisher exact test; P = 1 × 10−18; Table III). There is also a striking similarity between metabolites increased most in response to CBF3 overexpression (galactinol, raffinose, Fru, Glc, Man, and Pro; Cook et al., 2004) and those identified in our analyses. Thus, natural variation of nonacclimated and acclimated freezing tolerance in Arabidopsis could, at least in part, be mediated through the CBF-transcriptional pathway. There is currently little understanding of the basis for natural variation in nonacclimated freezing tolerance and the involvement of CBF. Indeed, increased expression of CBF genes in the absence of a cold stimulus was previously unknown. However, CBF involvement in acclimated tolerance is well established and is further supported by the recent identification of a CBF2 promoter deletion as the likely candidate for a QTL explaining approximately 20% of the variation in acclimated freezing tolerance between Cvi and Ler (Alonso-Blanco et al., 2005). Although this was the largest QTL, it also indicates that the majority of the observed variation was under alternative genetic control. It is also worth noting that the difference between the acclimated freezing tolerance of Ler and Cvi is only small in comparison to the total variation in acclimated freezing tolerance we observed (Fig. 1).
In agreement with a significant but limited role of the CBF pathway, only about 5% to 15% of the transcript changes we identified are established to be under the control of CBF. In addition, this overlap was only seen for transcripts up-regulated by CBF overexpression, but there was no significant overlap among down-regulated transcripts. Genes affected by CBF overexpression but not regulated during acclimation may indicate that some CBF regulated genes do not remain changed in the long term or may represent pleiotropic effects of ectopic overexpression. As our analyses support important roles for the down-regulation of many pathways, these changes are likely to be mediated by pathways independent of CBF. We identified 27 transcription factors, in addition to the CBFs and PAP1/2 that correlated positively with acclimated freezing tolerance, and 24 that showed negative correlations (Supplemental Table I). Negative regulators of freezing tolerance have received little attention. Here we identify two auxin-induced AUX/indole-3-acetic acid (Dharmasiri and Estelle, 2004) and two cytokinin-induced response regulator genes (Hwang and Sheen, 2001) among the negatively correlating transcription factors. This is in agreement with the strong down-regulation of hormone-related genes at all time points following cold treatment (Hannah et al., 2005) and in all accessions during cold acclimation (Table II). While positively correlating pathways have been identified by other methods, combining the power of natural genetic diversity with profiling analyses is advantageous especially to uncover possible roles for negatively correlating metabolic or transcriptome networks, thus helping to uncover so far unknown roles for regulatory mechanisms in complex adaptive responses that are under environmental selection.
MATERIALS AND METHODS
Plant Material
Nine Arabidopsis (Arabidopsis thaliana) accessions were greenhouse grown as described (Rohde et al., 2004; Hannah et al., 2005); a 16-h photoperiod at a minimum of 200 μmol m−2 s−1 and a day/night temperature of 20°C/18°C were used. Nonacclimated plants were harvested for all measurements at the common developmental stage of bolting (0.5–1 cm inflorescence growth). At the same time, replicate plants were transferred to a 4°C growth chamber with a 16-h photoperiod at 90 μmol m−2 s−1 for an additional 14 d (Rohde et al., 2004). Samples were harvested before and after cold treatment after 8 h of light. The experiment was repeated to give three independent biological replicates.
Expression Analysis
Whole rosettes pooled from 10 plants were harvested. Extraction, labeling, hybridization, and scanning were as described (Redman et al., 2004). Each biological replica was hybridized to a single Affymetrix ATH1 Genome array (ATH1) at the German Resource Center for Genome Research, Berlin (www.rzpd.de). The validation of ATH1 data using an independent platform, data normalization, and statistical testing of differential expression have been previously discussed (Hannah et al., 2005). Additionally, we must consider the use of the ATH1 array, designed for the Col-0 genome sequence, for multiple accessions. Polymorphisms between the different accessions could lead to differences in signal intensity but are less likely to affect signal changes. The low frequency of polymorphisms (exon = 0.003; 3′ untranslated region = 0.006; Nordborg et al., 2005) and the use of 11 probes/gene would minimize the effect on signal intensities. In addition, using multiple accessions for all analyses would make it unlikely for polymorphisms to introduce systematic bias. Data were analyzed using the bioconductor software project (Gentleman et al., 2004). Data quality was assessed using the affy (Gautier et al., 2004) and AffyPLM packages. Nonacclimated replicate 1 of C24 and Nd were outliers and excluded from all analyses. The GCRMA algorithm (ver. 1.1.0) was used to obtain expression estimates (Wu et al., 2004). An edited version of the probeset to Arabidopsis Genome Initiative locus code mapping of The Arabidopsis Information Resource (01/06/04) was used as described (Hannah et al., 2005). Additionally Arabidopsis Genome Initiative locus codes that had multiple probe matches were assigned to a single probeset either by taking the original design probeset or, in the small number of cases with no match, by selecting a match at random. These probesets matched 21,166 unique genes, of which 297 also matched a second closely homologous gene. All statistical analyses were made using linear models in the limma bioconductor package that uses an empirical Bayes approach, improving performance with low sample number (Smyth, 2004). First, a model was fitted that fully describes the systematic variation in the data (i.e. nine genotypes, two treatments) using a design matrix. This model was then used to extract comparisons of interest using a contrast matrix. These were the pairwise comparisons of acclimated and nonacclimated samples for each genotype and the comparisons of nonacclimated Te and Can to the summer annuals. These comparisons of interest were combined into F tests with significance defined as meeting the specified level in both the pairwise t test and F test. Linear regression was used for correlations with acclimated LT50. All P values were adjusted for multiple testing (see data analysis). Data from Vogel et al. (2005) was analyzed using the comparison mode of the Affymetrix MAS5 software as described (Hannah et al., 2005). Data are deposited under the accession number E-TABM-52 at Array-Express (www.ebi.ac.uk/arrayexpress).
Metabolite Analysis
In the first experiment, leaf discs were harvested from 30 individual plants. In experiments 2 and 3, six pooled samples from three plants were taken. Sugars and Pro were quantified independently as described (Rohde et al., 2004). Starch was measured using an enzymatic kit (Boehringher). For metabolite profiling, powdered samples (20 mg fresh weight) were extracted using 1 mL of chloroform:methanol:water (2:5:2, v/v/v) at −17°C. After centrifugation, the supernatant was dried and derivatized using 20 μL of 40 mg/mL methoxyamine hydrochloride in pyridine (30°C/90 min) followed by 180 μL of N-methyl-N-trimethylsilyltrifluoroacetamid at 37°C for 30 min. Derivatized samples were stored for 120 min at room temperature prior to injection to allow complete trimethylsilylation of amino moieties. Gas chromatography (GC)-time-of-flight analysis was performed using an Agilent 6890 GC coupled to a LECO Pegasus2 time-of-flight mass spectrophotometer (Leco) as described (Weckwerth et al., 2004). Briefly, nonacclimated samples were injected in splitless mode into tapered, deactivated split/splitless liners containing glasswool (Agilent) at 230°C. To prevent detector saturation and column overloading, acclimated samples were injected at a split ratio of 1:5. The GC was operated at constant helium flow of 1 mL/min with a 40-m RTX-5 column (0.25 mm id) with an integrated 10-m precolumn. After 2 min at 80°C, temperature was increased at 15°C/min to 330°C, which was held for 6 min.
Twenty spectra per second were recorded between mass-to-charge ratio 85 and 500. Peak identification and quantification were performed using the Pegasus software (Leco). Reference chromatograms at a minimum signal/noise threshold of 10:1 were used for automated chromatogram comparisons. Artifact peaks were removed from the result files. Areas were computed using reference unique ions as quantifiers. GC-mass spectrometry raw peak areas were normalized to the independent measure of Suc concentration and log2 transformed. The independent measures of Glc, Fru, raffinose, and Pro indicated good agreement (R2 = 0.83–0.93). Only data from experiment 1 is presented while, due to the reduced replication, experiments 2 and 3 were used for confirmation. Comparisons of known metabolites indicated that 0.83 and 0.85 were significantly (P < 0.05) correlated between experiments 1 and 2 and between experiments 1 and 3, respectively. As some data showed nonnormal distributions and/or unequal variances we used Wilcoxan tests to identify metabolite differences. Correlations were performed by linear regression using the median. Only metabolites detected in at least 10 samples were considered.
Data Analysis
Statistical comparisons of the overlap between different gene lists, such as cold-responsive genes among functional groups or transcriptional regulons, were made using a Fisher exact test in the R software (Hannah et al., 2005). Genetic variation of freezing tolerance was determined by a two-way ANOVA using the R software. Microsoft Excel, Access, and the R software were used to perform general analyses. Cluster validity was assessed using multiscale bootstrap resampling using the pvclust R package (Suzuki and Shimodaira, 2006) and is indicated by the approximately unbiased probability at selected nodes. This probability was proposed to be superior in terms of bias compared to ordinary bootstrap resampling (Suzuki and Shimodaira, 2006). FDR corrections for multiple testing across all genes/metabolites were used; the linear step-up (Benjamini and Hochberg, 1995) method for comparisons and the resampling based point estimate for correlations (Reiner et al., 2003). Unless an alternative value is stated the term significant is used to denote an FDR corrected P value <0.05.
Supplementary Material
Acknowledgments
We thank Aenne Eckhardt for running the GC-mass spectrometry samples, Thomas Altmann for providing seeds, the German Resource Center for Genome Research for ATH1 hybridizations, the R and Bioconductor community for software, Ellen Zuther for help with sugar measurements, Anat Reiner for advice on the FDR corrections, and the Mapman team at the Max-Planck-Institut für Molekulare Pflanzenphysiologie for providing annotations.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Matthew A. Hannah (hannah@mpimp-golm.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso-Blanco C, Gomez-Mena C, Llorente F, Koornneef M, Salinas J, Martinez-Zapater JM (2005) Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol 139: 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57: 289–300 [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong XH, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101: 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9: 302–308 [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54: 767–781 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Ishitani M, Zhu JK (2002) An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc Natl Acad Sci USA 99: 7786–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah MA, Heyer AG, Hincha DK (2005) A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 1: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann MH (2002) Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae). J Biogeogr 29: 125–134 [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotke J, Kopka J, Gatzke N, Heyer AG (2004) Impact of soluble sugar concentrations on the acquisition of freezing tolerance in accessions of Arabidopsis thaliana with contrasting cold adaptation—evidence for a role of raffinose in cold acclimation. Plant Cell Environ 27: 1395–1404 [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuittinen H, Mattila A, Savolainen O (1997) Genetic variation at marker loci and in quantitative traits in natural populations of Arabidopsis thaliana. Heredity 79: 144–152 [DOI] [PubMed] [Google Scholar]
- Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Puhakainen T, Welling A, Vihera-Aarnio A, Ernstsen A, Junttila O, Heino P, Pavla ET (2002) Cold acclimation in silver birch (Betula pendula): development of freezing tolerance in different tissues and climatic ecotypes. Physiol Plant 116: 478–488 [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Napp-Zinn K (1985) Arabidopsis thaliana. In A Halevy, ed, CRC Handbook of Flowering. CRC, Boca Raton, FL, pp 492–503
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J (2004) CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101: 3985–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart NJ, Gil P, Chen W, Han B, Chang H-S, Wang X, Zhu T (2003) Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiol 132: 893–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD (2004) Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38: 545–561 [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19: 368–375 [DOI] [PubMed] [Google Scholar]
- Repo T, Zhang G, Ryyppo A, Rikala R, Vuorinen M (2000) The relation between growth cessation and frost hardening in Scots pines of different origins. Trees-Structure and Function 14: 456–464 [Google Scholar]
- Rohde P, Hincha DK, Heyer AG (2004) Heterosis in the freezing tolerance of crosses between two Arabidopsis thaliana accessions (Columbia-0 and C24) that show differences in non-acclimated and acclimated freezing tolerance. Plant J 38: 790–799 [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Sorensen TR, Stracke R, Torjek O, Altmann T, Mitchell-Olds T, Weisshaar B (2003) Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res 13: 1250–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Smallwood M, Bowles DJ (2002) Plants in a cold climate. Philos Trans R Soc Lond B Biol Sci 357: 831–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004) Linear models and empirical Bayes for assessing differential expression in microarray experiments. Stat App Gen Mol Biol 3: Article3 [DOI] [PubMed]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2005) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis thaliana. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J (2004) A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci USA 101: 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Palta JP, Bamberg JB, Weiss LS, Harbage JF (1993) Inheritance of freezing resistance in tuber-bearing Solanum species: evidence for independent genetic control of nonacclimated freezing tolerance and cold acclimation capacity. Proc Natl Acad Sci USA 90: 7869–7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Takagi T, Nakamura M, Hayashi H, Inatsugi R, Yano R, Nishida I (2003) The leaf-order-dependent enhancement of freezing tolerance in cold-acclimated Arabidopsis rosettes is not correlated with the transcript levels of the cold-inducible transcription factors of CBF/DREB1. Plant Cell Physiol 44: 922–931 [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teutonico RA, Yandell B, Satagopan JM, Ferreira ME, Palta JP, Osborn TC (1995) Genetic-analysis and mapping of genes-controlling freezing tolerance in oilseed Brassica. Mol Breed 1: 329–339 [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Thorlby G, Fourrier N, Warren G (2004) The SENSITIVE TO FREEZING2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a beta-glucosidase. Plant Cell 16: 2192–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al (2005) Extension of the visualization tool MAPMAN to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Wanner LA, Junttila O (1999) Cold-induced freezing tolerance in Arabidopsis. Plant Physiol 120: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, McKown R, Marin AL, Teutonico R (1996) Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckwerth W, Wenzel K, Fiehn O (2004) Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4: 78–83 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Wu ZJ, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909–917 [Google Scholar]
- Zhu J, Shi H, Lee BH, Damsz B, Cheng S, Stirm V, Zhu JK, Hasegawa PM, Bressan RA (2004) An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc Natl Acad Sci USA 101: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, Lee B-h, Zhan X, Manabe Y, Sokolchik I, Zhu Y, Dong C-H, Zhu J-K, et al (2005) HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc Natl Acad Sci USA 102: 9966–9971 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zuther E, Buchel K, Hundertmark M, Stitt M, Hincha DK, Heyer AG (2004) The role of raffinose in the cold acclimation response of Arabidopsis thaliana. FEBS Lett 576: 169–173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.