Abstract
Hydrolytic deamination of cytosines in DNA creates uracil and, if unrepaired, these lesions result in C to T mutations. We have suggested previously that a possible way in which cells may prevent or reduce this chemical reaction is through the binding of proteins to DNA. We use a genetic reversion assay to show that a restriction enzyme, PspGI, protects cytosines within its cognate site, 5′-CCWGG (W is A or T), against deamination under conditions where no DNA cleavage can occur. It decreases the rate of cytosine deamination to uracil by 7-fold. However, the same protein dramatically increases the rate of deaminations within the site 5′-CCSGG (S is C or G) by ∼15-fold. Furthermore, a similar increase in cytosine deaminations is also seen with a catalytically inactive mutant of the enzyme showing that endonucleolytic ability of the protein is dispensable for its mutagenic action. The sequences of the mutants generated in the presence of PspGI show that only one of the cytosines in CCSGG is predominantly converted to thymine. Our results are consistent with PspGI ‘sensitizing’ the cytosine in the central base pair in CCSGG for deamination. Remarkably, PspGI sensitizes this base for damage despite its inability to form stable complexes at CCSGG sites. These results can be explained if the enzyme has a transient interaction with this sequence during which it flips the central cytosine out of the helix. This prediction was validated by modeling the structure of PspGI–DNA complex based on the structure of the related enzyme Ecl18kI which is known to cause base-flipping.
INTRODUCTION
Water is the most abundant molecule in living organisms and it damages DNA. It can deaminate the DNA bases cytosine, adenine and guanine converting them to uracil, hypoxanthine and xanthine, respectively. In addition, it causes depurination and depyrimidination creating abasic sites in DNA (1). If uncorrected, such DNA lesions can cause mutations, block replication and lead to cell death. Cells contain numerous DNA repair mechanisms to repair or bypass these lesions (2), suggesting that hydrolytic damage to the genetic material is a significant problem for cells over evolutionary time periods. The benefits of repairing these lesions are expected to be greater in thermophilic organisms where the rates of these reactions are several orders of magnitude higher than in mesophiles such as humans (3).
We have suggested previously that organisms may also have developed protective mechanisms that prevent or reduce hydrolytic damage to DNA (4). Specifically, we suggested that a possible mechanism for the protection of DNA against hydrolytic deamination of cytosines is its shielding with DNA-binding proteins. Protein–DNA complexes do contain bound water molecules, but many of these are immobile and are unlikely to initiate an attack on DNA bases unless they are properly positioned and oriented. We demonstrated the ability of proteins to protect DNA in this manner using a protein from Bacillus spores called SspC that binds DNA non-specifically. It reduced the rate of cytosine deamination in double-stranded DNA at 70°C by a factor of ∼10 (4).
As a way of generalizing this observation to proteins found in vegetative cells, we chose to study a protein from the hyperthermophilic organism Pyrococcus sp. strain GI-H. This organism grows at 85°C and the protein we selected for study is the restriction endonuclease PspGI (5). This enzyme binds the DNA sequence CCWGG (W is A or T) and cleaves before the first cytosine in the sequence in a Mg2+-dependent fashion. When Mg2+ ions are replaced with Ca2+ the enzyme binds the sequence, but does not cleave it (6). Thus, it is possible to study the biochemical effects of the binding of this enzyme at specific DNA sequences without triggering catalysis.
We chose a restriction enzyme for these studies in part because restriction endonucleases are known to possess high degree of discrimination between the cognate and non-cognate sequences (7). Thus, in contrast to SspC which binds DNA non-specifically, PspGI should protect DNA—if at all—at a limited number of sites. Furthermore, PspGI was suitable for these studies because we had already established a genetic system to study deamination of cytosines in CCWGG sequences to thymine (8). To perform this assay in vitro, it is necessary to heat plasmid DNA containing CCWGG to high temperatures (4) and, hence, the known stability of PspGI at high temperatures was an important factor in its selection for this study.
This genetic assay uses a plasmid that contains a defective kanamycin-resistance gene (kan). The plasmid is incubated at high temperatures with or without the protein of interest to cause cytosine deaminations and the DNA is subsequently electroporated into an Escherichia coli host that is defective in the repair of uracils in DNA (genotype—ung). In these cells the uracils are copied as if they were thymines and this results in C to T mutations. When cytosines in a specific codon of kan (which lies within a CCAGG site) are mutated to thymines, a functional kan gene is restored and these cells are scored as kanamycin-resistant colonies (phenotype-KanR) among the transformants (8–16). Thus if a protein protects DNA against cytosine deamination, a smaller increase in the KanR frequency is observed when the protein is included during the high temperature incubation (4).
Using this kan-reversion assay we did find that PspGI protects CCAGG sequences against cytosine deamination. Surprisingly, we also found that the enzyme and one of its mutants is capable of making cytosines in a related DNA sequence substantially more susceptible to deamination. These observations and their implications to PspGI structure and evolution are discussed below.
MATERIALS AND METHODS
Bacterial strains, plasmids and proteins
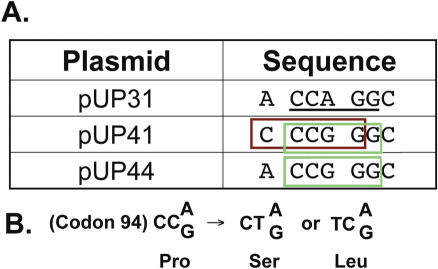
E.coli strains GM31 (dcm-6 thr-1 hisG4 leuB6 rpsL ara-14 supE44 lacY1 tonA31 tsx-78 galK2 galE2 xyl-5 thi-1 mtl-1) and BH156 (=GM31 ung-1 tyrA::Tn10) have been described previously (17). Plasmids pUP31 and pUP41 contain an active β-lactamase gene and an inactive kanamycin-resistance allele and have also been described previously (15,16). These plasmids contain mutation in codon 94 of kan gene (Figure 1). Plasmid pUP44 was constructed from pUP41 using USE mutagenesis (18). The mutagenic oligomer used was CTA93 (GACTGGCTGCTACCGGGCGAAGTG). The purification of wild-type and mutant PspGI proteins was performed as described previously (6).
Figure 1.
Sequence context of the mutable cytosines in the kan alleles. (A) The sequence of the plasmids used in this work around the proline codon (CCA or CCG) is shown. In plasmids pUP31, pUP41 and pUP44 the kan gene has the proline codon at position 94. The CCWGG site in pUP31 is underlined. The CCSGG sites in the different plasmids are indicated by green or brown boxes. (B) The effect of C to T mutation in codon 94 is shown. The mutations change a proline codon to either a leucine or serine codon and this restores kanamycin resistance.
DNA protection assay
One microgram of pUP31, pUP41 or pUP44 DNA was incubated with different amounts of PspGI in a 40 μl reaction containing the binding buffer (20 mM Tris–HCl, pH 7.9 at 25°C, 50 mM NaCl, 10 mM CaCl2 and 100 μg/ml BSA). Typically incubations were performed at 75°C for 4 h and the reactions were stopped by add SDS to 0.2%. In some experiments, other temperatures were used for the incubation. The DNA was extracted with phenol–chloroform, ethanol precipitated and redissolved in 10 μl TE (10 mM Tris–HCl, pH 7.8 and 1 mM EDTA). It was electroporated into the E.coli strain BH156 (or GM31) and transformants were selected on plates with 50 μg/ml carbenicillin or kanamycin. The frequency of kanamycin-resistant (KanR) revertants was calculated as the ratio (Number of KanR colonies/ml)/(Number of carbenicillin-resistant cells/ml).
DNA-binding assays with PspGI
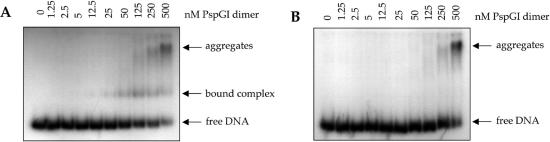
The region of the plasmids pUP31 and pUP41 containing codon 94 of kan was amplified using the primers 5′-GTCAAGACCGACCTGTCCGGTG-3′ and 5′-GTATGCAGCCGCCGCATTG-3′. The resulting 230 bp PCR products were used for gel retardation assays. PspGI was incubated with 5 nM 32P-labeled PCR product from either pUP31 (part A) or pUP41 (part B) in binding buffer [10 mM Tris–HCl, 100 mM NaCl, 5 mM CaCl2, 0.1 mg/ml BSA, 10% (v/v) glycerol (pH 8.5) in a total volume of 10 μl]. The reactions also contained 0.1 μg of poly(dI–dC) as a non-competitive inhibitor (Amersham Biosciences). After incubation for 15 min at ambient temperature, the samples were applied to a 6% polyacrylamide gel. After electrophoresis for 3 h at 80 V (10 V/cm) in 20 mM Tris–acetate and 5 mM CaCl2 (pH 8.5), gels were subjected to autoradiography using an instant imager (Packard Instrument Co.). The concentration of PspGI in each reaction is shown at the top of the lane (Figure 6).
Figure 6.
Sequence dependence of binding of PspGI to DNA. Gel electrophoretic mobility shift experiment to compare complex formation of PspGI and DNA with two different recognition sequences. Radioactively labeled PCR products containing either a ACCAGGC (A) or CCCGGGC (B) was incubated with various concentrations of PspGI (lanes 1–10). Free and bound DNA were separated by electrophoresis under non-denaturing conditions and detected using an instant imager.
Modeling the structure of PspGI bound to DNA
The model was obtained by aligning the PspGI sequence with the known protein structures using the fold-recognition MetaServer (19), and using the alignments reported by different servers as multiple alternative starting points for comparative modeling and recombination of fragments, followed by the optimization of the sequence–structure fit according to the ‘FRankenstein monster’ approach (20). The uncertain regions (insertions and terminal extensions) were remodeled de novo with ROSETTA (21).
All fold-recognition servers generated reliable matches between the PspGI sequence and the structures of EcoRII (22) and/or Ecl18kI (23) enzymes. We selected the Ecl18kI–DNA complex as the principal template to model PspGI structure and regarded EcoRII structure only as a secondary guide, as the latter structure is of lower resolution, lacks DNA and appears to be in a latent form. The final model of PspGI was constructed after several rounds of iterative optimization of sequence alignment (PspGI versus Ecl18kI) and evaluation of the resulting intermediate models by the COLORADO3D server (24). The PspGI model may be downloaded from ftp://genesilico.pl/iamb/models/R.PspGI/.
RESULTS
Protection of CCAGG sequences by PspGI against cytosine deamination
PspGI requires Mg2+ in the reaction buffer to cleave DNA and when this metal is replaced with Ca2+ the enzymebinds to its cognate sequence without cleaving it (6). We used this property to create stable protein–DNA complexes and incubated them at high temperatures to study the ability of PspGI to protect DNA against cytosine deamination. Specifically, plasmid pUP31 carrying a defective kan gene was incubated with different amounts of wild-type (WT) PspGI in a buffer containing Ca2+ at 75°C. This kan allele contains the cognate sequence for PspGI, CCAGG, overlapping codon 94 and a C to T change at either cytosine restores the phenotype to KanR (Figure 1). This is one of the 14 PspGI sites in pUP31. To prevent the repair of uracils created during the incubation, all the DNA samples were transformed into ung cells and the frequency of KanR revertants was determined in each case. The results are presented in Figure 2.
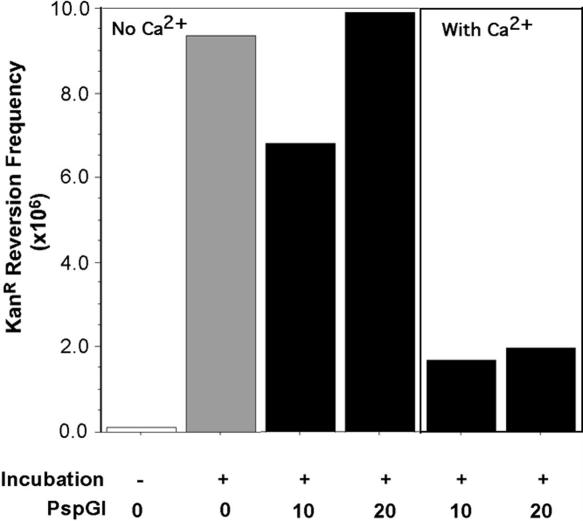
Figure 2.
Protection of cytosines in CCWGG by PspGI. The frequency of KanR revertants following DNA incubation in the presence of different amounts of PspGI is shown. The ratio, number of protein dimers:number of PspGI sites in DNA, is shown below the bar graph. Whether or not the DNA was incubated at 75°C is also indicated below the graph by plus (+) and minus (−) signs.
As expected, incubation of pUP31 DNA in the absence of PspGI substantially increased the reversion frequency. However, including PspGI in the reaction reduced this increase and at the higher concentrations of the enzyme the DNA was almost completely protected (Figure 2). Based on these results, we used the two highest concentrations of the protein, 10- and 20-fold molar excess of the protein over CCAGG sites, in subsequent experiments.
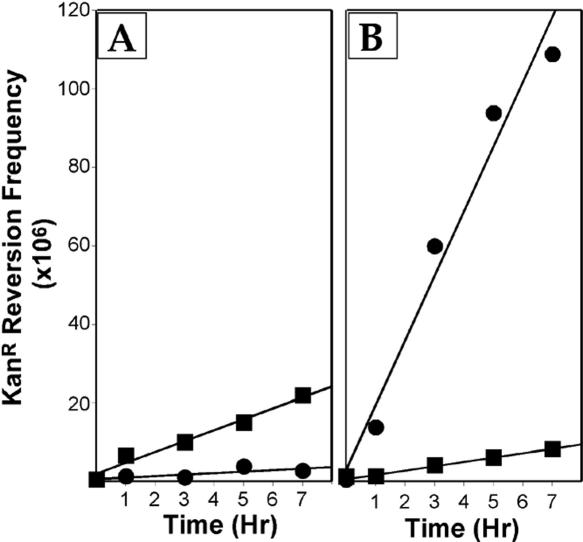
The restriction enzyme was able to protect DNA at 75°C for at least 7 h. During this time period, the rate of cytosine deamination in pUP31 was lowered by a factor of 7.4 in the presence of PspGI compared to the control (pseudo-first-order rates 1.1 × 10−10 s−1 and 7.8 × 10−10 s−1, respectively; Figure 3A). These results show that PspGI can provide substantial long-term protection of DNA against cytosine deamination at high temperatures.
Figure 3.
Kinetics of cytosine deamination in the presence of PspGI. The frequency of KanR revertants following incubation of DNA with or without PspGI is shown. Closed circles (•), PspGI at 10-fold molar excess; closed squares (▪), no PspGI added. The number of hours of incubation at 75°C is on the x-axis. The DNA used was pUP31 (A) or pUP41 (B).
PspGI does not bind tightly to its cognate sequence if a divalent metal is not included in the buffer (A. Pingoud, personal communication) and, thus, the enzyme should not protect the DNA under these conditions. To confirm this, we repeated the DNA protection assay described above in the absence of any divalent cation and found that the protective effects of the enzyme were reduced under these conditions. In fact, no protection of plasmid DNA against deamination could be detected at 20-fold excess of the protein over binding sites (Figure 4 and data not shown).
Figure 4.
Role of calcium in the protection of DNA by PspGI. The frequency of KanR revertants following incubation of pUP31 DNA in the presence or absence of calcium in the incubation buffer is shown. The incubation conditions for the various reactions are indicated below the bar graph in a manner similar to Figure 2.
Sensitization of a non-cognate site by PspGI
We wondered whether the protective effects of PspGI against deamination were sequence specific. To test this, plasmid pUP41 was used instead of pUP31 as the deamination target. The former plasmid is missing the PspGI site within which cytosine deamination must occur to obtain KanR revertants. The ACCAGGC sequence (PspGI site is underlined) in pUP31 is replaced with CCCGGGC in pUP41 (Figure 1) (15). This creates two overlapping CCSGG sites (S is C or G) in pUP41 at codon 94 (CCCGG and CCGGG; Figure 1). There are 18 other CCSGG sites in pUP41. We expected that PspGI would bind poorly to these CCSGG targets and thus not protect them against cytosine deamination.
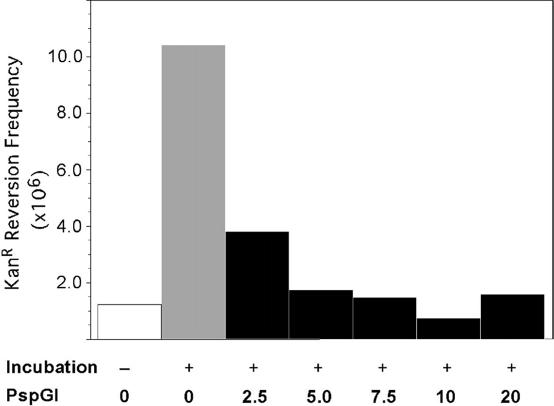
Surprisingly, the WT PspGI not only failed to protect pUP41 DNA, but also made it more susceptible to cytosine deaminations. When a 10-fold excess of WT PspGI (in terms of binding sites) was added to the reaction, the mean KanR frequency from six independent reactions increased by a factor of ∼19 compared to the incubated reactions lacking the protein (Figure 5). This increase is over and above the 8-fold increase in KanR revertants owing to incubation alone (Figure 5A). The effect of PspGI on the reversion frequency of pUP41 was time-dependent, and it increased the rate of cytosine deamination by 15-fold over a 7 h incubation (pseudo first-order rates without and with the protein of 3.1 × 10−10 and 4.4 × 10−9 s−1 respectively, Figure 3B). The increase in reversion frequency due to PspGI was eliminated if SDS was added to the reaction before the start of incubation or if the DNA was electroporated into ung+ E.coli (data not shown). These data, respectively, show that maintaining the native structure of PspGI was essential for its action and that PspGI helped convert to cytosines in pUP41 DNA to uracil.
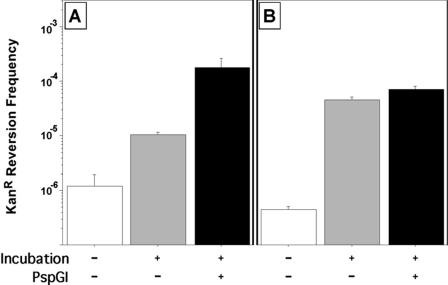
Figure 5.
Enhancement of cytosine deamination in CCSGG by PspGI. The frequency of KanR revertants following incubation of pUP41 DNA in the presence or absence of WT PspGI is shown. A 10-fold molar excess of the protein was incubated at 75°C for 4 h. The mean and standard error of the mean from six independent reactions are shown. Note that the Y-axis is logarithmic. Results with pUP41 (A) and pUP44 (B) are shown.
Binding of PspGI to CCAGG and CCSGG sequences
Although PspGI does not cleave DNA at CCSGG sites (5) we wondered whether it bound to these sites tightly to form non-productive complexes. To determine this, we amplified the DNA sequence containing codon 94 in pUP31 and pUP41 using PCR and performed a gel retardation assay in the presence of PspGI and CaCl2. The results are shown in Figure 6. PspGI forms a stable complex with DNA containing the target CCAGG site from pUP31 and this can be detected at PspGI concentrations at or above 5 nM (Figure 6A). In contrast, the equivalent DNA fragment from pUP41 does not form a stable complex at even 100 times higher concentration of PspGI (Figure 6B). These results show that PspGI does not bind tightly to CCSGG sequences, but do not preclude the possibility that it may bind to these sequences transiently.
Effect of catalytically inactive PspGI on deamination
Several mutants of PspGI that alter residues within its predicted catalytic site have been studied for their catalytic and DNA-binding properties (6). Some of these bind the cognate sequence (in the presence of Ca2+) but are catalytically inactive (in the presence of Mg2+). We chose the mutant, D138A, which is catalytically inactive but has KD for DNA binding at the cognate sequence that is only 2.5-fold higher than that of the wild-type (WT) protein (6).
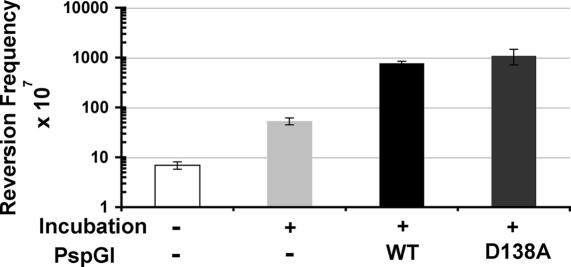
The effect of the D138A mutant on cytosine deaminations within CCSGG were studied using the plasmid pUP41. Similar to the WT protein, D138A protein also increased substantially the frequency of KanR revertants in pUP41. When a 10-fold molar excess of the protein was incubated with the plasmid, the KanR revertant frequency increased ∼20-fold over and above the increase due to heat alone and this was similar to the increase caused by the wild-type protein (Figure 7). These results show that the catalytic ability of PspGI was dispensible for its ability to sensitize cytosines in pUP41 for deamination.
Figure 7.
Effect of the D138A mutant of PspGI on cytosine deamination. The frequencies of KanR revertants following incubation of DNA with an excess of the mutant protein are shown. The incubation conditions for the different reactions are indicated below the bar graph in a manner similar to that in Figure 2.
Sequence requirements for the sensitization effect of PspGI on DNA
Codon 94 in the kan allele in pUP41 has two overlapping CCSGG sites (Figure 1). We wondered whether both these sites were needed for PspGI to sensitize the cytosines in codon 94 for deamination. To address this, the first CCSGG site was eliminated by changing the first cytosine in this sequence to adenine creating the plasmid pUP44 (Figure 1). When this plasmid was incubated with PspGI and the DNA subsequently transformed into E.coli, the KanR revertant frequency increased only slightly above the level seen with heat alone (Figure 5B). In three independent experiments (four reactions each), the deamination frequency of pUP44 increased 1–84% when PspGI was included in the reaction and this was substantially less than the magnitude of the increase seen under the same conditions for pUP41 (Figure 5A and data not shown). These results identify the cytosine at the third position of codon 93 (Figure 1) as being critical for the sensitization of the cytosines within codon 94 for deamination.
Sequence analysis of KanR revertants
Kanamycin-resistant revertants from pUP41 or its derivatives can be obtained by changing either the first or the second cytosine in codon 94 (Figure 1). We have found that different treatments of the plasmid result in different ratios of mutations at the first and the second C, and this creates a signature for each treatment (M. Samaranayake, C. Canugovi and A. S. Bhagwat, unpublished data). To determine whether the enhancement of KanR revertant frequency by PspGI also changed the pattern of C to T mutations within codon 94, several independent revertants were sequenced. The results are summarized in Table 1.
Table 1.
Mutations in KanR Revertants
| Revertants sequenced | First C | Second C | Ratioa | |
|---|---|---|---|---|
| No protein | 24 | 9b | 15c | 1.7 |
| WT PspGI | 52 | 3 | 49 | 16.3 |
Plasmid pUP41 was heated with or without WT PspGI. Following transformation of the DNA into ung E.coli, independent KanR revertants were obtained and sequenced. The location of mutations in independent KanR revertants is shown. ‘First C’ and ‘Second C’ refer to cytosines in codon 94 (CCG). With one exception, all the clones contained mutations at one of the two positions. The exception was a clone from the no protein control and contained C to T changes at both the positions. No mutations other than those listed here were seen in a 500 bp window around codon 94.
aIncludes two C to G and one C to A mutation.
bIncludes one C to A mutation.
cThe ratio—(Number of mutations at second C)/(Number of mutations at first C).
When plasmid pUP41 heated without PspGI was used for transformation, the mutations were modestly biased in favor of the second C in the CCG codon (Figure 1 and Table 1). We have observed previously a similar bias in pUP31 and other plasmids related to pUP41 that were incubated at 37°C (M. Samaranayake, C. Canugovi and A. S. Bhagwat, unpublished data). This bias increased drastically when the same plasmid was incubated in the presence of PspGI. In this case the mutations were overwhelmingly found in the second cytosine in CCG (Table 1). This cytosine is in the central C:G pair of the first 5′ CCSGG site overlapping codon 94 (enclosed in a brown box, Figure 1) and these data suggest that PspGI sensitizes this particular cytosine for deamination.
DISCUSSION
While studying the protective effects of DNA binding by restriction endonuclease PspGI at its cognate sequence CCAGG, we unexpectedly found that the enzyme promoted cytosine deaminations at CCSGG sites. PspGI reduced the pseudo-first-order rate constant for cytosine deamination to uracil at CCAGG by >7-fold, but increased the rate of this reaction at CCSGG by >15-fold. Thus, depending on the sequence context of cytosines, PspGI can alter the rate of cytosine deamination over two orders of magnitude. How does an enzyme that protects cytosines in one sequence against deamination can sensitize them for the same reaction in a closely related sequence?
The protection of CCAGG sequence by PspGI may be explained based on our understanding of the binding ofother restriction enzymes to their cognate sequence. PspGI could protect its cognate sequence by several related mechanisms. First, when restriction enzymes bind their cognate site they reduce the hydration of DNA and this may reduce the possibility of a hydrolytic attack. For example, both EcoRI and BamHI release substantial numbers of bound water molecules upon binding to their cognate sequences (25,26). Although the assay used to measure this effect does not distinguish between water molecules bound to DNA and those bound to the enzyme, it is reasonable to assume that at least some of the released water is from DNA. A second reason for the protective effects of PspGI may be that it restricts the access of free water to DNA. Most restriction enzymes are known to wrap themselves around DNA when bound to their cognate sites and make specific contacts to the DNA backbone and bases (27). This must reduce access of free water to DNA bases and reduce the rate of hydrolytic deamination of cytosines. Finally, binding of PspGI to its cognate sequence probably reduces the degrees of freedom within the DNA and prevents thermal motions that may tend to open the bases to the solvent.
However, the sharp increase in cytosine deamination caused by PspGI at a CCSGG site is unprecedented and is not easily explained. This is because CCSGG is not the cognate binding site for the enzyme and PspGI does not form stable complexes at this sequence (Figure 6). In addition, the CCSGG sites at codon 94 must compete with 13 CCAGG sites (to which the protein binds with much greater affinity) and 18 other CCSGG sites within the plasmid for binding. Finally, the target plasmid pUP41 is 6800 bp in size and provides a vast excess of non-specific DNA as competitor for binding to PspGI. Thus the binding at the CCSGG sites at codon 94 should be infrequent and short-lived, but somehow must cause a change in DNA structure at CCSGG sites that persists for some time and promotes cytosine deamination. In other words, PspGI cannot be causing a change such as modest local unwinding or kinking of DNA at CCSGG. Although restriction enzymes are known to cause such changes in DNA (27), these secondary structures would be quickly eliminated as soon as the enzyme leaves.
There are several reasons to think that endonucleolytic activity of PspGI is not the explanation for the increased cytosine deamination owing to PspGI in pUP41. We used a catalytically defective mutant of PspGI (D138A) and substituted the normal cofactor Mg2+ with Ca2+ to prevent DNA cleavage by the enzyme during the incubation. Under these conditions, the frequency of cytosine deaminations increased to the same level as caused by the WT enzyme (Figure 7). Furthermore, PspGI is not known to cleave CCSGG sites and, hence, under these conditions it is highly unlikely that the WT or the mutant protein cleaves DNA at or near CCSGG sites. Even if some cleavage were to occur at this site, the resulting linear DNA would transform E.coli poorly and would be unlikely to give rise to colonies. Lastly, if cleavage were to occur, the open ends of such DNA would be processed in E.coli resulting in deletion mutations and not base substitutions.
There is limited amount of information about how DNA secondary structure affects cytosine deamination. It is known that cytosines in single-stranded DNA deaminate at 140-fold higher rate than the corresponding double-stranded form at 37°C (28) and those in C•C and C•T mispairs have a rate of deamination intermediate between single- and double-stranded state (29). As cytosines in single-stranded DNA are freely accessible to the solvent, they are likely to have the highest rate of deamination among different secondary structures. The rate of cytosine deamination at codon 94 in the presence of PspGI is about one-fourth of the rate in single-stranded DNA [4.4 × 10−9 s−1 instead of 2.0 × 10−8 s−1; (3,28)] indicating that the susceptible cytosine in codon 94 has substantial single-stranded character.
One type of secondary structure change in DNA at CCSGG site that would be metastable and expose the cytosines to attack is the formation of a cruciform structure. If the susceptible cytosines were to lie in the loop region of a cruciform, it would be much more susceptible to deamination. However, analysis of the sequence surrounding codon 94 using MFOLD (30) does not predict the existence of such a cruciform structure. Furthermore, it is difficult to see how PspGI could cause the formation of cruciform structure while being bound at the same sequence.
Alternately, if a cytosine within CCSGG were to be flipped out of the double helix by PspGI, it would become readily susceptible to deamination. As the C to T mutations occur overwhelmingly at the second cytosine in codon 94 (Table 1), according to this hypothesis this particular cytosine would have to be flipped out by the enzyme. The first of two CCSGG sites (enclosed in a brown box in Figure 1) is essential for PspGI to increase deamination (Figure 5B), and this cytosine is the central base in this CCSGG site. These considerations suggest that PspGI flips out the cytosine in the central base pair in CCSGG. This implies that PspGI will also flip out the cytosine in the central base of the second CCSGG site at codon 94 (enclosed in a green box in Figure 1). However, this has no effect on the KanR reversion frequency because a C to T change at this position does not change the coded amino acid (CCG to CCA change). Thus, flipping of cytosine in the central base pair in CCSGG is consistent with our data.
These data are consistent with two models of how base-flipping by PspGI may sensitize CCCGG sequences for deamination of the central cytosine. The models are not mutually exclusive. In one, the flipped out cytosine either stays extrahelical for extended periods of time (several seconds) after the enzyme leaves either because of a large energy barrier preventing the return of the base into the double helix or because of repeated binding and dissociation of the enzyme (subsecond timescale). Currently, little information is available regarding the half-lives of a flipped out bases in naked DNA. Most discussions regarding ‘spontaneous’ base flipping are based on numbers obtained from NMR studies of imino proton exchanges of DNA bases with a general base [e.g. see (31)] and these studies use the loss of hydrogen bonding between the N1 of purines and N3 of pyrimidines as proxy for a complete base flipping reaction. It is very likely that half-life of the flipped out state of the base is much longer than the open state (i.e. just the loss of N1–N3 hydrogen bond) of a DNA base pair.
The second model for how PspGI may enhance cytosine deamination envisions an additional role for the enzyme in the process. In this case, the enzyme would continue to bind CCCGG sequences in DNA for an extended amount of time (several seconds, but not minutes; Figure 6) and would form an open complex with DNA that allows increased access of water to the flipped out bases. It is known that restriction enzymes form more solvent accessible complexes with non-specific DNA (32,33) and the same may be true regarding the interaction of PspGI at CCCGG sequences. Furthermore, CCCGG is a ‘star’ site for PspGI and restriction enzymes are known to pause at star sites (34). Thus, a combination of increased residence time for PspGI at CCCGG sites with the central bases flipped out and an increase in solvent accessibility may also explain the enhancement in cytosine deamination reported here.
After this work was completed, the crystal structure of the restriction enzyme Ecl18kI with DNA was published (23). Ecl18kI recognizes the sequence CCNGG, cleaves DNA before the first cytosine and is related to PspGI (34% sequence similarity). In the crystal, the enzyme is bound to CCAGG sequence and shows that Ecl18kI flips both the adenine and thymine in the central base pair out of the helix. Both the extra-helical bases are sandwiched between arginine and tryptophan residues (Figure 8). This is the first example of flipping of a base within its recognition sequence by a restriction enzyme. The only other example of base flipping by these enzymes is HinP1I, which flips an adenine outside its canonical recognition sequence (35).
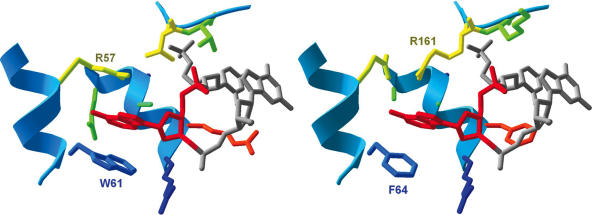
Figure 8.
Binding pocket for the central base in Ecl18kI and PspGI. The amino acid residues nearest to the central nucleoside (adenosine here) in the Ecl18kI–DNA complex (left) and the corresponding structure in the model for PspGI (right). Identical amino acid residues in the two proteins are shown in dark blue, chemically similar residues are in light blue, dissimilar residues are in green to yellow to orange. Secondary structures are shown in blue. DNA is shown in gray, and the central adenosine in the target sequence is in red. The flipped-out base in the PspGI model is held by F64 (homologous to W61 in Ecl18kI) and R161 (non-homologous to R57 in Ecl18kI, but with the side chain at an equivalent position). There are no base-specific interactions formed by amino acid side chains either with the Watson–Crick or the Hoogsteen edges, suggesting that the binding pockets of both Ecl18kI and PspGI can accommodate all four DNA bases.
Base flipping observed in the Ecl18kI co-crystal structure supports indirectly our conclusions regarding cytosine flipping by PspGI. To obtain more direct support for our hypothesis, we modeled a PspGI dimer bound to the same DNA sequence as the Ecl18kI co-crystal (CCAGG) based upon the known structures of EcoRII and Ecl18kI (22,23). The resulting model is a significant refinement over an earlier partial model which included only the catalytic domain, had no DNA and lacked N-terminal extension involved in protein–protein and protein–DNA contacts (6). The new model (Figure 8) shows that, similar to Ecl18kI, PspGI can also accommodate the central bases in the recognition sequence in a pocket formed by two α-helices. In both the structures (Figure 8), the flipped out base is buffeted by an aromatic side chain on one side and an arginine on the other. There are no base-specific contacts between the extra-helical bases and the protein, providing a simple explanation for how both the enzymes can accommodate a cytosine or a guanine in the pocket in a manner similar to how the thymine and adenine is found in the Ecl18kI structure. We expect that PspGI, similar to Ecl18kI, should also flip out the A and the T in CCWGG sequences. However, this should not sensitize the adjacent cytosine bases for deamination.
Although our results are consistent with base flipping observed in the Ecl18kI co-crystal structure, they provide several additional details that go beyond the structure. First, in this work the DNA was in aqueous solution and the experiments were performed using moderate salt concentrations. Thus structural artifacts sometimes associated with crystallization are not likely to be present in our experiments. Second, similar to Ecl18kI, PspGI also creates 5-nt overhangs. Bochtler et al. (23) argue that Ecl18kI flips out the central base pair as a way of contracting the distance between the scissile phosphodiester linkages effectively allowing it to cleave DNA on opposite strands at a distance of 4 nt in B DNA. The data presented here support this idea and make it likely that other restriction enzymes that leave 5 nt overhangs will also flip out the central base pair.
Third, the data presented here strongly suggest that CCSGG, a site not cleaved by PspGI, undergoes base flipping. In contrast, the Ecl18kI co-crystal has the canonical sequence CCAGG in its DNA. This leads to several observations and conclusions. It shows that flipping of the central bases (presumably the guanine opposite the extrahelical cytosine is also flipped) is not sufficient for PspGI to cleave at CCSGG sequences. It is possible that the flipping of bases in a non-canonical sequence destabilizes the protein–DNA complexes aborting cleavage. Additional studies are needed to determine whether this is correct or PspGI avoids cleaving DNA at CCSGG sites using some other mechanism.
The ability of PspGI to flip DNA bases in CCSGG also suggests that it may have evolved from an ancestor similar to Ecl18kI that cleaves both CCWGG and CCSGG sequences. Previously, evolutionary analysis of CCXGG (X is S, W or N) family of restriction enzymes had revealed that SsoII (recognition sequence CCNGG) is the closest known relative of PspGI (6). The work presented here suggests that the common ancestor of these two enzymes recognized and cleaved at CCNGG sequences. It would be fascinating to explore how the ability to cleave CCSGG sequences was lost by the PspGI branch of the family during its evolution.
Flipping of the bases in the central base pair of CCSGG by PspGI could be a serious problem for its host organism Pyrococcus sp. strain GI-H. In this archaeon, the CCWGG sequences are presumably protected by the corresponding modification methyltransferase, M.PspGI (5). However, if the CCSGG sequences are unmethylated in the archaeal DNA, flipping of bases within these sequences by PspGI endonuclease would make them greatly susceptible to damage. This would create a large increase in deamination, oxidation and other types of chemical damage to the exposed bases and would sharply increase the endogenous rate of mutations. This is unlikely to be sustainable over long time periods and probably does not happen in this organism. We suggest that one way in which the organism could avoid this mutational catastrophe is to methylate CCSGG sequences in addition to CCWGG sequences. Currently, the sequence specificity of M.PspGI is unknown and it is possible that it methylates both CCWGG and CCSGG sequences. Alternately, the organism may have another methyltransferase (similar to Dcm in E.coli) that protects these sequences from PspGI endonuclease.
In summary, we found that PspGI protects the two cytosines in its canonical sequence CCWGG against deamination, while making the cytosine in the central base pair in CCSGG sensitive to hydrolytic attack. These data are consistent with the flipping of the central base (pair) by PspGI and this has interesting implications for the evolution of PspGI and how it couples base sequence recognition with catalysis.
Acknowledgments
We would like to thank Shuang-yong Xu (New England Biolabs) for providing initial samples of PspGI. We would also like to thank Alfred Pingoud (Justus-Liebig University), John SantaLucia (Wayne State University) and Anjum Sohail (Wayne State University) for their help in planning some of the experiments and their comments on the manuscript. A.S.B. would like to thank Mats Ljungman (University of Michigan School of Medicine) for his hospitality during the writing of this manuscript. J.M.B. thanks Agnieszka Obarska (IIMCB) for running ROSETTA. This work was supported by grants from the National Institutes of Health to A.S.B (GM57200 and CA097899) and from Deutsche Forschungsgemeinschaft (Pi151/3-1) to V.P. J.M.B. was supported by the Fogarty International Center (grant R03 TW007163-01). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington, DC: ASM Press; 2005. [Google Scholar]
- 3.Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 4.Sohail A., Hayes C.S., Divvela P., Setlow P., Bhagwat A.S. Protection of DNA by alpha/beta-type small, acid-soluble proteins from Bacillus subtilis spores against cytosine deamination. Biochemistry. 2002;41:11325–11330. doi: 10.1021/bi026332t. [DOI] [PubMed] [Google Scholar]
- 5.Morgan R., Xiao J., Xu S. Characterization of an extremely thermostable restriction enzyme, PspGI, from a Pyrococcus strain and cloning of the PspGI restriction–modification system in Escherichia coli. Appl. Environ. Microbiol. 1998;64:3669–3673. doi: 10.1128/aem.64.10.3669-3673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pingoud V., Conzelmann C., Kinzebach S., Sudina A., Metelev V., Kubareva E., Bujnicki J.M., Lurz R., Luder G., Xu S.Y., et al. PspGI, a type II restriction endonuclease from the extreme thermophile Pyrococcus sp.: structural and functional studies to investigate an evolutionary relationship with several mesophilic restriction enzymes. J. Mol. Biol. 2003;329:913–929. doi: 10.1016/s0022-2836(03)00523-0. [DOI] [PubMed] [Google Scholar]
- 7.Halford S.E., Johnson N.P. The EcoRI restriction endonuclease with bacteriophage lambda DNA. Equilibrium binding studies. Biochem. J. 1980;191:593–604. doi: 10.1042/bj1910593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyszynski M., Gabbara S., Bhagwat A.S. Cytosine deaminations catalyzed by DNA cytosine methyltransferases are unlikely to be the major cause of mutational hot-spots at Sites of cytosine methylation in E.coli. Proc. Natl Acad. Sci. USA. 1994;91:1574–1578. doi: 10.1073/pnas.91.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhagwat A.S. DNA-cytosine deaminases: from antibody maturation to antiviral defense. DNA Repair. 2004;3:85–89. doi: 10.1016/j.dnarep.2003.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yebra M.J., Bhagwat A.S. A cytosine methyltransferase converts 5-methylcytosine in DNA to thymine. Biochemistry. 1995;34:14752–14757. doi: 10.1021/bi00045a016. [DOI] [PubMed] [Google Scholar]
- 11.Bandaru B., Gopal J., Bhagwat A.S. Overproduction of DNA cytosine methyltransferases causes methylation and C to T mutations at non-canonical sites. J. Biol. Chem. 1996;271:7851–7859. doi: 10.1074/jbc.271.13.7851. [DOI] [PubMed] [Google Scholar]
- 12.Lutsenko E., Bhagwat A.S. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat. Res. 1999;437:11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 13.Beletskii A., Grigoriev A., Joyce S., Bhagwat A.S. Mutations induced by bacteriophage T7 RNA Polymerase and their effects on the composition of T7 genome. J. Mol. Biol. 2000;300:1057–1065. doi: 10.1006/jmbi.2000.3944. [DOI] [PubMed] [Google Scholar]
- 14.Sharath A.N., Weinhold E., Bhagwat A.S. Reviving a dead enzyme: cytosine deaminations promoted by an inactive DNA methyltransferase and an S-adenosylmethionine analogue. Biochemistry. 2000;39:14611–14616. doi: 10.1021/bi001610e. [DOI] [PubMed] [Google Scholar]
- 15.Beletskii A., Bhagwat A.S. Transcription-induced cytosine-to-thymine mutations are not dependent on sequence context of the target cytosine. J. Bacteriol. 2001;183:6491–6493. doi: 10.1128/JB.183.21.6491-6493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beletskii A., Bhagwat A.S. Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol. Chem. 1998;379:549–551. [PubMed] [Google Scholar]
- 17.Lutsenko E., Bhagwat A.S. The role of the Escherichia coli mug protein in the removal of uracil and 3,N(4)-ethenocytosine from DNA. J. Biol. Chem. 1999;274:31034–31038. doi: 10.1074/jbc.274.43.31034. [DOI] [PubMed] [Google Scholar]
- 18.Deng W.P., Nickoloff J.A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 19.Kurowski M.A., Bujnicki J.M. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosinski J., Cymerman I.A., Feder M., Kurowski M.A., Sasin J.M., Bujnicki J.M. A ‘FRankenstein's monster’ approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins. 2003;53:369–379. doi: 10.1002/prot.10545. [DOI] [PubMed] [Google Scholar]
- 21.Simons K.T., Kooperberg C., Huang E., Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 22.Zhou X.E., Wang Y., Reuter M., Mucke M., Kruger D.H., Meehan E.J., Chen L. Crystal structure of type IIE restriction endonuclease EcoRII reveals an autoinhibition mechanism by a novel effector-binding fold. J. Mol. Biol. 2004;335:307–319. doi: 10.1016/j.jmb.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 23.Bochtler M., Szczepanowski R.H., Tamulaitis G., Grazulis S., Czapinska H., Manakova E., Siksnys V. Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease. EMBO J. 2006;25:2219–2229. doi: 10.1038/sj.emboj.7601096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasin J.M., Bujnicki J.M. COLORADO3D, a web server for the visual analysis of protein structures. Nucleic Acids Res. 2004;32:W586–589. doi: 10.1093/nar/gkh440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch T.W., Sligar S.G. Macromolecular hydration changes associated with BamHI binding and catalysis. J. Biol. Chem. 2000;275:30561–30565. doi: 10.1074/jbc.M000266200. [DOI] [PubMed] [Google Scholar]
- 26.Robinson C.R., Sligar S.G. Changes in solvation during DNA binding and cleavage are critical to altered specificity of the EcoRI endonuclease. Proc. Natl Acad. Sci. USA. 1998;95:2186–2191. doi: 10.1073/pnas.95.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pingoud A., Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frederico L.A., Kunkel T.A., Shaw B.R. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 29.Frederico L.A., Kunkel T.A., Shaw B.R. Cytosine deamination in mismatched base pairs. Biochemistry. 1993;32:6523–6530. doi: 10.1021/bi00077a005. [DOI] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao C., Jiang Y.L., Stivers J.T., Song F. Dynamic opening of DNA during the enzymatic search for a damaged base. Nature Struct. Mol. Biol. 2004;11:1230–1236. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 32.Viadiu H., Aggarwal A.K. Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol. Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 33.Winkler F.K., Banner D.W., Oefner C., Tsernoglou D., Brown R.S., Heathman S.P., Bryan R.K., Martin P.D., Petratos K., Wilson K.S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeltsch A., Alves J., Wolfes H., Maass G., Pingoud A. Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochemistry. 1994;33:10215–10219. doi: 10.1021/bi00200a001. [DOI] [PubMed] [Google Scholar]
- 35.Horton J.R., Zhang X., Maunus R., Yang Z., Wilson G.G., Roberts R.J., Cheng X. DNA nicking by HinP1I endonuclease: bending, base flipping and minor groove expansion. Nucleic Acids Res. 2006;34:939–948. doi: 10.1093/nar/gkj484. [DOI] [PMC free article] [PubMed] [Google Scholar]