Abstract
In tobacco chloroplast transcripts 34 nt are efficiently edited to U. No common consensus region is present around all editing sites; however, sites can be grouped in clusters that share short common sequences. Transgene transcripts carrying either the wild-type −31/+22 or −31/+60 sequence near NTrpoB C473, an editing site within tobacco rpoB transcripts, or three different mutated sequences, were all highly edited in vivo. Endogenous transcripts of rpoB, psbL and rps14, all of which contain common sequences S1, S2 and S3 5′ to NTrpoB C473, NTpsbL C2 and NTrps14 C80, were less edited in transgenic plants that over-express transcripts from NTrpoB C473 transgenes. Extent of reduction of endogenous editing differed between transgenic lines expressing mutated −31/+22 regions, depending on the abundance of the transgene transcripts. The −20/−5 sequence contains critical 5′ sequence elements. Synthetic RNA templates with alterations within this 5′ region were less efficiently edited in vitro than wild-type templates, by either tobacco or maize chloroplast extracts. The tobacco chloroplast extract supports both RNA editing and processing of 3′ transcript termini. We conclude that within the −20/−5 region, sequences common to editing sites in the transcripts of rpoB, psbL and rps14 are critical for efficient NTrpoB C473 editing.
INTRODUCTION
RNA editing, a form of RNA processing, occurs in both nuclear and organelle transcripts of diverse organisms. In vascular plants, ∼30 C targets of editing typically exist in chloroplast transcripts, while >400 such targets have been observed in plant mitochondria (1–4). Tobacco chloroplasts are a particularly good model system for editing because deliberate alteration in editing substrates can be assayed in vivo in chloroplast transgenic plants or in vitro with chloroplast extracts (5–7). Tobacco chloroplasts have 34 known editing sites within the 155 939 bp organelle genome, and all are modifications from cytidine to uridine. Of the 34 identified editing sites, 32 sites are known to be efficiently edited, with 70–100% of transcripts modified from C-to-U at each edited position (8). In vascular plant chloroplasts, identified editing sites are almost exclusively within coding regions and occur most frequently at the second position of a codon (2). Editing of all but one of the C targets identified in tobacco results in a change of encoded amino acid (2,8). Edited codons commonly encode amino acids that are conserved among orthologous proteins of other plants (9–11). Defects in editing of some transcripts result in plants with severe phenotypes, producing dysfunctional proteins (12–15). Editing in plant organelles is likely a mechanism for the correction of genomic T-to-C mutations rather than for creation of protein diversity (16,17). The low number of C-to-U modifications, accompanied by the high extent of editing of C targets, suggests the existence of a highly efficient and specific editing mechanism within chloroplasts.
In vivo and in vitro editing studies have focused on the sequence elements responsible for directing editing in chloroplasts as well as in mitochondria. Both organelles edit C-to-U and may share similar mechanisms for editing (18). Regions critical for editing are primarily located in nearby regions 5′ of the editing sites, and in vivo studies have identified a number of editable substrates that carry only 20–40 nt 5′ and 10–20 nt 3′ to the C editing target.
The editing site NTrpoB C473 is within the tobacco rpoB transcript; the C at position 473 from the A of the initiation codon is edited. This editing site has been referred to as rpoB-2 previously, but because of the number of additional species in which chloroplast editing has been characterized, a previous nomenclature system (2) for chloroplast C editing targets has become unwieldy. We propose here to name editing sites by initials of genus and species, gene name, then nucleotides from the A of the closest gene's initiation codon. C473 in tobacco rpoB is at the second position of the codon, altering the encoded amino acid from serine to leucine.
The cis-requirements for editing for NTrpoB C473 were examined previously in vivo by expressing transgenes carrying a small portion of the rpoB gene surrounding the C editing target (5,19–21). A 27 nt sequence flanking the editing site NTrpoB C473 is sufficient for editing; however, a more highly edited template contained 92 nt around the editing site. The lower amount of editing observed in the smaller 27 nt template is most likely due to the loss of important nucleotides in the reduced 3′ and 5′ regions around the edited C, compared to the larger substrate. Therefore, we have created transgenes and transcripts with 54 nt around the edited C to better define the important cis-acting region in vivo and in vitro.
Although no consensus sequence is common to all editing sites, groups of sites with common sequences can be gathered into clusters of sites that may also share sequence-dependent specificity factors. Over-expression of sequences flanking NTrpoB C473 or NTndhF C290 in tobacco chloroplasts results in a reduction in editing of a small group of endogenous editing sites which contain some short common sequence elements (20). All of the known editing sites within tobacco can be grouped into clusters based on short common sequences. Some of these clusters exhibit similar changes in efficiency of editing depending on tissue type (8). Upon over-expression of a template containing a region surrounding NTrpoB C473 in tobacco chloroplast transgenic plants, endogenous rpoB, psbL and rps14 transcripts exhibit less editing at NTrpoB C473, NTpsbL C2 and Ntrps14 C80 than in wild-type plants. These three editing sites carry three common sequence elements of 2–3 nt that we have termed S1, S2 and S3. We have constructed in vitro templates to examine the importance of S1, S2 and S3 in editing of NtrpoB C473 in both tobacco and maize chloroplast extracts. These studies indicate that all three sequences are important for efficient editing of C473 in rpoB transcripts. We have also explored the effect of 5′ and 3′ flanking sequences on editing efficiency in vivo and in vitro. Furthermore, we report the production of a maize chloroplast extract that is capable of editing the tobacco C473 editing site.
MATERIALS AND METHODS
Construction of plastid transformation vectors
The editing site and adjacent bases were amplified by PCR from tobacco leaf genomic DNA and specific-sequence alterations were generated by mutagenic PCR. Five different editing templates were constructed. Editing templates were flanked by NcoI and XbaI restriction sites. Transformation constructs were then created by the integration of the editing templates into the vector pLAA24A (22). Restriction enzyme digestion at sites NcoI and XbaI were used to remove the uidA coding sequence from pLAA24A and insert the NTrpoB C473 gene fragment, creating constructs for bombardment.
Transformation and tissue culture
Standard methods were used to create chloroplast transgenic plants (5,7,19,21,23). Young tobacco seedlings were bombarded with plasmid-coated tungsten particles. After bombardment plants were selected on regeneration media containing 500 mg/l spectinomycin (24). A number of initial transformants were created for each construct, and one line for each construct was maintained for further analysis. Plants were assayed for homoplasmicity after selection through Southern blotting. All plants with integrated constructs remained homoplasmic throughout this investigation, except for R54. Despite continued rounds of selection, line R54 never achieved homoplasmicity and was analyzed as a heteroplasmic plant.
Expressed transcripts from all constructs include an ATG codon because of the use of the NcoI restriction site. Translation beginning at this AUG would be out of frame of rpoB and would proceed just 16 amino acids before reaching a stop codon.
DNA blot analysis
Total DNA was isolated from transgenic and wild-type leaves from shoots grown on RMOP media. DNA (1 μg) was digested using BamHI, electrphoresed on 1% agarose and blotted onto positively charged nylon (Amersham) using a turboblotter (Schleicher and Schuell). Oligonucleotides (PC1.1 and PCα1.2) were used to amplify a 350 nt genomic probe from wild-type genomic DNA overlapping the insertion site. The probe was random labeled using the DECAprime II kit and [α-32P]dCTP and hybridized to the DNA blot for 24 h at 65°C.
S1 nuclease assay
A DNA probe was constructed by PCR using oligonucleotides T7_5′_500s and 500sreverse_Lg. The PCR product was designed to hybridize with the transgenic transcript and overlap the 3′ end terminator sequence from tobacco rps16. The DNA probe was restricted with NcoI (Invitrogen) and the 3′ end of the antisense strand was labeled using a Klenow fill reaction. Labeled probe and 1, 10 or 25 μg of RNA were hybridized overnight. S1 nuclease (500 U/ml) (Promega) was added to the nuclease reaction for 1 h at 37°C and the products were electrophesed on a 5% polyacrylamide gel.
Immunobloting
Immunoblotting was performed as in Hegeman et al. (21). Total leaf protein was obtained from shoots grown on RMOP medium using homogenization buffer containing 50 mM Tris–HCl, 1 mM EDTA, 1× Protease Inhibitor cocktail (Complete, Roche) and 0.1% (v/v) Triton X-100. Protein was quantified using Bio-Rad Protein Assay kit and a BSA standard curve. Total proteins (20 μg) were boiled in SDS–PAGE, electrophoretically separated onto 10% acrylamide gels and transferred to nitrocellulose membranes (Pierce). Membranes were blocked overnight in blocking buffer (5% dried milk powder, 1%TBS-T) after which the primary antibody was added to 1:500 dilution from crude serum. The washed blots were incubated in secondary antibody (horseradish peroxidase conjugated goat anti-rabbit; Amersham) diluted to 1:50 000 and proteins were visualized using SuperSignal West Dura Extended Duration Substrate as per manufacturer's specifications (Pierce Biotechnology).
Editing analysis
Total RNA was isolated using Trizol (Invitrogen) for transgenic plants and wild-type leaves. Contaminating DNA was removed using Turbo DNAse (Ambion) and cDNA was synthesized by reverse transcription (Omniscript; Qiagen) using degenerate hexamers. Transgenic transcripts were amplified using PPrrn2 and either Trps16lg for transcripts with 3′ end I or Trps16sh for amplification of transcripts with both 3′ ends. Amplified transcripts were then assayed for editing extent using the poisoned primer extension assay as described previously (11,21,25).
Substrates for analysis in vitro
For substrates equivalent to the transgenic transcripts, DNA substrates were produced from PCR amplifications using primers T7_5′_500s and either Trps16sh or Trps16lg. For substrates with sequence alterations in the −31 to +22 region around NTrpoB C473, the respective mutagenic PCR primers were used. The bacterial sequences SK and KS were added to flank the region of rpoB to prevent amplification from endogenous nucleic acids. A T7 sequence was added to the 5′ end of the substrate also by PCR amplification. RNA substrates were then produced using the PCR products as template by in vitro transcription using the T7 MEGAshortscript kit (Ambion). RNAs were then purified using the RNA clean-up kit-5 (Zymo Research).
Editing reactions in vitro
The editing reactions were performed as described previously (25). RNA (0.1 fmol) was added to 80 μg of tobacco, competent, chloroplast extract (25) in assay conditions. Maize extracts were prepared from 7 to 10 day old maize plants grown in the same conditions as tobacco (25). Leaves were homogenized and plastids isolated using a Percoll (Amersham Biosciences) gradient. Intact chloroplasts were lyzed using Triton X-100, and dialyzed in Dialysis Buffer (25). Conditions for the maize in vitro assay were identical to the conditions used for tobacco except only 20 μg of chloroplast extract was used. Editing of RNA substrates was analyzed using the poisoned primer extension assay (11,21,25).
In vitro processing assay
0.1 fmol of randomly- or 5′-labeled RNA was incubated in tobacco chloroplast extract for 2 h under the in vitro editing assay conditions described previously (25). RNA was purified by phenol:chloroform extraction and precipitated. Resuspended RNAs were separated on 6% poly-acrylamide gels.
Semi-quantitative RT–PCR
cDNA was synthesized for both short and long transcripts by reverse transcription (Sensiscript; Qiagen) from 50 ng of total RNA isolated from transplastomic plants and the Trps16sh primer. PCR amplification of the cDNA templates utilized the primers Trps16sh and PPrrn2. RT–PCR products from different rounds of PCR were then separated on 3% agarose gels. After 22 cycles differing quantities of RT–PCR products could be distinguished corresponding to the varying amounts of initial transcript. After 40 cycles all bands were of similar intensity and reactions without reverse transcriptase showed no specific amplification.
RESULTS
Production of transgenic plants for further analysis of an rpoB editing site in vivo
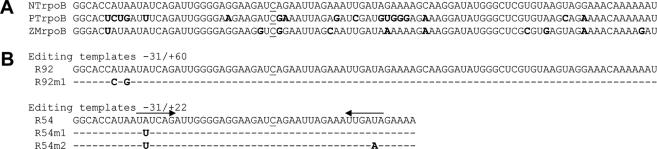
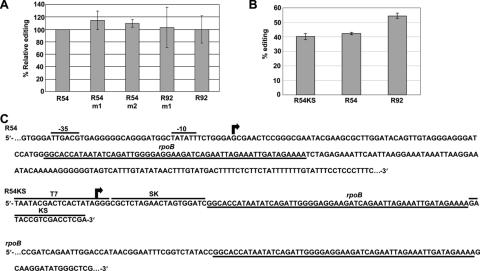
Previously, transgenic plants were analyzed that contained 27 nt surrounding the NTrpoB C473 editing site. Approximately 25% of the transcripts carrying the wild-type tobacco sequence or a sequence altered at either −7 or +2 from the C target were edited, on average (19). However, transcripts carrying a T rather than the wild-type A at −20 were poorly edited; only 3 of 221 individually analyzed transcripts exhibited editing. Furthermore, a homologous sequence from black pine (which contains T rather than A at −20) was also not edited. Because a homologous 92 nt region from maize rpoB had been observed previously to be highly edited (∼50%), we produced transgenic plants carrying a sequence larger than 27 nt, but smaller than 92 nt to further define sequence requirements for editing. A 54 nt region (−31 to +22) from tobacco was inserted into vector pLAA24A, as well as a 92 nt tobacco sequence (−31/+60) for use as a control analogous to the maize region expressed previously in tobacco (19). Several mutated versions were also produced. R92m1 carries 2 nt found in the black pine sequence at −23 and −25. R54m1 contains the −20 T change found inimical to editing in the 27 nt transgene. Because the region −16 to −21 was observed to be complementary to the +12 to +17 sequence, the +12 T was changed to A as a potential compensatory mutation for the −20A to T change (Figure 1).
Figure 1.
RNA substrates were created with regions of sequence around the editing site NTrpoB C473 to study template requirements of editing in vivo. (A) The sequence 31 nt 5′ to 60 nt 3′ around the editing site at position 473 from the initiation codon of Nicotiana tobacum, NTrpoB, was aligned with Pinus thunbergii PTrpoB and Zea mays ZMrpoB sequences. Boldface characters indicate nucleotides different from NTrpoB sequence. The position of the edited nucleotide is indicated by an underline. (B) The sequences of created templates are represented with differences at nucleotides that are divergent between NTrpoB and PTrpoB. Boldface characters indicate nucleotides that differ from the wild-type NTrpoB sequence. Dashes indicate positions where wild-type sequence is present in the template. Complementary sequences present around the editing site are indicated by arrows.
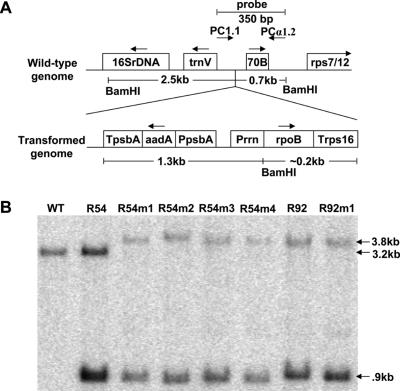
The transformation vector was introduced to young tobacco leaves by biolistic delivery, and plastid genome composition was determined in regenerating plants by Southern blotting (Figure 2). A single R54-containing shoot did not reach homoplasmy despite lengthy efforts and this line was therefore examined as a heteroplasmic plant. Single transformation events of the four other lines achieved homoplasmy after repeated selection on antibiotic medium (Figure 2).
Figure 2.
Creation of transplastomic plants. (A) Diagram representing the wild-type tobacco chloroplast genome and insertion of the transformation cassette. The probe used in the Southern blot is indicated at the top right of the diagram, and spans the wild-type insertion site. (B) Southern blots containing BamHI digested DNA from transplastomic plant leaves were probed with a labeled 350 bp PCR product. Bands at 3.2 kb are due to the untransformed genome. The 3.8 and 0.9 kb bands indicate an integrated transgene.
Regenerating shoots were treated with auxin and grown on rooting medium. Only R54m2 could not be induced to root, all other plants were taken to soil (Figure 3). R54 as a heteroplasmic plant exhibited sensitivity to light and leaves became bleached in exposure to levels of light equal to the normal growing conditions for the other plants. R54 plants did not survive in soil even in low light conditions. R54m1 plants thrived on soil but had severe leaf abnormalities. Leaves appeared malformed, with defects in venation, long trichomes and short internodes. Plants were severely stunted and did not flower. R92 plants were stunted and flowers were male sterile. R92m1 plants grew normally with no other defects observed from the transgene. Progeny resulting from an outcross using male pollen from wild-type plants and chloroplast transformant flowers with R92 were male sterile, but did not display any other significant phenotypes compared to wild-type tobacco plants (data not shown), suggesting that mutant phenotypes have resulted from the bombardment and tissue culture for plant regeneration.
Figure 3.
Phenotypes of transplastomic plant lines. R54 and R54m2 could not grow on soil and were photographed at their last developmental stage in which they would thrive. R54 displayed a bleached phenotype when grown on sucrose containing media under low light intensity. Transplastomic R54m2 shoots grown on regeneration media are shown. All other lines were grown in soil and are shown from two different perspectives. Viewed from the side, left, it is clear many lines display stunted phenotypes characterized by short internodes. From the top view, right, abnormal leaf morphology is evident in plant line R54m1.
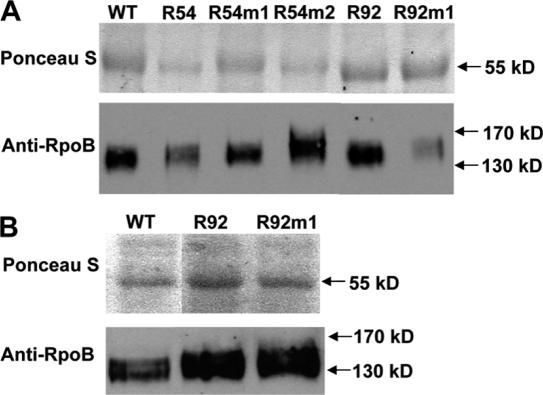
Total protein was isolated from plants and RpoB protein levels determined through immunoblotting. There was no correlation between RpoB protein levels and severity of phenotype, and only R92m1 appeared to have reduced amounts of RpoB protein compared to total protein (Figure 4A). The transplastomic progeny of R92 and R92m1 plants did not display reduced RpoB protein levels (Figure 4B). We have not investigated the developmental phenotypes further, as our primary interest was to analyze the effect of the transgenes on editing of related sequences. We cannot conclude whether or not the rpoB transgene stimulated tissue culture mutations or is directly responsible for any of these phenotypes without obtaining a number of additional independently generated transgenic lines.
Figure 4.
RpoB protein levels in transplastomic plants. To verify equal SDS–PAGE sample loading, blots were stained with Ponceau S to compare levels of the abundant Rubisco large subunit. The 120 kDa RpoB protein was detected using primary antisera raised against a RpoB peptide. (A) Total and RpoB protein levels were determined in protein preparations isolated from leaves of transplastomic plants growing on regeneration media. (B) Determination of total and RpoB protein levels isolated in preparations from transplastomic plants resulting from a cross between wild-type pollen on to stigmas from transformed plants grown in soil.
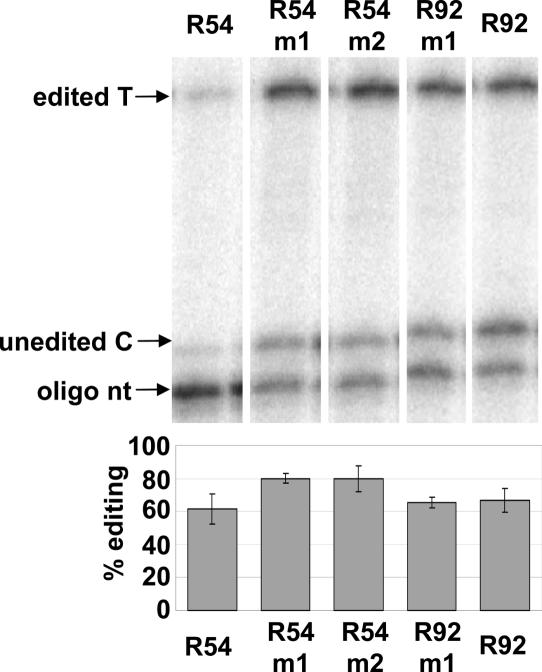
Assay of editing extent of transgene transcripts
Editing of transgene transcripts in leaves of the five transgenic lines were assessed by poisoned primer extension. R92m1 transcripts, which carry altered −23 and −25 nt were 65% edited compared to 67% for the R92 construct, the template with the same region of wild-type sequence. The −23 and −25 nt appear to be of little importance for the editing process. Surprisingly, even the lines carrying the −20A to T change that prevented editing in transgene transcripts of shorter length (−20 to +6) (19), exhibited editing over 60% (Figure 5). In fact, editing of the two lines containing −20A to T changes exhibited somewhat higher editing than the R54 line containing wild-type sequence. Evidently either the addition of 11 nt at the 5′ end or 15 nt 3′ to the edited C has affected the editing efficiency of the transgene transcripts in comparison to the transgenes carrying only 27 nt of chloroplast sequence.
Figure 5.
Poisoned primer extension reactions comparing editing in transgene transcripts between transplastomic plants. Primers PPrrn2 and Trps16sh, which amplify the region from −73 to +57 around the editing site in the transgene, yielded a 131 bp fragment by RT–PCR. Percent editing was calculated from band intensity for duplicate reactions. Error bars represent 1 SD from the mean.
Further characterization of transgene transcripts
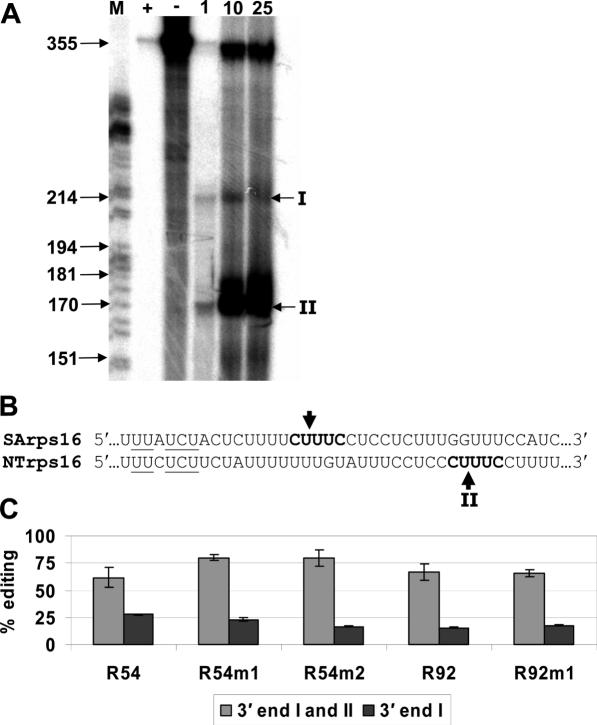
In order to understand how the increased size of the transcripts carrying 54 nt rather than 27 nt of rpoB sequence has enhanced editing efficiency, it was necessary to determine the exact RNA species produced in vivo. The 5′ end of the transcript is determined by the rrn16 promoter, which initiates transcription downstream of eubacterial-like −35/−10 promoter elements (24). The 3′ end of the transcript is determined by the terminator sequence, Trps16, from the 3′-untranslated region (3′-UTR) of tobacco rps16 (22). The 3′ end of the rps16 gene had not been mapped previously in tobacco; however, it had been determined in white mustard (26). In white mustard, a nuclease has been purified and implicated in cleavage at a recognition sequence creating the 3′ end (27,28). We aligned the Trps16 sequence from tobacco with white mustard and found the tobacco sequence carries a sequence similar to the white mustard nuclease recognition element (Figure 6). We performed 3′ end mapping in the tobacco transgenic plants and found that two RNA species are the products of the transcribed transgene in tobacco. One of the two, 3′ end I, might be a precursor to the more abundant 3′ end II. 3′ End II also matches a region of similar sequence where rps16 in white mustard is cleaved to form the mature 3′ end.
Figure 6.
The 3′ ends of transgenic transcripts were determined using S1 nuclease protection mapping. (A) The 355 nt antisense probe was mixed with S1 nuclease (lane +), without S1 nuclease (lane −), and 1, 10 and 25 μg (lanes 1, 10, 25) of total RNA from leaves of transplastomic shoots. Two 3′ ends were observed (I and II). Lane M contains a DNA sequencing reaction, which served as a molecular weight standard. (B) The 3′-UTR sequences from rps16 of Sinapsis alba (SArps16) and N.tobacum (NTrps16) were aligned. Arrows indicate mapped cleavage sites and underlined characters represent a 7mer protein-binding region from Nickelsen and Link (27). (C) The percentage of edited transgenic transcripts calculated by poisoned primer extension reactions with either 3′ end I or both 3′ ends. Error bars represent 1 SD from the mean.
After determining that two RNA species were the product of in vivo transcription of the transgene, we wanted to assess whether transcripts of both sizes were edited to the same extent. Through selective RT–PCR, it was possible to amplify the transcripts with 3′ end I only, unlike the data shown in Figure 5, in which primers were used so that transcripts of both sizes were assayed simultaneously. Of the two RNA species within transgenic plants, the smaller RNAs accumulated more edits in every construct than the longer precursor (Figure 6). This could either result from an inhibition of editing by the extra 3′ sequence, or because the longer transcripts are processed quickly to the smaller size, before significant editing has occurred.
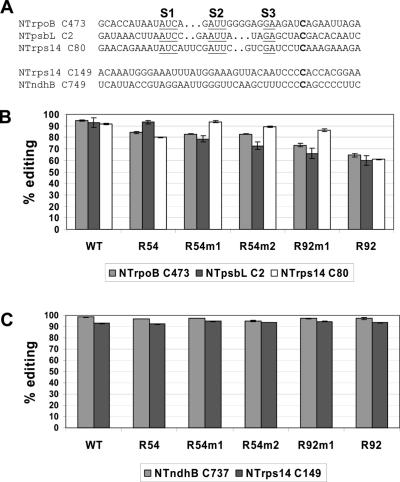
Editing of endogenous transcripts in transgenic plants carrying NTrpoB C473 transgenes
Previously we described editing of chloroplast transcripts in tobacco chloroplast transgenics that were over-expressing a 92 nt maize rpoB gene fragment encompassing the maize chloroplast sequence homologous to NTrpoB C473. Editing of the endogenous site NTrpoB C473 as well as sites NTpsbL C2 and NTrps14 C80, are less edited in these transgenic plants (20). All three sequences were observed to share common elements (Figure 7A) and were therefore described as a ‘cluster’ of editing sites. We analyzed the editing extent of endogenous transcripts of all three cluster members in the five new transgenic lines. All transgenic plants exhibit some reduction, varying from 10 to 30%, of endogenous NTrpoB C473 editing (Figure 7B). In every homoplasmic plant, a reduction of endogenous NTrpoB C473 editing was accompanied by a 10–32% reduction of endogenous NTpsbL C2 editing. A reduction in editing extent of cluster member NTpsbL C2 was not observed in R54. Endogenous NTrps14 C80 editing is reduced 12 and 32% in the wild-type sequence-containing R54 and R92 plants, respectively. Reduced Rps14 C80 endogenous editing correlates with plants with large, >20%, reductions in endogenous NTrpoB C473 and NTpsbL C2. Constructs with differences at −20 had little effect on NTrps14 C80 editing. NTrps14 C80 is less sensitive to over-expression of NTrpoB C473 substrates than is NTpsbL C2 editing. Editing extent of another editing cluster of endogenous sites that share common sequences with each other, NTrps14 C149 and NTndhB C737, but that do not carry S1, S2 or S3, are unaffected by expression of transgenes carrying NTrpoB C473 (Figure 7C).
Figure 7.
Percentage of sites edited in endogenous transcripts within transgenic plants. (A) Sequence alignment with induced spacing, of a cluster of three editing sites. Cluster members: NTrpoB C473, NTpsbL C2 and NTrps14 C80. Underlined characters represent common sequence elements: (S1), (S2) and (S3). (B) Editing in sites that contain S1, S2 and S3 sequences. (C) Editing in two sites that do not contain S1, S2 and S3 sequences. Error bars represent 1 SD from the mean.
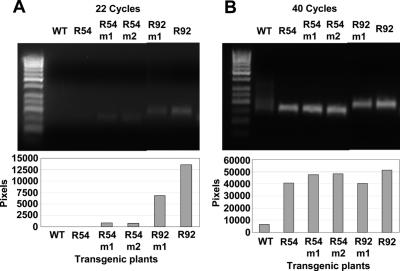
A possible reason for differences in extent of inhibition of editing of related endogenous sequences between different rpoB transgenic lines could be different levels of transgene transcripts. The reduction in endogenous transcript editing is thought to be due to competition among transcripts for a limited quantity of editing factors. Transgene transcripts presumably must be expressed at a sufficient level to engender this competition effect. We assayed the abundance of transgene transcripts in the five lines by semi-quantitative PCR. R92, the transgenic line which exhibited most reduction of editing of cluster members, also exhibited the highest transcript levels (Figure 8). R54, which exhibited no reduction in endogenous editing, had very low transcript levels, presumably due its heteroplasmic state. The other three lines exhibited intermediate amounts of transcripts and less reduction in editing of endogenous transcripts of cluster members than in R92 (Figure 8).
Figure 8.
Semi-quantitative RT–PCR of transgene transcripts from transplastomic plants. (A) At 22 cycles of RT–PCR, bands of different intensities correspond to varying transcript abundances. (B) After 40 cycles of RT–PCR all bands are at equivalent intensities.
Analysis of chimeric transcripts in vitro
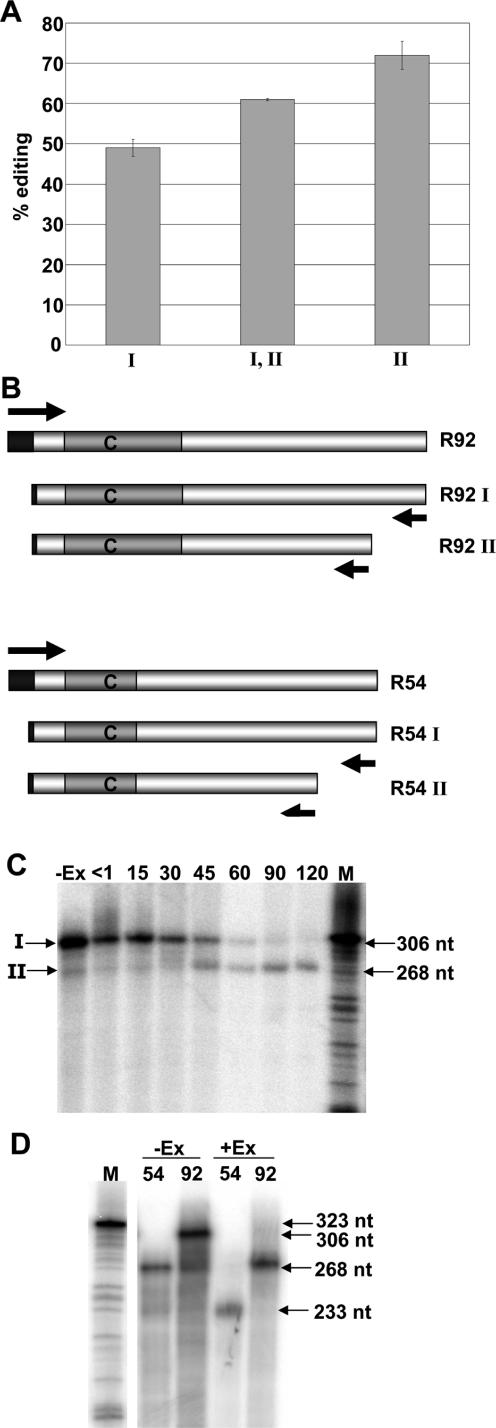
Analyzing the requirements for editing of transcripts incubated in chloroplast extracts in vitro allows more rapid examination of a large number of mutated RNA substrates than is possible with chloroplast transgenic plants, which require considerable labor and months of time for regeneration. We carried out further analysis of the cis-requirements for editing of rpoB using an in vitro editing system. Previously, the RNAs that have been assayed for editing efficiency in vitro have either been synthetic RNAs comprised only of chloroplast sequence and a 3′ KS amplification sequence (6,29,30) or have been surrounded by SK and KS amplification sequences following transcription in vitro by T7 polymerase (25). In order to mimic the transgene transcripts for in vitro analysis, a T7 promoter was placed 5′ of the 92 nt rpoB sequence, which was followed by the sequence corresponding to the longer 3′ end (3′ end I). A second substrate was created that carried the short 3′ end (3′ end II). Both were incubated with tobacco chloroplast extract, and editing efficiency was assessed. Transcripts with the shorter 3′ ends were more highly edited than those with the longer 3′ ends (Figure 9A), but the difference in editing extent between the long and short transcripts was less than in vivo. When a primer was used that would amplify both long and short transcripts from extract initially incubated with the longer transcript, a higher editing efficiency was obtained. This suggested that the chloroplast extract used for editing in vitro might be capable of 3′ end processing.
Figure 9.
In vitro editing of substrates corresponding to transgenic transcripts were incubated in chloroplast extract under in vitro editing conditions. Editing percentages were calculated by comparing poisoned primer extension reaction intensity and error bars represent one standard deviation from the mean. (A) Lanes (I) and (I, II), Substrates with either 3′ end I or both 3′ ends were amplified through selective RT–PCR from an initial RNA template equivalent to 3′ end I, respectively. Lane (II), substrate was amplified with 3′ end II from an RNA template with 3′ end II. (B) Diagram of DNA substrates created to express RNA templates corresponding to the transgenic transcripts. Arrows indicate primers used for PCR amplification. Bars represent DNA substrates, closed bars symbolize T7 sequence used for transcription in vitro and gray bars indicate the region of rpoB. (C) Incubating 5′ end-labeled RNA substrates under in vitro editing conditions. Lane −Ex, without chloroplast extract for 120 min. Lanes <1 through 120, with tobacco chloroplast extract for points indicated up to 120 min. Bands that correspond to S1 nuclease mapped ends I and II are indicated to the left of the figure. (D) Internally labeled RNA substrates for R54 and R92 were incubated with tobacco chloroplast extract, +Ex, and without extract, −Ex, for 120 min. (C and D) Lane M, sequencing reactions serving as a molecular weight standard with molecular weights in nucleotides are indicated to the right.
To assess the processing capacity of the chloroplast extracts used for editing analysis, RNA substrates equivalent to R92 with 3′ end I were amplified by PCR and transcribed in vitro. Substrates were then radiolabeled at their 5′ ends and incubated in chloroplast extracts to determine whether the longer RNA can be processed in vitro. The RNA was cleaved over time and was nearly fully digested after 2 h (Figure 9). RNA substrates equivalent to R54 and R92 were randomly labeled to show if they were equally cleaved and that no other cleavage products were formed (Figure 9). Only substrates with 3′ ends equivalent to in vivo 3′ ends I and II were observed. Thus, chloroplast extracts used for assaying editing also exhibit 3′ RNA processing activity. Since most transcripts have been processed by 2 h of incubation (Figure 9), most of the transcripts assayed with the primer that amplifies both short and long transcripts are actually the shorter transcripts.
RNA substrates were constructed with the same 5′ and 3′ ends as in vivo transgenic transcripts to compare editing in vitro versus in vivo. The extent of editing within transcripts with sequence differences versus substrates carrying wild-type sequence correlates with the differences observed in vivo (Figure 10A). To determine whether the presence of KS and SK sequence surrounding a chloroplast sequence affects editing efficiency of wild-type sequences in vitro, we compared a substrate with a KS sequence 3′ to the 54 nt rpoB sequence versus substrates carrying the rps16 3′ end II (Figure 10B). The insignificant difference between substrates carrying the 54 nt sequence and either the 5′ rrn16/MCS 5′ end and the rps16 3′ end versus the SK and KS 5′ and 3′ ends led us to utilize the convenient SK/KS sequences for further assessment of editing efficiency of substrates in chloroplast extracts. The 5′ and 3′ environment of the NTrpoB C473 region is necessarily different in the transgene and in vitro substrates than in actual rpoB transcripts (Figure 10C).
Figure 10.
(A) Relative in vitro editing of substrates analogous to transgenic transcripts created with 3′ end II. Editing is expressed as % of the wild-type R54 editing because substrates were assayed by two different extracts, one editing the constructs from 50 to 80% and the other from 22 to 36%. (B) In vitro editing of the R54KS substrate with bacterial sequences SK and KS around the editing site and substrates equivalent to the transgene transcript expressed in transplastomic plants. (C) Diagram of editing templates. Arrows represent the +1 position of transcription initiation. Underlined nucleotides signify the common rpoB sequence around the editing site.
We produced a number of RNA substrates to assess the effect of alterations within the conserved elements on editing efficiency and to identify any other important sequences. DNA templates were created by PCR with bacterial T7 promoter and SK and KS sequences flanking the rpoB region and then transcribed in vitro to produce the substrate RNA. All substrates carried the −31/+22 chloroplast sequence present in the transgenes tested in vivo.
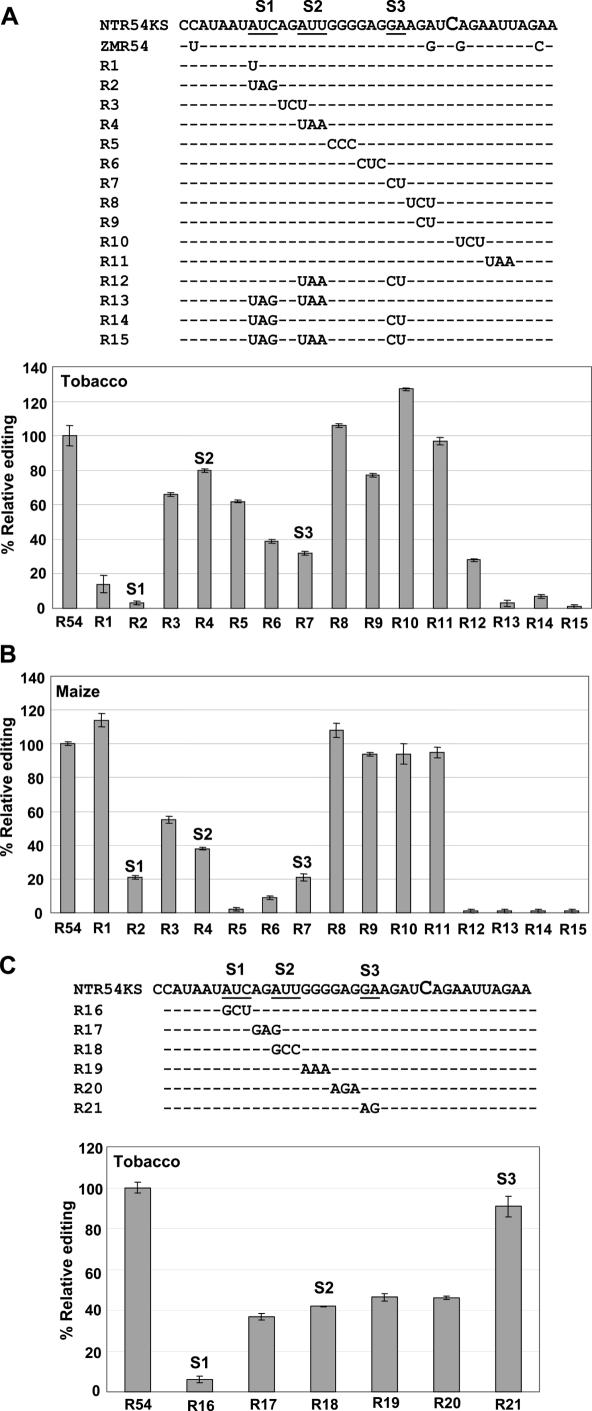
Substrates R1–R11 were created to determine the critical elements within −20/+6 region (Figure 11A). Together these substrates cover the −20/+6 region found to be important in vivo. The editing efficiency of substrates R1–R7 and R9 in tobacco extracts was significantly reduced compared to the wild-type substrates. Of the substrates with reduced editing efficiencies, R4 and R9 had the smallest reduction (20%) in relative editing, and alterations were either not in an important region or not a sufficient sequence change to alter processing by the editing apparatus. Substrate R8 contains the same alterations as R9 but was not reduced in its editing efficiency, suggesting that the −3 and −2 nt contained in R9 are not part of the critical sequence element. Curiously, R10 was a better substrate than R54KS perhaps indicating that the sequence restraints within coding regions result in endogenous sequences that do not always represent the optimal sequences for editing. The increase in editing is only ∼30% and may be due to enhancement of RNA structure rather than representing a positive alteration in part of an important sequence element.
Figure 11.
Effect of substrate sequence alterations of S1, S2 and S3. (A) Top: Alignment of RNA templates differing from wildtype sequence by purine to pyrmidine changes or vice versa. Bottom: RNA templates were incubated with tobacco or maize (B) chloroplast extracts. (A and B) The percentage of edited substrates, as calculated by poisoned primer extension. Percentage relative editing equals the percent editing of the substrate divided by the percent editing of the wild-type substrate. Data in (A) were derived from three experiments in which wild-type R54KS was edited at either 30, 34 or 72%. Data in (B) were derived from three experiments in which R54KS editing was either 30, 60 or 64%. (C) Upper panel, alignment if RNA templates differing from wild-type sequence by purine to purine or pyrimidine to pyrimidine changes. Lower panel, RNA templates were incubated with tobacco chloroplast extracts and relative editing calculated after one experiment with R54KS editing of 73%. Error bars represent one standard deviation 1 SD from the mean from two replicates.
Sequence changes in S1, S2 and S3 affect editing efficiency
A single −20 A→U change nearly completely abolished editing, in contrast to the results obtained in vivo with the −31/+22 transgene (Figure 5), but in agreement with the low editing efficiency when only −20/+7 chloroplast sequence is included in the transgene (19). Evidently the presence of the SK sequence 5′ to the chloroplast sequence results in greater sensitivity of the transcripts to editing perturbation by alteration of the −20 nt than does the presence of a few nucleotides derived from the rrn16 promoter.
Substrates R2, R4, R7 and R12–R15 were created with differences in S1, S2, S3 to test their importance for editing NTrpoB C473 (Figure 11A). R4 showed small, ∼20%, reductions in editing relative to wild type. This can be compared to the 65% reduction in R7 and abolition of editing in R2. Substrates carrying a combination of altered common elements, R12–R15, were created to study the effect of multiple changes. The effects of changes in S1, S2 and S3 are evident in cumulative reductions of editing in substrates with combinations of altered elements compared to wild-type substrates. All substrates with differences in S1 could not be edited by tobacco chloroplast extracts.
Substrates R1–R15 all had sequence changes from purine to pyrimidine and vice versa. Substrates R3–R5 only exhibited minor 20–40% reductions in editing compared to wild type (Figure 11A). To test whether purine to purine and pyrimidine to pyrimidine mutations might disrupt editing more significantly, such mutations were created within the previously defined critical region in substrates R16–R21 (Figure 11C). Again nucleotide changes in S1, as in R16, had the greatest effect on editing. R17–R19 had more severe effects than substrates R3–R5. R18, carrying a change in S2 is a much poorer editing substrate than R4 or R54KS and confirms that S2 is a critical sequence. Overall, purine to purine and pyrimidine to pyrimidine base changes had a larger impact on editing than purine to pyrimidine and pyrimidine to purine. In spite of this, R21 had no effect compared to R7 although they both alter S3. S3 is flanked by GA nucleotides and the mutations in R21 happen to create two S3 sequences shifted 1 nt 5′ and 3′ of the endogenous position from the editing site. The sequence of R21 is evidently not sufficiently altered to reduce the editability of the substrate.
When maize rpoB is aligned with tobacco rpoB, ZMrpoB C467 is at the same position as NTrpoB C473, and the surrounding sequence is very similar to that in tobacco. Tobacco chloroplasts can edit a template expressing a −31/+61 region around ZMrpoB C467 (data not shown). Therefore, it is likely that tobacco and maize contain factors that can recognize similar cis-acting elements around NTrpoB C473. The same substrates tested in vitro using tobacco extracts were tested in vitro using maize extracts. Substrates R2–R7, which have sequence alterations in the −20/−5 region, exhibited major reductions in editing efficiency. The same region of sequence is therefore critical for editing in maize as in tobacco. All substrates with differences in S1, S2 and S3 were less efficient editing substrates in maize extracts compared to wild type (Figure 11B). As in the tobacco extracts, changes in S1 and S3 affected editing of the substrate more than S2. In contrast, the single −20 T to A change that severely affected editing in tobacco had little effect on editing in maize extracts. The editing efficiencies of the substrates with mutations within the −20/−5 region were reduced to a greater extent in maize compared to tobacco. Also, substrates R5 and R6 had the largest reductions in editing compared to R2, which had the largest reduction in tobacco. These results indicate that, although the RpoB C467/C473 editing factors in the two species may be similar, some evolutionary divergence has probably occurred that has resulted in differential preference for sequences surrounding the editing site.
DISCUSSION
With the addition of maize extracts described here, assays for chloroplast editing in vitro are available in four species including tobacco, pea and Arabidopsis as well (6,25,31). In vitro systems have been particularly useful in studying the cis-acting elements for editing sites due to their relative speed, flexibility of species and cost advantages. Studies in vivo have presently been limited to tobacco because of the technical difficulty of chloroplast transformation. The maize in vitro assay described here should facilitate identification of editing factors in an organism with better genetic resources. This is also the first monocot in vitro editing system and should allow comparisons of editing between dicot and monocot species.
The same substrates have not been assayed previously both in vivo and in vitro to test the biological relevance of the in vitro system. Here we describe the assay of comparable templates for NTrpoB C473 in vivo and in vitro, which allows a direct comparison between systems. Five substrates with sequence differences but the same approximate flanking sequences have been compared to wild-type substrates in vivo and in vitro. All five substrates showed similar in vivo and in vitro editing efficiencies, indicating that data from the in vitro system are applicable to editing in intact plant chloroplasts. Chloroplast extracts are therefore likely to be complete in their complement of critical editing factors.
Transcripts containing −31 to +22 nt surrounding NTrpoB C473 were more efficiently edited in vivo than transcripts with only −20 to +6 nt. Thus, substrates reduced to the immediate flanking and adequate region for editing do not represent the full complement of cis-acting elements that influence editing. Sequence elements that enhance editing are probably present outside of the −20/+6 region around the editing site. Nevertheless, the −20 to +6 region is sufficient to specify editing the proper C target in vivo.
Substrates have been constructed and tested in vivo or in vitro with changes in the nucleotides within the −20/+6 minimal region (Figure 12). Changes in the sequence from −20/−5 had large reductions in the editing efficiency of the corresponding substrate and the critical sequence element is contained within that region. Differences in S1, S2 and S3 consistently affected editing of the templates when assayed in extracts from tobacco and maize. Consistent with our hypothesis that elements conserved between cluster members are important site recognition features, S1, S2 and S3 are critical for RNA substrate editing in both tobacco and maize extracts. A substrate that contains a −20 A to U change is very poorly edited in tobacco but well edited in maize. This suggests that in maize the −20 nt is not critical. The editing factor in maize apparently sufficiently differs from the tobacco factor that it can tolerate a change in the S1 sequence whereas the tobacco factor cannot. In addition, substrates with changes within the −20/−5 region have different reductions in editing efficiency between tobacco and maize. Although editing factors in maize recognize the same region of sequence, they have diverged to the extent that different nucleotides are critical for editing.
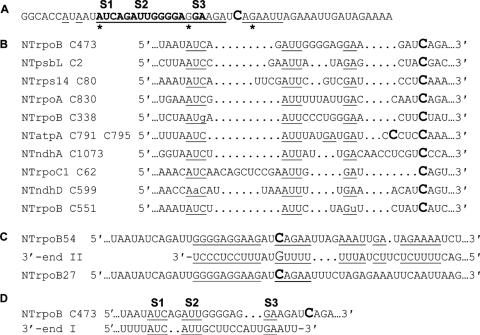
Figure 12.
(A) Nucleotides that have been altered within in vivo and in vitro substrates the editing site NTrpoB C473. Underlined characters represent positions where substrates with sequence alterations have been assayed in vivo or in vitro. The asterisk indicates nucleotides that were changed in a substrate containing a −20 to +6 region of rpoB expressed in transplastomic plants by Reed et al. (19). Boldface characters represent nucleotides that when are altered within substrates are edited less efficiently than wild type. (B) Alignment of editing sites that contain S1, S2 and S3 sites. (C) Alignment of sequence of substrates containing 54 and 27 nt regions around NTrpoB C473 expressed in vivo and sequence from the substrates 3′ end II. Underlined characters represent regions of complementarity. (D) An alignment of sequence from the 3′ end I and wild-type sequence around the NTrpoB C473 site. Underlined sequences represent S1, S2 and S3 and similar sequences.
Of the 34 editing sites within tobacco chloroplasts 11 share elements similar to S1, S2 and S3, although not all are reduced by over-expression of NTrpoB C473 (Figure 12). One explanation for the lack of a competition effect is the existence of different trans-factors for some or all of these sites. Another possible reason that sites with common cis-acting elements are differently affected by over-expression of NTrpoB C473 is that the same sequestered factor may be important for editing of multiple sites, but may not always be limiting. This seems to be the case even among sites whose editing is reduced by over-expression, as there are significant differences in the strength of competition between different members of the RpoB C467 cluster. Some sites may have stronger affinities for a particular trans-factor than other sites. Therefore, all 11 sites may share important cis-acting elements, but the possible relationship of such sites will not be obvious by over-expression of a single RNA editing site in a transgenic plant.
The presence of S1, S2 and S3 in the three genes whose editing is altered in transgenic plants could only be identified by allowing significant gaps, suggesting critical sequence elements could be irregularly spaced from the edited site. This complicates understanding how editing sites are specifically targeted, since any common distance from a cis-acting element to the editing site might differ among sites. Possibly, either a processive mechanism specifies a particular nucleotide for editing that is different between sites from a cis-acting element, or there are elements that require particular spacing that may serve roles in locating the editing components to the particular C to be edited. Any processive mechanism within the NTrpoB C473 cluster would have to differ significantly between sites due to different sequences around the editing site, and the position of 5′ and 3′ Cs around editing sites. Sensitivity of editing site substrates to altered spacing has been observed in mitochondria, suggesting a potential for a ‘molecular ruler’ determining the target for editing by its distance from a cis-acting element (32,33). In the NTrpoB C473 cluster, S3 is 4–5 nt away from the edited nucleotide and could be important for editing site targeting. Substrate R21 is not reduced in its editing efficiency and suggests that small perturbations in the precise position of this sequence do not affect editing, at least in rpoB transcripts (Figure 11). In mitochondria, editing sites separated by only a short length of sequence were found to share cis-acting elements (34,35). Chloroplast sites NTatpA C791 and NTatpA C795 contain two S3 elements 5 and 6 nt upstream of the edited sites, respectively, and share the same upstream elements S1 and S2 (Figure 12), but their editing efficiency is unaffected by over-expression of NTrpoB C473.
Some editing sites in plant mitochondria and chloroplasts have been shown to be sensitive to changes in nucleotides immediately adjacent to the editing site (30,33). However, an inserted ZMrpoB C467 editing site from maize sequence, flanked by 92 bases, differs from the tobacco sequence by a 3′ adjacent G and is edited as well as the tobacco sequence (5,21). Endogenous editing of NTrpoB C473 and cluster member NTpsbL C2 are reduced when the NTrpoB C473 gene fragment is over-expressed, although the NTpsbL C2 editing site differs in its immediate 5′ and 3′ nucleotides (20). If NTpsbL C2 shares factors involved in the editing mechanism, then the editing complex for NTrpoB C473 may be insensitive to differences in nucleotides flanking the editing site.
The −20A nucleotide has been determined to be critical in vivo using a 27 nt (−20, +6) template and in vitro using a 54 nt (−31, +22) template surrounded by SK and KS sequences. In vivo, however, a substrate containing a 54 nt region around NTrpoB C473 with the −20U alteration can be more highly edited than a wild-type substrate. A substrate analogous to the in vivo transgenic transcript is also well edited in vitro compared to wild type. Therefore, the reduction in editing caused by the −20U alteration depends upon the particular sequences 5′ to the −20 nt. Since two independent observations show a critical role for the −20A, it is likely it is important for editing. Possibly the importance of the region 5′ to −20 relates to the existence of complementary sequence in the mature 3′ end, which could be involved in a secondary structure near the editing site (Figure 12). The extent of the complementarity would differ between the 54 nt region around NTrpoB C473 and the 27 nt region, and would not be present in the in vitro substrate that carries SK and KS flanking sequences. Possibly this region of complementarity can compensate for the −20U mutation in the in vivo R54m1 and R54m2 transcripts. Other unknown RNA secondary structures or cis-elements might also result in the compensation for the −20U alteration.
The presence or absence of critical nucleotides near the C editing target does not solely determine the extent to which an RNA substrate is edited in vivo or in vitro. RNA editing sites are sensitive to not only to the amount of local sequence around the edited site, but can also be affected by sequence features significantly distal to the editing site. In vivo and in vitro RNA substrates with 3′ ends containing 44 nt more rps16 sequence, 111 nt away from the edited nucleotide, are edited less efficiently. The endogenous NTrpoB C473 editing site is located far from the 3′ end of its transcript, and possibly the edited machinery adapted to edit this site may have done so without the bulky stem–loops and other elements that normally characterize plastid transcript 3′ ends. Another possibility is that there is sequence in the longer 3′ end that has the ability to sequester editing factors. Indeed, near the end of the longer rps16 3′ end present in the transgene transcripts, there is some sequence similar to the common elements known to be critical for NTrpoB C473 editing (Figure 12). Substrate editing is therefore sensitive to the presence of nearby cis-acting elements, more distant enhancing elements and flanking sequence. These sensitivities could be responsible for the early difficulty in creation of an in vitro system, and suggest why only a limited set of editing sites have been described that are edited in chloroplast extracts in vitro at high efficiency.
Table 1.
Oligonucleotides (Integrated DNA Technologies, Coralville, IA) used in experiments reported here
| Name | Sequence 5′–3′ | Purpose |
|---|---|---|
| 500f | GATCCCCATGGGGCACCATAATATCAGATTGGGGAGG | Transgene construction |
| 500r | CCGTCTAGATTTTCTATCAATTTCTAATTCTGATCTTC | Transgene construction |
| 501f | GATCCCCATGGGGCACCATAATTTCAGATTGGGGAGG | Transgene construction |
| 502f | GATCCCCATGGGGCACCATAATTTCAGATTGGGGAGG | Transgene construction |
| 502r | CCGTCTAGATTTTCTTTCAATTTCTAATTCTGATCTTC | Transgene construction |
| 505f | GATCCCCATGGGGCACCATAATATCAGATTGGGGA | Transgene construction |
| 505r | GATCCCCATGGGGCACCCTGATATCAGATTGGGGA | Transgene construction |
| 506f | CCGTCTAGAATTTTTTGTTTCCTACTTACACGAGCCCA | Transgene construction |
| 500sreverse_Lg | TGTCCATTTTTCGGGGTCTCAAAGGGGCGTGGAAA | S1 nuclease mapping |
| T7_5′_500s | TAATACGACTCACTATAGGGGCGAACTCCGGGCGAATA | Transcript production |
| PC1.1 | TCTTGAACAACTTGGAGCCGGGCC | Southern probe |
| PCα1.2 | GAGGATAGCAAGTTCCAAATTCTGTCTCGG | Southern probe |
| PPrrn2 | AATACGAAGCGCTTGGATACAGTTGTAGGGA | PCR transgenic transcript |
| Trps16sh | TCCTTAATTTATTTCCTTAATTGAATTTCTCTAGA | PCR 3′ end II |
| Trps16lg | AATTCAATGGAAGCAATGATAAAAAAATACAAATA | PCR 3′ end I |
| FRpoB2 | ACTCCAGGTTCCTCGGGGTAAA | PCR endogenous rpoB |
| RrpoB2 | TTGCGGAGTAAATGGGCTTCTAA | PCR endogenous rpoB |
| RpoB-2(C) | GGCACCATAATATCAGATTGGGGAGGAAG | PPE NTrpoB C473 |
| FpsbL | TACCGTCTTTTTTTTGGGATC | PCR endogenous psbL |
| RpsbL | ATTTTGTTCGTTCGGGTTTGA | PCR endogenous psbL |
| PsbL(G) | AACATTTTGTTCGTTCGGGTTTGATTGTGT | PPE NTpsbL C2 |
| FRps14 | CAGAGGGAGAAGAAGAGGC | PCR endogenous rps14 |
| RRps14 | GCTCCTGGCAACAAACAT | PCR endogenous rps14 |
| Rps14-1(C) | GGAACAGAAATATCATTCGATTCGTCGATCC | PPE NTrps14 C80 |
| Rps14-2(A) | CGATGAAGGCGTGTAGGTGCACTATTCC | PPE NTrps14 C149 |
| FndhB2 | TACGGTCTAATGAGGCTACTA | PCR endogenous ndhB |
| RndhB2 | TCCCAATATCATGCTAAGAA | PCR endogenous ndhB |
| ndhB-6(T) | TCATTACCGTAGGAATTGGGTTCAAGCTTT | PPE NTndhB C737 |
| RpoB54ForSK | CTAGAACTAGTGGATCGGCACCATAATATCAGATTGGGGA | R54KS in vitro substrate |
| RpoB54RevKS | TATCTTTTCTATCAATTTCTAATTCTGATCTTCCTCCCCA | R54KS in vitro substrate |
| R54R2For | CTAGAACTAGTGGATCGGCACCATAATTAGAGATTGGGGA | R2 and R14 in vitro substrate |
| R54R7For | CTAGAACTAGTGGATCGGCACCATAATATCAGTAAGGGGA | R7 in vitro substrate |
| R54R5Rev | TATCTTTTCTATCAATTTCTAATTCTGAAGTTCCTCCCCA | R9 in vitro substrate |
| R54R7Rev | TATCTTTTCTATCAATTTCTAATTCTGATCTAGCTCCCCA | R7 and R15 in vitro substrate |
| R54R6For | ATCGGCACCATAATATCAGTAAGGGGAGCTAGATCAGAAT | R12 in vitro substrate |
| R54R7For | ACTAGTGGATCGGCACCATAATTAGAGTAAGGGGAGGAAG | R13 in vitro substrate |
| R54R9For | ACTAGTGGATCGGCACCATAATTAGAGTAAGGGGAGCTAG | R15 in vitro substrate |
| R54R1For | CTAGAACTAGTGGATCGGCACCATAATTTCAGATTGGGGA | R1 in vitro substrate |
| T7SK | TAATACGACTCACTATAGGGCGCTCTAGAACTAGTGGATC | in vitro substrate construction |
| R54R3For | CGCTCTAGAACTAGTGGATCggcaccataatatcTCTttggggaggaagatCagaattag | R3 in vitro substrate |
| R54R3Rev | TCGAGGTCGACGGTATCttttctatcaatttctaattctGatcttcctccc | R3, R16, R17, R18 in vitro substrate |
| R54R5For | CGCTCTAGAACTAGTGGATCggcaccataatatcagattCCCgaggaagatCaga | R5 in vitro substrate |
| R54R5Rev | TCGAGGTCGACGGTATCttttctatcaatttctaattctGatcttcctcGGGaat | R5 in vitro substrate |
| R54R6For | CGCTCTAGAACTAGTGGATCggcaccataatatcagattgggCTCgaagatCaga | R6 in vitro substrate |
| R54R6Rev | TCGAGGTCGACGGTATCttttctatcaatttctaattctGatcttcGAGcccaat | R6 in vitro substrate |
| R54R8For | CGCTCTAGAACTAGTGGATCggcaccataatatcagattggggaggaTCTtCaga | R8 in vitro substrate |
| R54R8Rev | TCGAGGTCGACGGTATCttttctatcaatttctaattctGaAGAtcctccccaat | R8 in vitro substrate |
| R54R10For | CGCTCTAGAACTAGTGGATCggcaccataatatcagattggggaggaagatCTCT | R10 in vitro substrate |
| R54R10Rev | TCGAGGTCGACGGTATCttttctatcaatttctaatAGAGatcttcctccccaat | R10 in vitro substrate |
| R54R11For | CGCTCTAGAACTAGTGGATCggcaccataatatcagattggggaggaagatCagaT | R11 in vitro substrate |
| R54R11Rev | TCGAGGTCGACGGTATCttttctatcaatttctTTAtctGatcttcctccccaat | R11 in vitro substrate |
| R54R16For | TAGAACTAGTGGATCggcaccataatGCTagattggggaggaagatCaga | R16 in vitro substrate |
| R54R17For | TAGAACTAGTGGATCggcaccataatatcGAGttggggaggaagatCaga | R17 in vitro substrate |
| R54R18For | TAGAACTAGTGGATCggcaccataatatcagGCCggggaggaagatCaga | R18 in vitro substrate |
| R54R19For | TAGAACTAGTGGATCggcaccataatatcagattAAAgaggaagatCagaat | R19 in vitro substrate |
| R54R19Rev | GAGGTCGACGGTATCttttctatcaatttctaattctGatcttcctc | R19 in vitro substrate |
| R54R20For | TAGAACTAGTGGATCggcaccataatatcagattgggAGAgaagatCagaattag | R20 in vitro substrate |
| R54R20Rev | TCGAGGTCGACGGTATCttttctatcaatttctaattctGatcttc | R20 in vitro substrate |
| R54R21For | TAGAACTAGTGGATCggcaccataatatcagattggggagAGagatCagaattagaa | R21 in vitro substrate |
| R54R21Rev | TCGAGGTCGACGGTATCttttctatcaatttctaattctGatctCTctcc | R21 in vitro substrate |
Acknowledgments
We thank Barbara Eaglesham for maintaining transplastomic plants in tissue culture and Pal Maliga for providing the chloroplast transformation vector. This work was supported by NSF MCB 0344007 and NIH GM 50723 grants to M.R.H and a NIH graduate traineeship to M.L.H. Funding to pay the Open Access publication charges for this article was provided by NSF MCB 0344007.
Conflict of interest statement. None declared.
REFERENCES
- 1.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsudzuki T., Wakasugi T., Sugiura M. Comparative analysis of RNA editing sites in higher plant chloroplasts. J. Mol. Evol. 2001;53:327–332. doi: 10.1007/s002390010222. [DOI] [PubMed] [Google Scholar]
- 3.Notsu Y., Masood S., Nishikawa T., Kubo N., Akiduki G., Nakazono M., Hirai A., Kadowaki K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 4.Giege P., Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed M.L., Hanson M.R. A heterologous maize rpoB editing site is recognized by transgenic tobacco chloroplasts. Mol. Cell. Biol. 1997;17:6948–6952. doi: 10.1128/mcb.17.12.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirose T., Sugiura M. Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: development of a chloroplast in vitro RNA editing system. EMBO J. 2001;20:1144–1152. doi: 10.1093/emboj/20.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudhuri S., Carrer H., Maliga P. Site-specific factor involved in the editing of the psbL mRNA in tobacco plastids. EMBO J. 1995;14:2951–2957. doi: 10.1002/j.1460-2075.1995.tb07295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chateigner-Boutin A.L., Hanson M.R. Developmental co-variation of RNA editing extent of plastid editing sites exhibiting similar cis-elements. Nucleic Acids Res. 2003;31:2586–2594. doi: 10.1093/nar/gkg354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz-Linneweber C., Regel R., Du T.G., Hupfer H., Herrmann R.G., Maier R.M. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol. Biol. Evol. 2002;19:1602–1612. doi: 10.1093/oxfordjournals.molbev.a004222. [DOI] [PubMed] [Google Scholar]
- 10.Maier R.M., Zeltz P., Kossel H., Bonnard G., Gualberto J.M., Grienenberger J.M. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 11.Peeters N.M., Hanson M.R. Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA. 2002;8:497–511. doi: 10.1017/s1355838202029424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki Y., Kozaki A., Ohmori A., Iguchi H., Nagano Y. Chloroplast RNA editing required for functional acetyl-CoA carboxylase in plants. J. Biol. Chem. 2001;276:3937–3940. doi: 10.1074/jbc.M008166200. [DOI] [PubMed] [Google Scholar]
- 13.Schmitz-Linneweber C., Kushnir S., Babiychuk E., Poltnigg P., Herrmann R.G., Maier R.M. Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase alpha-subunit mRNA. Plant Cell. 2005;17:1815–1828. doi: 10.1105/tpc.105.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zito F., Kuras R., Choquet Y., Kossel H., Wollman F.A. Mutations of cytochrome b6 in Chlamydomonas reinhardtii disclose the functional significance for a proline to leucine conversion by petB editing in maize and tobacco. Plant Mol. Biol. 1997;33:79–86. doi: 10.1023/a:1005734809834. [DOI] [PubMed] [Google Scholar]
- 15.Bock R., Kossel H., Maliga P. Introduction of a heterologous editing site into the tobacco plastid genome: the lack of RNA editing leads to a mutant phenotype. EMBO J. 1994;13:4623–4628. doi: 10.1002/j.1460-2075.1994.tb06784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Covello P.S., Gray M.W. On the evolution of RNA editing. Trends Genet. 1993;9:265–268. doi: 10.1016/0168-9525(93)90011-6. [DOI] [PubMed] [Google Scholar]
- 17.Smith H.C., Gott J.M., Hanson M.R. A guide to RNA editing. RNA. 1997;3:1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 18.Takenaka M., Brennicke A. In vitro RNA editing in pea mitochondria requires NTP or dNTP, suggesting involvement of an RNA helicase. J. Biol. Chem. 2003;278:47526–47533. doi: 10.1074/jbc.M305341200. [DOI] [PubMed] [Google Scholar]
- 19.Reed M.L., Peeters N.M., Hanson M.R. A single alteration 20 nt 5′ to an editing target inhibits chloroplast RNA editing in vivo. Nucleic Acids Res. 2001;29:1507–1513. doi: 10.1093/nar/29.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chateigner-Boutin A.L., Hanson M.R. Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis-elements. Mol. Cell. Biol. 2002;22:8448–8456. doi: 10.1128/MCB.22.24.8448-8456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegeman C.E., Halter C.P., Owens T.G., Hanson M.R. Expression of complementary RNA from chloroplast transgenes affects editing efficiency of transgene and endogenous chloroplast transcripts. Nucleic Acids Res. 2005;33:1454–1464. doi: 10.1093/nar/gki286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoubenko O.V., Allison L.A., Svab Z., Maliga P. Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Res. 1994;22:3819–3824. doi: 10.1093/nar/22.19.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed M.L., Lyi S.M., Hanson M.R. Edited transcripts compete with unedited mRNAs for trans-acting editing factors in higher plant chloroplasts. Gene. 2001;272:165–171. doi: 10.1016/s0378-1119(01)00545-5. [DOI] [PubMed] [Google Scholar]
- 24.Svab Z., Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl Acad. Sci. USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegeman C.E., Hayes M.L., Hanson M.R. Substrate and cofactor requirements for RNA editing of chloroplast transcripts in Arabidopsis in vitro. Plant J. 2005;42:124–132. doi: 10.1111/j.1365-313X.2005.02360.x. [DOI] [PubMed] [Google Scholar]
- 26.Neuhaus H., Scholz A., Link G. Structure and expression of a split chloroplast gene from mustard (Sinapis alba): ribosomal protein gene rps16 reveals unusual transcriptional features and complex RNA maturation. Curr. Genet. 1989;15:63–70. doi: 10.1007/BF00445753. [DOI] [PubMed] [Google Scholar]
- 27.Nickelsen J., Link G. RNA-protein interactions at transcript 3′ ends and evidence for trnK-psbA cotranscription in mustard chloroplast. Mol. Gen. Genet. 1991:89–96. doi: 10.1007/BF00282452. [DOI] [PubMed] [Google Scholar]
- 28.Nickelsen J., Link G. The 54 kDa RNA-binding protein from mustard chloroplasts mediates endonucleolytic transcript 3′ end formation in vitro. Plant J. 1993;3:537–544. doi: 10.1046/j.1365-313x.1993.03040537.x. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto T., Obokata J., Sugiura M. Recognition of RNA editing sites is directed by unique proteins in chloroplasts: biochemical identification of cis-acting elements and trans-acting factors involved in RNA editing in tobacco and pea chloroplasts. Mol. Cell. Biol. 2002;22:6726–6734. doi: 10.1128/MCB.22.19.6726-6734.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto T., Obokata J., Sugiura M. A site-specific factor interacts directly with its cognate RNA editing site in chloroplast transcripts. Proc. Natl Acad. Sci. USA. 2004;101:48–52. doi: 10.1073/pnas.0307163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakajima Y., Mulligan R.M. Nucleotide specificity of the RNA editing reaction in pea chloroplasts. J. Plant Physiol. 2005;162:1347–1354. doi: 10.1016/j.jplph.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 32.Choury D., Farre J.C., Jordana X., Araya A. Different patterns in the recognition of editing sites in plant mitochondria. Nucleic Acids Res. 2004;32:6397–6406. doi: 10.1093/nar/gkh969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuwirt J., Takenaka M., JA V.D.M., Brennicke A. An in vitro RNA editing system from cauliflower mitochondria: editing site recognition parameters can vary in different plant species. RNA. 2005;11:1563–1570. doi: 10.1261/rna.2740905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Merwe J.A., Takenaka M., Neuwirt J., Verbitskiy D., Brennicke A. RNA editing sites in plant mitochondria can share cis-elements. FEBS Lett. 2006;580:268–272. doi: 10.1016/j.febslet.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Hermann M., Bock R. Transfer of plastid RNA-editing activity to novel sites suggests a critical role for spacing in editing-site recognition. Proc. Natl Acad. Sci. USA. 1999;96:4856–4861. doi: 10.1073/pnas.96.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]